Abstract
Signal transduction by fibroblast growth factor (FGF) receptors in Drosophila depends upon the intracellular protein Dof, which has been proposed to act downstream of the receptors and upstream of Ras. Dof is the product of a fast-evolving gene whose vertebrate homologs, BCAP and BANK, are involved in signaling downstream of the B-cell receptor. Mapping functional domains within Dof revealed that neither of its potential interaction motifs, the ankyrin repeats and the coiled coil, is essential for the function of Dof. However, we have identified a region within the N terminus of the protein with similarity to BCAP and BANK, which we refer to as the Dof, BCAP, and BANK (DBB) motif, that it is required for FGF-dependent signal transduction and is necessary for efficient interaction of Dof with the FGF receptor Heartless. In addition, we demonstrate that Dof is phosphorylated in the presence of an activated FGF receptor and that tyrosine residues could contribute to the function of the molecule.
Communication between cells is an important feature of metazoans that regulates the development and maintenance of distinct tissues within an organism. Many of the molecules involved in the pathways that cells use to sense their environment are highly conserved. For example, the Drosophila homolog of Grb2, Drk, which links receptor tyrosine kinases to the mitogen-activated protein (MAP) kinase cascade (31, 37), shows 65% identity with its human counterpart. However, there are also molecules, such as Argos, a negative regulator of epidermal growth factor signaling in Drosophila (14, 40), that appear to be specific to a particular species or phylum.
Most receptor tyrosine kinases are able to activate the MAP kinase cascade by recruiting Grb2, but in the case of the vertebrate fibroblast growth factor (FGF) receptors, a myristylated protein known as FRS2, or SNT, which contains an N-terminal phosphotyrosine binding (PTB) domain, is required to couple the receptor to the MAP kinase cascade (23, 41). The PTB domain of the FRS2/SNT proteins is unusual in that it interacts with a nonphosphorylated region of the FGF receptors close to the membrane (10, 25, 32, 43). Upon activation of the receptor, FRS2/SNT is rapidly phosphorylated and acts as a binding site for other molecules, such as Grb2 or Shp2, which leads to activation of the MAP kinase cascade and the phosphatidylinositol (PI) 3-kinase pathway (17, 18, 23, 33, 34, 41). Interestingly, the function of FRS2/SNT appears to be unique to vertebrates. The gene most closely related to FRS2/SNT (Flybase no. CG13398) in Drosophila shows a high degree of divergence compared to the vertebrate FRS2/SNT genes. In Drosophila, the formation of the mesoderm and the tracheal tissues depends upon signaling through the FGF receptors Heartless and Breathless, respectively (reviewed in references 16, 26, 28, and 42). Neither of these processes is affected if the function of the gene related to FRS2/SNT is removed (A. Michelson, personal communication). Furthermore, the juxtamembrane region of the vertebrate FGF receptors, which is the region that interacts with FRS2/SNT, is not conserved in the Drosophila FGF receptors.
The activities of the Drosophila FGF receptors depend upon a gene known as downstream-of-FGF receptor (dof), heartbroken (hbr), or stumps (sms) (20, 27, 39). dof is essential for the morphogenesis of both the mesoderm and the tracheae. It is required for the accumulation of the activated form of MAP kinase within these tissues and has been proposed to be an adaptor molecule that could relay the activated state of the Drosophila FGF receptors to components that act downstream of the receptors. The gene encodes two transcripts predicted to produce proteins of >1,000 amino acids that differ slightly at the N terminus (39). These proteins contain two ankyrin repeats and a coiled coil but not structural domains, such as SH2 or PTB domains, typically present in molecules that function as adaptors or docking proteins in signal transduction pathways.
The proteins most closely related to Dof that are found in vertebrates are BCAP and BANK (30, 45). These proteins are involved in signaling; however, unlike Dof, they are not implicated in FGF signaling but in the events downstream of the B-cell receptor. Both proteins regulate the release of calcium within B cells (44, 45). BANK interacts directly with the IP-3 receptor (45), while BCAP is thought to be an adaptor protein that mediates the activation of PI 3-kinase and phospholipase C γ2 by the B-cell antigen receptor (30). Recently, BCAP has been shown to be essential for the activation and maturation of B cells (44). Thus, Dof, BCAP, and BANK are all involved in signaling, although they appear to have undergone rapid change during the course of evolution and may have evolved in parallel with FRS2/SNT. It is not clear how the structures of these molecules relate to their functions, except that the N-terminal part of BANK associates with the IP-3 receptor. We describe here the domains of Dof required for the function of the protein in vivo.
MATERIALS AND METHOD
Sequence analysis
Throughout the course of this work, we refer to the annotated Drosophila genome (11), the mosquito genome pages at ENSEMBL, EBI (19), and GenBank (6). Sequence comparison was carried out with BLAST or a program that counted the identities shared by two protein sequences (details available on request). Protein sequences were aligned using CLUSTAL W, and the output was formatted with BOXSHADE. To determine the most highly conserved regions of the Ag. and Dm. Dof homologs, the protein sequences (accession numbers Q8T5J9 and O96757) were aligned with CLUSTAL W. Then, the insertions and deletions within the proteins were determined, and regions representing the insertions in the Anopheles sequence were removed from the alignment. Subsequently, the number of identities shared by the homologs within a window of 15 amino acids was determined for each position of Drosophila Dof (program available on request). The data were plotted using Microsoft Excel.
In vitro mutagenesis
Mutagenesis was carried out by PCR or by the pAlter system (Promega), with the exception that we used the Escherichia coli strains JM109 and ES455. The presence of the desired mutation was confirmed by DNA sequence analysis.
Plasmids
A detailed description of the constructs used is included as supplemental material.
Drosophila genetics
Standard procedures were followed to produce transgenic flies. Insertions were assigned to a particular chromosome based on the segregation of the transgenes from dominant markers on the second and third chromosome balancers and sex linkage for the X chromosome. The tracheal assay was carried out essentially as previously described (39). For the mesoderm assay, an X chromosome with multiple P[w+ twi-Gal4] insertions, constructed by Nick Brown (35), was mobilized using a third-chromosome transposase source. Insertions of the P[w+ twi-Gal4] element on a dof1 e11 chromosome were recovered and balanced using a TM3 chromosome carrying a P[ry+ ftz-lacZ] insertion. Homozygous dof mutant embryos were identified in a 4- to 6-h embryo collection by staining them with an antibody directed against β-galactosidase. To determine the localization of the Dof mutant proteins, males from stocks containing chromosomes with upstream activation sequence insertions were crossed to virgins with the third-chromosome P[w+ GAL4-prd.F] insertion (7).
Immunohistochemistry
Standard protocols were followed to collect, fix, and stain embryos. The primary antibodies used were rabbit anti-Even-Skipped (1:5,000) (13) (kindly provided by M. Frasch), rabbit anti-Dof (1:200) (39), polyclonal mouse anti-β-galactosidase (1:500) (Sigma G4644), and 2A12 (1:20) (kindly provided by Nipam Patel). Western blotting was carried out using 1.25 μg of the mouse anti-FLAG M5 monoclonal antibody (Sigma)/ml and a dilution (1:5) of the mouse anti-phosphotyrosine monoclonal antibody 4G10 (courtesy of Frank Sprenger). Biotinylated wheat germ agglutinin (Molecular Probes) was used at 0.01 mg/ml and visualized with Avidin-Alexa 647 (Molecular Probes) at a dilution of 1/500.
Yeast methods
Protein interaction experiments were carried out in the yeast strain PJ69-4a (21) (kindly provided by P. James). The techniques and constructs were as described previously (4).
Drosophila cell culture
Drosophila Schneider S2 cells were maintained at 22°C in Schneider's Drosophila medium (Gibco-BRL) supplemented with 10% (vol/vol) fetal calf serum (Sigma). Transient transfections were as described previously (4). For the production of stable cell lines, cells were transfected with 5′Flag-Dof.pAT-Hygro and lambda-Btl.phsT-Neo by means of a modified calcium phosphate procedure (9). Geneticin (G418; Roche) was used at 1 mg/ml, and hygromycin (Roche) was used at 200 μg/ml to establish stably transfected lines. To induce the expression of λ-Btl from the heat shock promoter, cells were incubated at 37°C for 30 min and then allowed to recover for 4 h at 22°C before analysis. For whole-cell lysate assays, a 60-mm-diameter plate of confluent cells was lysed in 400 μl of boiling sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. The sample was heated at 95°C for 3 min and then sonicated for 20 s prior to 7.5% SDS-PAGE analysis. For immunoprecipitation, a 60-mm-diameter plate of confluent cells was washed twice with ice-cold 1× Tris-buffered saline (25 mM Tris-HCl, pH 7.4, 137 mM NaCl, 3 mM KCl) and lysed on ice for 20 min using 1 ml of lysis buffer (10% glycerol, 50 mM HEPES, pH 7.5, 150 mM NaCl, 1% Triton X-100, 1.5 mM MgCl2, 1 mM EGTA, 200 μM Na3VO4, 10 mM NaF) supplemented with a protease inhibitor cocktail (P8340; Sigma). The lysates were transferred to a microcentrifuge tube and precleared by centrifugation at 20,150 × g for 15 min at 4°C. Immunoprecipitation was carried out for 2 h at 4°C with 300 μl of lysate, 1 μl of rabbit anti-Dof antiserum (39), and 15 μl of protein G-Sepharose (Amersham-Pharmacia) that had been blocked for 20 min with lysis buffer containing 5% bovine serum albumin and then washed with lysis buffer. Protein complexes bound to the protein G Sepharose were washed three times with 1 ml of wash buffer (10% glycerol, 20 mM HEPES, pH 7.5, 0.1% Triton X-100, 150 mM NaCl, 1 mM Na3VO4) each time; 40 μl of 1× SDS-PAGE sample buffer containing 2-mercaptoethanol was added, and the samples were heated at 95°C for 3 min to denature the proteins. The proteins were separated by SDS-7.5% PAGE. Western blotting, using 1% bovine serum albumin solution to saturate the nitrocellulose membrane, followed standard procedures. Immunoreactive proteins were visualized using a horseradish peroxidase-catalyzed chemiluminescence reaction (Amersham-Pharmacia).
RESULT
Sequence analysis of proteins related to Dof
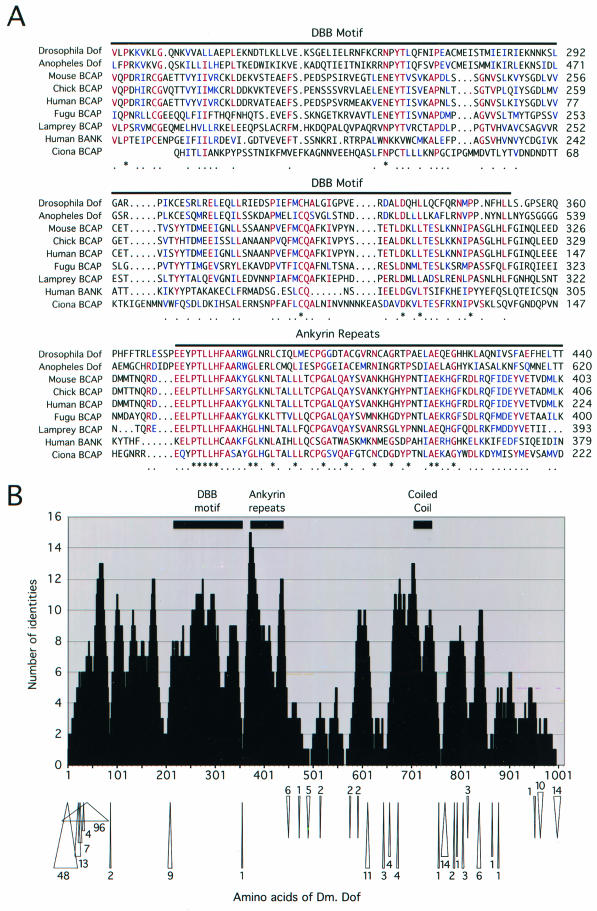
When Dof was initially characterized, the sequence of the protein did not resemble any other in the database. Two vertebrate proteins that function in B-cell signaling, BCAP and BANK, which share structural domains with Dof, have since been described (30, 45). These proteins possess a C-terminal coiled coil and have a high degree of similarity to Dof over the ankyrin repeats. Mouse BCAP shows 47% identity over 55 amino acids with this region of Dof, while BANK shows 43% identity. Interestingly, the similarity among Dof, BCAP, and BANK extends beyond the ankyrin repeats toward the N terminus. Mouse BCAP and human BANK show 19 and 17% identity, respectively, with amino acids 216 to 353 of Dof (Fig. 1A). We have designated this the Dof, BCAP, and BANK (DBB) region.
FIG.1.
Sequence analysis of Dof and related proteins. (A) Aligment over the DBB motif and the ankyrin repeats of Drosophila Dof (accession number O96757), Anopheles Dof (accession number Q8T5J9), mouse BCAP (accession number Q9EQ32), chicken BCAP (accession number AAG48583), human BCAP (accession number XP_058343), fugu BCAP (ENSEMBL SINFRUP00000078060), lamprey BCAP (accession number AAN64296), human BANK (accession number Q8WYN5), and Ciona intestinalis BCAP (a translation of accession number AK115423). The alignment begins at positions 216 of Drosophila Dof, 395 of Anopheles Dof, 182 of mouse BCAP, 3 of human BCAP, 180 of the predicted fugu BCAP sequence, 178 of lamprey BCAP, 170 of human BANK, and 1 of the incomplete Ciona BCAP sequence. Six or more identical or similar residues shared by the sequences are shown in red and blue, respectively, and are marked below the alignment by a dot. An asterisk denotes a position that is absolutely conserved. (B) Alignment of A. gambiae Dof and Drosophila Dof was made using CLUSTAL W, and regions representing insertions in the Anopheles sequence were removed. Then, the number of identities found within a window of 15 amino acids was plotted for each position of the Drosophila protein. The locations of the DBB motif, the ankyrin repeats, and the coiled coil within Drosophila Dof are indicated at the top of the chart. The positions of insertions and deletions within the Anopheles sequence are shown below the chart as arrows pointing toward and away from the sequence of the Drosophila protein, respectively. The numbers represent the lengths of the insertions and deletions.
The completion of the Anopheles gambiae genome has revealed a homolog of Dof (accession number Q8T5J9) that shows 37% identity and 50% similarity over 924 amino acids of the Drosophila protein (Fig. 1A). A number of expressed sequence tags derived from this gene, such as AJ285928 and AJ281683, indicate that it is expressed. The degree of identity shared by the Drosophila and Anopheles Dof homologs is considerably lower than 56%, which is the average degree of conservation between clear Drosophila and Anopheles orthologs (46), so dof may represent an example of a fast-evolving gene. However, the regions of Dof that are similar to the vertebrate proteins BCAP and BANK are also among the most highly conserved parts of the Anopheles and Drosophila Dof homologs (Fig. 1B; see also Table S1 in the supplemental material) and have not been interrupted by amino acid insertions during evolution. Interestingly, the sequence between the DBB motif and ankyrin repeats diverges (Fig. 1), suggesting that the DBB region may represent an independent domain of the protein. Within the C-terminal half, the coiled coil shows the highest degree of similarity and is the most extensive block of conservation. Notably, the coiled-coil motif of Dof also shows 48% identity and 84% similarity with the hypothetical open reading frame of a partial Bombyx cDNA (accession number AU000659), suggesting that the motif may be important functionally. Thus, the sequences and organizations of three distinct domains of Dof appear to have been conserved among the homologs in insects and the most closely related vertebrate proteins. Furthermore, these domains are the regions with the highest degree of conservation among the vertebrate BCAP proteins (see Table S1 in the supplemental material) and as such define the general structure and features of this family of proteins.
In vitro mutagenesis of Dof
To determine if specific domains of Dof mediate different cellular functions, we generated by in vitro mutagenesis a series of mutants of the protein encoded by transcript II of dof. We chose to analyze the protein encoded by this transcript, which will be referred to as Dof below, since it has been shown that it can rescue the defects in the development of the tracheae and mesoderm of homozygous dof mutant embryos (39). To assay the in vivo functions of the mutant Dof proteins, we used the Gal4 upstream activation sequence system (7) to express transgenes encoding the mutant protein in wild-type and homozygous dof mutant embryos.
Characterization of the functions of the mutant Dof proteins in vivo
FGF receptor-dependent signal transduction regulates transcriptional activity and morphogenesis within the tracheae and the mesoderm. We tested Dof mutant constructs for FGF-dependent morphogenesis by examining the tracheae of homozygous dof mutant embryos expressing the mutant forms of Dof under the control of btl-Gal4. To determine their abilities to activate transcription of FGF-dependent genes, we monitored the expression of Even-skipped in the mesoderm. In this case, the constructs were expressed in dof mutant embryos under the control of twi-Gal4.
Differences in the behaviors of the mutant Dof proteins in these biological assays may not simply reflect differences in the functions of the proteins but could also reflect differences in the protein stabilities. We assessed whether the mutant Dof transgenes could produce stable proteins by expressing FLAG-tagged versions in Schneider S2 cells in tissue culture. Western blots probed with antibodies against the FLAG tag showed that in this system, all mutant proteins were expressed. They showed only slight variations in abundance, except for Dof[1-522], which was significantly more abundant than the full-length form, and Dof[168-1012], which appeared to be much less stable (see Fig. S1 in the supplemental data). When the proteins were expressed ectopically in the ectoderm of embryos by using prd-Gal4 and were visualized with an antiserum directed against Dof, we were able to detect expression of all mutant proteins (see below for more details), with the exception of the large C-terminal truncations (Dof[1-277] and Dof[1-446]). The fact that Dof[1-446] can be immunoprecipitated from Schneider S2 cells but is undetectable in formaldehyde-fixed embryos using the Dof antibody suggests that formaldehyde destroys the epitopes remaining in this part of the molecule. Furthermore, in yeast cells, the truncation Dof[1-446] fused to the Gal4 DNA binding domain must be stable, since it can interact with the kinase domain of Htl in the yeast two-hybrid assay (see below). Thus, we included these transgenes in further functional tests.
The DBB domain is necessary for the function of Dof
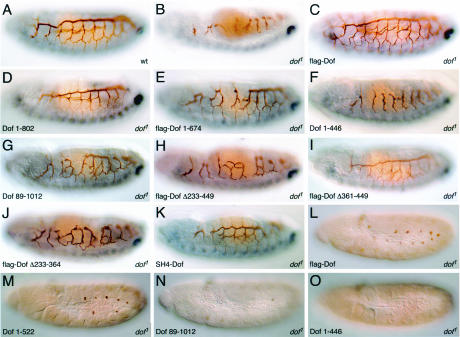
In homozygous dof or btl mutant embryos, differentiation of the lumen of the tracheae can be recognized using the antibody 2A12, as shown in Fig. 2B. In comparison to the wild type (Fig. 2A), the tubular structures lack connections to one another; in particular, the dorsal and lateral trunks are absent and the prominent visceral branches do not grow out normally. The morphogenesis of the tracheae in dof mutants can be completely rescued by expression of Dof (38), and this is not compromised by the addition of a FLAG epitope tag to the amino terminus of the protein (Fig. 2C).
FIG. 2.
Abilities of different mutant Dof proteins to rescue defects in the development of the mesoderm and tracheae. (A to K) Tubular network of the tracheae, revealed using the monoclonal antibody 2A12, is shown in brown. Even-skipped, visualized as a blue stain, was used to identify the homozygous mutant embryos by the absence of pericardial cells expressing Eve in the mesoderm, which can be seen in the wild-type embryo in panel A as a dorsal row of faint out-of-focus spots. (A) Stage 15 wild-type embryo; (B) homozygous dof1 mutant embryo. (C to K). Tracheal development of homozygous dof1 mutant embryos in which mutant forms of Dof were overexpressed using a btl-Gal4 transgene. The embryos chosen for flag-Dof[1-674] (E), Dof[1-446] (F), Dof[89-1012] (G), flag-Dof[Δ361-449] (I), and SH4-Dof (K) show the typical rescues obtained with these constructs, while those chosen for flag-Dof[Δ233-449] (H) and flag-Dof[Δ233-364] (J) show the best rescue of tracheal development obtained with these constructs. (L to O) Abilities of the mutant Dof proteins flag-Dof (L), Dof[1-522] (M), Dof[89-1012] (N), and Dof[1-446] (O) to promote differentiation of the mesoderm in stage 11 embryos were determined by monitoring the expression of Even-skipped, shown in brown, in parasegments 3 to 13. The embryos shown are representative, although a large degree of variation in the number of Even-skipped positive clusters was observed for the mutant Dof[89-1012].
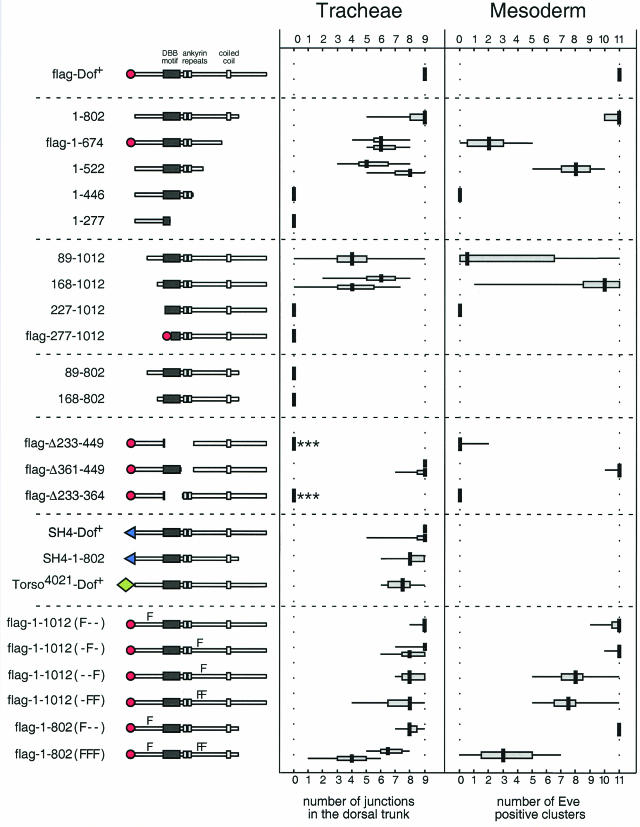
A truncated form of Dof that lacks the last 210 amino acids (Dof[1-802]) (Fig. 2D) is able to rescue morphogenesis of the tracheae completely. This was consistently observed with independently derived transgenic lines, suggesting that these C-terminal amino acids are not essential for this function in vivo. With transgenes encoding shorter proteins lacking the last 338 amino acids, or the last 481 amino acids, and which do not contain the coiled-coil motif, we still observed a partial rescue of the dof tracheal phenotype that was variable between embryos (Fig. 2E and 3). The formation of the dorsal trunk was least efficiently rescued by the transgenes, particularly towards the anterior of the embryo. We used this feature as a semiquantitative measure to compare different transgenic constructs (Fig. 3). In a few cases, complete rescue was observed with these constructs, indicating that the last 481 amino acids, which include the coiled coil, are not absolutely essential for the transmission of the signal from the FGF receptor. In contrast, transgenes encoding proteins with more extensive C-terminal deletions (data summarized in Fig. 3) had no activity in this assay.
FIG. 3.
Functions of mutant Dof proteins in in vivo assays. The Dof constructs are shown schematically on the left. The amino acids present in each construct are noted. A flag epitope tag is represented by a red circle, while the membrane-targeting SH4 domain of human Src is shown as a blue triangle. The extracellular portion and transmembrane domain of the Torso mutant 4021 is shown as a green diamond (also see Fig. 6 for details). To assess the ability of a transgene to rescue tracheal development, we counted the junctions formed by the dorsal trunk in five stage 15 embryos. In the case of the mesoderm, we determined the number of Eve-positive clusters formed in five stage 11 embryos. The degree of variability of the data is depicted as a box-and-whiskers plot. A thick vertical line indicates the median value, a horizontal shaded box represents the interquartile range between the first and third quartiles, and a thin horizontal line represents the entire range of the data. For the mutants flag-Dof[1-674], Dof[1-522], Dof[168-1012], flag-Dof[Δ361-449], SH4-Dof, flag-Dof <Y486F>, and flag-Dof[1-802]<Y97F, Y486F, Y515F>, two independent transgenic lines were assessed in the tracheae. In most cases, the absence of a dorsal trunk reflected a complete failure of the transgene to rescue the homozygous dof1 mutant phenotype (cf. Fig. 2). The asterisks shown for the mutants flag-Dof[Δ233-449] and flag-Dof[Δ233-364] indicate the formation of some lateral trunk (Fig. 2H and J).
Dof constructs lacking the first 88 or 168 amino acids at the N terminus partially rescued development of the tracheae (Fig. 2G). With both of these N-terminal truncations, the degree of rescue was more variable than with the C-terminal truncations with biological activity. Surprisingly, rescue with Dof[89-1012] was worse than with Dof[168-1012]. However, the breakpoint at amino acid 89 appears to have a nonspecific deleterious effect on the protein, since we have also observed that the 89-to-1012 truncation has less activity than the 168-to-1012 truncation in yeast two-hybrid experiments irrespective of the interaction being tested (A. Battersby, unpublished results). Notably, the N-terminal amino acids deleted in these mutants are not absolutely essential for signal transmission. However, further deletions, extending into the DBB motif (up to amino acid 227 or 277), resulted in complete failure of the constructs to rescue tracheal morphogenesis (data summarized in Fig. 3).
When we examined the activities of proteins with combined amino- and carboxy-terminal truncations, namely, Dof[89-802] and Dof[168-802], we found that they had no activity in the assays. Since neither of these deletions alone abolished the activity of the protein, this indicates that the amino and carboxy termini of the protein could have overlapping functions. None of the mutant Dof transgenes in these experiments had a dominant-negative effect on the function of the wild-type protein in the heterozygous sibling embryos.
The experiments described above suggested that the region of Dof containing the ankyrin repeats and the DBB motif, which has the most similarity to the vertebrate proteins BCAP and BANK, may be important for the function of the protein. To investigate the importance of these domains, we examined the activities of a number of transgenes with internal deletions. A protein that lacked the DBB motif and the ankyrin repeats, flag-Dof[Δ233-449], was severely compromised in its ability to support tracheal morphogenesis (Fig. 2H). Formation of a normal dorsal trunk was never observed in these embryos; however, occasional misdirected outgrowth of visceral branches and partial formation of the lateral trunk distinguished them from homozygous dof mutant embryos. A transgene in which just the ankyrin repeats were deleted could fully rescue tracheal morphogenesis (Fig. 2I). Thus, surprisingly, even though the ankyrin repeats are the most highly conserved part of the protein, they are not essential for transmission of the signal from the FGF receptor in this assay. By contrast, deletion of just the DBB region had the same phenotype as the larger deletion (Fig. 2J), suggesting that the DBB domain provides a critical function of Dof.
We also assayed the constructs for the ability to participate in FGF signaling in the mesoderm by expressing them in homozygous dof mutant embryos and determining the number of Even-skipped-expressing cell clusters. In wild-type embryos, Even-skipped is expressed in the dorsal mesoderm in clusters comprised of two or three cells, which include the pericardial cells (2, 3, 13). Mutations in the genes encoding the FGF receptor Htl (5, 15, 36) and Dof (20, 27, 39) result in the loss of these Even-skipped-positive cells. The activities of the Dof mutants in this assay were similar to those in the tracheae (summarized in Fig. 3), although the degree of rescue of Even-skipped-positive cells with the N-terminally truncated forms of Dof was more variable than that observed in the tracheal assay, and the activity of flag-Dof[1-674] in this assay was poor in comparison with that in the tracheal assay. Thus, in summary, for any Dof mutant that was capable of rescuing tracheal morphogenesis, we could also detect activation of Even-skipped within the mesoderm. The mutants we have analyzed have allowed us to identify a domain within the N-terminal half of the protein that is critical for efficient transmission of the FGF signal in both situations.
Subcellular localization of the mutant Dof proteins
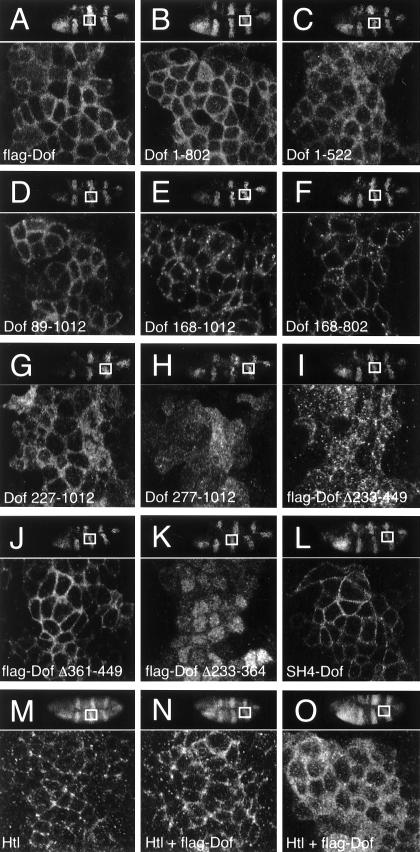
We examined whether the truncations and deletions within Dof altered the subcellular localization of the protein. The wild-type protein is observed in a punctate pattern within the cytoplasm of mesodermal, glial, and tracheal cells and is slightly enriched at the cell cortex (39). An identical or similar subcellular distribution was also observed for full-length Dof with an N-terminal FLAG epitope tag upon expression of the protein in the ectoderm with prd-Gal4 and most of the mutant Dof proteins (Fig. 4A to C, E, and F). However, both flag-Dof[277-1012] and flag-Dof[Δ233-449] (Fig. 4H and I) were distributed in a punctate fashion throughout these cells, whereas flag-Dof[Δ361-449] (Fig. 4J) appeared to be more tightly associated with the cell cortex than the wild-type protein and comparable in distribution to full-length Dof protein fused to the C terminus of the SH4 domain of human Src (Fig. 4L). Surprisingly, flag-Dof[Δ233-364] appeared to be enriched within nuclei and only present at low levels in the cytoplasm (Fig. 4K; for double labeling with markers for the nuclear envelope and the plasma membrane, see Fig. S2 in the supplemental material).
FIG. 4.
Subcellular localization of mutant Dof proteins and influence of Dof on localization of ectopically expressed Htl. (A to L) The stabilities and subcellular localization of mutant forms of Dof were assessed using a prd-Gal4 transgene to express each construct in ectodermal cells. The expression and subcellular localization of the mutant proteins were followed by staining fixed embryos with an anti-Dof antiserum. (M to O) A transgene encoding Htl was expressed using prd-Gal4 either alone (M) or in combination with a transgene encoding Dof with an N-terminal flag epitope tag (N and O). The subcellular distribution of Htl (M and N) was visualized by staining the embryos with an anti-Htl antiserum, and the localization of Dof (O) was determined as described above. All embryos depicted are of the same age (stage 10). The orientations of the embryos are shown in the top right corner of each panel; the white boxes indicate the regions (39 by 35 μm) that were photographed at high magnification. The proteins ectopically expressed are indicated in each panel.
There are several molecules involved in signal transduction, such as Corkscrew, the Drosophila SHP-2 homolog, and Raf, which become active when recruited to the plasma membrane. To investigate whether high levels of Dof at the plasma membrane are sufficient to rescue the defects observed in homozygous dof mutant embryos, we expressed variants of Dof with an N-terminal SH4 epitope tag. When targeted to the plasma membrane in this way (Fig. 4L), both the full-length Dof protein and the C-terminal truncation lacking the last 210 amino acids gave a good rescue (Fig. 2K and 3). However, we found that targeting Dof to the membrane was not sufficient to induce signaling independent of an active receptor, since the SH4-Dof fusion proteins did not rescue homozygous btl-dof double-mutant embryos, which lack both the FGF receptor Btl and Dof. Similar results were also obtained with Torso4021-Dof chimeras (Fig. 3).
Interaction of Dof with the FGF receptors
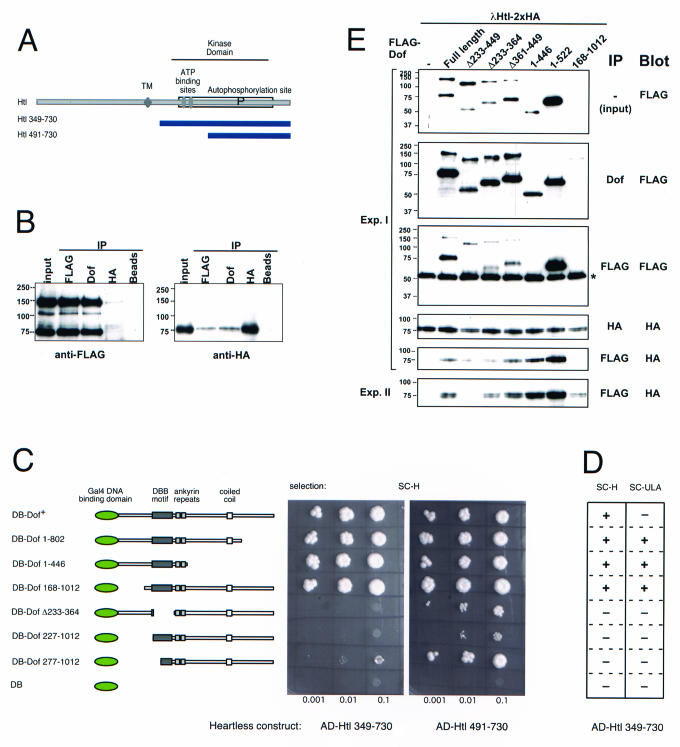
In a yeast two-hybrid screen, in which we used Dof[1-802] as bait (4), we found a number of proteins that interact with Dof, including the FGF receptor Heartless (Fig. 5A). Dof[1-802] can interact with a fragment covering most of the intracellular domain of Heartless, as well as a fragment lacking most of the smaller lobe of the kinase domain. We confirmed the interaction by coimmunoprecipitation from lysates of Schneider S2 cells expressing Dof and the activated FGF receptor (Fig. 5B).
FIG. 5.
Interaction of the FGF receptor Heartless with mutant forms of Dof. (A) Diagram of Heartless (accession number Q07407) showing the transmembrane domain and the ATP binding and autophosphorylation sites. The two blue bars represent the longest and shortest regions encoded by heartless clones isolated in a yeast two-hybrid screen with Dof (the fusion of the Gal4 activation domain with Htl corresponds to amino acids 349 and 491, respectively). TM, transmembrane region. (B) Coimmunoprecipitation of Dof with the FGF receptor Htl. Lysates from Schneider S2 cells coexpressing a constitutively active, hemagglutinin (HA)-tagged form of Htl (lambda-Htl) and a Flag-tagged full-length Dof protein were used for immunoprecipitation (IP) with antibody against FLAG, Dof, or HA or control beads without antibodies. The immunoprecipitates were analyzed by SDS-PAGE together with the whole-cell lysate (input). Western blots were stained with antibodies against Flag to visualize Dof (left) and against HA to visualize the FGF receptor (right). In addition to the band corresponding to the full-length Flag-tagged Dof molecule at ∼130 kDa, a breakdown product at 70 kDa is usually seen. (C and D) Growth of yeast clones coexpressing Dof mutants fused to the Gal4 DNA binding domain (DB) and fragments of Htl fused to the Gal4 activation domain (AD). The different Dof mutants tested are shown schematically on the left (see Fig. 3 for details). Serial dilutions of cell suspensions were spotted onto plates containing the indicated selection media (His selection, SC-H, or the more stringent Ade selection, SC-ULA) and grown for 3 days. In panel D, the results of the comparison of the behavior of the long fragment of Htl under the two selection conditions are shown schematically. +, strong growth at all three densities; −, no detectable growth at the lower densities and minimal or no growth at the highest density. (E) Western blots of whole-cell lysates (input) and immunoprecipitations from Schneider S2 cells expressing a constitutively active HA-tagged FGF receptor (λHtl) and Flag-tagged mutated versions of Dof. Protein complexes were immunoprecipitated from the lysates using antibody against Dof, Flag, or HA, and the precipitated proteins were detected on the Western blots using antibody against Flag or HA. The asterisk marks the immunoglobulin G heavy chain of the Flag antibody used for immunoprecipitation. In addition to a protein of approximately the size predicted for each of the constructs, in each lane there is also a smaller fragment representing a breakdown product. The sizes of these smaller fragments are in all cases consistent with a cleavage of Dof at the same position between the ankyrin repeat and the coiled-coil region. The five top panels are from the same experiment. The bottom panel is from a separate experiment to show the variation in the amount of DofΔ233-449 associated with the receptor. The expression levels in this experiment were the same as in experiment (Exp.) I. Note that although Dof[168-1012] is not detectable in the whole-cell lysate at this exposure, a certain amount is precipitable and (in experiment II) is able to coprecipitate the receptor.
We used the yeast two-hybrid assay to investigate which regions of Dof are required for the interaction with Heartless. When we tested the Dof deletion constructs in this assay, we found that the first 446 or the last 844 amino acids of Dof were able to interact with both fragments of Htl (Fig. 5C). However, the constructs with more extensive N-terminal deletions or constructs lacking the DBB motif were unable to interact with the long form of Htl, and their interaction with the short form of Htl was significantly reduced. This resembled results from coimmunoprecipitation of the Dof deletion constructs with the activated receptor from Schneider S2 cell lysates (Fig. 5E). Deletions that did not affect the DBB region had minimal effects on the interaction (note that even Dof[168-1012], although expressed at very low levels, is able to coprecipitate the receptor, albeit at similarly low levels). By contrast, constructs lacking the DBB region alone or in combination with the ankyrin repeat showed strongly reduced or even abolished binding, with a certain degree of experimental variation.
The long fragment of Htl interacts with the C-terminally truncated DB-Dof[1-802] under both low- and high-stringency selection conditions. Surprisingly, the full-length form of Dof could interact only under the less stringent selection conditions (Fig. 5D), indicating that the C terminus of Dof can weaken the interaction of Dof with Heartless (4).
Thus, in summary, the integrity of the region that lies between amino acids 168 and 364, which spans the DBB domain, is critical for the efficient interaction of Dof with the cytoplasmic domain of an FGF receptor in Drosophila. Whether the ability of the DBB deletion mutants to interact weakly with the activated receptor in Schneider S2 cells and the large lobe of the kinase domain in the yeast assay, but not with the whole kinase domain in yeast, is a reflection of a difference in affinity for the activated and nonactivated kinase is an interesting problem that remains to be elucidated.
We investigated whether Dof influences the distribution of Htl protein expressed ectopically in the ectodermal cells. In the absence of Dof, we observed Htl largely in a punctate pattern at the membranes of these cells (Fig. 4M), and coexpression of Dof did not change this distribution significantly (Fig. 4N), nor did the presence of the FGF receptor have any gross effect upon the distribution of Dof in these cells (compare Fig. 4A and O). Thus, the subcellular localizations of Htl and Dof are independent of each other and are not dependent upon an interaction between the proteins.
Dof is phosphorylated by the FGF receptors
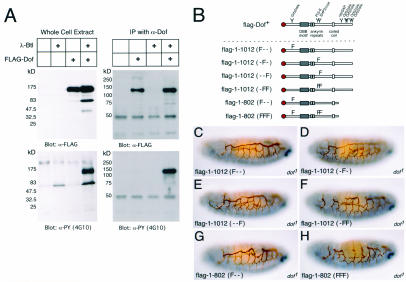
To investigate whether Dof could be phosphorylated in the presence of activated FGF receptors, we expressed the protein in Drosophila S2 cells and examined the effect of adding an activated form of the FGF receptor Btl. A phosphorylated protein with an apparent molecular mass of roughly 130 kDa was present specifically in cells expressing both the activated FGF receptor and Dof (Fig. 6A, left) (the predicted molecular mass for Dof with the Flag tag is 112.8 kDa). We confirmed that this protein was indeed Dof by immunoprecipitation with an anti-Dof antiserum followed by Western blotting with an anti-phosphotyrosine antibody (Fig. 6A, right). Thus, these experiments indicate that Dof is phosphorylated in the presence of an active FGF receptor, although not whether the phosphorylation is mediated directly by the FGF receptors.
FIG. 6.
Dof phosphorylation in response to FGF receptor activation and the effect of specific tyrosine mutations on the function of Dof. (A) Drosophila Schneider S2 cell lines were stably transformed as indicated above each lane with different combinations of an activated form of the FGF receptor Btl under the control of a heat shock promoter (λ-Btl) and Dof with an N-terminal FLAG epitope tag under the control of the actin5C promoter (FLAG-Dof). The presence of Dof and proteins with phosphorylated tyrosine residues in whole-cell lysates of these cell lines, previously induced to express the FGF receptor (left), or immunoprecipitates (IP) of the induced lysates with an antiserum directed against Dof (α-Dof) (right) was monitored by Western blotting with antibodies directed against the FLAG epitope tag (M5) (α-FLAG) and phosphotyrosine (4G10) (α-PY). The sizes of the proteins detected are indicated on the left. Dof is seen at an apparent molecular mass of ∼130 kDa. Often, an additional band representing a breakdown product is seen at ∼70 kDa. (B) Schematic representations of Dof constructs. The name of a signaling protein and a Y indicates the position of a tyrosine in Dof that is a potential binding site for this molecule. An F indicates a specific mutation that results in a protein containing a phenylalanine residue rather than a tyrosine; different combinations of mutations in amino acids 97, 486, and 515 are shown. (C to H) Tracheal development of homozygous dof mutant embryos expressing the constructs flag-Dof <Y95F> (C), flag-Dof <Y486F> (D), flag-Dof <Y515F> (E), flag-Dof <Y486F, Y515F> (F), flag-Dof[1-802] <Y97F> (G), and flag-Dof[1-802] <Y97F, Y486F, Y515F> (H). The embryos are stained for 2A12 in brown and Even-skipped in blue to distinguish the tracheae and the genotype, respectively, as described in the legend to Fig. 2.
The FGF receptors require Dof to generate high levels of diphosphorylated MAP kinase (20, 27, 39). The analysis of the truncated forms of Dof described above indicates that the last 210 amino acids are not essential for the function of the molecule. Thus, we focused our attention upon the potential binding sites for Drk (Grb2), PI 3-kinase, and Corkscrew (SHP2) that are present at positions 97, 486, and 515 of Dof, respectively (Fig. 6B).We constructed a number of transgenes with different combinations of mutations that result in the production of proteins containing phenylalanine residues in place of these tyrosines (Fig. 6B). When expressed in Schneider S2 cells, these constructs are stably expressed at levels comparable to those of the full-length wild-type protein (see Fig. S3 in the supplemental material). The level of phosphorylation of these constructs is reduced compared to the full-length wild-type protein, but even in Dof1-802(FFF), phosphorylation is not completely abolished (see Fig. S3 in the supplemental material). This suggests that tyrosines other than those that lie within recognizable consensus binding sites for known adapter proteins can also be phosphorylated. Comparison of the phosphorylation states of the deletion mutants (see Fig. S3 in the supplemental material) indicates that these tyrosines lie in the region between amino acids 522 and 802, which contains seven tyrosines.
In the tracheal assay, the mutation of individual sites had little effect on the activity of Dof, irrespective of whether the full-length or the truncated form was used (data summarized in Fig. 3 and 6C, D, E, and G). However, the combination of these mutations gave a less reliable rescue (Fig. 6F), and the mutation of all three tyrosines in a transgene that lacked the C terminus reduced the activity in this assay further (Fig. 6H). In the mesoderm assay, the single mutations of the potential Drk and PI 3-kinase binding sites also had little effect (Fig. 3). However, constructs in which the potential Corkscrew binding site was mutated were compromised in the ability to rescue the expression of Even-skipped, and a transgene lacking the C terminus and with mutations in all three tyrosines had much less activity. Taken together, these results indicate that the most important of the potential phosphorylation sites is Y515, suggesting that one function of Dof may be to recruit Corkscrew/SHP-2 upon the activation of an FGF receptor.
DISCUSSION
Dof has been proposed to mediate the transmission of a signal from an activated receptor to other components of the cell, including the MAP kinase cascade (20, 27, 39). We can envisage three different ways in which Dof could function, all of which are consistent with the physical interaction of Dof with the FGF receptors.
First, Dof could be required for the transport of the FGF receptors to the cell surface. However, the detection of Heartless at the peripheries of cells in the absence of Dof argues against such a function. Second, Dof may have a role in the activation of the receptors. It could, for example, facilitate or stabilize conformational changes or autophosphorylation of the receptors. We have no data that would specifically support this model, but it is not ruled out by any of our results. Finally, Dof is phosphorylated in the presence of an activated FGF receptor and could be involved in transmission of the signal from the receptors.
Although Dof is a large protein, the only motifs that can be identified in the primary sequence are two ankyrin repeats and a coiled coil (39). Comparison of the Drosophila protein with its Anopheles homolog and the most closely related vertebrate proteins, BCAP and BANK (30, 45), suggests that dof may be an example of a fast-evolving gene used for FGF signaling in Drosophila but which has acquired a novel function with the development of the immune system in higher vertebrates. Dof shares a number of distinct parts with these proteins, namely, the ankyrin repeats, the coiled coil, and the region adjacent to the ankyrin repeats, which we have called the DBB motif. Surprisingly, despite the conservation of these domains, only the DBB domain appears to be indispensable for FGF signaling in Drosophila. In this respect, it is interesting that the association of BANK with the IP-3 receptor, which stimulates the release of calcium from intracellular stores upon activation of the B-cell receptor, also depends upon the N-terminal part of the protein but not upon the ankyrin repeats that are present within this region (45). However, two caveats apply to the in vivo assays that we used to test the function of Dof. First, there are aspects of FGF signaling besides those examined in our assays, such as feedback regulation (8, 16, 29) and the response to oxygen deprivation (22), which could be affected by the Dof mutations. Second, we determined the consequence of a particular deletion based on the overexpression of the mutant protein, and this may have masked certain physiological requirements for particular domains of the protein. Thus, we may not have identified all of the functional domains of Dof; nevertheless, this approach has revealed a critical part of the protein.
The most important region for the function of Dof corresponded to the DBB motif, which is critical for FGF signaling and for the efficient interaction of Dof with the receptor. Dof mutants with deletions that disrupted the DBB motif had only miminal biological activity and were no longer capable of interacting efficiently with the FGF receptor. These observations suggest that the DBB domain interacts directly with the FGF receptors. This is unlikely to be the only function of the DBB motif, since this domain is conserved in BCAP and BANK, which are expressed in B cells and macrophages and are required for the function of the B-cell receptor and thus are unlikely to interact with FGF receptors. Indeed, we have found that the DBB domain in both Dof and BCAP is required to mediate self-association in yeast cells, indicating that this domain may have a more general role in mediating protein-protein interactions (4).
Constructs lacking this domain still provide above-background biological activity, suggesting that perhaps other parts of the molecule participate in receptor binding, allowing residual signal transmission in the absence of the DBB. Conversely, the DBB region in itself is clearly not sufficient for transmission of the signal, showing that other essential functions reside elsewhere in the molecule. The smallest N-terminal fragment of Dof with biological activity was Dof[1-522]. A mutant with a more extensive C-terminal truncation, Dof[1-446], had no biological activity but was still able to interact with the cytoplasmic domain of the FGF receptor Htl. Together, these findings imply that in addition to the DBB domain there are essential properties of Dof that are located within the 76 amino acids between residues 446 and 522. Intriguingly, there are two tyrosine residues in this region that are potential binding sites for PI 3-kinase and Corkscrew (39). The mutation of the potential Corkscrew binding site had a clear effect upon the activation of Even-skipped within the mesoderm, suggesting that this binding site contributes to the function of the molecule.
In summary, we have shown that two parts of Dof are important for its function. The DBB motif is necessary for the efficient interaction of Dof with the receptor, and tyrosines between the ankyrin repeat and the coiled-coil region also contribute to the function of Dof, possibly by recruiting Csw. Thus, similar to FRS2 in vertebrate FGF signal transmission, Dof uses a protein-protein interaction domain to interact with the receptor and acts as a substrate for phosphorylation, and it can therefore recruit other signaling molecules. It is intriguing that although the FGF signaling pathway must have existed in the common precursor of insects and vertebrates, different molecules have taken on this role in the two lineages, while their respective homologs in the other lineage have diverged in sequence and function.
Supplementary Material
Acknowledgments
We thank N. Brown, M. Frasch, R. Herbst, P. James, A. Michelson, D. Montell, N. Patel, K. Saigo, M. Simon, F. Sprenger, and T. Kurosaki for providing reagents used in this work. We are also very grateful to the E. coli Genetic Stock Center (http://cgsc.biology.yale.edu/) for the strain ES455. Flybase (http://flybase.bio.indiana.edu/) was used as a source for information throughout this work. A. Michelson and F. Sprenger were kind enough to read the manuscript, which benefited as a result of their comments and suggestions.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (LE 546/2-2 and LE 546/3-1).
Footnotes
Present address: School of Biological Sciences, University of Wales—Bangor, Bangor LL57 2UW, United Kingdom.
REFERENCE
- 1.Allard, J. D., H. C. Chang, R. Herbst, H. McNeill, and M. A. Simon. 1996. The SH2-containing tyrosine phosphatase corkscrew is required during signaling by sevenless, Ras1 and Raf. Development 122:1137-1146. [DOI] [PubMed] [Google Scholar]
- 2.Azpiazu, N., P. A. Lawrence, J.-P. Vincent, and M. Frasch. 1996. Segmentation and specification of the Drosophila mesoderm. Genes Dev. 10:3183-3194. [DOI] [PubMed] [Google Scholar]
- 3.Bate, M. and A. Martinez Arias. 1993. The development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 4.Battersby, A., A. Csiszar, M. Leptin, and R. Wilson. 2003. Isolation of proteins that interact with the signal transduction molecule Dof and identification of a functional domain conserved between Dof and vertebrate BCAP. J. Mol. Biol. 329:479-493. [DOI] [PubMed] [Google Scholar]
- 5.Beiman, M., B. Z. Shilo, and T. Volk. 1996. Heartless, a Drosophila FGF receptor homolog, is essential for cell migration and establishment of several mesodermal lineages. Genes Dev. 10:2993-3002. [DOI] [PubMed] [Google Scholar]
- 6.Benson, D. A., I. Karsch-Mizrachi, D. Lipman, J. Ostell, B. A. Rapp, and D. A. Wheeler. 2002. GenBank. Nucleic Acids Res. 30:17-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brand, A. H., and N. Perrimon. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401-415. [DOI] [PubMed] [Google Scholar]
- 8.Casci, T., J. Vinos, and M. Freeman. 1999. Sprouty, an intracellular inhibitor of Ras signaling. Cell 96:655-665. [DOI] [PubMed] [Google Scholar]
- 9.Cerbas, L., R. Moss, and P. Cerbas. 1994. Transformation techniques for Drosophila cell lines. Methods Cell Biol. 44:161-179. [DOI] [PubMed] [Google Scholar]
- 10.Dhalluin, C., K. Yan, O. Plotnikova, K. W. Lee, L. Zeng, M. Kuti, S. Mujtaba, M. P. Goldfarb, and M. M. Zhou. 2000. Structural basis of SNT PTB domain interactions with distinct neurotrophic receptors. Mol. Cell 6:921-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flybase Consortium. 2002. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 30:106-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forbes, A., and R. Lehmann. 1999. Cell migration in Drosophila. Curr. Opin. Genet. Dev. 9:473-478. [DOI] [PubMed] [Google Scholar]
- 13.Frasch, M., T. Hoey, C. Rushlow, H. Doyle, and M. Levine. 1987. Characterization and localization of the even-skipped protein of Drosophila. EMBO J. 6:749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freeman, M., C. Klämbt, C. S. Goodman, and G. M. Rubin. 1992. The argos gene encodes a diffusible factor that regulates cell fate decisions in the Drosophila eye. Cell 69:963-975. [DOI] [PubMed] [Google Scholar]
- 15.Gisselbrecht, S., J. B. Skeath, C. Q. Doe, and A. M. Michelson. 1996. heartless encodes a fibroblast growth factor receptor (DFR1/DFGF-R2) involved in the directional migration of early mesodermal cells in the Drosophila embryo. Genes Dev. 10:3003-3017. [DOI] [PubMed] [Google Scholar]
- 16.Hacohen, N., S. Kramer, D. Sutherland, Y. Hiromi, and M. A. Krasnow. 1998. sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell 92:253-263. [DOI] [PubMed] [Google Scholar]
- 17.Hadari, Y. R., N. Gotoh, H. Kouhara, I. Lax, and J. Schlessinger. 2001. Critical role for the docking-protein FRS2 alpha in FGF receptor-mediated signal transduction pathways. Proc. Natl. Acad. Sci. USA 98:8578-8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hadari, Y. R., H. Kouhara, I. Lax, and J. Schlessinger. 1998. Binding of Shp2 tyrosine phosphatase to FRS2 is essential for fibroblast growth factor-induced PC12 cell differentiation. Mol. Cell. Biol. 18:3966-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hubbard, T., D. Barker, E. Birney, G. Cameron, Y. Chen, L. Clark, T. Cox, J. Cuff, V. Curwen, T. Down, et al. 2002. The Ensembl genome database project. Nucleic Acids Res. 30:38-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imam, F., D. Sutherland, W. Huang, and M. A. Krasnow. 1999. stumps, a Drosophila gene required for fibroblast growth factor (FGF)-directed migrations of tracheal and mesodermal cells. Genetics 152:307-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarecki, J., E. Johnson, and M. A. Krasnow. 1999. Oxygen regulation of airway branching in Drosophila is mediated by branchless FGF. Cell 99:211-220. [DOI] [PubMed] [Google Scholar]
- 23.Kouhara, H., Y. R. Hadari, T. Spivak-Kroizman, J. Schilling, D. Bar-Sagi, I. Lax, and J. Schlessinger. 1997. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell 89:693-702. [DOI] [PubMed] [Google Scholar]
- 24.Lee, T., N. Hacohen, M. Krasnow, and D. J. Montell. 1996. Regulated Breathless receptor tyrosine kinase activity required to pattern cell migration and branching in the Drosophila tracheal system. Genes Dev. 10:2912-2921. [DOI] [PubMed] [Google Scholar]
- 25.Lin, H. Y., J. Xu, I. Ischenko, D. M. Ornitz, S. Halegoua, and M. J. Hayman. 1998. Identification of the cytoplasmic regions of fibroblast growth factor (FGF) receptor 1 which play important roles in induction of neurite outgrowth in PC12 cells by FGF-1. Mol. Cell. Biol. 18:3762-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metzger, R. J., and M. A. Krasnow. 1999. Genetic control of branching morphogenesis. Science 284:1635-1639. [DOI] [PubMed] [Google Scholar]
- 27.Michelson, A. M., S. Gisselbrecht, E. Buff, and J. B. Skeath. 1998. Heartbroken is a specific downstream mediator of FGF receptor signalling in Drosophila. Development 125:4379-4389. [DOI] [PubMed] [Google Scholar]
- 28.Montell, D. J. 1999. The genetics of cell migration in Drosophila melanogaster and Caenorhabditis elegans development. Development 126:3035-3046. [DOI] [PubMed] [Google Scholar]
- 29.Ohshiro, T., Y. Emori, and K. Saigo. 2002. Ligand-dependent activation of breathless FGF receptor gene in Drosophila developing trachea. Mech. Dev. 114:3-11. [DOI] [PubMed] [Google Scholar]
- 30.Okada, T., A. Maeda, A. Iwamatsu, K. Gotoh, and T. Kurosaki. 2000. BCAP: the tyrosine kinase substrate that connects B cell receptor to phosphoinositide 3-kinase activation. Immunity 13:817-827. [DOI] [PubMed] [Google Scholar]
- 31.Olivier, J. P., T. Raabe, M. Henkemeyer, B. Dickson, G. Mbamalu, B. Margolis, J. Schlessinger, E. Hafen, and T. Pawson. 1993. A Drosophila SH2-SH3 adaptor protein implicated in coupling the sevenless tyrosine kinase to an activator of Ras guanine nucleotide exchange, Sos. Cell 73:179-191. [DOI] [PubMed] [Google Scholar]
- 32.Ong, S. H., G. R. Guy, Y. R. Hadari, S. Laks, N. Gotoh, J. Schlessinger, and I. Lax. 2000. FRS2 proteins recruit intracellular signaling pathways by binding to diverse targets on fibroblast growth factor and nerve growth factor receptors. Mol. Cell. Biol. 20:979-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ong, S. H., Y. R. Hadari, N. Gotoh, G. R. Guy, J. Schlessinger, and I. Lax. 2001. Stimulation of phosphatidylinositol 3-kinase by fibroblast growth factor receptors is mediated by coordinated recruitment of multiple docking proteins. Proc. Natl. Acad. Sci. USA 98:6074-6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ong, S. H., Y. P. Lim, B. C. Low, and G. R. Guy. 1997. SHP2 associates directly with tyrosine phosphorylated p90 (SNT) protein in FGF-stimulated cells. Biochem. Biophys. Res. Commun. 238:261-266. [DOI] [PubMed] [Google Scholar]
- 35.Seher, T. C., and M. Leptin. 2000. Tribbles, a cell-cycle brake that coordinates proliferation and morphogenesis during Drosophila gastrulation. Curr. Biol. 10:623-629. [DOI] [PubMed] [Google Scholar]
- 36.Shishido, E., N. Ono, T. Kojima, and K. Saigo. 1997. Requirements of DFR1/Heartless, a mesoderm-specific Drosophila FGF-receptor, for the formation of heart, visceral and somatic muscles, and ensheathing of longitudinal axon tracts in CNS. Development 124:2119-2128. [DOI] [PubMed] [Google Scholar]
- 37.Simon, M. A., G. S. Dodson, and G. M. Rubin. 1993. An SH3-SH2-SH3 protein is required for p21Ras1 activation and binds to Sevenless and Sos proteins in vitro. Cell 73:169-177. [DOI] [PubMed] [Google Scholar]
- 38.Sprenger, F., M. M. Trosclair, and D. K. Morrison. 1993. Biochemical analysis of Torso and D-Raf during Drosophila embryogenesis: implications for terminal signal transduction. Mol. Cell. Biol. 13:1163-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vincent, S., R. Wilson, C. Coelho, M. Affolter, and M. Leptin. 1998. The Drosophila protein Dof is specifically required for FGF signaling. Mol. Cell. 2:515-525. (Erratum, Mol. Cell 3:540, 1999.) [DOI] [PubMed] [Google Scholar]
- 40.Vinos, J., and M. Freeman. 2000. Evidence that Argos is an antagonistic ligand of the EGF receptor. Oncogene 19:3560-3562. [DOI] [PubMed] [Google Scholar]
- 41.Wang, J. K., H. Xu, H. C. Li, and M. Goldfarb. 1996. Broadly expressed SNT-like proteins link FGF receptor stimulation to activators of Ras. Oncogene 13:721-729. [PubMed] [Google Scholar]
- 42.Wilson, R., and M. Leptin. 2000. Fibroblast growth factor receptor-dependent morphogenesis of the Drosophila mesoderm. Phil. Trans. R. Soc. Lond. B 355:891-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu, H., K. W. Lee, and M. Goldfarb. 1998. Novel recognition motif on fibroblast growth factor receptor mediates direct association and activation of SNT adaptor proteins. J. Biol. Chem. 273:17987-17990. [DOI] [PubMed] [Google Scholar]
- 44.Yamazaki, T., K. Takeda, K. Gotoh, H. Takeshima, S. Akira, and T. Kurosaki. 2002. Essential immunoregulatory role for BCAP in B cell development and function. J. Exp. Med. 195:535-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yokoyama, K., I. Su Ih, T. Tezuka, T. Yasuda, K. Mikoshiba, A. Tarakhovsky, and T. Yamamoto. 2002. BANK regulates BCR-induced calcium mobilization by promoting tyrosine phosphorylation of IP(3) receptor. EMBO J. 21:83-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zdobnov, E. M., C. von Mering, I. Letunic, D. Torrents, M. Suyama, R. R. Copley, G. K. Christophides, D. Thomasova, R. A. Holt, G. M. Subramanian, et al. 2002. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science 298:149-159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.