Abstract
Nuclear export of the transcription factor Swi6 during the budding yeast Saccharomyces cerevisiae cell cycle is known to require phosphorylation of the Swi6 serine 160 residue. We show that Clb6/Cdc28 kinase is required for this nuclear export. Furthermore, Cdc28 combined with the S-phase cyclin Clb6 specifically phosphorylates serine 160 of Swi6 in vitro. Nuclear import of Swi6 occurs concomitantly with dephosphorylation of serine 160 in late M phase. We show that Cdc14 phosphatase, the principal effector of the mitotic exit network, can trigger nuclear import of Swi6 in vivo and that Cdc14 dephosphorylates Swi6 at serine 160 in vitro. Taken together, these observations show how Swi6 dephosphorylation and phosphorylation are integrated into changes of Cdc28 activity governing entry and exit from the G1 phase of the cell cycle.
In the G1 phase of the eukaryotic cell cycle, initiation of a new round cycle of division only occurs when environmental and physiological conditions are suitable. In Saccharomyces cerevisiae, this regulatory step, called Start, is marked by the expression of a large number of biosynthetic and regulatory genes under the regulation of the MBF and SBF transcription factors (18, 48). SBF and MBF are heteromeric complexes; Swi6 and Swi4 are combined in SBF, while Swi6 is complexed with Mbp1 in MBF. Swi4 and Mbp1 impart sequence-specific DNA binding (1, 20, 24, 34), while the Swi6 subunit provides both positive and negative regulatory activity (1, 10, 24, 45). Swi6 has several functional domains, including two regions with transcriptional activation activity, a C-terminal region for interaction with Swi4 and Mbp1, and N- and C-terminal regions for responding to Cln3 (44, 58). This multifunctionality is reflected at the structural level, where Swi6 has a modular organization with discrete structural units connected by unstructured linkers (44).
Like many other cell cycle events, passage through Start is marked by oscillations in the activity of cyclin-dependent kinases. SBF-regulated genes include CLN1 and CLN2, which encode G1 cyclins (28, 31) whose activity triggers degradation of Sic1 (41, 55). As Sic1 is an inhibitor of Clb/Cdc28 kinase, its degradation is important for the transition from Cln-dependent G1 to Clb-dependent S and M stages of the cell cycle. At the same time, MBF activity in G1 produces the S-phase cyclins Clb5 and Clb6 (11, 22, 42). With the elimination of Sic1, S-phase Clb/Cdc28 acts through multiple mechanisms to stimulate formation of prereplicative complexes (reviewed in reference 19) and, by phosphorylation of Cdc6, Orc, and Mcm, to prevent rereplication from activated origins (reference 29 and references therein).
Clearly, stimulation of the transcriptional activity of SBF and MBF is a pivotal regulatory step in initiating this cascade of events. However, the regulatory mechanisms governing SBF and MBF activity are complex, genetically redundant, and not fully understood. Swi6 is envisaged as the regulatory subunit, but its levels are only weakly cell cycle dependent and only SWI4 displays significant cell cycle-regulated expression (6). SBF and MBF become associated with their promoters in early G1 (7, 15, 21). Although Swi6 does not bind DNA, Swi4 binding to DNA requires interaction with Swi6 to release an autoinhibitory effect of the C terminus of Swi4 on its N-terminal DNA binding domain (2).
However, DNA binding alone is insufficient to account for regulation of SBF and MBF, as several other regulatory factors, some of which act through Swi6, are still required for activation of transcription (9, 12, 16, 25, 50, 57, 58). Of these, the primary activator is the Cln3/Cdc28 kinase complex (21, 50, 53, 58). Intriguingly, phosphorylation of Swi6 is not required for Cln3/Cdc28 to exert its regulatory activities on cell size and on the execution of Start (58). Nevertheless, phosphorylation of Swi6 has long been suspected as a possible regulatory mechanism (51). Indeed, Swi6 is subject to multiple phosphorylation events (17, 25, 45, 60), but only phosphorylation at serine 160 is cell cycle dependent. Phosphorylation at serine 160 is required for nuclear export of Swi6 from the end of G1 to M phase, while dephosphorylation in late mitosis is needed for nuclear import. Serine 160 is located in a Cdc28 consensus motif, but the kinase responsible for its phosphorylation has not been identified (46). Export of Swi6 to the cytoplasm requires the karyopherin Msn5 and is required for an uncharacterized licensing step that is required for subsequent SBF- but not MBF-dependent gene expression (36). The timing of nuclear import in late M phase coincides with the elimination of mitotic cyclin activity by the phosphatase Cdc14 (56). Cdc14 activity is in turn regulated by the FEAR pathway and the mitotic exit network (49; reviewed in reference 3).
Given this plethora of regulatory processes, phosphorylation at serine 160 remains the only cell cycle-specific change in Swi6 that has been linked with a change in cell cycle behavior. Therefore, to understand further Swi6 regulation, we have investigated which kinase phosphorylates Swi6 at serine 160 to control its cellular localization. We show that Cdc28 and the S-phase cyclin Clb6 specifically phosphorylate Swi6 at serine 160 in vitro and also direct nuclear export of Swi6 in vivo. We propose that Clb6 not only facilitates DNA replication but, by stimulating export of Swi6, curtails further G1 activity. We also show that the mitotic exit network effector Cdc14 can stimulate nuclear import of Swi6. Therefore, in addition to regulating the final stages of mitosis and cytokinesis, the mitotic exit network also primes G1 by regulating nuclear import of a key, G1-specific transcription factor, Swi6.
MATERIALS AND METHOD
Strains, culture, and media
SWI6 sequences were synthesized by PCR of yeast genomic DNA and cloned into the hexahistidinyl tagging plasmid pET22b. PCR fragments encoding CLB1, CLB2, CLB3, CLB4, CLB5, CLB6, CLN1, CLN2, and CLN3 were cloned and expressed as glutathione S-transferase (GST)-cyclin-6-His fusions in pGEX-KG. E. Randle kindly provided pMAL-2c (New England Biolabs) expressing MBP-Cdc14 and MBP-Cdc14 C283A. Cloned gene expression in Escherichia coli BL21(DE3) was induced by 3 h of growth in LB broth containing 0.3 μM isopropyl β-d-thiogalactopyranoside. pESC-GST.28 has an in-frame fusion of GST and CDC28 coding sequences cloned 3′ to the galactose-inducible promoter of pESC-URA (Stratagene). pESC-GST.28-13 was similarly produced by PCR amplification from cdc28-13 genomic DNA.
S. cerevisiae MGY2 contains pESC-GST.28 and MGY15 contains pESC-GST.28-13, and both have chromosomal CDC28::LEU2 deletions to enforce selection of pESC-GST.28 and pESC-GST.28-13 in rich YEP medium with no further plasmid selection. Cyclin-defective derivatives of MGY2 were made by transformation with linear knockout cassettes made by PCR (5). Expression of GST-Cdc28 and GST-Cdc28-13 was induced by addition of 2% galactose to cells growing in YEP-sucrose for 6 h at 30°C or 25°C. 3′ fusions of genomic SWI6 with green fluorescent protein (GFP) or 13Myc were constructed by using PCR-tagging cassettes (23). pCDC14yexEMBL expressing galactose-inducible Cdc14 was from S. Jensen.
Protein purification and β-galactosidase assays
Frozen pellets of yeast cells were thawed, resuspended in cold lysis buffer (50 mM Tris, pH 7.5, 250 mM NaCl, 1% NP-40, 1 mM EDTA, 1 mM dithiothreitol, 10 mM NaF, 50 mM β-glycerophosphate, 1 mM sodium orthovanadate, and Complete protease inhibitor cocktail [Roche]), passed twice through a French pressure cell, and centrifuged at 18,000 x g at 4°C for 45 min. In the “low-salt” purification, the supernatant was mixed with glutathione-Sepharose beads at 4°C. After 1 h, a column of beads was washed with 50 mM Tris (pH 7.5)-250 mM NaCl-1% NP-40-1 mM dithiothreitol. GST-Cdc28 protein was eluted in 50 mM Tris (pH 7.5)-100 mM NaCl-0.1% NP-40-1 mM dithiothreitol-15 mM reduced glutathione. In the “high-salt” purification procedure, (NH4)2SO4 was added to 50% saturation (52). Precipitation was for 1 h on ice. The pellet was dissolved in lysis buffer, and GST-Cdc28 was purified by affinity binding to GST-Sepharose as described above. Eluted protein was dialyzed overnight against 50 mM Tris (pH 7.5)-100 mM NaCl-1 mM dithiothreitol-10% glycerol and stored at −80°C.
For proteins expressed in E. coli, GST fusion proteins were purified by affinity to glutathione-Sepharose beads (14), hexahistidinyl-tagged proteins were purified by affinity to nickel-agarose beads (Qiagen), and maltose binding protein (MBP) fusion proteins were purified by affinity binding to amylose resin (New England Biolabs). Swi6-driven gene expression was assayed in extracts from cells carrying pTR339, which is a triple-SCB lacZ reporter plasmid (constructed by L. Johnston) transposed in HIS3 with Tn1000TRP1. β-Galactosidase was assayed spectroscopically at 420 nm with o-nitrophenyl β-d-galactopyranoside and quantified in arbitrary units normalized for protein concentration and time of reaction.
Swi6/cyclin binding
Binding of Swi6 to glutathione-Sepharose beads loaded with GST-cyclin fusions was assayed as described by Hagemeier et al. (14). After electrophoresis through sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE), separated proteins were transferred to a Protran nitrocellulose membrane (Schleicher and Schuell). Anti-Swi6 rabbit polyclonal antibodies were produced against the full-size six-His-tagged Swi6 protein by G. Banks and A. Spanos (unpublished data). Secondary antibodies were a goat anti-rabbit-peroxidase conjugate (Sigma) that were revealed by chemiluminescence (Amersham).
Cdc28 kinase assays
Kinase assays in 30 μl of kinase buffer (50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM dithiothreitol) contained 1 to 10 μg of substrate. Following addition of kinase, 50 μM ATP and 0.1 μl of [γ-32P]ATP (Amersham, 370 MBq/ml, 3,000 Ci/mmol) were added. The assays were then incubated at 30°C for 15 min. For cyclin stimulation of kinase activity, kinase and cyclins were mixed together on ice for 10 min before addition to the reaction mixture. Reactions were inactivated by addition of 10 μl of 4× Laemmli buffer and heating at 100°C for 2 min. After SDS-PAGE, radiolabeling was visualized by autoradiography. Quantification of radiolabel in protein bands was performed with a Storm 860 PhosphorImager (Amersham) and ImageQuant software.
Cdc14 phosphatase assays
Full-length Swi6-six-His bound to Ni-agarose beads (Qiagen) was washed twice with kinase buffer and resuspended in an equal volume of kinase buffer; 200 μl of beads was suspended in 400 μl of kinase buffer containing 100 μM ATP and 5 μl of [γ-32P]ATP (Amersham, 370 MBq/ml, 3,000 Ci/mmol) and 20 μl of the low-salt Cdc28 kinase preparation. After 60 min at 30°C, the beads were washed four times with 4 ml of kinase buffer and resuspended in an equal volume of 50 mM Tris-HCl, pH 7.5. Dephosphorylation assays were carried out for 60 min at 30°C in 100 μl of phosphatase buffer (50 mM Tris-HCl, pH 8.0, 1 mM EDTA, and 1 mM dithiothreitol) containing 3 to 5 μg of 32P-labeled phosphorylated Swi6 attached to Ni-agarose beads and 1.5 μg of MBP-Cdc14 or MBP-Cdc14 C283A. Dephosphorylation by 2 U of calf intestine alkaline phosphatase (Boehringer Mannheim) was also assayed in the manufacturer's own phosphatase buffer for 60 min at 30°C. After washing three times with phosphatase buffer, the treated Swi6-Ni-agarose beads were resuspended in 100 μl of SDS-PAGE sample buffer and heated at 100°C for 2 min; 50 μl of the supernatant was analyzed on an SDS-10% PAGE gel. Proteins were visualized by staining with Coomassie blue, and their radioactive content was detected by autoradiography.
Cell biology protocols and fluorescence microscopy
Immunolocalization of Swi6-13Myc was performed essentially as detailed before (35) with 9E10 primary antibody and secondary tetramethylrhodamine isothiocyanate-conjugated anti-mouse immunoglobulin (Jackson ImmunoResearch Laboratories). Fluorescence microscopy was performed with a Photometrics CH350L liquid-cooled charge-coupled device camera on an Olympus IX70 inverted microscope with a 100× objective. Cell images were captured and manipulated with SoftWoRx software (Applied Precision Inc., Issaquah, Wash.) and Adobe Photoshop version 6.0 (Adobe Systems Inc.).
SWI6-GFP pCDC14yexEMBL cells were cultivated overnight in YNB selective medium with 2% sucrose as the carbon source. The cells were reinoculated in 2% sucrose YEP rich medium and cultivated for 1.5 h before 15 μg of nocodazole per ml was added for a further 2 h of incubation. The culture was divided and 2% galactose or 2% dextrose was added. After a final 2 h of cultivation, the cells were harvested and examined for DNA content and GFP fluorescence as described above.
RESULT
Phosphorylation of Swi6 by Cdc28 at serine 160
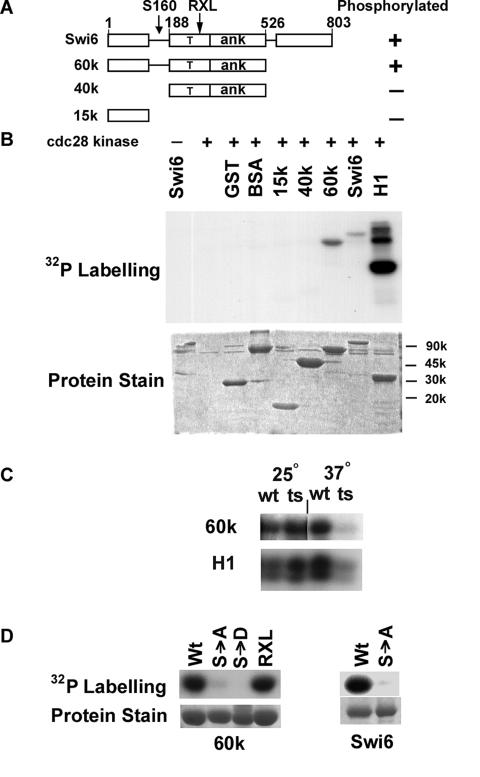
Unphosphorylated full-length six-His-Swi6 and deletion derivatives were prepared from E. coli and used as substrates for kinase assays with Cdc28 (Fig. 1A). Cdc28 was prepared under low-salt conditions from MGY2 yeast to favor copurification of accessory proteins such as cyclins that are essential for kinase activity. Purified Cdc28 was able to phosphorylate full-length Swi6 and control histone H1 (Fig. 1B). To confirm the Cdc28 specificity of this kinase preparation, the activities of purified wild-type Cdc28 and thermosensitive Cdc28-13 were compared (Fig. 1C). Levels of phosphorylation of Swi6 and histone H1 by Cdc28 and Cdc28-13 at 25°C were similar. In contrast, at 37°C the activity of Cdc28-13 was markedly decreased compared to wild-type protein. Thus, Cdc28 is the major kinase that phosphorylates Swi6 in our preparations.
FIG. 1.
Cdc28 phosphorylates Swi6 at serine 160. (A) Modular structure of Swi6 with amino acid residues numbered showing protein fragments used as substrates for Cdc28 kinase. Phosphorylation is indicated by + or −. The arrows indicate the location of serine 160 in a flexible linker region and an RXL motif at residues 256 to 258. T, transcriptional activation region; ANK, ankyrin repeat domain. (B) Upper panel, autoradiogram showing phosphorylation of the substrates indicated by low-salt, cyclin-proficient preparations of Cdc28. Lower panel, Coomassie blue staining of input proteins. BSA, bovine serum albumin; H1, histone H1. (C) Phosphorylation of the Swi6-60k fragment and histone H1 by wild-type (wt) Cdc28 and thermosensitive (ts) Cdc28-13 at 25°C and 37°C. (D) Comparison of phosphorylation by Cdc28 of wild-type and mutant forms of Swi6-60k (left panel) and full-length Swi6 (right panel). S→A, serine 160 mutated to alanine; S→D, serine 160 mutated to aspartate; RXL, R256A L257A L258A triple mutant. Protein stain shows input protein substrates visualized by Coomassie blue staining.
To localize the site(s) of in vitro phosphorylation, approximately equal amounts of full-length Swi6 and Swi6 fragments containing different domains were tested as substrates for Cdc28 (Fig. 1A and 1B). Cdc28 phosphorylated full-length Swi6 and Swi6-60k, which terminates after the ankyrin repeat module (Fig. 1A). However, Cdc28 showed little activity towards a central 40-kDa fragment of Swi6, or to an N-terminal 15-kDa domain, or to GST and bovine serum albumin controls. Given the modular structure of Swi6 (Fig. 1A), a flexible linker region between residues 126 and 188 is the only part of the 60-kDa fragment not represented in either the 15-kDa or 40-kDa fragment. Therefore, by elimination, this linker region of Swi6 either contains the target for Cdc28 kinase activity or is needed for that activity to be targeted elsewhere.
The N-terminal linker region of Swi6 includes serine 160, which was earlier identified as the only residue of Swi6 to undergo cell cycle-dependent phosphorylation (46). As a test of whether serine 160 is required for phosphorylation by Cdc28 in vitro, S160 mutant derivatives of the full-length and 60k forms of Swi6 were tested for phosphorylation by Cdc28. Compared to normal Swi6, Cdc28 had little kinase activity towards full-size S160A Swi6 or towards the S160A and S160D forms of Swi6-60k. (Fig. 1D). Thus, Cdc28 phosphorylates Swi6 in vitro at the same S160 residue which, in vivo, shows cell cycle changes in phosphorylation.
An RXL motif is a feature for substrate recognition by certain cyclins (8, 40) and such a motif is present in Swi6 at residues 256 to 258. However, wild-type and RXL>AAA forms of Swi6-60k were phosphorylated with equal efficiency by purified Cdc28 (Fig. 1D).
Cyclin specificity of Swi6 phosphorylation
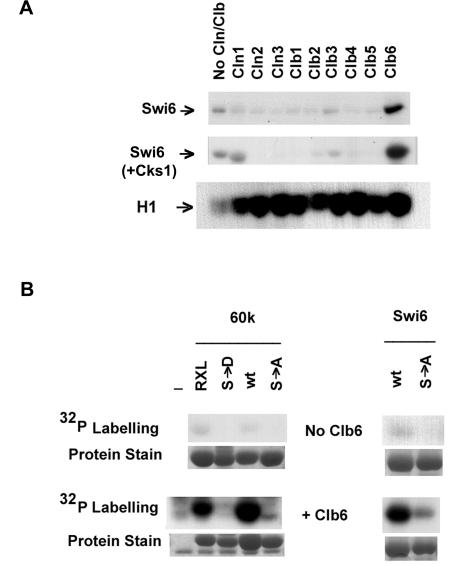
To determine the cyclin specificity for Swi6 phosphorylation, Cdc28 was prepared under high-salt conditions that reduce the levels of copurifying cyclins (52). With no additional cyclin, the high-salt preparation of Cdc28 showed relatively low levels of kinase activity towards Swi6, Swi6-60k, and histone H1 (Fig. 2). All nine Cdc28 cyclins were then prepared from E. coli and added individually to the high-salt kinase preparation to test for stimulation of phosphorylation of Swi6. The addition of affinity-purified GST-Clb6 greatly stimulated Cdc28 kinase activity towards Swi6 and Swi6-60k (Fig. 2). In contrast, no stimulation of phosphorylation of Swi6 occurred after addition of purified Cln1, Cln2, Cln3, Clb1, Clb2, Clb3, Clb4, and Clb5. Nevertheless, these cyclins were all active, as they all increased Cdc28 kinase activity towards the nonspecific substrate histone H1 (Fig. 2A). Incidentally, the cyclin-dependent increases in kinase activity seen in Fig. 2 further indicate that it was Cdc28 and not a contaminating kinase that was active in our preparations.
FIG. 2.
Phosphorylation of Swi6 at serine 160 depends on Clb6. (A) High-salt, cyclin-depleted preparations of Cdc28 were tested for phosphorylation of full-length Swi6 and histone H1 in the absence or presence of additional cyclins or Cks1 as indicated. (B) High-salt, cyclin-depleted preparations of Cdc28 were tested for phosphorylation of wild-type and mutant forms of full-length Swi6 and the Swi6-60k fragment in the absence or presence of additional Clb6. Abbreviations are as described in the legend for Fig. 1D. The protein stain shows input protein substrates visualized by Coomassie blue staining.
Optimal kinase activity of Cdc28 has been shown to require Cks1, at least in conjunction with G1 cyclins (39). To eliminate the possibility that cyclins other than Clb6 were unable to stimulate phosphorylation of Swi6 because of limiting Cks1 activity, kinase assays with Cdc28 and individual cyclins were repeated with additional affinity-purified Cks1 (Fig. 2A). However, even with Cks1, Swi6 was only phosphorylated efficiently by Clb6-Cdc28. Cks1 did not affect the phosphorylation of histone H1 (data not shown). Whether Cks1 was added or not, the addition of some cyclins other than Clb6 reduced the ability of Cdc28 to phosphorylate Swi6 but not histone H1. We interpret the reduction of activity to competitive binding of non-Swi6 specific cyclin that displaces the residual endogenous Clb6 that copurified with Cdc28.
The initial characterization of Cdc28 activity with no additional cyclin showed that serine 160 was the target for phosphorylation of Swi6 (Fig. 1D). To test whether the increased kinase activity brought about by Clb6 is also directed to serine 160, high-salt preparations of Cdc28 with and without additional Clb6 were assayed for kinase activity on wild-type and S160 mutant forms of Swi6. Clb6 stimulated the phosphorylation of wild-type full-length and 60k Swi6 but there was little phosphorylation when serine 160 was replaced with alanine or aspartate (Fig. 2B). Clb6-Cdc28 also phosphorylated RXL>AAA Swi6-60k (Fig. 2B). We conclude that Clb6 specifically directs phosphorylation of Swi6 by Cdc28 at serine 160 but the RXL256-258 motif of Swi6 is not necessary for this event.
Δclb6 mutants have reduced ability to phosphorylate Swi6
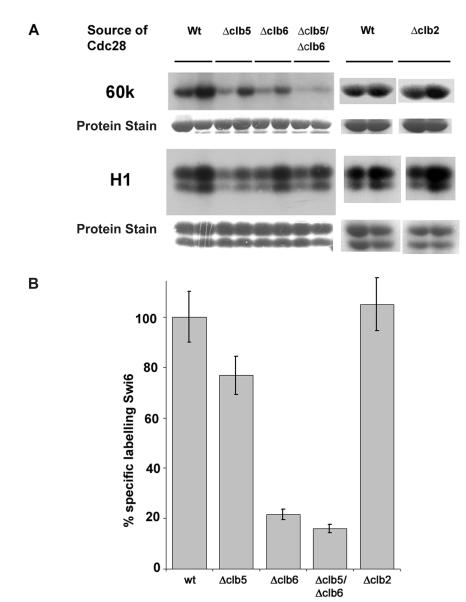
In a further test of cyclin specificity, Cdc28 was produced from derivatives of S. cerevisiae MGY2 in which specific cyclin genes were deleted. This permitted an investigation of the cyclin requirement for Swi6 phosphorylation with the endogenous cyclins that copurify with Cdc28 under low-salt conditions. CLB6 was deleted from the genome of the Cdc28-producing strain, and the resulting purified kinase was assayed for activity towards Swi6p-60k and histone H1. Since Clb6 and Clb5 display a degree of redundancy in their in vivo roles in S phase (22, 42), the effects of the clb5 and clb5 clb6 defects on phosphorylation of Swi6 were also examined (Fig. 3). Similarly, as there appears to be some in vivo interchangeability between Clbs (27), the effect of clb2 deletion was also tested. Two concentrations of kinase were used both to ensure that phosphorylation was assayed under nonsaturating conditions and to compensate for differing input levels of histone phosphorylation activity of individual cyclin/kinase preparations. These data were quantified by normalizing the levels of phosphorylation of Swi6 relative to that of histone H1.
FIG. 3.
Clb5 and Clb6 are needed for optimal phosphorylation of Swi6. (A) Low-salt, cyclin-containing preparations of GST-Cdc28 were made from wild-type and Δclb5, Δclb6, Δclb5 Δclb6, and clb2 MGY2 yeast cells as indicated. Two concentrations of Cdc28 preparations were used to ensure that assays were performed under nonsaturating conditions. The amounts of input affinity-purified Cdc28 were between 25 and 150 ng. Protein staining indicates input substrate visualized by Coomassie blue staining. (B) Quantification of phosphorylation of Swi6 compared to histone H1. The level of phospholabeling of Swi6 by each kinase preparation was normalized against the level of phosphorylation of histone H1 by phosphorimaging; 100% represents the ratio of Swi6 to histone H1 labeling from wild-type extracts.
Compared to wild-type cells, phosphorylation of Swi6-60k was reduced by almost 80% with Cdc28 prepared from a Δclb6 strain. Deletion of CLB5 had a smaller effect, reducing activity by only 25%. A double Δclb5 Δclb6 deletion reduced the purified in vivo activity of Cdc28p towards Swi6p-60k by more than 85% of the wild-type level. In contrast, deletion of CLB2 had no detectable effect on the ability of Cdc28 to phosphorylate Swi6 (Fig. 3). Thus, Clb6 appears to be the major determinant of Swi6 phosphorylation in this assay, although Clb5 may have a minor role.
Clb6 and Swi6 interact
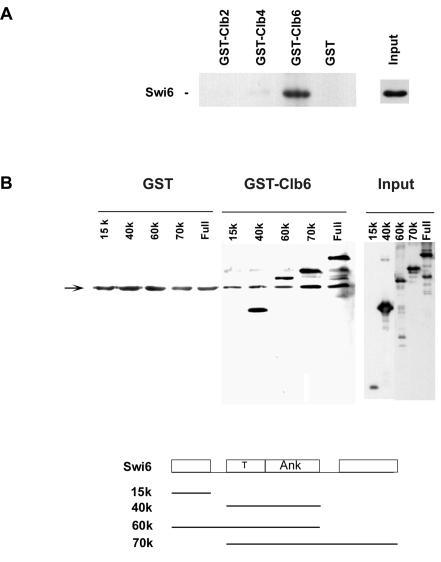
Substrate recognition of the cyclin/Cdk complex has been largely attributed to the cyclin subunit. Accordingly, purified Swi6 was tested for binding to GST-cyclin fusions immobilized on glutathione-Sepharose beads. Swi6 was retained by GST-Clb6 but not by GST-Clb2, GST-Clb4, or GST beads (Fig. 4A). Similarly, Swi6 was not bound by GST-Cln1 (data not shown). These GST-cyclin fusion proteins were nevertheless active, as they were also used in kinase assays such as those presented in Fig. 2. The regions of Swi6 required for binding to GST-Clb6 were mapped in a similar binding experiment with full-size and modular fragments of Swi6. Binding was detected with all fragments carrying the central transcriptional activation and ankyrin repeat domain, whether or not the serine 160 residue was present (Fig. 4B). Binding was not detected with an N-terminal 15-kDa fragment of Swi6 (Fig. 4B) even after prolonged exposure in the enhanced chemiluminescence procedure to enhance detection of this smaller fragment (data not shown). Thus, Clb6 binding requires features within Swi6 residues 188 to 526. Despite this in vitro interaction, attempts to detect an in vivo interaction between Swi6 and Clb6 by coimmunoprecipitation proved unsuccessful, possibly because of the transient nature of enzyme-substrate interactions and the short half-life of cyclins.
FIG. 4.
Swi6 interacts with Clb6. (A) Full-length Swi6 was assayed for binding to glutathione-Sepharose beads carrying GST, GST-Clb2, GST-Clb4, and GST-Clb6. (B) Deletion mapping of Swi6 for binding to Clb6. The fragments of Swi6 diagrammed in the line drawing were assayed for binding to glutathione-Sepharose beads carrying GST and GST-Clb6. T, transcriptional activation region; ANK, ankyrin repeat domain. Bound proteins were visualized by immunoblotting with polyclonal anti-Swi6 antibodies. Nonspecific bands are indicated by the arrow.
Clb6 is required for nuclear export of Swi6
Given the in vitro evidence for Swi6 phosphorylation at serine 160 by Clb6/Cdc28, clb6 mutants might be expected to have reduced or delayed nuclear export of Swi6 because the phosphoserine 160 signal for nuclear export would be absent or at least reduced. Cellular localization of Swi6 was therefore monitored with indirect immunofluorescence against an endogenously expressed Swi6 C-terminal 13myc fusion protein. The activity of Swi6-13myc was first confirmed by its ability to support SBF-lacZ reporter gene expression at similar levels to untagged cells: 0.602 (± 0.043) arbitrary units of β-galactosidase were expressed in wild-type untagged W303-1a SWI6 cells compared to 0.478 (± <0.001) units in the SWI6-13myc derivative. Compared to CLB+ SWI6-13myc cells, reporter gene expression was increased by 2.3-fold in Δclb6 SWI6-13myc cells to 1.152 (± 0.065) in accord with earlier reports of increased G1-specific gene expression in clb6 mutants (4).
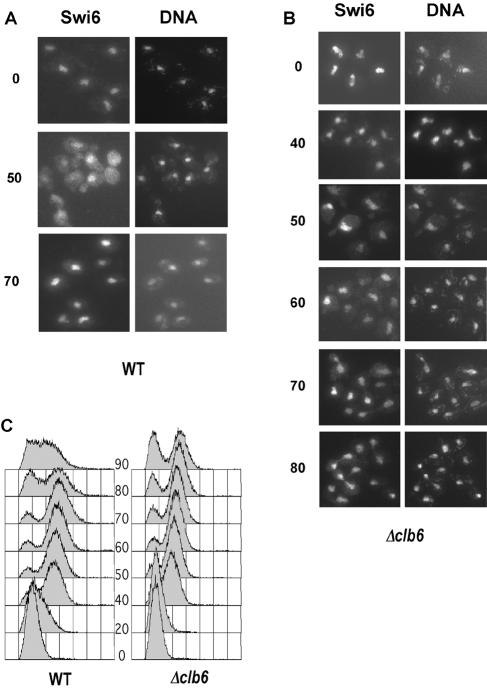
The effect of CLB6 deletion on Swi6 localization was observed during cell cycle progression after α-factor block and release of wild-type and Δclb6 mutants expressing Swi6-13myc fusion protein. Swi6 localization was monitored at 10-min intervals, but for clarity, only pertinent time points are shown in Fig. 5. In wild-type cells, Swi6-13myc was nuclear during the arrest point and became cytoplasmic 50 min after release. At this stage of the cell cycle, the Swi6 signal became diffuse throughout the cell and showed indistinct nuclear localization compared to the DAPI-stained nuclei; 70 min after release from α-factor arrest, Swi6 had become predominantly nuclear once again, in agreement with earlier studies (36, 46). In Δclb6 cells, cell cycle progression was slightly slower than in wild-type cells because of a delay in entry into S phase (Fig. 5C). Importantly, Swi6 localization in the Δclb6 mutant coincided mainly with DAPI staining of the nuclei, indicating that Swi6 was predominantly nuclear at all times. However, a low-level cytoplasmic signal was seen in Δclb6 cells at 60 min, but in observations of several hundred cells, we never saw the diffuse and almost uniform cytoplasmic distribution of Swi6 that was seen in wild-type cells. The reduction in nuclear export in Δclb6 mutants therefore indicates that Clb6/Cdc28 activity is required in vivo for nuclear trafficking of Swi6.
FIG. 5.
Clb6 is required for nuclear export of Swi6. Immunolocalization of Swi6-myc in wild-type cells (A) and Δclb6 cells (B) after release from α-factor arrest. Numbers denote minutes after release from arrest. DNA was visualized by DAPI staining. (C) DNA content of cells following release from α-factor arrest, determined by flow cytometry.
Dephosphorylation and nuclear import of Swi6 by Cdc14
In vivo, dephosphorylation of Swi6 is required for nuclear import (46). The timing of import is in the later stages of mitosis because Swi6 is cytoplasmic in a metaphase arrest imposed by nocadazole (46, 51) (Fig. 6C) and phosphorylated Swi6 accumulates in the late mitotic arrest of a thermosensitive cdc15 kinase mutant (46). As the phosphatase, Cdc14, acts after Cdc15 in the final stages of mitotic exit (reviewed in reference 3), we asked whether Cdc14 might direct dephosphorylation and nuclear localization of Swi6.
FIG. 6.
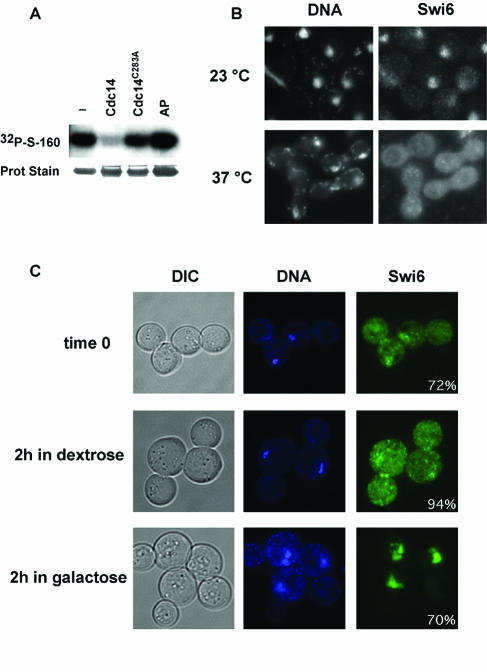
Dephosphorylation and nuclear import of Swi6 after Cdc14 activity. (A) Radioactive Swi6 labeled at serine 160 was treated with the phosphatase indicated and the remaining radioactivity was visualized by autoradiography. AP, alkaline phosphatase. (B) Immunolocalization of Swi6-13myc and nuclear staining in cdc14-1 SWI6-13myc cells at 23°C or after 165 min of incubation at 37°C. (C) SWI6-GFP cells carrying pCDC14yexEMBL, expressing Cdc14 under the control of the GAL1,10 galactose-inducible promoter, were grown for 2 h in 2% sucrose and nocodazole-containing medium to produce a metaphase arrest (time zero). The culture was divided and incubation was continued a further 2 h with nocodazole in the presence of dextrose or galactose to repress or induce Cdc14 expression. Inset values quantify the percentage of cells of the culture as shown in the images (n > 100).
To test if Swi6 can be a substrate for Cdc14, it was first phosphorylated in vitro at serine 160 by Cdc28 as described above. Wild-type, affinity-purified MBP-Cdc14 clearly dephosphorylated Swi6 at serine 160, but no dephosphorylation was detected with mutationally inactive MBP-Cdc14 C283A (Fig. 6A). Although Cdc14 has a broad range of substrates, the conditions used here employed molar ratios of phosphatase to substrate that were established as nonsaturating (data not shown) and which were similar (±2-fold) to those used to demonstrate Cdc14-specific dephosphorylation of the known substrates Swi5 and Sic1 (56). Likewise, no dephosphorylation occurred with alkaline phosphatase treatment of Swi6 (Fig. 6A), reminiscent of the failure of alkaline phosphatase to act on Swi5 (56) even though the same preparation of Cdc14 was active against the nonspecific substrate p-nitrophenylphosphate (data not shown).
As Cdc14 can dephosphorylate the serine 160 residue required for nuclear import, we examined whether Cdc14 was required for relocalization of Swi6 to the nucleus in vivo. In an initial experiment we asked whether Swi6 nuclear import was blocked during the thermal arrest of a cdc14-1 thermosensitive strain. A culture of early-logarithmic-phase cdc14-1 Swi6-13myc cells was split, and incubation was continued at either 25°C or 37°C for a further 165 min. At the permissive temperature Swi6 was largely nuclear, as indicated by the colocalization of Swi6-13myc immunofluorescence and DAPI staining, although a fraction of presumably M-phase cells displayed a more diffuse distribution typifying cytoplasmic Swi6 (Fig. 6B). At the restrictive temperature the cdc14-1 cells arrested as large dumbbells, where, in more than 90% of cells, Swi6 was distributed throughout the cell and did not colocalize with the nuclear DAPI staining (Fig. 6B). Thus, in the absence of Cdc14, Swi6 is largely cytoplasmic.
Although this experiment might indicate a requirement for Cdc14 in Swi6 nuclear import, it does not distinguish between inhibition of a Cdc14-dependent process and inhibition of an independent process through general arrest in cell cycle progression. To address this question, SWI6-GFP cells were held in a metaphase arrest and Swi6-GFP localization was followed after ectopic expression of Cdc14 from a galactose-inducible CDC14 plasmid. (As noted previously, Swi6-GFP behaves similarly to Swi6 in cellular localization studies [46].) The cells were blocked in metaphase with nocodazole to produce a population of cells arrested as large dumbbells with predominantly cytoplasmic Swi6 (Fig. 6C). Incubation of one half of the culture continued in dextrose and nocodazole medium to maintain metaphase arrest under repressing conditions for Cdc14 expression. After 2 h, Swi6 remained cytoplasmic in more than 90% of the cells. In the other half of the culture, galactose was added to induce Cdc14 expression for 2 h during the ongoing metaphase arrest. Under these conditions, Swi6 changed from being predominantly cytoplasmic to being nuclear in 70% of the cells, even though the cells remained arrested as large dumbbells. Thus, in vivo Cdc14 activity can stimulate nuclear import of Swi6.
DISCUSSION
We show that the major cell cycle kinase Cdc28 and its Clb6 cyclin subunit specifically phosphorylate Swi6 at serine 160 and direct export of Swi6 from the nucleus. We also show that dephosphorylation of Swi6 at serine 160 and nuclear import are controlled by Cdc14, a phosphatase active in the final stages of M phase.
The pleiotropic and essential activity of Cdc28 and the redundancy of cyclin function have historically made it difficult to identify the substrate specificities of this kinase. Nevertheless, we conclude that Clb6/Cdc28 activity is responsible for phosphorylation at serine 160 of Swi6 because of the thermosensitive phosphorylation of Swi6 and histone H1 by Cdc28-13, because of the cyclin-specific stimulation of this activity, and because phosphorylation occurred at a putative Cdc28 consensus site. Phosphorylation at serine 160 had earlier been identified in vivo as the only site in Swi6 subject to cell cycle-dependent phosphorylation (46). The same phosphorylation at serine 160 in vivo and in vitro indicates that the availability of this kinase site is independent of interaction of Swi6 with Mbp1 or Swi4. Note that this specificity of in vitro phosphorylation occurred despite the presence of other putative Cdc28 kinase sites and other sites subject to non-cell-cycle-dependent phosphorylation in vivo (45).
In addition, by purifying all nine cyclins of Cdc28, we show that the specific phosphorylation of serine 160 is directed largely by Clb6-Cdc28 and not by other G1 or mitotic cyclin-Cdc28 kinase complexes. The specificity of this approach is emphasized by identical assays with a different substrate, where Ndd1 was phosphorylated by Clb2 rather than Clb6 (6). Clb6 specificity was confirmed with Cdc28 purified from cells depleted for different cyclins. This approach also suggested a minor contribution of Clb5 to Swi6 phosphorylation. However, as we were unable to detect this in other assays and as there is a near overlap of error bars of kinase activity in wild-type and Δclb5 extracts, we do not consider Clb5 to be a major source of Swi6 phosphorylation.
A specific interaction was also detected between Clb6 and Swi6 in vitro. The interaction occurred with the central section of Swi6 encompassing the ankyrin repeat domain and an associated transcriptional activation region (13). However, an RXL motif in this region was not required even though this motif has been equated with cyclin-substrate binding in other systems (8, 40). Nevertheless, ankyrin repeats have a well-established role in protein-protein interactions (reviewed in reference 43), and Clb2 is known to interact with the structurally related ankyrin repeats of Swi4 (47).
Prior to this work, extensive characterization of S-phase cyclin activities focused mainly on their effects at replication origins (19, 26, 29, 30, 61), but a direct demonstration of the substrate specificity of the Clb6-Cdc28 kinase has remained elusive. Thus, for the first time, we provide direct evidence for Clb6/Cdc28 substrate specificity.
To relate the in vitro phosphorylation of serine 160 by Clb6/Cdc28 to in vivo events, we demonstrated that Clb6 and presumably its associated kinase, Cdc28, are required for nuclear export of Swi6. Nevertheless, a small amount of cytoplasmic Swi6 was seen in Δclb6 mutants (Fig. 5), and Cdc28 purified from Δclb6 cells also had a residual ability to phosphorylate Swi6 (Fig. 3). Thus, in accord with the apparent in vivo redundancy of Clb activity, it is possible that other cyclins may have a minor activity which in vivo is sufficient to stimulate nuclear export.
We also examined how Swi6 dephosphorylation and nuclear import of Swi6 might be governed by Cdc14. Cdc14 is a phosphatase that acts as an antagonist of Clb/Cdc28 activity in the final phases of mitosis (32, 49, 56). Our finding that arrested cdc14-1 cells had predominantly cytoplasmic Swi6 is consistent with earlier observations that phosphorylated Swi6 accumulated during a cdc15 arrest (46). Moreover, we show that Cdc14 can also stimulate nuclear import of Swi6 in metaphase-arrested cells and, at least in vitro, is capable of dephosphorylating the serine 160 residue of Swi6. Together these results implicate Cdc14 in nuclear import of Swi6. Although a direct effect of Cdc14 on Swi6 dephosphorylation is likely, we cannot exclude the intervention of another phosphatase that is directly or indirectly controlled by Cdc14.
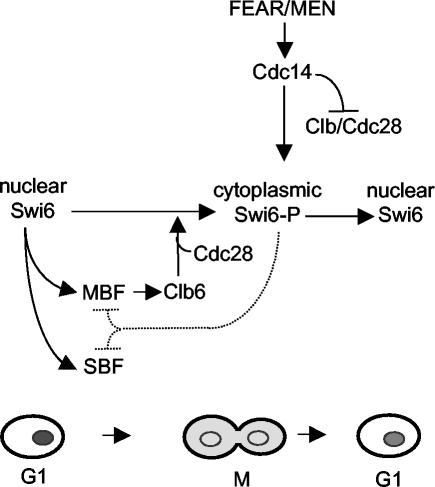
The idea of nuclear trafficking as a mechanism of Swi6 regulation was first proposed by Taba et al. (51). In this context, Clb6 can be seen as a negative regulator that curtails G1 by promoting nuclear export of Swi6 in addition to other mechanisms of antagonizing G1 Cdc28 activity (4). This conclusion is supported by the inhibitory effects of Clb6 overexpression and the stimulatory effects of CLB6 deletion on SBF- and MBF-dependent gene expression (4). Likewise, the effects of CLB6 overexpression and swi6 deletion are similar in that both deregulate CDC6 expression (4, 33). We propose that Swi6 phosphorylation by Clb6/Cdc28 generates a negative regulatory feedback mechanism. After passing Start, Clb6 is synthesized by Swi6 acting in MBF. The subsequent phosphorylation of Swi6 by Clb6 then helps to reduce further G1-specific gene expression by localizing Swi6 to the cytoplasm (Fig. 7).
FIG. 7.
Scheme for cell cycle regulation of nuclear export and import of Swi6. Swi6 is represented by grey shading in the lower cartoon. The broken line indicates reduction in MBF and SBF activity by loss of Swi6 from the nucleus. See the Discussion for further explanation.
Localization of Swi6 to the cytoplasm was initially envisaged to result from a stimulation of nuclear export (46). However, this idea is not consistent with a more recent proposal that nuclear trafficking of Swi6 is an essential step in licensing Swi6 for optimal SBF activity after reentry to the nucleus (36). In particular, the increased SBF activity in clb6 mutants would not be expected as reduced export of Swi6 would reduce licensing and hence attenuate G1-specific gene expression. These conflicting ideas can be reconciled, if as suggested (36), there is continuous nucleocytoplasmic shuttling of Swi6. In this model, phosphorylation of serine 160 would inhibit reentry into the nucleus rather than stimulating nuclear export. This would result in an apparent nuclear localization of Swi6 in a Δclb6 mutant throughout the cell cycle, but having been licensed by nuclear shuttling, the Swi6 or Swi6S160A would be transcriptionally active. Clearly, the question of whether phosphorylation triggers nuclear export or prevents import requires further examination, but in either case Clb6 would play a determining role. It is interesting that the S160A mutation that leads to nuclear localization of Swi6 does not reveal any major changes in SBF- or MBF-driven transcription (46). As Δclb6 mutants show increased G1-specific transcription (4), Clb6 may have other roles in controlling Swi6-dependent transcription in addition to cellular localization of Swi6.
Our conclusion that Cdc28 has cell cycle-dependent activity towards Swi6 appears to contradict two earlier experiments which were used as evidence against Swi6 phosphorylation by Cdc28 (46). In one, Swi6 was not phosphorylated when protein synthesis was inhibited after release from a G1 arrest. It was argued that Cdc28 was unlikely to phosphorylate Swi6 at serine 160, as Cdc28 is active whether or not protein synthesis is permitted. However, if Clb6 specifies Cdc28 activity against Swi6, this cyclin would first have to be synthesized in G1 to allow Swi6 to be phosphorylated. Thus, this earlier result is completely consistent with our findings that Clb6/Cdc28 phosphorylates Swi6.
In a second experiment, cdc28-4 mutants were released from a hydroxyurea block into the restrictive temperature, where they arrested at M phase. Under these conditions, it was concluded that Cdc28 did not phosphorylate Swi6 because phosphorylation, albeit reduced, still occurred, even though the thermosensitive Cdc28-4 kinase was unable to perform an essential step in mitosis (46). It is important to stress that this result does not actually exclude Cdc28 from being the kinase that phosphorylates Swi6 at serine 160. Although the conditions used do impair an essential step at G2/M (38), they do not exclude the possibility of there being sufficient residual Cdc28 activity to phosphorylate a substrate implicated in a nonessential step of the cell cycle. Note that similar arguments have to be made to explain why thermosensitive CDC28 mutants normally arrest in G1 even though Cdc28 has multiple essential targets throughout the cell cycle (38).
Unfortunately, assays of possible residual activities of thermosensitive Cdc28 kinase at the restrictive temperature are problematic. Cdc28-13 assayed at 37°C has no activity against histone H1 or Sic1 even though cdc28-13 cells are able to pass the Cdc28-dependent step in mitosis and arrest only in G1 (37, 59). Moreover, no activity towards histone H1 can be detected with Cdc28-4 even at the permissive temperature (37). Given these reservations, our observations of altered Swi6 cellular localization in Δclb6 mutants strongly indicate that Cdc28 coupled with Clb6 is required for driving and/or maintaining Swi6 in the cytoplasm. Importantly, after submission of this report, the specific inhibition of Cdc28-as1 in vivo was shown to block phosphorylation of Swi6 (54). Although neither the phosphorylated residue nor the specific cyclin were identified, this observation is in complete agreement with the conclusions made here that Swi6 is a substrate for the Cdc28 kinase.
In summary, we show how phosphorylation and nuclear export of Swi6 are integrated with changes in cyclin-dependent kinase in the transition from Cln-dependent G1 to the subsequent Clb-dependent phase of the cell cycle. Similarly, with the elimination of Clb kinase activity at the end of M phase, Cdc14 also triggers nuclear import of Swi6 in preparation for SBF- and MBF-driven transcription in the ensuing G1 phase (Fig. 7).
Acknowledgments
We are indebted to Lee Johnston, Eliot Randle, Sanne Jensen, and Ian Taylor for plasmid and yeast stocks and purified proteins. We thank Lee Johnston, Sanne Jensen, Anthony Johnson, and Marisa Segal for help, advice, and encouragement.
G. Wells was supported by an MRC studentship and M. Geymonat was supported by the Association for International Cancer Research, grant 99-008.
REFERENCE
- 1.Andrews, B. J., and L. A. Moore. 1992. Interaction of the yeast Swi4 and Swi6 cell cycle regulatory proteins in vitro. Proc. Natl. Acad. Sci. USA 89:11852-11856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baetz, K., and B. Andrews. 1999. Regulation of cell cycle transcription factor Swi4 through auto-inhibition of DNA binding. Mol. Cell. Biol. 19:6729-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardin, A. J., and A. Amon. 2001. Men and sin: what's the difference? Nat. Rev. Mol. Cell. Biol. 2:815-826. [DOI] [PubMed] [Google Scholar]
- 4.Basco, R. D., M. D. Segal, and S. I. Reed. 1995. Negative regulation of G1 and G2 by S-phase cyclins of Saccharomyces cerevisiae. Mol. Cell. Biol. 15:5030-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berben, G., J. Dumont, V. Gilliquet, P. A. Bolle, and F. Hilger. 1991. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast 7:475-477. [DOI] [PubMed] [Google Scholar]
- 6.Breeden, L., and G. E. Mikesell. 1991. Cell cycle-specific expression of the SWI4 transcription factor is required for the cell cycle regulation of HO transcription. Genes Dev. 5:1183-1190. [DOI] [PubMed] [Google Scholar]
- 7.Cosma, M. P., S. Panizza, and K. Nasmyth. 2001. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol. Cell 7:1213-1220. [DOI] [PubMed] [Google Scholar]
- 8.Cross, F. R., and M. D. Jacobson. 2000. Conservation and function of a potential substrate-binding domain in the yeast Clb5 B-type cyclin. Mol. Cell. Biol. 20:4782-4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Como, C. J., H. Chang, and K. T. Arndt. 1995. Activation of CLN1 and CLN2 G1 cyclin gene expression by BCK2. Mol. Cell. Biol. 15:1835-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dirick, L., T. Moll, H. Auer, and K. Nasmyth. 1992. A central role for SWI6 in modulating cell cycle Start-specific transcription in yeast. Nature 357:508-513. [DOI] [PubMed] [Google Scholar]
- 11.Epstein, C. B., and F. R. Cross. 1992. CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev. 6:1695-1706. [DOI] [PubMed] [Google Scholar]
- 12.Epstein, C. B., and F. R. Cross. 1994. Genes that can bypass the CLN requirement for Saccharomyces cerevisiae cell cycle START. Mol. Cell. Biol. 14:2041-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foord, R., I. A. Taylor, S. G. Sedgwick, and S. J. Smerdon. 1999. X-ray structural analysis of the yeast cell cycle regulator Swi6 reveals variations of the ankyrin fold and has implications for Swi6 function. Nat. Struct. Biol. 6:157-165. [DOI] [PubMed] [Google Scholar]
- 14.Hagemeier, C., A. J. Bannister, A. Cook, and T. Kouzarides. 1993. The activation domain of transcription factor PU.1 binds the retinoblastoma (RB) protein and the transcription factor TFIID in vitro: RB shows sequence similarity to TFIID and TFIIB. Proc. Natl. Acad. Sci. USA 90:1580-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrington, L. A., and B. J. Andrews. 1996. Binding to the yeast SwI4,6-dependent cell cycle box, CACGAAA, is cell cycle regulated in vivo. Nucleic Acids Res. 24:558-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho, Y., M. Costanzo, L. Moore, R. Kobayashi, and B. J. Andrews. 1999. Regulation of transcription at the Saccharomyces cerevisiae start transition by Stb1, a Swi6-binding protein. Mol. Cell. Biol. 19:5267-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho, Y., S. Mason, R. Kobayashi, M. Hoekstra, and B. Andrews. 1997. Role of the casein kinase I isoform, Hrr25, and the cell cycle-regulatory transcription factor, SBF, in the transcriptional response to DNA damage in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:581-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horak, C. E., N. M. Luscombe, J. Qian, P. Bertone, S. Piccirrillo, M. Gerstein, and M. Snyder. 2002. Complex transcriptional circuitry at the G1/S transition in Saccharomyces cerevisiae. Genes Dev. 16:3017-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelly, T. J., and G. W. Brown. 2000. Regulation of chromosome replication. Annu. Rev. Biochem. 69:829-880. [DOI] [PubMed] [Google Scholar]
- 20.Koch, C., T. Moll, M. Neuberg, H. Ahorn, and K. Nasmyth. 1993. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science 261:1551-1557. [DOI] [PubMed] [Google Scholar]
- 21.Koch, C., A. Schleiffer, G. Ammerer, and K. Nasmyth. 1996. Switching transcription on and off during the yeast cell cycle: Cln/Cdc28 kinases activate bound transcription factor SBF (Swi4/Swi6) at start, whereas Clb/Cdc28 kinases displace it from the promoter in G2. Genes Dev. 10:129-141. [DOI] [PubMed] [Google Scholar]
- 22.Kuhne, C., and P. Linder. 1993. A new pair of B-type cyclins from Saccharomyces cerevisiae that function early in the cell cycle. EMBO J. 12:3437-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 24.Lowndes, N. F., A. L. Johnson, L. Breeden, and L. H. Johnston. 1992. SWI6 protein is required for transcription of the periodically expressed DNA synthesis genes in budding yeast. Nature 357:505-508. [DOI] [PubMed] [Google Scholar]
- 25.Madden, K., Y. J. Sheu, K. Baetz, B. Andrews, and M. Snyder. 1997. SBF cell cycle regulator as a target of the yeast PKC-MAP kinase pathway. Science 275:1781-1784. [DOI] [PubMed] [Google Scholar]
- 26.Masumoto, H., S. Muramatsu, Y. Kamimura, and H. Araki. 2002. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature 415:651-655. [DOI] [PubMed] [Google Scholar]
- 27.Nasmyth, K. 1996. At the heart of the budding yeast cell cycle. Trends Genet. 12:405-412. [DOI] [PubMed] [Google Scholar]
- 28.Nasmyth, K., and L. Dirick. 1991. The role of SWI4 and SWI6 in the activity of G1 cyclins in yeast. Cell 66:995-1013. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen, V. Q., C. Co, and J. J. Li. 2001. Cyclin-dependent kinases prevent DNA rereplication through multiple mechanisms. Nature 411:1068-1073. [DOI] [PubMed] [Google Scholar]
- 30.Noguchi, E., P. Shanahan, C. Noguchi, and P. Russell. 2002. CDK phosphorylation of Drc1 regulates DNA replication in fission yeast. Curr. Biol. 12:599-605. [DOI] [PubMed] [Google Scholar]
- 31.Ogas, J., B. J. Andrews, and I. Herskowitz. 1991. Transcriptional activation of CLN1, CLN2, and a putative new G1 cyclin (HCS26) by SWI4, a positive regulator of G1-specific transcription. Cell 66:1015-1026. [DOI] [PubMed] [Google Scholar]
- 32.Pereira, G., C. Manson, J. Grindlay, and E. Schiebel. 2002. Regulation of the Bfa1p-Bub2p complex at spindle pole bodies by the cell cycle phosphatase Cdc14p. J. Cell Biol. 157:367-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piatti, S., C. Lengauer, and K. Nasmyth. 1995. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 14:3788-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Primig, M., S. Sockanathan, H. Auer, and K. Nasmyth. 1992. Anatomy of a transcription factor important for the start of the cell cycle in Saccharomyces cerevisiae. Nature 358:593-597. [DOI] [PubMed] [Google Scholar]
- 35.Pringle, J. R., A. E. Adams, D. G. Drubin, and B. K. Haarer. 1991. Immunofluorescence methods for yeast. Methods Enzymol. 194:565-602. [DOI] [PubMed] [Google Scholar]
- 36.Queralt, E., and J. C. Igual. 2003. Cell cycle activation of the Swi6p transcription factor is linked to nucleocytoplasmic shuttling. Mol. Cell. Biol. 23:3126-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reed, S. I., J. A. Hadwiger, and A. T. Lorincz. 1985. Protein kinase activity associated with the product of the yeast cell division cycle gene CDC28. Proc. Natl. Acad. Sci. USA 82:4055-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reed, S. I., and C. Wittenberg. 1990. Mitotic role for the Cdc28 protein kinase of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 87:5697-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reynard, G. J., W. Reynolds, R. Verma, and R. J. Deshaies. 2000. Cks1 is required for G1 cyclin-cyclin-dependent kinase activity in budding yeast. Mol. Cell. Biol. 20:5858-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russo, A. A., P. D. Jeffrey, A. K. Patten, J. Massague, and N. P. Pavletich. 1996. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature 382:325-331. [DOI] [PubMed] [Google Scholar]
- 41.Schneider, B. L., Q. H. Yang, and A. B. Futcher. 1996. Linkage of replication to start by the Cdk inhibitor Sic1. Science 272:560-562. [DOI] [PubMed] [Google Scholar]
- 42.Schwob, E., and K. Nasmyth. 1993. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 7:1160-1175. [DOI] [PubMed] [Google Scholar]
- 43.Sedgwick, S. G., and S. J. Smerdon. 1999. The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem. Sci. 24:311-316. [DOI] [PubMed] [Google Scholar]
- 44.Sedgwick, S. G., I. A. Taylor, A. C. Adam, A. Spanos, S. Howell, B. A. Morgan, M. K. Treiber, N. Kanuga, G. R. Banks, R. Foord, and S. J. Smerdon. 1998. Structural and functional architecture of the yeast cell cycle-transcription factor swi6. J. Mol. Biol. 281:763-775. [DOI] [PubMed] [Google Scholar]
- 45.Sidorova, J. M., and L. L. Breeden. 1997. Rad53-dependent phosphorylation of Swi6 and down-regulation of CLN1 and CLN2 transcription occur in response to DNA damage in Saccharomyces cerevisiae. Genes Dev. 11:3032-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sidorova, J. M., G. E. Mikesell, and L. L. Breeden. 1995. Cell cycle-regulated phosphorylation of Swi6 controls its nuclear localization. Mol. Biol. Cell 6:1641-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siegmund, R. F., and K. A. Nasmyth. 1996. The Saccharomyces cerevisiae Start-specific transcription factor Swi4 interacts through the ankyrin repeats with the mitotic Clb2/Cdc28 kinase and through its conserved carboxy terminus with Swi6. Mol. Cell. Biol. 16:2647-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stegmeier, F., R. Visintin, and A. Amon. 2002. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell 108:207-220. [DOI] [PubMed] [Google Scholar]
- 50.Stuart, D., and C. Wittenberg. 1995. CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes Dev. 9:2780-2794. [DOI] [PubMed] [Google Scholar]
- 51.Taba, M. R., I. Muroff, D. Lydall, G. Tebb, and K. Nasmyth. 1991. Changes in a SWI4,6-DNA-binding complex occur at the time of HO gene activation in yeast. Genes Dev. 5:2000-2013. [DOI] [PubMed] [Google Scholar]
- 52.Thuret, J. Y., J. G. Valay, G. Faye, and C. Mann. 1996. Civ1 (CAK in vivo), a novel Cdk-activating kinase. Cell 86:565-576. [DOI] [PubMed] [Google Scholar]
- 53.Tyers, M., G. Tokiwa, and B. Futcher. 1993. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 12:1955-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ubersax, J. A., E. L. Woodbury, P. N. Quang, M. Paraz, J. D. Blethrow, K. Shah, K. M. Shokat, and D. O. Morgan. 2003. Targets of the cyclin-dependent kinase Cdk1. Nature 425:859-864. [DOI] [PubMed] [Google Scholar]
- 55.Verma, R., R. S. Annan, M. J. Huddleston, S. A. Carr, G. Reynard, and R. J. Deshaies. 1997. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science 278:455-460. [DOI] [PubMed] [Google Scholar]
- 56.Visintin, R., K. Craig, E. S. Hwang, S. Prinz, M. Tyers, and A. Amon. 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell 2:709-718. [DOI] [PubMed] [Google Scholar]
- 57.Wijnen, H., and B. Futcher. 1999. Genetic analysis of the shared role of CLN3 and BCK2 at the G(1)-S transition in Saccharomyces cerevisiae. Genetics 153:1131-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wijnen, H., A. Landman, and B. Futcher. 2002. The G1 cyclin Cln3 promotes cell cycle entry via the transcription factor Swi6. Mol. Cell. Biol. 22:4402-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wittenberg, C., and S. I. Reed. 1988. Control of the yeast cell cycle is associated with assembly/disassembly of the Cdc28 protein kinase complex. Cell 54:1061-1072. [DOI] [PubMed] [Google Scholar]
- 60.Zhu, H., J. F. Klemic, S. Chang, P. Bertone, A. Casamayor, K. G. Klemic, D. Smith, M. Gerstein, M. A. Reed, and M. Snyder. 2000. Analysis of yeast protein kinases with protein chips. Nat. Genet. 26:283-289. [DOI] [PubMed] [Google Scholar]
- 61.Zou, L., and B. Stillman. 2000. Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol. Cell. Biol. 20:3086-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]