Abstract
Pseudomonas aeruginosa is a pathogenic Gram-negative bacterium that affects patients with cystic fibrosis and immunocompromised individuals. This bacterium coexpresses two unique forms of lipopolysaccharides (LPSs) on its surface, the A- and B-band LPS, which are among the main virulence factors that contribute to its pathogenicity. The polysaccharides in A-band LPSs are synthesized in the cytoplasm and translocated into the periplasm via an ATP-binding-cassette (ABC) transporter consisting of a transmembrane protein, Wzm, and a cytoplasmic nucleotide-binding protein, Wzt. Most of the biochemical studies of A-band PSs in Pseudomonas aeruginosa are focused on the stages of the synthesis and ligation of PS, leaving the export stage involving the ABC transporter mostly unexplored. This difficulty is compounded by the fact that the subunit composition and structure of this bi-component ABC transporter are still unknown. Here we propose a simple but powerful method, based on Förster Resonance Energy Transfer (FRET) and optical micro-spectroscopy technology, to probe the structure of dynamic (as opposed to static) protein complexes in living cells. We use this method to determine the association stoichiometry and quaternary structure of the Wzm-Wzt complex in living cells. It is found that Wzt forms a rhombus-shaped homo-tetramer which becomes a square upon co-expression with Wzm, and that Wzm forms a square-shaped homo-tetramer both in the presence and absence of Wzt. Based on these results, we propose a structural model for the double-tetramer complex formed by the bi-component ABC transporter in living cells. An understanding of the structure and behavior of this ABC transporter will help develop antibiotics targeting the biosynthesis of the A-band LPS endotoxin.
Introduction
ATP-binding cassette (ABC) transporters form a superfamily of membrane transport proteins that is common to all living organisms.1 ABC transporters translocate molecules across the cell membrane by utilizing energy released through the hydrolysis of ATP. In general, it is assumed that the core of an ABC transporter consists of two transmembrane domains (TMDs) and two or more nucleotide-binding domains (NBDs).2–4 TMDs form the translocation pathway for transporting substrate across the membrane, while the NBDs are involved in ATP binding and hydrolysis and contain the conserved ABC sequence motifs. Most TMDs and NBDs in bacterial ABC transporters are expressed in separate polypeptides, while many of eukaryotes’ TMDs and NBDs are expressed in a single polypeptide.5
Pseudomonas aeruginosa coexpresses two unique forms of lipopolysaccharide (LPS) on the outer leaflet of its outer membrane, the common antigen A-band LPS and serologically variable O antigen B-band LPS.6 For both forms, polysaccharides (PSs) initially synthesized in the cytoplasm have to be translocated to the periplasm prior to reaching their final destination, the outer leaflet of the outer membrane, to form LPSs. The B-band PSs are translocated to the periplasmic face by Wzx.7 The TMD protein Wzm and the NBD protein Wzt form a bi-component ABC transporter8–10 in this bacterium that translocates A-band PSs6 to the periplasm. Genetic studies showed that the disruption of either wzt or wzm gene hinders PS export across the inner membranes, resulting in accumulation of only the A-band PS in mutants’ cytoplasm. 11 In homology to other bacterial systems, Wzt has one NBD located at its N terminus, while the PS binding site is thought to be located at its C terminus12, 13 (supplemental Fig. S1). Wzm is thought to provide a passage for PS during the translocation.
Pseudomonas aeruginosa affects patients with cystic fibrosis and immunocompromised individuals,14, 15 and LPSs represent one of the main virulence factors which contribute to its pathogenicity.16, 17 The exact mechanism of PS export by ABC transporters to its functional destination in the outer leaflet is still unknown,8, 16 and the problem can be traced back to the difficulty of determining the spatial assembly of subunits into functional ABC transporters.
Förster Resonance Energy Transfer (FRET) – a process through which energy is transferred non-radiatively from an optically excited donor (D) fluorophore to a non-excited acceptor (A) fluorophore18, 19 – is a suitable tool to study macromolecular interactions in living cells. FRET has been used for detecting conformational changes in macromolecules,20–24 tracking complex formation,25 probing heteromerization of more than two proteins,26 analyzing chromatin compaction,27 and monitoring dynamic protein interactions.28 More quantitatively, FRET has been used for determining the average size of the oligomers from average FRET efficiency values of populations of interacting molecules29–31 as well as the average fraction of associated versus unassociated monomers in a population of homo-oligomerizing proteins.29, 32
Advances in the FRET stoichiometry theory33 together with the advent of optical micro-spectroscopy technology has led to the development of a FRET imaging method for the determination of the stoichiometry and relative disposition of the protomers (i.e., quaternary structure) within a protein complex in living cells.34 In contrast to standard average-based methods, this method relies on the analysis of distributions of apparent FRET efficiencies, Eapp, across the image pixels of individual cells expressing proteins of interest. The most probable quaternary structure of the complex is identified from the number of peaks in the distribution of FRET efficiencies and their mutual relationships. Such peaks collectively create a unique fingerprint of a specific oligomer (quaternary) structure,35 or a “FRET spectrum” of the complex.
Although pixel-level FRET provides invaluable information for stable (i.e., long-lived) oligomeric complexes, it is difficult to use in the following situations: (i) when the number of proteins in a cell is very small; and (ii) when proteins form dynamic complexes that rapidly associate and dissociate, thereby leading to uniform distributions of complexes with same proportion of donors and acceptors. In both of these cases, the FRET efficiency histograms present few or even single peaks; this may easily leave one with the incorrect impression that the protein in question forms dimers, although it may in fact form higher order oligomers.
In our investigations of various oligomeric proteins, we have observed that although the Eapp distributions of most cells expressing dynamic protein oligomers may present single peaks, the FRET efficiency values corresponding to the maxima of those peaks do not take on arbitrary values, but cluster themselves about certain values, with each value corresponding to a single donor/acceptor configuration within an oligomer. Based on this observation, we developed a novel method that we used to determine the quaternary structure of the Wzm and Wzt homo-oligomers separately as well as that of their functional hetero-oligomer, the P. aeruginosa ABC transporter. Using CHO cells as a very competent medium for expression of an ABC transporter, we show that Wzt forms a rhombus-shaped homo-tetramer, which changes its shape into a square upon co-expression with Wzm. In its turn, Wzm forms a square-shaped homo-tetramer whose geometry remains unchanged upon co-expression with Wzt. Based on these results, we propose two slight variations of a hetero-octameric model of the quaternary structure of the Wzm-Wzt transporter from P. aeruginosa.
The new insights provided by the present study and, in general, by the proposed technology, will help understand infectious diseases caused by P. aeruginosa and possibly other Gram-negative bacteria, and to develop effective treatments.
Materials and Methods
Gene cloning and plasmid constructs
To construct pcDNA3.1 (+) gfp2-wzm, pcDNA3.1 (+) yfp-wzm, pcDNA3.1 (+) gfp2-wzt, and pcDNA3.1 (+) yfp-wzt, the plasmids pcDNA3.1 (+) gfp2-M2 and pcDNA3.1 (+) yfp-M2 (i.e., gfp2 or yfp fused to human muscarinic acetylcholine receptor gene M2, with alpha7 membrane signal peptide from nicotinic receptor) (a gift from Professor James W. Wells, University of Toronto), were used in inverse PCR reactions to remove the M2 gene (leaving the alpha7 signal peptide) by using the following two primers: 5`- AAC ATG GGC GCT ACA AGG TAA TCT AGA GGG CCC- 3` and 5`- CTT GTA CAG CTC GTC CAT GCC GAG CGT GAT CCC G- 3`. The wzm gene was PCR-amplified from Pseudomonas aeruginosa genomic DNA (P. aeruginosa PAO1 strain was a gift from Professor Robert E.W. Hancock, University of British Columbia). The PCR primers are the following: 5`- /Phos/ ATG CTT CTT GGC TTG TCT CGT TCC TTG TGG GGC- 3` and 5`- ATC CGG TCT AGA CTA GAG TTC ATC CAC CAT TTC-3`. Also, the wzt gene was PCR-amplified from genomic DNA with the following two primers: 5`- /Phos/ ATG GGA CAG ATA CGC GTA TCC GGC CTC GGC AAG G - 3` and 5`- GGG CCC TCT AGA TCA TGG AGT GCT CTC CGC GGA AGT G -3`. Underlined sequences represent the XbaI sites. All PCR products were gel purified (Qiagen), digested with XbaI, and then gel purified and ligated to produce the desired plasmids. pcDNA3.1 (+) gfp2-wzt, pcDNA3.1 (+) yfp-wzt plasmids were used in another inverse PCR to remove the alpha7 mammalian signal peptide by using the following primers: 5`- /Phos/ AAG GGC GAG GAG CTG TTC ACC GGG GTG GTG CCC A-3` and 5`- /Phos/ GGC GGT ACC AAG CTT AAG TTT AAA CGC TAG CCA G-3`. Then, gel-purified PCR products were self-ligated to produce the desired plasmids.
To construct the untagged wzm gene in plasmid pcDNA3.1 (pcDNA3.1 (+)-wzm), pcDNA3.1 (+) yfp-wzm was used in an inverse PCR reaction to remove the yfp gene while leaving the signal peptide intact by using the following primers: 5`- /Phos/ GTA CAG CTT CCT TTG GAA CTC TCC TTG CAG GGA C-3`; and 5`- /Phos/ ATG CTT CTT GGC TTG TCT CGT TCC TTG TGG GGC-3`.
To construct the untagged wzt gene in plasmid pcDNA3.1 (pcDNA3.1 (+)-wzt), pcDNA3.1 (+) yfp-wzt was used in an inverse PCR reaction to remove the yfp gene by using the following primers: 5`- /Phos/ GGC GGT ACC AAG CTT AAG TTT AAA CGC TAG CCA G-3`; and 5`- /Phos/ ATG GGA CAG ATA CGC GTA TCC GGC CTC GGC AAG G-3`. All engineered plasmids were checked by DNA sequencing.
Expression of fluorescently-tagged Wzm and Wzt in mammalian cells
The fusion proteins GFP2-Wzm, GFP2-Wzt, YFP-Wzm and YFP-Wzt were transiently expressed in CHO cells, singly or in combination, by transfection with pcDNA3.1 (+) gfp2-wzm, pcDNA3.1 (+) yfp-wzm, pcDNA3.1 (+) gfp2-wzt, or pcDNA3.1 (+) yfp-wzt using Lipofectamine 2000 (Invitrogen, USA), according to the manufacturer’s directions and as recently described by Pisterzi et al.30 Briefly, CHO-S cells were maintained in DMEM with 10% fetal bovine serum and 1% non-essential amino acids. Cells were seeded in 6-well plates at approximately 40% confluency and cultured overnight. Plasmid DNA and Lipofectamine were diluted in Opti-MEM medium (Invitrogen, USA), then added to cells. We used 2 µg DNA for a single-plasmid transfection reaction and 1 to 4 µg of each plasmid in co-transfection reactions. Cells were cultured for 36 hours. The cell growth medium was removed and 250 µl of Opti-MEM was added to each well of the 6-well plates. For imaging and FRET analysis, the cells were manually removed from the dishes using cell scrapers. Control reactions included single-plasmid transfections and mock transfections (i.e., no DNA).
Optical micro-spectroscopy
Spectrally resolved fluorescence images were acquired at room temperature using a two photon microscope with spectral resolution constructed in-house.34 The excitation light was provided by an ultrashort-pulse (modelocked) Ti: Sapphire laser (KM Labs, Boulder, CO), running at 80 MHz, and tunable from ~750 to 820 nm. The laser light was focused with an infinity-corrected Plan Apochromat objective (×100 magnification, NA=1.4, oil immersion; Nikon Instruments, Melville, NY) and raster-scanned across the sample using galvanometric scanners (Nutfield Technology, Hudson, NH). A non-descanned detection scheme was used, in which the emitted fluorescence was projected through a transmission grating onto a cooled electron-multiplying CCD (EMCCD) camera with single-photon sensitivity (Andor, iXon 897). In this manner, full spectral information was obtained from each sample voxel on a time scale much shorter than that which would correspond to molecular diffusion. This is a critical feature when using a two-photon microscope for molecular imaging in live cells.
Selection of FRET pairs
GFP2 was used as a donor of energy, since it has a single-photon excitation maximum at ~400 nm36 and hence a two-photon excitation at ~800 nm.34 This choice matches the center wavelength of our Ti: Sapphire laser which was set to ~800 nm and a full-width half maximum (FWHM) of ~30 nm. In addition, GFP2 (by contrast to the more popular variant eGFP) has a large Stokes shift, which allows one to avoid acceptor direct excitation in a FRET experiment. YFP was used as an acceptor, since its excitation spectrum overlaps perfectly with the donor emission.29 Moreover, YFP has a two-photon excitation maximum at ~1020 nm and it therefore cannot be directly excited by the laser light used to excite GFP2. To prevent nonspecific oligomerization the A206K mutation37 was included in our fluorescent protein sequences.
Emission spectra
CHO cells expressing either GFP2-tagged or YFP-tagged proteins were placed on a microscope slide and covered with a cover slip. Spectral images of the GFP2-expressing cells were obtained by using laser light with an average power of ~25 mW at the entrance of the microscope. Emission spectra obtained from several cells expressing GFP2 were averaged and normalized with respect to their maximum emission intensities to obtain the normalized spectrum of the GFP2 donor (D). To obtain the emission spectrum of YFP, CHO cells expressing YFP were excited with laser light with a red-shifted spectrum (centered about 820 nm) and twice the average power, which increased the excitation rate of YFP several-fold;38 this was necessary since YFP does not absorb efficiently at 800 nm. Spectral images were obtained from cells showing high levels of YFP expression (as judged from their overall emission). Since the emission intensities of the cells expressing YFP were much lower than those expressing GFP2, this suggested that the emission spectrum of YFP was contaminated by cell auto-fluorescence in the lower wavelengths of the spectrum. To correct for this spurious contribution, the YFP spectrum was fitted to a sum of three Gaussians: a broad Gaussian corresponding to the cellular auto-fluorescence, as determined from mock-transfected cells, and two closely-spaced Gaussians corresponding to the maximum plus a shoulder normally seen in the spectrum of YFP. We corrected the YFP spectrum by subtracting the Gaussian which fits the auto-fluorescence part of the spectrum from the measured spectrum. The corrected YFP spectra were averaged over many cells expressing YFP, and the resulting spectrum was normalized with respect to its maximum emission intensity to obtain the normalized (or elementary) acceptor (A) spectrum.
FRET efficiency determination
CHO cells were transfected to co-express GFP2-tagged and YFP-tagged proteins to determine whether they self-associate to form oligomers. Spectral images obtained from cells co-expressing GFP2-tagged and YFP-tagged proteins were unmixed to obtain separate donor (denoted by kDA) and acceptor (kAD) images, as described previously.29, 34 The apparent FRET efficiency (Eapp) distribution in a given optical section of the imaged cell was determined for each image pixel using kDA and kAD and the equation:
where wA and wD are the integrals of the measured elementary spectra of A and D, respectively. QD and QA are the quantum yields of D and A, respectively,34, 35 and their values (QD = 0.55 and QA = 0.61) were obtained from the literature.29, 36 All the computations were performed using the Matlab program (The MathWorks, Inc., USA). Pseudo-FRET efficiencies for pixels showing only background noise were avoided by rejecting all intensities not exceeding 2.5% of the maximum emission intensity in the Eapp calculations.
Distributions of FRET efficiencies
The number of image pixels that fell into a particular range of Eapp values was plotted against the corresponding Eapp values to obtain the distribution of FRET efficiencies in the cell, which is referred to here as “the Eapp histogram.” About 90% of the cells showed single peaks and the rest showed multiple peaks in their Eapp histograms. The use of histograms for representing the FRET data allows one to discriminate between FRET caused by specific interactions between the protein of interest and unwanted stochastic FRET that is caused by simple molecular agglomeration, if any.39 About 5% of all the imaged cells and for which Eapp histograms were determined were not considered in our analysis. These fall into two categories. The cells in the first category, which represents a minute fraction of the cells not considered for analysis, showed single peaks in their histogram with their peak position occurring at very large values of Eapp compared to the rest of the cells, and were therefore considered outliers. This occurred in cells that appeared to express unusually high concentrations of Wzt tagged with YFP. We believe this was caused by direct excitation with laser light. The second category of spurious signals came from cells with emission intensities that exceeded the threshold value of 2.5% of the maximum intensity for those individual cells, but their overall intensities were barely exceeding the background noise level. Subsequent analysis of the histograms that passed all the quality checks was dependent on whether they showed single or multiple peaks, and was performed as described in the next section.
Estimation of the protein expression level
From the fluorescence of the donor in the presence of acceptor (kDA) and the fluorescence of acceptor in the presence of donor (kAD), we computed the fluorescence of the donor in the absence of acceptor using the formula FD = kDAWD + kADWAQD / QA, where all the symbols are as defined above. Then we calculated the average donor fluorescence in the absence of FRET. In order to obtain an order of magnitude estimate of the donor concentration inside the cells, we used the following expression, derived from the expression for the number of photons generated by a two-photon absorption process:38
where C is the molar concentration, F is the average fluorescence intensity of the donors in the absence of energy transfer, σ is the two-photon absorption cross-section, Q is the quantum yield, td is the pixel dwell time of the scanning system, NA is the numerical aperture of the microscope objective, P is the average excitation light power, f is the repetition rate of the laser pulse, τ is the laser pulse duration (of the order of 10 to 100 fs), and λex is the excitation wavelength, while the subscripts s and x stand for standard solution and for the unknown concentration (of donor-tagged molecules). The average fluorescence intensity of a fluorescent standard consisting of an aqueous solution of YFP (10 µM concentration) was determined using a separate microscope Zeiss Axio Observer (Zeiss, Thornwood, NY) equipped with an OptiMiS™ module (Aurora Spectral Technologies, Bayside, WI) and an ultrashort-pulse laser (MaiTai™, Spectra Physics, Santa Clara, CA) tuned to 960 nm. The solution of YFP was prepared as described elsewhere.40 The absorption cross sections and the quantum yields were obtained from the literature.41, 42
Analysis of Eapp histograms
Of all the cells analyzed that co-expressed either GFP2-Wzt and YFP-Wzt or GFP2-Wzm and YFP-Wzm, a large majority (see Results section) exhibited single peaks located at various positions along the horizontal axis (i.e., Eapp), while some showed multiple, and often overlapping peaks in their Eapp histograms. The Eapp distributions for cells presenting only one peak were fitted to single Gaussian functions. Some cells, which presented broad Eapp distributions with dominant peaks, were also considered for analysis, with their dominant peaks fitted to Gaussian functions. The peak positions from individual histograms were binned and collected in “meta-histograms” representing the total number of peak positions obtained for each Eapp value (in intervals of 0.02). The five or three clearly distinct peaks in the meta-histograms (see for instance Figs. 3C and 4C) were simulated using five or three Gaussian functions respectively, with the location on the horizontal axis of their maxima predicted by either a rhombus-shaped tetramer model (as depicted in, e.g., Fig. 3D) or a square tetramer (see Fig. 4D). Details about the derivations of the expressions for the peak positions for these models are given elsewhere.34, 35 The fitting of the simulated curves to the experimental ones consisted of minimizing the mismatch between experimental and simulated data by adjusting a number of parameters, as described next.
The exact value of the pair-wise FRET efficiency (Ep), which determines the position of all peaks in a histogram (according to expressions given in Figs. 3D and 4D), depends on the distance between individual donors and acceptors within the protein complex, relative to the Förster radius corresponding to the FRET pair used. Ep was therefore considered to be an adjustable parameter in the data fitting process, which determined the positions of all five Gaussians predicted by the rhombus-shaped tetramer model (Fig. 3D) or all three Gaussians predicted by the square-shaped tetramer model (Fig. 4D).
The width of each Gaussian in the meta-histograms accounts for variations from cell to cell due to pH, viscosity, or other factors that may alter the properties of the fluorescent probes and their rotational diffusion rates; the widths of the peaks in the meta-histogram were therefore adjusted independently in the fitting process.
Finally, the amplitudes of the peaks depend on the number of instances each donor-acceptor configuration is observed in all the cells. For a large population of cells imaged, the average D to A concentration ratio should approach unity if the transfection ratio varies randomly from cell to cell. This would allow one to control the amplitudes for the individual peaks in the meta-histogram. However, it is possible that we underestimated instances when high FRET efficiencies were present in Eapp histograms, due to the fact that peaks corresponding to higher values of FRET efficiencies were usually broader and had lower amplitudes, which may have caused them to be masked by peaks corresponding to lower values of FRET efficiencies. Therefore, the peak amplitudes were used as independently adjustable parameters, and no significance was ascribed to them in the present analysis.
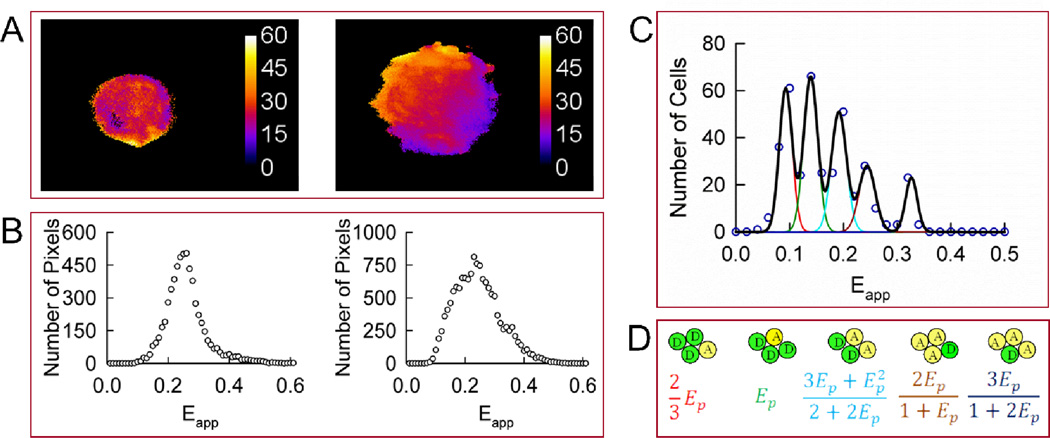
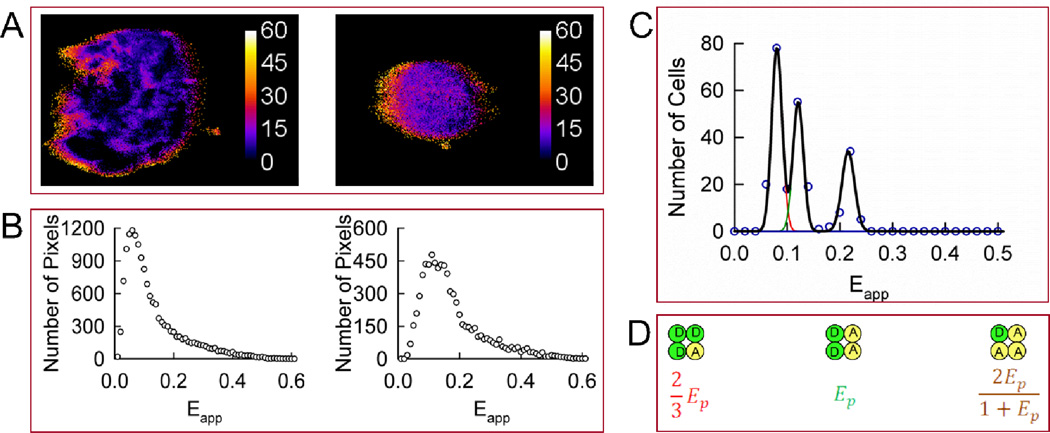
FIGURE 3.
Homo-oligomerization of Wzt expressed in CHO cells in the absence of Wzm. A. Apparent FRET efficiency (Eapp) maps of representative cells expressing Wzt. B. Distributions of FRET efficiencies (Eapp histograms) across the pixels composing the FRET efficiency maps shown in panel A. C. The peak positions of 380 cells showing single or dominant peaks in their corresponding Eapp histograms were binned and used to form meta-histograms (circles), and then fitted simultaneously to five correlated Gaussians shown individually (red, green, cyan, brown, and blue lines) or as a sum (black line). The five peaks in the meta-histogram correspond to different configurations of a rhombus-shaped tetramer shown in panel D. The peak positions of the five simulated Gaussians were determined from a single adjustable parameter, the pair-wise FRET efficiency (Ep) (see equations in panel D). The best fit was obtained for Ep ± S.D. equal to 0.139 ±0.013. D. A rhombus-shaped tetramer model used to interpret the results in panel C. Ep is the pair-wise FRET efficiency between a single donor (D) and a single acceptor (A).
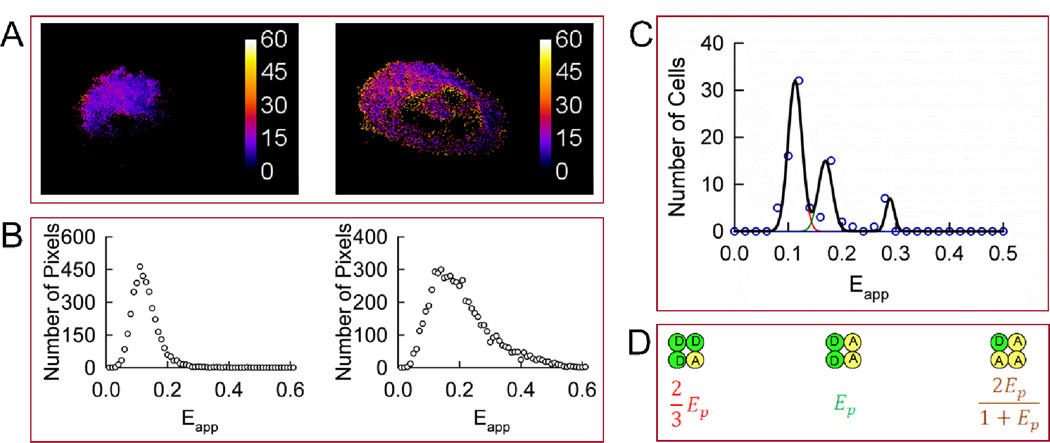
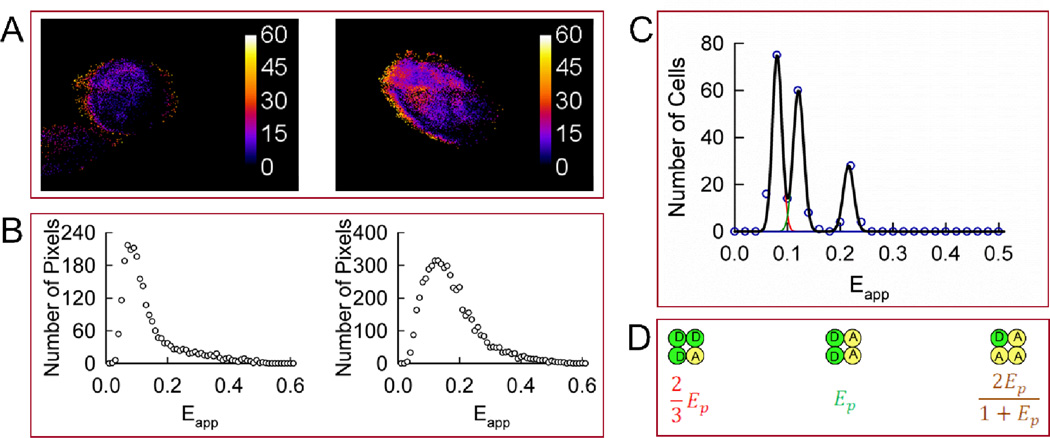
FIGURE 4.
Homo-oligomerization of Wzt expressed in CHO cells in the presence of untagged Wzm. A. Apparent FRET efficiency (Eapp) maps of representative cells expressing Wzt in the presence of Wzm. B. Distributions of FRET efficiencies (Eapp histograms) across the pixels composing the FRET efficiency maps shown in panel A. C. Meta-histogram obtained by accumulating the peak positions of 87 cells presenting single or dominant peaks in their Eapp distributions (circles) and theoretical best-fit to three correlated Gaussians shown individually (red, green, and brown lines) or as a sum (black line). The three peaks in the meta-histogram correspond to different configurations of a square-shaped tetramer shown in the same order in panel D. The best-fit was obtained for Ep ± S.D. equal to 0.17 ± 0.016. D. Square-shaped tetramer model used to interpret the results in panel C. Ep is the pair-wise FRET efficiency between a single donor (D) and a single acceptor (A).
Results
Cytoplasmic Wzt and membrane-bound Wzm interact at the plasma membrane
From many bioinformatics servers, including NCBI (ncbi.nlm.nih.gov) and ExPASy (expasy.org), we found that the Wzm and Wzt proteins possess no domains or motifs that can be targeted for modification by eukaryotic cellular machinery or exhibit any signal for organelle localization. Hence, we chose Chinese hamster ovary (CHO) cells as a model system to study the interaction of Wzm and Wzt proteins. CHO cells are readily transfected to transiently express proteins of interest using standard techniques. Moreover, the significantly larger size of CHO cells compared to most bacteria makes them suitable for pixel-level FRET studies.
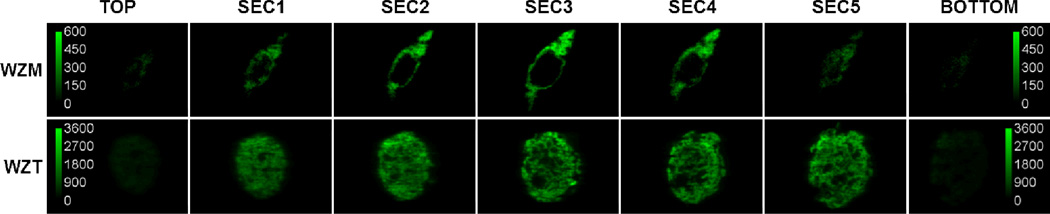
Before probing the quaternary structure of Wzm and Wzt, we wanted to determine their cellular localization. We fused the fluorescent protein GFP236 to their N termini and expressed them individually in CHO cells, which were then subjected to two-photon excitation using our home-made two-photon microscope with spectral resolution (or optical microspectroscopy system).34 Fluorescent images of different optical sections of the CHO cells expressing GFP2-Wzm or GFP2-Wzt fusion proteins were collected at ~508 nm, the wavelength at which the intensity of the GFP2 emission spectrum is at its maximum (Fig. 1). The consecutive sections of these cells are 1.5 µm apart (i.e., close to the limit of the axial resolution of the two-photon microscope). The fluorescence in the CHO cell expressing GFP2-Wzm originates mainly from annular regions, consistent with the localization of this protein at the level of the plasma membrane (Fig. 1, upper panels), while the fluorescence in the CHO cell expressing GFP2-Wzt is distributed throughout the cytoplasm (Fig. 1, bottom panels). The dark pockets in some sections are probably cellular organelles. From these images, we concluded that Wzm is targeted to the cellular membrane whereas Wzt localizes in the cytoplasm.
FIGURE 1.
Localization of Wzm and Wzt proteins in optical sections of Chinese hamster ovary (CHO) cells as detected by a two-photon microscope with spectral resolution. The top row displays fluorescence images (at 508 nm) of seven sections from top to bottom of a CHO cell expressing Wzm protein fused to the green fluorescent protein variant GFP2. The bottom row displays fluorescence images (at 508 nm) of sections, from top to bottom, of a CHO cell expressing Wzt protein fused to GFP2. Consecutive sections of these cells were 1.5 µm apart, which is roughly within the axial resolution of the two-photon microscope. Vertical scale bar indicates fluorescence intensity, in arbitrary units.
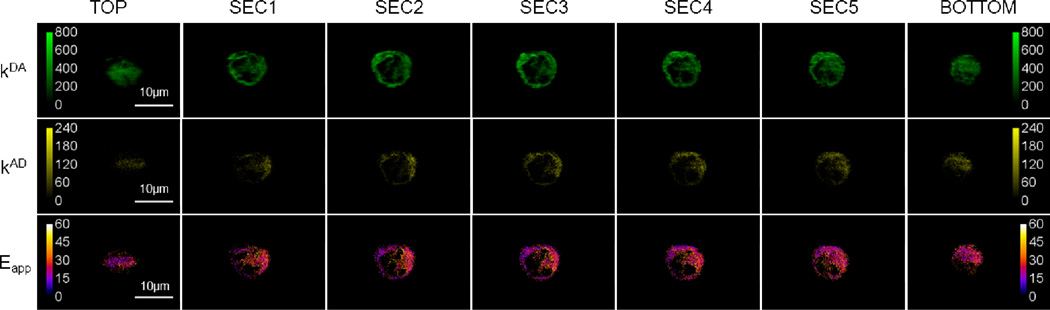
In order to probe the mutual interaction of Wzm and Wzt, we expressed fusion proteins GFP2-Wzm and YFP-Wzt – where YFP was the yellow variant of the green fluorescent protein GFP43 – separately or in combination in CHO cells, and imaged them using a two-photon microscope with spectral resolution.34 The cells expressing only GFP2-Wzm or only YFP-Wzt were imaged to obtain elementary spectra of GFP2 and YFP, respectively. In the cells co-expressing both fusion proteins, GFP236 was directly excited by laser light and acted as a donor (D) of energy, while YFP43 acted as an acceptor (A) in a FRET process18, 19, 44 and was almost completely insensitive to excitation by laser light under the conditions that excite GFP2. Spectral images obtained from cells co-expressing GFP2-Wzm and YFP-Wzt were unmixed (see the Matherial and Methods section) to obtain the fluorescence of donors in presence of acceptors (kDA) and the fluorescence of acceptors in presence of donors (kAD). From kDA and kAD, the apparent FRET efficiency (Eapp) for every image pixel was determined as described briefly in the Matherial and Methods section, and in more detail elsewhere.29, 34 Typical results shown in Fig. 2 indicate that Wzt and Wzm interact in living cells to form complexes at the plasma membrane, as evidenced by FRET occuring mostly at the plasma membrane but also in regions possibily occupied by intracellular membranes (such as the ER, Golgi, and transport vesicles). Analysis of these data alone is insufficient to extract detailed structural information regarding the complexes. However, such data do provide critical evidence for the interaction between Wzm and Wzt at the level of the plasma membrane, which will be used below in elucidating the structure of the entire hetero-oligomer.
FIGURE 2.
Mapping the spatial distribution of complexes of Wzm and Wzt proteins in thin (1.5 µm) optical sections through a CHO cell co-expressing Wzm proteins tagged with GFP2 as a donor, and Wzt proteins tagged with YFP as an acceptor. Two-dimensional spatial distribution maps of fluorescence of donors in presence of acceptors (kDA) and the fluorescence of acceptors in presence of donors (kAD) were obtained from spectral unmixing, while the apparent FRET efficiency (Eapp) map was calculated from kDA and kAD for every pixel (see text for details).
The quaternary structure of Wzt in the absence of Wzm
CHO cells were transfected to express the fusion proteins GFP2-Wzt and YFP-Wzt, separately or in combination at varied plasmid ratios, and imaged as described above. The apparent FRET efficiency (Eapp) for every image pixel was determined and a distribution of FRET efficiencies (or the Eapp histogram) for each cell imaged was obtained by plotting the number of pixels that fell into a particular range of Eapp values against the corresponding Eapp value (Fig. 3). Of all the cells co-expressing GFP2-Wzt and YFP-Wzt that were analyzed, roughly 90% exhibited single peaks located at various positions along the horizontal axis (i.e., Eapp), while about 10% showed multiple peaks in their Eapp histograms. The two-dimensional Eapp maps for representative cells expressing Wzt proteins and their corresponding Eapp distributions are shown in Fig. 3A–B. The Eapp distributions for cells presenting only one peak were fitted to single Gaussian functions. Some cells, which presented broad Eapp distributions with dominant peaks, were also considered for analysis, with their dominant peaks fitted to Gaussian functions. The best-fit curves provided individual peak positions, that is, Eapp values corresponding to the distribution maxima (Fig. 3B). The Eapp distributions presenting multiple peaks were analyzed separately (see below).
Peak positions from single-peaked histograms acquired from 380 cells expressing Wzt in the absence of Wzm were binned and collected in “meta-histograms” representing the total number of peak positions obtained for each Eapp value (in intervals of 0.02). As seen from Fig. 3C, the meta-histogram showed five distinct peaks, which means that the peak positions obtained for individual histograms did not vary over a continuum of Eapp values but rather clustered around five distinct values each corresponding to a different tetrameric configuration in individual cells. The five peaks in the meta-histograms (Fig. 3C) were simulated using five Gaussian functions, with the location on the horizontal axis of their maxima accurately predicted by a rhombus-shaped tetramer model 34, 35 depicted in Fig. 3D. According to this model, the positions of all five peaks in the meta-histogram may be computed from a single Eapp value, which is termed the “pair-wise FRET efficiency” (Ep) and, incidentally, corresponds to the second peak in the meta-histogram shown in Fig. 3C. (see Fig. 3D for mathematical expressions corresponding to each peak position). The fitting process consisted of minimizing the mismatch between experimental and simulated data by adjusting Ep as well as the amplitudes and widths of the Gaussian functions. A detailed justification of this fitting approach is provided in the Materials and Methods section above.
The quaternary structure of Wzt in the presence of untagged Wzm
To investigate, whether the presence of Wzm affects the stoichiometry and quarternary structure of Wzt, CHO cells were transfected to coexpress GFP2-Wzt, YFP-Wzt and untagged Wzm, and imaged to obtain Eapp histograms for each cell. As in the experiments described above, the Eapp histograms of about 90% of the cells showed single peaks, while the rest showed broader distributions. Typical results for cells expressing Wzt in the presence of untagged Wzm are presented in Fig. 4.
The peak positions for the singly-peaked histograms and broad histograms showing dominant peaks were collected, binned, and plotted against Eapp. The meta-histogram thus obtained and its best-fit to a sum of three Gaussians are shown in Fig. 4C, while the three configurations of a square tetramer model used to simulate the histogram are shown in Fig. 4D. In contrast to the model describing Wzt homo-oligomers in the absence of Wzm, the square-shaped tetramer correctly described this experimental meta-histogram.
The quaternary structure of Wzm in the absence or in the presence of untagged Wzt
To determine the quaternary structure of Wzm, CHO cells were transfected to express fusion proteins GFP2-Wzm and YFP-Wzm in the absence or presence of untagged Wzt and were imaged to obtain pixel-level distributions of Eapp for each individual cell. Here again, the Eapp histograms of most cells expressing Wzm in the absence of Wzt presented single peaks, while some presented multiple peaks. Typical data for the cells expressing Wzm proteins are shown in Fig. 5. Tails towards high Eapp values were seen in almost all the histograms, suggesting the presence of small and poorly defined peaks at high Eapp values. The meta-histogram obtained by collecting and binning the peak positions of the single-peaked Eapp histograms or the dominant peaks in broad and multi-peaked histograms and its theoretical best-fit to a sum of three Gaussians is shown in Fig. 5C. The peak positions of the individual Gaussians in the meta-histogram correspond to the apparent FRET efficiencies of different configurations of square-shaped tetramers as shown in Fig. 5D.
FIGURE 5.
Homo-oligomerization of Wzm expressed in CHO cells in the absence of Wzt. A. Apparent FRET efficiency (Eapp) maps of representative cells expressing Wzm. B. Distributions of FRET efficiencies (Eapp histograms) across the pixels composing the FRET efficiency maps shown in panel A. Most histograms presented single peaks, although those with peaks at lower values also present tails at higher Eapp values. C. Meta-histogram obtained from the peak positions of 240 cells presenting single or dominant peaks in their Eapp distributions (circles) and their theoretical best-fit to three Gaussians shown individually (red, green and brown lines) or as a sum (black line). The three Gaussians correspond to different configurations of square-shaped tetramers shown in the same order in panel C. The best fit was obtained for Ep ± S.D. equal to 0.121±0.010. D. Square-shaped tetramer model and its corresponding expressions for FRET efficiencies used to interpret the results in panel C. Ep is the pair-wise FRET efficiency between a single donor (D) and a single acceptor (A).
Similarly, the Wzm oligomeric structure in the presence of Wzt was also investigated, and typical results are shown in Fig. 6. Analysis of the meta-histogram obtained from such cells revealed that Wzm forms square-shaped tetramers in the presence of untagged Wzt (Fig. 6C).
FIGURE 6.
Homo-oligomerization of Wzm expressed in CHO cells in the presence of untagged Wzt. A. Apparent FRET efficiency (Eapp) maps of representative cells expressing Wzm in the presence of Wzt. B. Distributions of FRET efficiencies (Eapp histograms) across the pixels composing the FRET efficiency maps shown in panel A. The majority of cells presented single peaks, with minor tails at higher Eapp values. C. Meta-histogram obtained from the peak positions of 210 cells presenting single or dominant peaks in their Eapp distributions (circles) and theoretical best-fit to three correlated Gaussians shown individually (red, green and brown lines) or as a sum (black line). The three Gaussians correspond to different configurations of a square-shaped tetramer shown in the same order in panel D. The best-fit was obtained for Ep ± S.D. equal to 0.121±0.010. D. Square-shaped tetramer model and its corresponding expressions for FRET efficiencies used to interpret the results in panel C. Ep is the pair-wise FRET efficiency between a single donor (D) and a single acceptor (A).
Confirmation of the quaternary structure from analysis of broad Eapp distributions
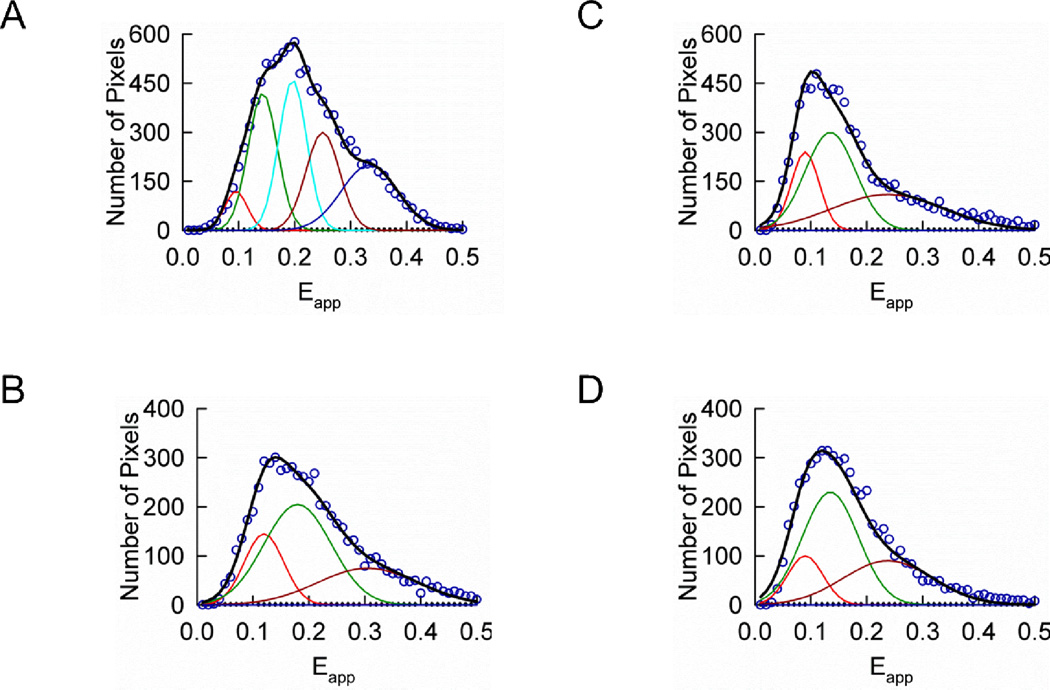
We simulated, using either the rhombus-shaped tetramer or the square-shaped tetramer model, the distribution of FRET efficiencies for individual cells showing multiple peaks (~10% of the total population of cells). The Ep values for individual cells were allowed to vary between Ep−S.D. and Ep+S.D. as determined from the meta-histogram analysis (Figs. 3C, 4C, 5C and 6C). The theoretical best-fits of the cells showing multiple peaks in their Eapp distributions are shown in Fig. 7. The different Gaussians correspond to the different configurations of the tetramers depicted in Figs. 3D and 4D, respectively. Variations in the FRET efficiency due to the relative orientation of the donor and acceptor transition dipoles, as well as smearing of the FRET distributions due to the point spread function of the microscope, influence the width of individual Gaussians. Because of this, here again, the widths of the Gaussians were adjusted independently of each other in the process of data fitting. Following this procedure, an agreement between the quaternary structure models and the experimental results was obtained as follows: the rhombus-shaped tetramer for Wzt in the absence of Wzm (Fig. 7A), and the square tetramer for Wzt in the presence of Wzm (Fig. 7B), Wzm in the absence of Wzt (Fig. 7C), and Wzm in the presence of Wzt (Fig. 7D). This is similar to the results of the analysis of meta-histograms obtained from single-peaked histograms. As for the observation that the Eapp histograms of most cells exhibited single peaks (corresponding to single combinations of donors and acceptors in the cell) while some exhibited multiple peaks (corresponding to multiple D-A combinations), an explanation will be attempted in the DISCUSSION section.
FIGURE 7.
Determination of the quaternary structure of Wzt and Wzm expressed in CHO cells showing multiple peaks in their Eapp histograms. A. Eapp histogram of a cell expressing Wzt in the absence of untagged Wzm (circles) and theoretical best-fit (for Ep = 0.143) to five Gaussians shown individually (red, green, cyan, brown, and blue lines) corresponding to different configurations of the rhombus shaped tetramer shown in Fig. 3D or as a sum (black line). B. Eapp histogram of a cell expressing Wzt in the presence of untagged Wzm (circles) and theoretical best-fit (for Ep = 0.180) to three Gaussians shown individually (red, green and brown lines) corresponding to different configurations of a square shaped tetramer shown in Fig. 4D or as a sum (black line). C. Eapp histogram of a cell expressing Wzm in the absence of untagged Wzt (circles) and theoretical best-fit (for Ep = 0.135) to three Gaussians shown individually (red, green and brown lines) corresponding to different configurations of square shaped tetramer of Fig. 5D or as a sum (black line). D. Eapp histogram of a cell expressing Wzm in the presence of untagged Wzt (circles) and theoretical best-fit (for Ep = 0.135) to five Gaussians shown individually (red, green and brown lines) corresponding to different configurations of square shaped tetramer of Fig. 6D or as a sum (black line).
Reliability of the pixel-level FRET method
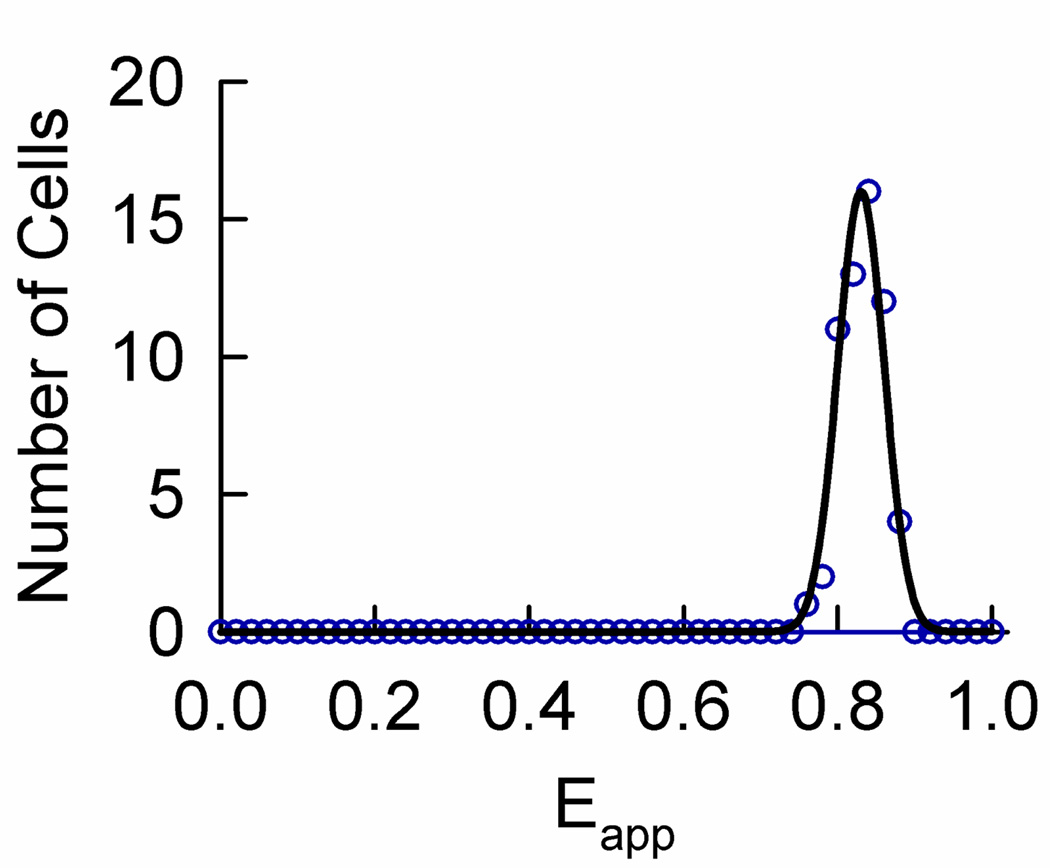
As seen above, determinations of the quaternary structure of Wzt in the absence or presence of Wzm, and of that of Wzm in the absence or presence of Wzt were based primarily on FRET efficiency meta-histograms (obtained from the peak position of single-peaked histograms or multi-peaked histograms exhibiting dominant peaks) and their best-fit to different theoretical models. Analysis of the smaller number of histograms presenting multiple peaks provided additional support to the quaternary structure model. Since all the results presented above relied heavily on the assumption that one peak in the meta-histogram corresponds to one oligomeric configuration, it was important to assess the validity of this assumption. To this end, we transfected CHO cells with plasmids encoding an artificial tetrameric construct consisting of three Venus molecules (a yellow variant of GFP, here serving as the FRET acceptor), and a Cerulean molecules (a cyan variant, serving as the FRET donor); this construct is denoted herein by VCVV. The plasmids were a generous gift from Dr. Steven Vogel of NIH and were described and characterized in detail elsewhere. 45 Analysis of cells expressing this construct was carried out the same way as for Wzm- and Wzt-expressing cells. As expected, the individual histograms of all cells expressing VCVV presented a single peak. By collecting and binning all those peaks, we obtained the meta-histogram shown in Fig. 8, which shows a clearly identifiable single peak. This is in agreement with our assumption that one tetrameric configuration corresponds to one peak in the meta-histogram, and demonstrates that our method of analysis does not introduce spurious peaks in the meta-histogram. We conclude that the multiple peaks obtained for Wzm and Wzt do originate from different combinations of donors and acceptors within a rhombus or square tetramer.
FIGURE 8.
Meta-histogram obtained from peak positions of individual Eapp histograms for CHO cells expressing an artificial tetramer (circles) consisting of three acceptors (Venus) and one donor (Cerulean).45 The solid line represents the theoretical best-fit to a single Gaussian.
Order-of-magnitude estimate of the two hetero-tetramers concentration
Using a reference solution consisting of 10 µM YFP and the average fluorescence intensity of donors corrected for loss through FRET (FD), we first estimated the molar concentrations for Wzm and Wzt separately. For instance, for the cells shown in Fig. 4 (for Wzt) and Fig. 6 (for Wzm), the molar concentrations of donor-tagged molecules were on the order of 10 nM. Then, using an approximate radius value of 10 µm for a CHO cell, we estimated the Wzm or Wzt concentrations to be on the order of a few hundred thousand molecules per cell. For all the cells imaged in this study, the numbers varied between a hundred thousand and one million molecules per CHO cell. For a P. aeruginosa cell, shaped as a rod of diameter of about 700 nm and length of 2 µm, this is the equivalent of a few tens to a few hundreds of molecules per cell. Given the quaternary structure we just determined, we estimate the number of Wzt and Wzm tetramers to be on the order of a few to a few tens per cell, which does not appear to be a particularly high expression level and might be close to physiological levels.
Discussion
Why the individual histogram peaks are broader than those of meta-histograms
Visual comparison between the individual peaks in the meta-histograms shown in Fig. 3–6 and the individual peaks of the multi-peaked histograms (Fig. 7) revealed that the widths of the individual peaks in multi-peaked histograms were vastly wider than those of the meta-histograms. The width of each Gaussian in the meta-histograms accounts for variations from cell to cell due to pH, viscosity, or other factors that may alter the properties of the fluorescent probes and their rotational diffusion rates, while the distributions of Eapp values accounting for the widths of the peaks in Eapp histograms showing single peaks or individual peaks of Eapp histograms showing multiple peaks were caused by two main factors: (a) changes in relative orientations of the individual transition dipoles of donors and acceptors undergoing rotational diffusion during the process of measurement, and (b) mixing of signals between adjacent tetramers (or possibly between tetramers and free monomers) due to image-blurring effects caused by the point spread function of the instrument. Numerical simulations suggest that the second effect should be stronger for higher concentrations of molecules.39 Both of these effects add significant blur to individual histograms, which is cleared away by the method based on generation of meta-histograms introduced in this paper. This allowed us to clearly resolve the fine structure of the Eapp histograms – as if we were using an imaging method with super-resolution – and thus determine the structure of the Wzm and Wzt oligomers.
Proposed structural model
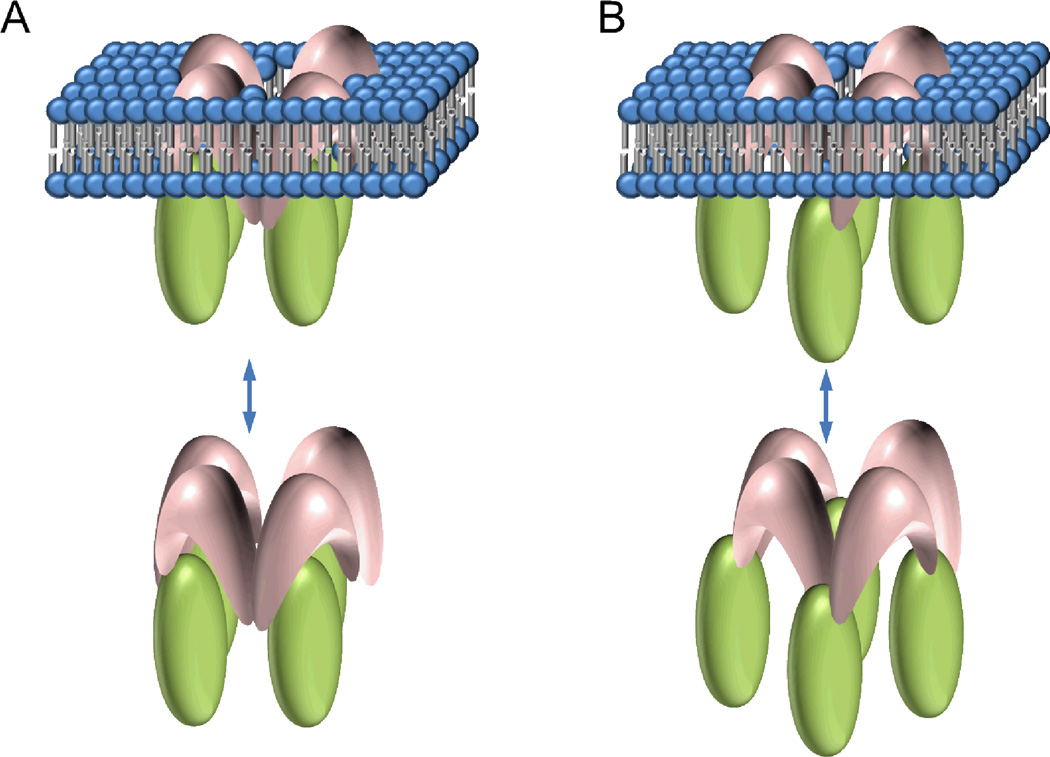
The FRET analysis described above indicates that the cytoplasmic Wzt protein forms a rhombus-shaped tetramer in the absence of the transmembrane domain Wzm, but changes its shape into a square-shaped tetramer in the presence of Wzm. By contrast, Wzm forms a square-shaped tetramer whether Wzt is present or not. In addition, we have demonstrated that the Wzm and Wzt oligomers interact with one another to form a hetero-oligomer. Based on these results, we are now ready to formulate a model for the supramolecular assembly of the Pseudomonas aeruginosa ABC transporter for polysaccharides, which consists of one Wzm homo-tetramer on top of one Wzt homo-tetramer, creating a hetero-dimer of homo-tetramers, or a hetero-octamer. Some ambiguity remains with regard to whether each protomer in the Wzm tetramer interacts with a single protomer in the Wzt tetramer and thus forms a head-to-head structure, or a Wzm protomer interacts with two adjacent Wzt protomers thereby forming an interlaced structure (Fig. 9). The two configurations differ from each other by a 45° rotation of the Wzt and Wzm tetramers relative to each other and around a common axis perpendicular to the membrane. Further biochemical work will be necessary to determine the specific interactions sites in order to choose one configuration over the other.
FIGURE 9.
Two versions of the hetero-octameric model proposed for the quaternary structure of the Wzm-Wzt ABC transporter. A. Four units of Wzm interacting with four units of Wzt face-to-face. B. Four units of Wzm are interlaced with four units of Wzt.
Considerations regarding the stability of the complexes
Additional information regarding this octamer may be gleaned from the observation that, while most of the cells presented single-peaked Eapp histograms, a small fraction presented multiple peaks. In other words, while for most cells, the proportion of donors and acceptors was on average the same in each homo-tetramer (of Wzm or Wzt), for some cells all three (for square tetramer) or five (for rhombus tetramers) peaks were present. This difference in composition suggests that the observed structures are dynamic and that the individual monomers may associate or dissociate reversibly. The fact that the histograms of some cells presented all the peaks suggests that the association/dissociation process is slow and those cells had not expressed the protein tetramers for a length of time sufficient to allow full randomization of their composition.
Comparison to other studies
The discovery that Wzm and Wzt form interacting tetramers is not an unusual finding in the study of ABC transporters. Indeed, several ABC transporters have been reported to associate into either dimers or multimers. For example, the human multidrug resistant half transporter MRP1,46 human cystic fibrosis transmembrane conductance regulator (CFTR)47 and the yeast Pdr5p full transporter48 have been reported to form dimers. Recently, the multidrug resistant half-transporter human breast cancer protein, ABCG2, was purified as a tetramer.49 Similarly, human ABCA1 transporter exists in intact fibroblasts as a homo-tetramer or even higher oligomerization states.50 Finally, the bi-component ABC transporter protein BtuCD involved in vitamin B12 uptake in E. coli – consisting of the homo-dimeric transmembrane protein BtuC and a homo-dimeric nucleotide-binding protein, BtuD – appears to form a hetero-tetramer 51 and serves as a paradigm for all other bacterial ABC transporters. All these studies were done in vitro and most of them were limited to the determination of the size of oligomers. Unlike those approaches, the spectrally-resolved FRET used in this study allowed us to determine the stoichiometry of a bi-component bacterial ABC transporter in living cells. We determined the oligomer size of each component as well as their quaternary structure in vivo and, based on these observations, proposed a quaternary structure model which is different from the model proposed for the transporter BtuCD in E. coli.
Significance of the present results
At this point the question that remains is how would tetramerization of Wzt or Wzm be relevant to substrate or even its translocation? Unlike the ABC transporters mentioned in the previous paragraph, which mostly translocate drugs or ions, the ABC transporter formed by Wzm and Wzt translocates a relatively large substrate, the A-band PS. The A-band PS sugar chain is a homopolymer of D-Rhamnose of approximately 70 monomers,52 giving it the length of ~28 nm, assuming an extended form of the polymer with a 0.4 nm per monomer. In fact, recent atomic force microscopy (AFM) studies have shown that the A-band PS can be up to ~36 nm long.53 In light of this, higher order oligomerization of Wzm and Wzt might be required for the translocation of the PS substrate. The Wzt tetramer could provide multiple binding sites for the long or bulky PS substrate. In analogy, the biochemical studies on the homo-tetrameric human ABCA1 led to the proposal that multiple binding sites exist for its substrates.50 Wzm is supposed to form a channel to facilitate the passage of PS across the membrane. It is conceivable therefore that Wzm being a tetramer can form an accommodating channel for the translocation of the bulky PS, a scenario originally proposed by Cuthbertson et al.12 Also, the fact that the Wzt protein tetramer changes its shape upon binding to Wzm may suggest that these structural changes allow for PS or PS-Wzt orientation prior to translocation.
One of the next steps in these studies would be to determine the binding interfaces for Wzt as well as the side chains of the Wzm that face the channel. This could be done by attempting to orient the Wzm and Wzt subunits within the general quaternary structure and under the constraints of the relative distances between protomers set by the present study. This step would require knowledge of the tertiary structure, which could be achieved using X-ray crystallographic measurements, which are currently unavailable.
An understanding of the structure and function of this ABC transporter provides critical information for developing antibiotics targeting the biosynthesis of the A-band LPS endotoxin, a primary factor in P. aeruginosa pathogenesis.
Supplementary Material
Insight, innovation, integration.
Recently introduced methods for Förster Resonance Energy Transfer (FRET) imaging of living cells permit determination of the quaternary structure of protein complexes from the number and relative disposition of peaks in the distribution of FRET efficiencies across FRET image pixels. Unfortunately, when the proteins of interest form rapidly dissociating/associating oligomers, the FRET efficiency distributions exhibit few or even single peaks, causing severe underestimation of the oligomer size. We describe a FRET-based method that circumvents this problem and allows one to probe the stoichiometry and structure of dynamic protein complexes in living cells. We use this method to determine the quaternary structure of an ABC transporter complex involved in the export of lipopolysaccharides to the surface of the pathogenic bacterium Pseudomonas aeruginosa.
Acknowledgments
This work was partly supported by a Bradley Foundation Catalyst grant (#MIL103076) and National Science Foundation grants (PFI-1114305 and PHY-1058470) to V.R. and a National Institutes of Health grant (R01-GM088403) and a National Science Foundation grant (DMR-1006332) to L.M. We thank Professor Kalina Hristova from the Department of Material Science and Engineering, Johns Hopkins University for providing the YFP standard, giving us access to her OptiMiS system and for helpful advice and discussions, and Ms. Sarvenaz Sarabipour from Professor Hristova research group for YFP protein production and characterization. We also thank Professor Robert E.W. Hancock (University of British Columbia) for providing Pseudomonas aeruginosa PAO1 strain, Professor James W. Wells (University of Toronto) for providing the pcDNA3.1 (+) gfp2-M2 and pcDNA3.1 (+) yfp-M2 plasmids and Dr. Steven S. Vogel (National Institutes of Health) for providing the plasmid for the artificial tetramer construct used to test the FRET method used in the study.
REFERENCES
- 1.Oldham ML, et al. Structural insights into ABC transporter mechanism. Curr. Opin. Struct. Biol. 2008;18:726–733. doi: 10.1016/j.sbi.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rees DC, et al. ABC transporters: the power to change. Nat. Rev. Mol. Cell Biol. 2009;10:218–227. doi: 10.1038/nrm2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seeger MA, van Veen HW. Molecular basis of multidrug transport by ABC transporters. Biochim. Biophys. Acta. 2009;1794:725–737. doi: 10.1016/j.bbapap.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Vergani P, et al. CFTR channel opening by ATP-driven tight dimerization of its nucleotide-binding domains. Nature. 2005;433:876–880. doi: 10.1038/nature03313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Igarashi Y, et al. The evolutionary repertoires of the eukaryotic-type ABC transporters in terms of the phylogeny of ATP-binding domains in eukaryotes and prokaryotes. Mol. Biol. Evol. 2004;21:2149–2160. doi: 10.1093/molbev/msh226. [DOI] [PubMed] [Google Scholar]
- 6.Rocchetta HL, et al. Genetics of O-antigen biosynthesis in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 1999;63:523–553. doi: 10.1128/mmbr.63.3.523-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reeves PR, et al. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 1996;4:495–503. doi: 10.1016/s0966-842x(97)82912-5. [DOI] [PubMed] [Google Scholar]
- 8.Hug I, Feldman MF. Analogies and homologies in lipopolysaccharide and glycoprotein biosynthesis in bacteria. Glycobiology. 2011;21:138–151. doi: 10.1093/glycob/cwq148. [DOI] [PubMed] [Google Scholar]
- 9.Abeyrathne PD, et al. Functional characterization of WaaL, a ligase associated with linking O-antigen polysaccharide to the core of Pseudomonas aeruginosa lipopolysaccharide. J. Bacteriol. 2005;187:3002–3012. doi: 10.1128/JB.187.9.3002-3012.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagao K, et al. Lipid outward translocation by ABC proteins. FEBS Lett. 2010;584:2717–2723. doi: 10.1016/j.febslet.2010.04.036. [DOI] [PubMed] [Google Scholar]
- 11.Rocchetta HL, Lam JS. Identification and functional characterization of an ABC transport system involved in polysaccharide export of A-band lipopolysaccharide in Pseudomonas aeruginosa. J. Bacteriol. 1997;179:4713–4724. doi: 10.1128/jb.179.15.4713-4724.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuthbertson L, et al. ABC transporters involved in export of cell surface glycoconjugates. Microbiol. Mol. Biol. Rev. 2010;74:341–362. doi: 10.1128/MMBR.00009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuthbertson L, et al. Substrate binding by a bacterial ABC transporter involved in polysaccharide export. Proc. Natl. Acad. Sci. USA. 2007;104:19529–19534. doi: 10.1073/pnas.0705709104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bodey GP, et al. Infections caused by Pseudomonas aeruginosa. Rev. Infect. Dis. 1983;5:279–313. doi: 10.1093/clinids/5.2.279. [DOI] [PubMed] [Google Scholar]
- 15.Govan JR, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoiby N, et al. Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol. 2010;5:1663–1674. doi: 10.2217/fmb.10.125. [DOI] [PubMed] [Google Scholar]
- 17.Hogardt M, Heesemann J. Adaptation of Pseudomonas aeruginosa during persistence in the cystic fibrosis lung. Int. J. Med. Microbiol. 2010;300:557–562. doi: 10.1016/j.ijmm.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 18.Selvin PR. The renaissance of fluorescence resonance energy transfer. Nat. Struct. Biol. 2000;7:730–734. doi: 10.1038/78948. [DOI] [PubMed] [Google Scholar]
- 19.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3 ed. New York, NY, USA: Springer; 2006. [Google Scholar]
- 20.Pearce LL, et al. Role of metallothionein in nitric oxide signaling as revealed by a green fluorescent fusion protein. Proc. Natl. Acad. Sci. USA. 2000;97:477–482. doi: 10.1073/pnas.97.1.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parsons M, et al. Imaging protein-protein interactions in cell motility using fluorescence resonance energy transfer (FRET) Biochem. Soc. Trans. 2004;32:431–433. doi: 10.1042/BST0320431. [DOI] [PubMed] [Google Scholar]
- 22.Caorsi V, et al. FRET measurements on fuzzy fluorescent nanostructures. Microsc. Res. Tech. 2007;70:452–458. doi: 10.1002/jemt.20444. [DOI] [PubMed] [Google Scholar]
- 23.Hink MA, et al. Imaging protein-protein interaction in living cells. Plant Mol. Biol. 2002;50:871–883. doi: 10.1023/a:1021282619035. [DOI] [PubMed] [Google Scholar]
- 24.Heyduk T. Measuring protein conformational changes by FRET/LRET. Curr. Opin. Biotechnol. 2002;13:292–296. doi: 10.1016/s0958-1669(02)00332-4. [DOI] [PubMed] [Google Scholar]
- 25.An SJ, Almers W. Tracking SNARE complex formation in live endocrine cells. Science. 2004;306:1042–1046. doi: 10.1126/science.1102559. [DOI] [PubMed] [Google Scholar]
- 26.Carriba P, et al. Detection of heteromerization of more than two proteins by sequential BRET-FRET. Nat. Methods. 2008;5:727–733. doi: 10.1038/nmeth.1229. [DOI] [PubMed] [Google Scholar]
- 27.Lleres D, et al. Quantitative analysis of chromatin compaction in living cells using FLIM-FRET. J. Cell Biol. 2009;187:481–496. doi: 10.1083/jcb.200907029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Demarco IA, et al. Monitoring dynamic protein interactions with photoquenching FRET. Nat. Methods. 2006;3:519–524. doi: 10.1038/nmeth889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raicu V, et al. Protein interaction quantified in vivo by spectrally resolved fluorescence resonance energy transfer. Biochem. J. 2005;385:265–277. doi: 10.1042/BJ20040226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pisterzi LF, et al. Oligomeric size of the M2 muscarinic receptor in live cells as determined by quantitative fluorescence resonance energy transfer. J. Biol. Chem. 2010;285:16723–16738. doi: 10.1074/jbc.M109.069443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maurel D, et al. Cell-surface protein-protein interaction analysis with time-resolved FRET and snap-tag technologies: application to GPCR oligomerization. Nat. Methods. 2008;5:561–567. doi: 10.1038/nmeth.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen L, et al. The extracellular domain of fibroblast growth factor receptor 3 inhibits ligand-independent dimerization. Sci. Signal. 2010;3:ra86. doi: 10.1126/scisignal.2001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raicu V. Efficiency of resonance energy transfer in homo-oloigomeric complexes of proteins. J. Biol. Phys. 2007;33:109–127. doi: 10.1007/s10867-007-9046-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raicu V, et al. Determination of supramolecular structure and spatial distribution of protein complexes in living cells. Nat. Photonics. 2009;3:107–113. [Google Scholar]
- 35.Raicu V. In: Nanoscopy and Multidimensional Optical Fluorescence Microscopy. Diaspro A, editor. Boca Raton: CRC Press; 2010. [Google Scholar]
- 36.Zimmermann T, et al. Spectral imaging and linear un-mixing enables improved FRET efficiency with a novel GFP2-YFP FRET pair. FEBS Lett. 2002;531:245–249. doi: 10.1016/s0014-5793(02)03508-1. [DOI] [PubMed] [Google Scholar]
- 37.Zacharias DA, et al. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 38.Denk W, et al. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 39.Singh DR, Raicu V. Comparison between whole distribution- and average-based approaches to the determination of fluorescence resonance energy transfer efficiency in ensembles of proteins in living cells. Biophys. J. 2010;98:2127–2135. doi: 10.1016/j.bpj.2010.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li E, et al. Quantitative measurements of protein interactions in a crowded cellular environment. Anal. Chem. 2008;80:5976–5985. doi: 10.1021/ac800616u. [DOI] [PubMed] [Google Scholar]
- 41.Zipfel WR, et al. Nonlinear Magic: Multiphoton Microscopy in the Biosciences. Nat. Biotechnol. 2003;21:1369–1377. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- 42.Svoboda K, Yasuda R. Principles of two-photon excitation microscopy and its applications to neuroscience. Neuron. 2006;50:823–839. doi: 10.1016/j.neuron.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 43.Tsien RY. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 44.Raicu V, Popescu AI. Integrated Molecular and Cellular Biophysics. London, United Kingdom: Springer; 2008. [Google Scholar]
- 45.Koushik SV, et al. Anomalous surplus energy transfer observed with multiple FRET acceptors. Plos One. 2009;4:e8031. doi: 10.1371/journal.pone.0008031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenberg MF, et al. The structure of the multidrug resistance protein 1 (MRP1/ABCC1) - Crystallization and single-particle analysis. J. Biol. Chem. 2001;276:16076–16082. doi: 10.1074/jbc.M100176200. [DOI] [PubMed] [Google Scholar]
- 47.Zhang L, et al. Architecture of the cystic fibrosis transmembrane conductance regulator protein and structural changes associated with phosphorylation and nucleotide binding. J. Struct. Biol. 2009;167:242–251. doi: 10.1016/j.jsb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 48.Ferreira-Pereira A, et al. Three-dimensional reconstruction of the Saccharomyces cerevisiae multidrug resistance protein Pdr5p. J. Biol. Chem. 2003;278:11995–11999. doi: 10.1074/jbc.M212198200. [DOI] [PubMed] [Google Scholar]
- 49.Dezi M, et al. The multidrug resistance half-transporter ABCG2 is purified as a tetramer upon selective extraction from membranes. Biochim. Biophys. Acta. 2010;1798:2094–2101. doi: 10.1016/j.bbamem.2010.07.034. [DOI] [PubMed] [Google Scholar]
- 50.Denis M, et al. Characterization of oligomeric human ATP binding cassette transporter A1. Potential implications for determining the structure of nascent high density lipoprotein particles. J. Biol. Chem. 2004;279:41529–41536. doi: 10.1074/jbc.M406881200. [DOI] [PubMed] [Google Scholar]
- 51.Locher KP, et al. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science. 2002;296:1091–1098. doi: 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- 52.King JD, et al. Review: Lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Innate Immun. 2009;15:261–312. doi: 10.1177/1753425909106436. [DOI] [PubMed] [Google Scholar]
- 53.Ivanov IE, et al. Relating the physical properties of Pseudomonas aeruginosa lipopolysaccharides to virulence by atomic force microscopy. J. Bacteriol. 2011;193:1259–1266. doi: 10.1128/JB.01308-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.