Abstract
Xenopus transcription factor IIIA (TFIIIA) is phosphorylated on serine-16 by CK2. Replacements with alanine or glutamic acid were made at this position in order to address the question of whether phosphorylation possibly influences the function of this factor. Neither substitution has an effect on the DNA or RNA binding activity of TFIIIA. The wild-type factor and the alanine variant activate transcription of somatic- and oocyte-type 5S rRNA genes in nuclear extract immunodepleted of endogenous TFIIIA. The glutamic acid variant (S16E) supports the transcription of somatic-type genes at levels comparable to those of wild-type TFIIIA; however, there is no transcription of the oocyte-type genes. This differential behavior of the phosphomimetic mutant protein is also observed in vivo when using early-stage embryos, where this mutant failed to activate transcription of the endogenous oocyte-type genes. Template exclusion assays establish that the S16E mutant binds to the oocyte-type 5S rRNA genes and recruits at least one other polymerase III transcription factor into an inactive complex. Phosphorylation of TFIIIA by CK2 may allow the factor to continue to act as a positive activator of the somatic-type genes and simultaneously as a repressor of the oocyte-type 5S rRNA genes, indicating that there is a mechanism that actively promotes repression of the oocyte-type genes at the end of oogenesis.
The synthesis of 5S rRNA during Xenopus oogenesis and embryogenesis provides a paradigm for developmental control of transcription (79). The oocyte-type 5S rRNA genes, which number more than 20,000 per haploid genome, are transcribed during oogenesis and briefly during early embryogenesis. The 400 somatic-type genes are active at all stages of development. Formation of transcription initiation complexes on the internal promoters of the 5S rRNA genes requires the initial binding of transcription factor IIIA (TFIIIA), followed by the ordered addition of TFIIIC and TFIIIB (46). Despite minor differences in the sequences of the two types of 5S rRNA genes, TFIIIA binds to the internal promoters of both with equal affinity (52). TFIIIC, however, preferentially binds to and stabilizes the complex of TFIIIA on the somatic-type genes (44, 79). Thus, the differential transcription of the two 5S rRNA genes during early development could, at least in part, be due to the levels of transcription factors. Nonetheless, the principal mediator of 5S rRNA gene transcription from gastrulation onward is chromatin structure.
Histone H1 orchestrates the repression of oocyte-type genes in somatic cells (5, 21, 66, 78). This inhibition occurs after the midblastula transition (MBT), when adult H1A begins to replace the maternal histone H1 variant, H1M (20, 21, 40). Nucleosomes are arranged differently over the two types of 5S RNA genes in somatic cells (10, 30, 83) because histone H1 acts as an architectural determinant (58, 67). In the presence of the linker histone, a stable nucleosome is positioned on the oocyte 5S rRNA gene that prevents binding of TFIIIA, whereas on the somatic gene, essential promoter elements remain exposed to the transcription factor (67). It is not apparent whether histone modification is important in this case (37, 71, 72); however, this simple model of promoter accessibility controlled by histone H1 appears to be sufficient to explain the differential expression of the Xenopus 5S rRNA genes in somatic cells (35).
Whether there are any events during oogenesis that contribute to the differential activity of the two types of 5S rRNA genes is less clear. During oogenesis, the somatic- and oocyte-type 5S rRNA genes are transcribed with nearly the same efficiency (ratio of somatic to oocyte gene transcription [S/O ratio] of 4). After fertilization, when transcription resumes at the MBT of embryogenesis, the transcriptional efficiency of the oocyte-type 5S rRNA genes has already dropped more than 10-fold relative to that of the somatic genes (81). This S/O ratio of 50 to 100 is actually established in the egg at some time during hormone-dependent maturation, a process that includes germinal vesicle (GV) breakdown and progression to metaphase of meiosis II. The latter transcriptional bias can be mimicked in whole-cell extract (S150) prepared from mixed-stage oocytes, which has led to the suggestion that a cytoplasmic component may participate in the initial inactivation of the oocyte-type genes (54, 59).
We have found that TFIIIA is phosphorylated, preferentially on serine-16, by a CK2 activity in Xenopus oocytes (75). CK2 (formerly casein kinase II) is a ubiquitous serine/threonine protein kinase composed of two catalytic (α or α′) and two regulatory (β) subunits (reviewed in references 49, 53, and 61). In addition to this tetrameric structure, the individual α and β subunits have been found to associate with a significant number of other proteins. Recent evidence indicates that CK2 plays an essential role in cell growth, proliferation, and survival. In order to determine whether there is a functional consequence to the phosphorylation of TFIIIA by CK2, serine-16 and those at two other CK2 consensus sites in TFIIIA were changed to either alanine or glutamic acid. None of the mutations has a significant impact on the binding of TFIIIA to either the oocyte- or somatic-type genes or to 5S rRNA. However, TFIIIA in which serine-16 is replaced with glutamic acid, which acts as a putative mimetic of phosphoserine, cannot support transcription of oocyte-type 5S rRNA genes in nuclear extract, whereas transcription of somatic-type genes is unaffected. The mutant also exhibits this differential activity in developing embryos. Template exclusion assays demonstrate that transcription complexes containing the glutamic acid variant can form on the oocyte 5S rRNA genes but are inactive. The behavior of this mutant protein mirrors the transcriptional activity of embryos when zygotic transcription begins at the MBT, suggesting that phosphorylation of TFIIIA by CK2, during oocyte maturation, contributes to the initial differential expression of 5S rRNA genes.
MATERIALS AND METHOD
Plasmids and nucleic acids
Plasmid pTFLNK has the 5′-flanking sequence of Xenopus TFIIIA inserted between the EcoRI and SacI restriction sites of pGEM2 and was a generous gift from M. T. Andrews (University of Minnesota, Duluth). A 1,497-bp NdeI-BamHI restriction fragment, containing the entire TFIIIA coding sequence, was excised from plasmid pTA102 (18) and ligated into pTFLNK digested with the same enzymes to generate plasmid pTFA. Mutations in pTFA were made with a QuickChange site-directed mutagenesis kit (Stratagene). Serine-16 (TCT) was changed to alanine (GCA) or glutamic acid (GAA). Serine-314 (TCG) and serine-328 (TCT) were also changed to these amino acids with the same codons. Capped mRNA encoding TFIIIA and its variants was prepared by runoff transcription of pTFA linearized with BamHI with SP6 RNA polymerase (70). After digestion of the DNA template with RQ DNase I (Promega), the mRNA was purified by passage through a Microspin S-300 HR column (Amersham). Internally radiolabeled 5S rRNA was prepared by runoff transcription of pT75S linearized with DraI with T7 RNA polymerase (62). Plasmids pWXlo1 and pWXls1 contain a single copy of a Xenopus laevis oocyte-type or somatic-type 5S rRNA gene, respectively (75), and were used for in vitro transcription assays and DNase I footprinting. The X. borealis maxigene encodes a somatic 5S rRNA gene with an insertion at position 115 that yields a 140-nucleotide transcript (65). A 250-bp EcoRI restriction fragment containing a copy of the Xenopus tRNAMet1 gene (14) was cloned into pBS/SK+ to generate pWXlt1.
Preparation of oocytes and embryos
Oocytes were isolated from samples of ovary tissue and manually staged in modified high-salt Barth's solution (110 mM NaCl, 2 mM KCl, 1 mM MgSO4, 2 mM NaHCO3, 0.5 mM Na2HPO4, 15 mM Tris-HCl, pH 7.6). Stage VI oocytes were matured by incubation with 10 μg of progesterone per ml in OR2 buffer (82.5 mM NaCl, 2.5 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 1 mM Na2HPO4, 5 mM HEPES, pH 7.8) at room temperature until GV breakdown had occurred in the majority of the cells. Only oocytes displaying the white maturation spot were taken for experiments. For in vitro fertilization of Xenopus eggs, adult females were injected in the dorsal lymph sac with 250 U of human chorionic gonadotropin the night before spawning. In the morning, they were injected with 500 U of human chorionic gonadotropin. Testes were removed from an adult male, gently minced, added to 0.1× OR2 buffer, and mixed with eggs at room temperature. Approximately 30 min following fertilization, an equal volume of 2% cysteine (pH 7.8) was added and gently mixed until the jelly coat was removed from the majority of the embryos. The embryos were washed 5 times with 0.1× OR2 buffer and then placed at 18°C. GVs were isolated from oocytes that were initially swelled by incubation in TM buffer (5 mM Tris-HCl [pH 7.8], 10 mM MgCl2) for 1 h. Cells were transferred to J buffer (10 mM HEPES [pH 7.4], 70 mM NH4Cl, 7 mM MgCl2, 0.1 mM EDTA, 2.5 mM dithiothreitol [DTT], 10% glycerol), and GVs were forced from the whole cells by pressure applied with a spatula or forceps. Each GV was collected with 1 μl of J buffer and stored at −80°C.
In vivo phosphorylation of TFIIIA
Staged oocytes were prepared as already described and injected with 400 nCi of [γ-32P]ATP. Oocytes and eggs were then incubated in OR2 buffer at 18°C for 16 h. Three oocytes or GVs per stage were homogenized in 50 mM HEPES (pH 7.5)-5 mM DTT-1 mM phenylmethylsulfonyl fluoride containing a cocktail of protease inhibitors (Roche 1 836 170). Protein A-Sepharose bound with TFIIIA antibody was used to immunoprecipitate TFIIIA (in 50 mM HEPES [pH 7.5]-100 mM NaCl-0.1% NP-40-5 mM DTT) at 4°C for 2 h. Beads were washed five times with 1 ml of the same buffer before resuspension in loading buffer containing sodium dodecyl sulfate (SDS). The immunoprecipitated samples were analyzed by SDS-polyacrylamide gel electrophoresis, followed by autoradiography.
Binding assays
The binding of TFIIIA to 5S rRNA genes was measured with quantitative DNase I footprinting assays, and its binding to 5S rRNA was measured with mobility shift assays. Both procedures have been described earlier (62).
In vitro transcription assays
Transcription assays in GV extract were carried out as described previously (74). The amount of DNA template used and the order of addition are given in the figure legends. The activity of mutant forms of TFIIIA was measured in extract depleted of endogenous factor by immunoprecipitation with protein A-Sepharose beads that had been incubated with anti-TFIIIA antiserum as described in reference 65a. GV extract (90 μl) was incubated with 30 μl of beads suspended in J buffer with end-to-end rotation for 1 h at 4°C. The Sepharose beads were removed by low-speed (500 × g) centrifugation. The amount of antiserum needed to deplete the extract without interfering with general polymerase III (pol III) transcription was determined empirically. Samples of mutant and wild-type TFIIIA were diluted into J buffer and added to the depleted extract, which was then used in standard assay conditions (74).
Assays of transcription in embryos
Capped synthetic mRNA (7.2 ng) encoding wild-type or mutant TFIIIA was coinjected along with 78 nCi of [α-32P]UTP into the animal hemisphere of one-cell embryos just as the first cleavage furrow appeared (64). Injected embryos were allowed to develop in 0.1× OR2 buffer at room temperature and collected approximately 10 h after fertilization at the beginning of gastrulation (stage 10) (56). Three embryos injected with the same mRNA were homogenized, and total RNA was isolated as described by Rollins et al. (64) and analyzed by electrophoresis on 10% polyacrylamide gels containing 7 M urea, followed by autoradiography. In order to determine the levels of expression of the injected mRNAs, embryos were homogenized in protein dissociation buffer (1.5 M Tris-HCl [pH 8.9], 2% SDS, 1% DTT) and then incubated at 100°C for 2 min. The proteins were alkylated in 0.3 M iodoacetamide for 30 min at 37°C. One embryo equivalent was loaded onto an SDS-polyacrylamide gel and taken through a Western blot assay with polyclonal anti-TFIIIA antiserum.
Template exclusion assays
GV extract immunodepleted of endogenous TFIIIA was used for template exclusion assays. The total amount of DNA in each assay was kept constant at 500 ng by addition of plasmid (pUC19) DNA. The amount of each template DNA, the order of addition, and the times of incubation are given in the figure legends. In all cases, transcription was initiated by addition of ribonucleoside triphosphates. All other details of these in vitro assays are the same as those described above.
RESULT
Phosphorylation of TFIIIA
TFIIIA in Xenopus oocytes is phosphorylated on serine-16, which is located in a consensus sequence for protein kinase CK2 (75). A CK2 activity, in fact, is associated with TFIIIA and remains with the factor through several steps of purification. TFIIIA has two functions in Xenopus oocytes. In the nucleus, it is a positive regulator of transcription of 5S rRNA genes, necessarily being the first transcription factor to bind to the internal promoters of these genes (22). TFIIIA also forms a complex with 5S rRNA in the cytoplasm as a storage particle for the RNA until it is needed for ribosome biosynthesis (34, 60). In order to determine whether the phosphorylation of TFIIIA has some functional consequence, we first examined the levels of phosphorylation during oogenesis and whether this modification was limited to a certain subcellular fraction of TFIIIA.
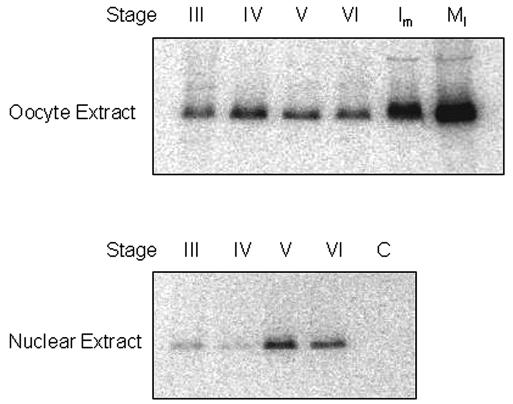
Staged oocytes were injected with [γ-32P]ATP and incubated overnight. TFIIIA was isolated by immunoprecipitation from extract prepared from whole oocytes or from GVs. Radiolabeled TFIIIA was visualized by electrophoresis, followed by autoradiography (Fig. 1). It is noteworthy that the mobility of the radiolabeled TFIIIA recovered by immunoprecipitation is greater than that of TFIIIA isolated from the 7S RNP complex. This difference, presumably due to phosphorylation, is under study. The first detectable phosphorylation of TFIIIA occurs during stage III, and it remains relatively constant up to, and including, stage VI; however, there is some redistribution of phosphorylated TFIIIA during oogenesis. Whereas much of the labeled TFIIIA is cytoplasmic during stages III and IV, it becomes increasingly localized to the nucleus during stage V. It needs to be kept in mind that only about 1% of the TFIIIA in oocytes is located in the nucleus, so the distribution of labeled factor in early-stage oocytes reflects the general distribution of total TFIIIA. However, the phosphorylated form of the factor accumulates in the nucleus during the later stages of oogenesis. Progesterone-induced maturation of oocytes causes GV breakdown, the progression from meiosis I to arrest in metaphase of meiosis II, and the activation of several protein kinases, including CK2. We detected an apparent fivefold increase in the amount of phosphorylated TFIIIA during in vitro maturation. However, during this period, the levels of TFIIIA actually decrease more than 10-fold (68), meaning that a substantial amount of TFIIIA ultimately becomes phosphorylated in unfertilized eggs. These labeling experiments are consistent with the known activation of CK2 that occurs during oocyte maturation (3, 41, 55).
FIG. 1.
Increased phosphorylation of TFIIIA upon oocyte maturation. Staged oocytes were injected with [γ-32P]ATP and kept at 18°C for 16 h; extract from either whole cells or GVs was then prepared. In the case of mature oocytes, the cells were either injected and then treated with progesterone (lane Im) or treated with progesterone and injected after GV breakdown (lane MI). TFIIIA was retrieved from the extracts by immunoprecipitation and analyzed by SDS-polyacrylamide gel electrophoresis, followed by autoradiography. The intensity of the bands was measured with a laser densitometer. Extract from injected stage IV oocytes incubated with protein A-Sepharose (lane C) served as a control for immunoprecipitation.
Amino acid substitutions at serine-16 do not affect the nucleic acid binding activity of TFIIIA
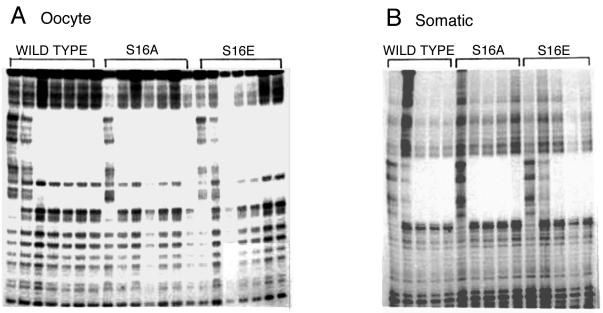
In order to examine whether phosphorylation of TFIIIA might influence one of its functions, we created a series of variants that potentially mimic either the phosphorylated (substitution with glutamic acid) or unphosphorylated (substitution with alanine) state of the protein. Serine-16, the primary site of phosphorylation, is located within the β-turn of the first zinc finger of TFIIIA. There is considerable evidence that much of the free energy of binding of TFIIIA to the 5S rRNA genes is derived from the first three zinc fingers (11, 12, 15, 17, 43, 48, 69). Thus, it seemed reasonable that phosphorylation or mutations at serine-16 might affect the binding of TFIIIA to either or both 5S rRNA genes. However, quantitative footprinting assays revealed no significant differences in the binding affinities of the wild-type and serine-16 mutant forms for either the oocyte- or somatic-type genes (Fig. 2). Since purified components were used in these assays, it was also possible to measure binding with TFIIIA expressed in Escherichia coli and phosphorylated in vitro with commercial CK2. We did not detect any difference in binding affinity between control TFIIIA and protein that was phosphorylated in vitro (results not shown). TFIIIA can be phosphorylated on serine-314 in addition to serine-16 in vitro, although we have not detected this modification in vivo (75). An alanine substitution at serine-314 had no impact on binding, whereas replacement with glutamic acid caused about a fourfold decrease in binding affinity for both 5S rRNA genes. We judge this not to be a significant change.
FIG. 2.
Substitutions at serine-16 do not affect the binding affinity of TFIIIA for either 5S rRNA gene. Wild-type TFIIIA or mutant TFIIIA with serine-16 replaced with alanine (S16A) or glutamic acid (S16E) was used in DNase I footprinting assays with the oocyte- or somatic-type 5S rRNA gene. Each sample contained 2 nM end-labeled, linearized plasmid and 0, 10, 15, 20, 30, 40, or 100 nM protein (oocyte gene) or 0, 10, 20, 30, or 40 nM protein (somatic gene).
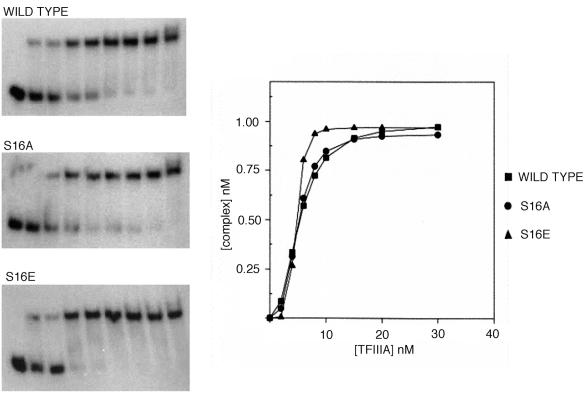
The collection of mutants was also tested for binding to 5S rRNA with mobility shift assays (Fig. 3). None of these variants exhibited any detectable perturbation in binding affinity relative to the wild-type protein. TFIIIA phosphorylated in vitro never exhibited more than a fourfold decrease in binding affinity for 5S rRNA (results not shown). The relative dissociation constants for the wild-type and mutant forms of TFIIIA are presented in Table 1. These data indicate that phosphorylation is unlikely to have an appreciable effect on the binding of TFIIIA to either DNA or RNA. The structure of a complex containing the first three zinc fingers of TFIIIA bound to its cognate DNA sequence has been determine by nuclear magnetic resonance spectroscopy (82), and a cocrystal structure of the first six zinc fingers bound to DNA has also been solved (57). In these structures, serine-16 is exposed and lies opposite the recognition helix that is positioned in the major groove. Thus, the absence of any effect by either mutagenesis or in vitro phosphorylation is consistent with these structures.
FIG. 3.
Substitutions at serine-16 do not affect the binding affinity of TFIIIA for 5S rRNA. Internally radiolabeled 5S rRNA (1 nM) was incubated with 0, 2, 4, 6, 8, 10, 15, 20, or 30 nM protein. Samples were analyzed by electrophoresis on 8% nondenaturing gels. Autoradiographs were scanned with a laser densitometer for the construction of binding isotherms.
TABLE 1.
Relative dissociation constants for wild-type and mutant forms of TFIIIAa
| Mutation | Avg relative dissociation constant ± SE
|
||
|---|---|---|---|
| Somatic-type gene | Oocyte-type gene | 5S rRNA | |
| S16A | 0.41 ± 0.04 | 0.48 ± 0.03 | 0.91 ± 0.04 |
| S16E | 1.08 ± 0.04 | 1.09 ± 0.14 | 0.97 ± 0.08 |
| S314A | 1.03 ± 0.22 | 0.95 ± 0.22 | 1.17 ± 0.34 |
| S314E | 4.56 ± 0.38 | 3.76 ± 0.79 | 0.51 ± 0.08 |
| S328A | NDb | ND | 0.79 ± 0.28 |
| S328E | ND | ND | 0.85 ± 0.09 |
Binding is expressed as the ratio of the apparent dissociation constant of mutant TFIIIA relative to that of wild-type TFIIIA.
ND, not determined
TFIIIA with glutamic acid substituted for serine-16 cannot support transcription of oocyte-type 5S rRNA genes
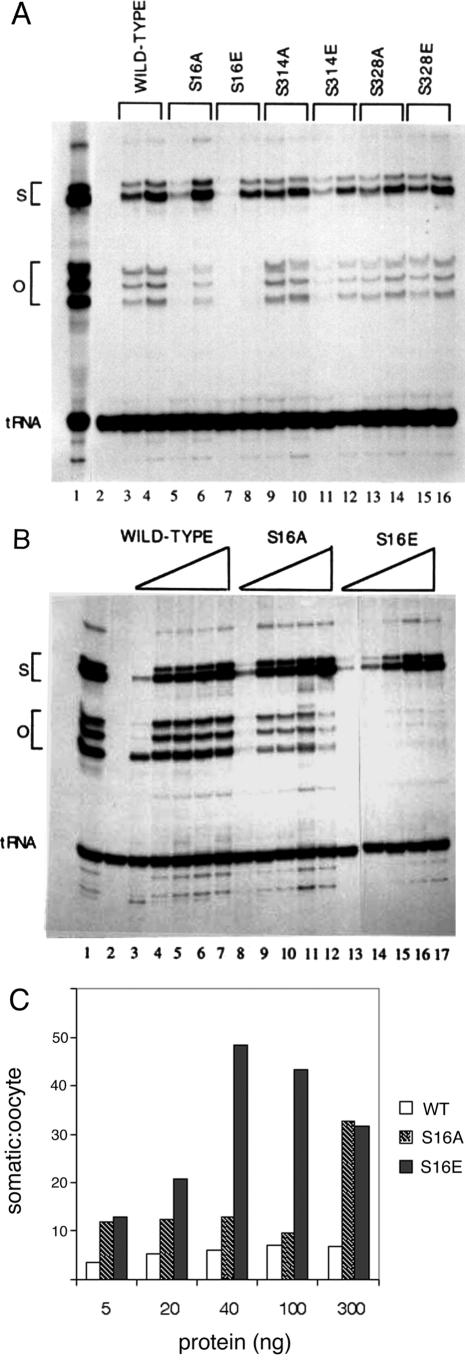
The ability of the TFIIIA variants to activate transcription was tested in GV extract that was immunodepleted of endogenous TFIIIA (Fig. 4A). In order to compare directly the transcription of oocyte-type and somatic-type genes, both templates were included in the assay mixture at a ratio of 4:1, respectively, to compensate for the difference in transcriptional efficiency in this extract. The somatic maxigene contains an insertion downstream of the internal promoter that generates transcripts that are sufficiently separated from the transcripts generated from the oocyte gene (65). TFIIIA or the indicated variants were added to immunodepleted extract, and the transcription products were analyzed on a denaturing polyacrylamide gel. Internally radiolabeled tRNA was added to each reaction mixture at the end of the assay to control for nucleic acid recovery and sample loading. A small decrease in the transcription of both genes for the S314E mutant (lane 11) may be due to the modest reduction in binding affinity measured for this protein (Table 1). Likewise, there is also a small decrease in the activity of the S16A mutant protein (lane 5). However, the most substantive effect is seen for the S16E mutant protein, which can activate transcription of the somatic, but not the oocyte, gene (lanes 7 and 8).
FIG.4.
TFIIIA with serine-16 replaced with glutamic acid cannot restore transcription of oocyte 5S rRNA genes in immunodepleted GV extract. Each transcription assay mixture includes 5 μl of extract and 100 ng of DNA (containing a 4:1 mixture of oocyte-type to somatic-type maxigene). After preincubation for 15 min, transcription was initiated by the addition of ribonucleoside triphosphates, including [α-32P]CTP. Reactions were stopped after 90 min, and radiolabeled tRNA was added in order to normalize the recovery of RNA from each sample, which were analyzed on a polyacrylamide gel containing 7 M urea. Lane 1 is an assay with GV extract prior to immunodepletion of endogenous TFIIIA, and lane 2 is depleted extract to which DNA template, but no TFIIIA, was added. (A) Lanes 3 to 16 contain pairs of assays containing 20 or 100 ng of the indicated variant of TFIIIA. (B) Lanes 3 to 7, 8 to 12, and 13 to 17 contain 5, 20, 40, 100, or 300 ng of the indicated variant of TFIIIA. S and O indicate the positions of transcripts from the somatic- and oocyte-type genes, respectively. (C) The autoradiograph in panel B was scanned with a laser densitometer. The S/O transcript ratios were normalized relative to the exogenous tRNA added to each assay and are presented as bar graphs. WT, wild type.
Additional transcription assays at higher concentrations of TFIIIA established that the mutant protein with glutamic acid at residue 16 does not support appreciable transcription of the oocyte-type gene even though transcription of the somatic gene is essentially at wild-type levels (Fig. 4B). The alanine substitution at this site somewhat diminishes, but clearly does not eliminate, transcription of the oocyte gene. Importantly, robust transcription of the somatic gene establishes that the glutamic acid substitution at position 16 does not significantly perturb the structure of the protein. The modest decrease in transcription obtained with the alanine variant suggests that the serine residue might normally participate in some type of interaction that facilitates transcription of the oocyte gene. Transcription assays at even higher concentrations of TFIIIA exhibited nonspecific “squelching” effects (29), so it was not possible to reach levels of the glutamic acid mutant protein that support transcription of the oocyte genes.
The autoradiograph in Fig. 4B was scanned with a laser densitometer in order to quantitate the amounts of transcription in these assays; the data are presented as bar graphs in Fig. 4C. The S/O ratio is approximately 4 to 5 for immunodepleted extract supplemented with wild-type TFIIIA. This is the same preference measured in unfractionated GV extract (80) and for 5S rRNA genes injected into oocyte nuclei (6, 26). In the case of the S16A mutant protein, this ratio is elevated slightly to about 10, except at the highest concentration of exogenous factor, where squelching effects appear to inhibit transcription of the oocyte, but not the somatic, genes. The modest increase in the S/O ratio suggests that the alanine substitution affects the transcription complex on the oocyte-type genes, perhaps by altering the interaction between TFIIIA and TFIIIC (4, 79). TFIIIA with the glutamic acid substitution shows a progressively larger S/O ratio as the amount of this variant is increased that plateaus at levels of around 50. Notably, this preference corresponds to that measured in S150 extract prepared from whole oocytes (52, 54, 59) and to the endogenous levels of transcription in embryos following the midblastula transition (73, 81).
We have supplemented immunodepleted GV extract with wild-type TFIIIA that was first incubated with purified CK2 and ATP (results not shown). There is some specific reduction in the transcription of the oocyte-type 5S rRNA genes in this case; however, the degree of repression was variable in these experiments. It is possible that the CK2 in the exogenous sample of TFIIIA acts on other pol III targets, such as TFIIIB, during the course of the transcription assay, potentially masking or offsetting any effects of phosphorylation on the former. In addition, there was no control for the amount of phosphatase activity in any given preparation of extract. For these reasons, subsequent experiments used the glutamic acid and alanine mutant forms of TFIIIA as opposed to factor phosphorylated in vitro.
Activity of TFIIIA variants in vivo
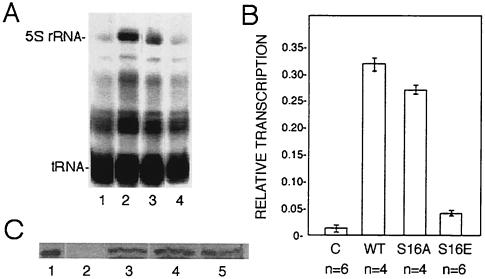
Single-cell Xenopus embryos injected with mRNA encoding TFIIIA have elevated levels of 5S rRNA by the gastrula stage, owing to the reactivation of the normally dormant oocyte-type genes (2, 64). We used this assay to examine the behavior of the serine-16 mutant forms of TFIIIA in vivo. When the first cleavage furrow became visible, embryos were injected with [α-32P]UTP and mRNA encoding wild-type or mutant TFIIIA. Embryos were collected at the beginning of gastrulation (stage 10) (56), and total RNA from three embryo equivalents was analyzed by electrophoresis on denaturing polyacrylamide gels (Fig. 5A). Injection of [α-32P]UTP alone (lane 1) shows the basal level of 5S rRNA synthesis, which is predominantly the somatic type (2). Injection of mRNA encoding wild-type TFIIIA results in significant activation of 5S rRNA synthesis (lane 2). This RNA was isolated from the gel and analyzed on a partially denaturing polyacrylamide gel that can resolve oocyte and somatic 5S rRNAs owing to differences in secondary-structure stability (73). The majority of the 5S rRNA comigrated with oocyte-type 5S rRNA (results not shown), in accord with previous experiments that showed that overexpression of TFIIIA in early embryos activates the normally repressed oocyte-type genes (2, 6, 64). Expression of the S16A mutant factor can also activate transcription of the oocyte-type genes, albeit somewhat less than the wild-type factor (lane 3). Strikingly, the S16E mutant factor does not stimulate transcription significantly beyond the basal level (lane 4). Autoradiographs of replicates of this experiment were scanned with a laser densitometer, and the amount of 5S rRNA synthesized in each case was normalized to a band corresponding to tRNA (Fig. 5B).
FIG. 5.
TFIIIA with serine-16 replaced with glutamic acid fails to reactivate transcription of oocyte-type 5S rRNA genes in early embryos. Embryos were injected when the first cleavage furrow began to appear with capped mRNA encoding wild-type TFIIIA or the indicated mutant TFIIIA along with [α-32P]UTP. (A) Embryos were collected at the beginning of gastrulation (stage 10), total RNA was isolated, and 3 embryo equivalents was analyzed by electrophoresis and autoradiography. Lanes: 1, embryos injected with [α-32P]UTP only; 2 to 4, embryos injected with mRNA encoding wild-type TFIIIA or S16A or S16E mutant TFIIIA, respectively. (B) Autoradiographs of replicates of this experiment were scanned with a laser densitometer. The amount of 5S rRNA in each lane relative to the indicated band of tRNA, which serves as an internal control, is presented. Error bars indicate the standard deviation of the mean, and n is the number of independent measurements. C, control; WT, wild type. (C) The amount of ectopically expressed TFIIIA in injected embryos was determined with a Western blot assay. Whole-cell extract corresponding to one embryo was analyzed in each lane. Lanes: 1, purified TFIIIA; 2, embryos injected with [α-32P]UTP only; 3 to 5, embryos injected with radiolabel and mRNA encoding wild-type TFIIIA or the S16A or S16E variant of TFIIIA, respectively.
Consistent with the transcription assays in immunodepleted nuclear extract, the embryo injection experiments show that the glutamic acid mutant form of TFIIIA cannot activate transcription of the oocyte-type 5S rRNA genes. Aliquots of the homogenate prepared from each sample of embryos were also examined by Western blot assay in order to establish that this effect is due to the activity of the protein, rather than its level of expression (Fig. 5C). The amount of TFIIIA synthesized from the injected mRNA is similar in each case and does not account for the differences in the amount of transcription.
The S16E mutant of TFIIIA forms an inactive transcription complex on oocyte 5S rRNA genes
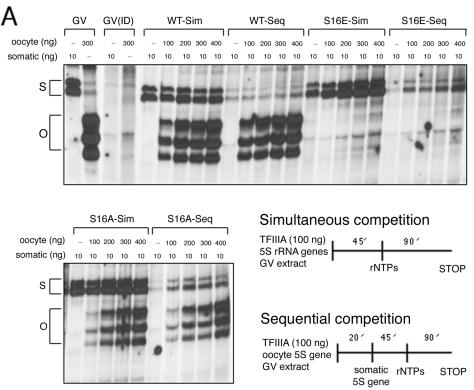
The DNA footprinting assays indicate that the binding affinity of the S16E mutant form of TFIIIA for the oocyte-type gene is similar to that of wild-type TFIIIA. Thus, the effect of this substitution on transcription is likely due to the perturbation of a protein-protein interaction within the transcription initiation complex. In order to examine this point more closely, we carried out a series of template exclusion assays with immunodepleted GV extract (Fig. 6). The competition assays were organized in two different ways. In those referred to as simultaneous (Sim), both oocyte and somatic genes were incubated together in extract supplemented with the indicated form of TFIIIA and transcription was initiated by the addition of ribonucleoside triphosphates. In the sequential (Seq) assays, the oocyte-type gene was incubated in the extract prior to the addition of the somatic-type gene. This was followed by a further incubation before initiation of transcription.
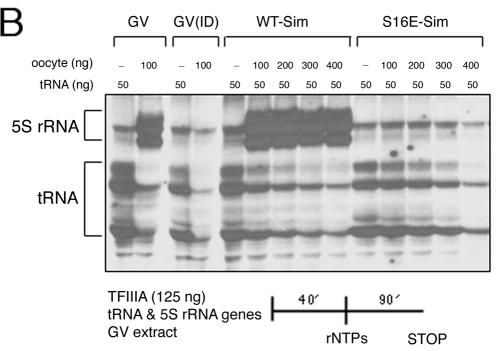
FIG. 6.
Template exclusion assays demonstrate that the S16E mutant form of TFIIIA supports the formation of an inactive complex on oocyte 5S rRNA genes. GV extract was immunodepleted of endogenous TFIIIA and then supplemented with wild-type (WT) or mutant TFIIIA. (A) A somatic 5S rRNA maxigene was challenged with increasing amounts of oocyte-type gene. The two DNA templates were added together at the beginning of the assay (Sim), or the oocyte-type gene was added to the extract first, followed after 20 min by the somatic-type gene (Seq). Transcription was initiated by the addition of ribonucleoside triphosphates (rNTPs). Transcripts were analyzed on a denaturing polyacrylamide gel. The positions of the somatic (S) and oocyte (O) transcripts are denoted by brackets. Lanes marked GV indicate assays in extract prior to immunodepletion, and GV(ID) indicates assays in depleted extract that was not supplemented with exogenous TFIIIA. (B) A tRNA gene was challenged with increasing amounts of oocyte-type gene to determine whether the S16E mutant form of TFIIIA recruits other pol III transcription factors. The two DNA templates were added together to extract, and after a 40-min incubation, the transcription assays were initiated by the addition of ribonucleoside triphosphates. Brackets denote the position of 5S rRNA and tRNA transcripts.
Wild-type TFIIIA behaves as expected. Both 5S rRNA genes are transcribed when they are incubated together in extract. As the amount of the oocyte gene is increased relative to the somatic gene in this assay, there is some decrease in transcription of the latter (WT-Sim). When an excess of the oocyte gene is incubated with extract prior to the addition of the somatic gene, transcription of the latter is completely blocked (WT-Seq), since limiting amounts of pol III transcription factors are recruited to the first template.
The activity of the S16A mutant form of TFIIIA is generally the same as that of wild-type TFIIIA. When both templates are added together, the amount of oocyte transcripts is proportional to the amount of template. When the oocyte gene is incubated prior to the somatic gene (S16A-Seq), transcription is predominantly from the former. However, unlike the case of wild-type TFIIIA, transcription of the somatic gene is not blocked entirely. This suggests that the stability of the initiation complex containing the alanine mutant protein may be modestly compromised during the time course of the assay, allowing for some assembly of new complexes on the promoters of the somatic genes. Similarly, in some replicates of the simultaneous assays in which the two genes were added together, transcription of the somatic gene was slightly greater with the alanine variant relative to wild-type TFIIIA, which likewise suggests that transcription complexes containing S16A on the oocyte-type genes are either less stable or form more slowly than those containing the wild-type factor. A similar phenomenon was noted in template exclusion experiments with S150 extract (59).
The results of the template exclusion assays with the glutamic acid mutant are similar to those with the alanine mutant with the important exception that in no case is there any transcription of the oocyte genes. The results of the sequential assay are particularly notable because they provide compelling evidence that, despite being quiescent, the oocyte-type genes must be recruiting one or more of the pol III transcription factors. This is consistent with our determination that the glutamic acid substitution does not affect the affinity of TFIIIA for either 5S rRNA gene. Moreover, it appears that this change in TFIIIA has converted it from an activator to a specific repressor of transcription of the oocyte-type 5S rRNA genes.
To determine whether other pol III factors besides the glutamic acid mutant form of TFIIIA are being assembled on the oocyte 5S rRNA gene, we performed a template exclusion assay with a tRNAMET gene, which only requires TFIIIB and TFIIIC to form a functional initiation complex for RNA pol III (Fig. 6). In the presence of the S16E mutant protein, increasing amounts of the oocyte gene diminished transcription of the tRNA gene. This result indicates that at least one other pol III transcription factor is recruited to the promoter of the 5S rRNA gene by the mutant form of TFIIIA. Because assembly of initiation complexes on 5S rRNA genes is ordered, with TFIIIC following TFIIIIA, and because the former stabilizes the binding of the latter, it is most likely that, at a minimum, TFIIIC is also present on the oocyte-type genes.
DISCUSSION
CK2 and control of pol III transcription
Of the general transcription factors used by RNA pol III, TFIIIB seems to be the most frequent target of protein kinases. ERK2 (24), p34cdc2 (31), and CK2 (27, 39) are three kinases identified thus far that can modify this factor. TFIIIB directly recruits pol III to the preinitiation complex, and phosphorylation of this factor can either activate (ERK2 and CK2) or repress (p34cdc2) transcription. There is evidence that TFIIIC may be phosphorylated; however, no putative kinases have been determined (13, 16, 33, 63, 76). La antigen, a termination and recycling factor that also influences the transcriptional efficiency of class III genes, is down-regulated upon phosphorylation by CK2 (23). Thus, at least three factors (TFIIIA, TFIIIB, and La) that control pol III transcription are targets of CK2.
CK2 activity is ubiquitous and has been implicated in the phosphorylation of an extraordinary number of targets, which has confounded efforts to determine the physiological roles of this kinase and its various isoforms (49, 53). Moreover, there is little information regarding the regulation of CK2 that might otherwise provide clues to it function(s). Recent studies indicate that stable interactions between the regulatory β subunits of CK2 and its targets may provided some degree of regulation (49), and indeed, CK2 forms stable complexes with TFIIIB (28, 39) and TFIIIA (75). Notwithstanding this limited understanding of regulation, there is considerable evidence to implicate CK2, which is closest phylogenetically to the cyclin-dependent and mitogen-activated kinases, in the control of cell growth and proliferation (1, 49, 53).
CK2 is necessary for efficient transcription of class III genes in yeast and mammalian cells, suggesting the conservation of a fundamental mechanism for regulation (27, 32, 39). Surprisingly, CK2 targets the TBP subunit of TFIIIB in yeast cells (27) but the Brf subunit in human cells (39). In the latter case, it was shown that phosphorylation of TFIIIB by CK2 facilitates its interaction with TFIIIC and thereby promotes formation of the transcription initiation complex. In yeast, the phosphorylation of TFIIIB by CK2 also enhances recruitment to the promoter; however, the involvement of TFIIIC in this instance has not been tested (28). The strong correlation between the elevated levels of pol III transcription (7) and CK2 activity in transformed and rapidly dividing cells (1, 49) likewise indicates that this kinase generally acts as a positive effector of transcription of class III genes. In the case of TFIIIA, phosphorylation by CK2 at the end of oogenesis seemingly allows the factor to promote (somatic type) and simultaneously to repress (oocyte type) transcription.
Another example of a dual role for CK2 in the regulation of pol III transcription comes from the human U6 genes (36). The kinase, which appears to be physically located near the U6 promoter, targets some component of the RNA polymerase itself, and this phosphorylation is required for transcription. Conversely, the kinase can also phosphorylate some component of the TFIIIB variant (TBP-Brf2-Bdp1) used by type 3 promoters, which lie upstream rather than within their respective genes. Phosphorylation in the latter case suppresses U6 transcription. These results underscore not only the essential role of CK2 in the transcription of pol III genes but also the complexity and diversity of regulation.
CK2 activity during oocyte maturation
A full-grown, stage VI Xenopus oocyte is arrested in meiosis I. Upon progesterone stimulation, GV breakdown occurs and the oocyte completes the first meiotic division and then arrests in metaphase of meiosis II (25). This hormone-induced maturation is mediated by components of a mitogen-activated protein kinase cascade, as well as cell cycle regulators. It is notable that CK2 has been implicated in the activity of two key regulatory kinases that are necessary for oocyte maturation: Mos, which is part of the progesterone signaling cascade (8, 9), and Cdc2, which, when complexed with cyclin B, forms maturation-promoting factor (MPF), a major regulator of the cell cycle (55).
The Mos/MEK1/p42 mitogen-activated protein kinase cascade is activated by progesterone and leads to the activation of MPF (reviewed in reference 25). In Xenopus oocytes, the β subunit of CK2 binds to Mos and suppresses the kinase activity of the latter (9). Cdc2 phosphorylates the β subunit of Xenopus oocyte CK2 (55); similarly, human and avian cells arrested in mitosis also exhibit a marked increase in the phosphorylation of both the α and β subunits of CK2 by Cdc2 (50, 51). It will be especially important to determine whether the phosphorylation of the β subunit of CK2 by Cdc2 influences its interaction with Mos, since this could explain the positive feedback effect Cdc2 has on Mos activity during maturation. Notwithstanding this relationship, the phosphorylation of CK2 by Cdc2 during oocyte maturation results in modest (2.5-fold) activation of the activity of the former and may release it from its complex with Mos, thereby increasing its effective concentration and possibly its ability to phosphorylate other substrates (3, 41, 55). Notably, these changes in CK2 activity occur at the same time that there is some reprogramming of the oocyte-type 5S rRNA genes, which becomes apparent when transcription resumes at the MBT (81).
Although CK2 appears to be predominantly nuclear in somatic cells, reports of its location in amphibian oocytes have been contradictory, with some indicating that there is a significant amount of the kinase in the cytoplasm (38, 41, 45, 47). We found CK2 activity associated with TFIIIA purified from 7S RNP particles (75), consistent with reports that this kinase is found with assorted cytoplasmic RNP complexes in amphibian oocytes (19, 41, 42). Furthermore, TFIIIA can be coimmunoprecipitated from oocyte extract with antibodies to the α subunit of CK2, indicating that a stable complex exists between the factor and the kinase, possibly through the catalytic subunit (M. Malik and P. W. Huber, unpublished results). These observations, along with the increased phosphorylation of TFIIIA that occurs at oocyte maturation (Fig. 1), indicate that, following GV breakdown, much of the factor available to the 5S rRNA genes will likely be phosphorylated. This could then account for the S/O ratio of 50 when transcription begins again at the MBT. Association of CK2 with nuclear TFIIIA has not been studied.
Inactivation of the oocyte-type genes at maturation
During the previtellogenic stages of oogenesis, TFIIIA, TFIIIB, TFIIIC, and RNA pol III are abundant and the oocyte and somatic genes are transcribed with nearly equal efficiencies (73, 81). Although the amount of TFIIIA declines more than 20-fold by the end of oogenesis, there are still ∼1010 molecules per oocyte (68). Most (>98%) of the TFIIIA in late-stage oocytes is located in the cytoplasm in a storage particle with 5S rRNA; however, sufficient amounts of the factor remain in the nucleus to support transcription of both somatic- and oocyte-type 5S rRNA genes. Nuclear extract prepared from late-stage oocytes (stages V and VI) exhibits an approximately fivefold preference for transcription of somatic 5S rRNA genes over oocyte-type gene transcription (59, 80) that matches the relative efficiency of transcription of cloned genes injected into oocyte nuclei (6, 26, 59). However, transcriptional efficiency in whole-cell (S150) extract prepared from predominantly late-stage oocytes is considerably different (52, 54, 59). In this case, transcription of the oocyte-type genes is specifically reduced, so that there is an approximately 50-fold bias in favor of the somatic genes. This level of discrimination is remarkably similar to that seen when transcription of the 5S rRNA genes resumes during early embryogenesis at the MBT (73, 81).
The transcriptional efficiency of other class III genes in S150 extract is comparable to that of somatic 5S rRNA genes, indicating that the activity of the oocyte-type genes is being specifically repressed (54). TFIIIA is not limiting in this extract (54, 59) and binds with the same affinity to the oocyte- and somatic-type 5S rRNA genes (52). Millstein et al. (54) showed that complexes assembled on the oocyte genes in S150 extract are transcriptionally silent and resist reactivation when injected into oocyte nuclei; conversely, active complexes form on the somatic genes. Most importantly, TFIIIA in this extract binds not only to the promoters of the active somatic-type genes but also to the promoters of the inactive oocyte-type genes. Addition of partially purified TFIIIC to S150 extract can selectively increase transcription of the oocyte-type genes, reducing the S/O ratio to approximately 5 (79). Since there is no comparable stimulation of tRNA genes or the somatic 5S rRNA genes, it appears that only the association of TFIIIC with the oocyte-type genes is affected in this extract. The simplest explanation for the strikingly different transcriptional activities of nuclear versus whole-cell extracts is that some component, presumably in the cytoplasm, can specifically mediate the repression of the oocyte-type genes. This activity, then, could account for the differential transcription of the two 5S rRNA genes observed at the MBT and suggests that there is a mechanism that actively promotes the repression of oocyte-type genes upon oocyte maturation. The activity of the S16E mutant form of TFIIIA exactly mirrors the transcriptional activity of S150 extract and suggests that CK2 is the agent responsible for the specific repression of oocyte-type 5S rRNA genes. Since TFIIIC binds to the binary complex of TFIIIA on the promoter, it is possible that phosphorylation of TFIIIA influences the interaction of the former with the oocyte-type genes. However, the present experiments cannot rule out effects on other interactions involving TFIIIA.
The intracellular distribution of CK2 in amphibian oocytes (41) and its increased activity at meiosis, owing to phosphorylation by Cdc2 (3, 51, 55), are consistent with this model of regulation. When Cdc2-cyclin B (MPF) was added to a reconstituted transcription system, the S/O ratio increased from 7 to 50 (77), which likely explains the apparent connection between Cdc2 activity and differential expression of the two 5S rRNA gene families. Our results suggest that the switch in 5S rRNA gene expression detected at the MBT results from phosphorylation events that occur during oocyte maturation. This mechanism for selective repression of the oocyte-type genes when levels of TFIIIA are still high (68) is not at odds with the accumulated evidence that the transcriptional activity of the 5S rRNA genes in somatic cells, where TFIIIA is limiting, is ultimately due to differences in nucleosome positioning. These two modes of transcriptional regulation operate in distinctly different circumstances, at different points in development, and in different cell types.
Acknowledgments
We are especially indebted to Matthew T. Andrews (University of Minnesota) for assistance with the embryo injection experiments, for long-term use of his microinjection equipment, gifts of plasmids, and general guidance. We thank him and members of our laboratory for comments on the manuscript.
This work was supported by a grant (GM38200) from the National Institutes of Health.
REFERENCE
- 1.Ahmed, K., D. A. Gerber, and C. Cochet. 2002. Joining the cell survival squad: an emerging role for protein kinase CK2. Trends Cell Biol. 12:226-230. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, M. T., and D. D. Brown. 1987. Transient activation of oocyte 5S RNA genes in Xenopus embryos by raising the level of the trans-acting factor IIIA. Cell 51:445-453. [DOI] [PubMed] [Google Scholar]
- 3.Belle, R., P. Cormier, R. Poulhe, J. Morales, D. Huchon, and O. Mulner-Lorillon. 1990. Protein phosphorylation during meiotic maturation of Xenopus oocytes: cdc2 protein kinase targets. Int. J. Dev. Biol. 34:111-115. [PubMed] [Google Scholar]
- 4.Bieker, J. J., P. L. Martin, and R. G. Roeder. 1985. Formation of a rate-limiting intermediate in 5S RNA gene transcription. Cell 40:119-127. [DOI] [PubMed] [Google Scholar]
- 5.Bouvet, P., S. Dimitrov, and A. P. Wolffe. 1994. Specific regulation of Xenopus chromosomal 5S rRNA gene transcription in vivo by histone H1. Genes Dev. 8:1147-1159. [DOI] [PubMed] [Google Scholar]
- 6.Brown, D. D., and M. S. Schlissel. 1985. A positive transcription factor controls the differential expression of two 5S RNA genes. Cell 42:759-767. [DOI] [PubMed] [Google Scholar]
- 7.Brown, T. R., P. H. Scott, T. Stein, A. G. Winter, and R. J. White. 2000. RNA polymerase III transcription: its control by tumor suppressors and its deregulation by transforming agents. Gene Expr. 9:15-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, M., and J. A. Cooper. 1997. The β subunit of CKII negatively regulates Xenopus oocyte maturation. Proc. Natl. Acad. Sci. USA 94:9136-9140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, M., D. Li, E. G. Krebs, and J. A. Cooper. 1997. The casein kinase II β subunit binds to Mos and inhibits Mos activity. Mol. Cell. Biol. 17:1904-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chipev, C. C., and A. P. Wolffe. 1992. Chromosomal organization of Xenopus laevis oocyte and somatic 5S rRNA genes in vivo. Mol. Cell. Biol. 12:45-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choo, Y., and A. Klug. 1993. A role in DNA binding for the linker sequences of the first three zinc fingers of TFIIIA. Nucleic Acids Res. 21:3341-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christensen, J. H., P. K. Hansen, O. Lillelund, and H. C. Thogersen. 1991. Sequence-specific binding of the N-terminal three-finger fragment of Xenopus transcription factor IIIA to the internal control region of a 5S RNA gene. FEBS Lett. 281:181-184. [DOI] [PubMed] [Google Scholar]
- 13.Clark, M. E., and A. Dasgupta. 1990. A transcriptionally active form of TFIIIC is modified in poliovirus-infected HeLa cells. Mol. Cell. Biol. 10:5106-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarkson, S. G., V. Kurer, and H. O. Smith. 1978. Sequence organization of a cloned tDNAmet fragment from Xenopus laevis. Cell 14:713-724. [DOI] [PubMed] [Google Scholar]
- 15.Clemens, K. R., P. Zhang, X. Liao, S. J. McBryant, P. E. Wright, and J. M. Gottesfeld. 1994. Relative contributions of the zinc fingers of transcription factor IIIA to the energetics of DNA binding. J. Mol. Biol. 244:23-35. [DOI] [PubMed] [Google Scholar]
- 16.Conesa, C., R. N. Swanson, P. Schultz, P. Oudet, and A. Sentenac. 1993. On the subunit composition, stoichiometry, and phosphorylation of the yeast transcription factor TFIIIC/τ. J. Biol. Chem. 268:18047-18052. [PubMed] [Google Scholar]
- 17.Del Rio, S., S. R. Menezes, and D. R. Setzer. 1993. The function of individual zinc fingers in sequence-specific DNA recognition by transcription factor IIIA. J. Mol. Biol. 233:567-579. [DOI] [PubMed] [Google Scholar]
- 18.Del Rio, S., and D. R. Setzer. 1991. High yield purification of active transcription factor IIIA expressed in E. coli. Nucleic Acids Res. 19:6197-6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deschamps, S., H. Jacquemin-Sablon, G. Triqueneaux, O. Mulner-Lorillon, M. Potier, J. P. Le Caer, F. Dautry, and M. le Maire. 1997. mRNP3 and mRNP4 are phosphorylatable by casein kinase II in Xenopus oocytes, but phosphorylation does not modify RNA-binding affinity. FEBS Lett. 412:495-500. [DOI] [PubMed] [Google Scholar]
- 20.Dimitrov, S., G. Almouzni, M. Dasso, and A. P. Wolffe. 1993. Chromatin transitions during early Xenopus embryogenesis: changes in histone H4 acetylation and in linker histone type. Dev. Biol. 160:214-227. [DOI] [PubMed] [Google Scholar]
- 21.Dworkin-Rastl, E., H. Kandolf, and R. C. Smith. 1994. The maternal histone H1 variant, H1M (B4 protein), is the predominant H1 histone in Xenopus pregastrula embryos. Dev. Biol. 161:425-439. [DOI] [PubMed] [Google Scholar]
- 22.Engelke, D. R., S.-Y. Ng, B. S. Shastry, and R. G. Roeder. 1980. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell 19:717-728. [DOI] [PubMed] [Google Scholar]
- 23.Fan, H., A. L. Sakulich, J. L. Goodier, X. Zhang, J. Qin, and R. J. Maraia. 1997. Phosphorylation of the human La antigen on serine 366 can regulate recycling of RNA polymerase III transcription complexes. Cell 88:707-715. [DOI] [PubMed] [Google Scholar]
- 24.Felton-Edkins, Z. A., J. A. Fairley, E. L. Graham, I. M. Johnston, R. J. White, and P. H. Scott. 2003. The mitogen-activated protein (MAP) kinase ERK induces tRNA synthesis by phosphorylating TFIIIB. EMBO J. 22:2422-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferrell, J. E., Jr. 1999. Xenopus oocyte maturation: new lessons from a good egg. Bioessays 21:833-842. [DOI] [PubMed] [Google Scholar]
- 26.Gargiulo, G., F. Razvi, and A. Worcel. 1984. Assembly of transcriptionally active chromatin in Xenopus oocytes requires specific DNA binding factors. Cell 38:511-521. [DOI] [PubMed] [Google Scholar]
- 27.Ghavidel, A., and M. C. Schultz. 1997. Casein kinase II regulation of yeast TFIIIB is mediated by the TATA-binding protein. Genes Dev. 11:2780-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghavidel, A., and M. C. Schultz. 2001. TATA binding protein-associated CK2 transduces DNA damage signals to the RNA polymerase III transcriptional machinery. Cell 106:575-584. [DOI] [PubMed] [Google Scholar]
- 29.Gill, G., and M. Ptashne. 1988. Negative effect of the transcriptional activator GAL4. Nature 334:721-724. [DOI] [PubMed] [Google Scholar]
- 30.Gottesfeld, J. M., and L. S. Bloomer. 1980. Nonrandom alignment of nucleosomes on 5S RNA genes of X. laevis. Cell 21:751-760. [DOI] [PubMed] [Google Scholar]
- 31.Gottesfeld, J. M., V. J. Wolf, T. Dang, D. J. Forbes, and P. Hartl. 1994. Mitotic repression of RNA polymerase III transcription in vitro mediated by phosphorylation of a TFIIIB component. Science 263:81-84. [DOI] [PubMed] [Google Scholar]
- 32.Hockman, D. J., and M. C. Schultz. 1996. Casein kinase II is required for efficient transcription by RNA polymerase III. Mol. Cell. Biol. 16:892-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoeffler, W. K., R. Kovelman, and R. G. Roeder. 1988. Activation of transcription factor IIIC by the adenovirus E1A protein. Cell 53:907-920. [DOI] [PubMed] [Google Scholar]
- 34.Honda, B. M., and R. G. Roeder. 1980. Association of a 5S gene transcription factor with 5S RNA and altered levels of the factor during cell differentiation. Cell 22:119-126. [DOI] [PubMed] [Google Scholar]
- 35.Howe, L., and J. Ausio. 1998. Nucleosome translational position, not histone acetylation, determines TFIIIA binding to nucleosomal Xenopus laevis 5S rRNA genes. Mol. Cell. Biol. 18:1156-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu, P., S. Wu, and N. Hernandez. 2003. A minimal RNA polymerase III transcription system from human cells reveals positive and negative regulatory roles for CK2. Mol. Cell 12:699-709. [DOI] [PubMed] [Google Scholar]
- 37.Jerzmanowski, A., and R. D. Cole. 1990. Flanking sequences of Xenopus 5S RNA genes determine differential inhibition of transcription by H1 histone in vitro. Mitotic phosphorylation of H1 decreases its inhibitory power. J. Biol. Chem. 265:10726-10732. [PubMed] [Google Scholar]
- 38.Jin, Y. J., and S. J. Burakoff. 1993. The 25-kDa Fk506-binding protein is localized in the nucleus and associates with casein kinase II and nucleolin. Biochemistry 90:7769-7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnston, I. M., S. J. Allison, J. P. Morton, L. Schramm, P. H. Scott, and R. J. White. 2002. CK2 forms a stable complex with TFIIIB and activates RNA polymerase III transcription in human cells. Mol. Cell. Biol. 22:3757-3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kandolf, H. 1994. The H1A histone variant is an in vivo repressor of oocyte-type 5S gene transcription in Xenopus laevis embryos. Proc. Natl. Acad. Sci. USA 91:7257-7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kandror, K. V., A. O. Benumov, and A. S. Stepanov. 1989. Casein kinase II from Rana temporaria oocytes. Intracellular localization and activity during progesterone-induced maturation. Eur. J. Biochem. 180:441-448. [DOI] [PubMed] [Google Scholar]
- 42.Kandror, K. V., and A. S. Stepanov. 1984. RNA-binding protein kinases from amphibian oocytes is a casein kinase II. FEBS Lett. 170:33-37. [DOI] [PubMed] [Google Scholar]
- 43.Kehres, D. G., G. S. Subramanyan, V. S. Hung, G. W. Rogers, Jr., and D. R. Setzer. 1997. Energetically unfavorable interactions among the zinc fingers of transcription factor IIIA when bound to the 5S rRNA gene. J. Biol. Chem. 272:20152-20161. [DOI] [PubMed] [Google Scholar]
- 44.Keller, H. J., P. J. Romaniuk, and J. M. Gottesfeld. 1992. Interaction of Xenopus TFIIIC with the TFIIIA-5S RNA gene complex. J. Biol. Chem. 267:18190-18198. [PubMed] [Google Scholar]
- 45.Krek, W., G. Maridor, and E. A. Nigg. 1992. Casein kinase II is a predominantly nuclear enzyme. J. Cell Biol. 116:43-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lassar, A. B., P. L. Martin, and R. G. Roeder. 1983. Transcription of class III genes: formation of preinitiation complexes. Science 222:740-748. [DOI] [PubMed] [Google Scholar]
- 47.Leiva, L., D. Carrasco, A. Taylor, M. Veliz, C. Gonzalez, C. C. Allende, and J. E. Allende. 1987. Casein kinase II is a major protein phosphorylating activity in the nuclei of Xenopus laevis oocytes. Biochem. Int. 14:707-717. [PubMed] [Google Scholar]
- 48.Liao, X., K. R. Clemens, L. Tennant, P. E. Wright, and J. M. Gottesfeld. 1992. Specific interaction of the first three zinc fingers of TFIIIA with the internal control region of the Xenopus 5S RNA gene. J. Mol. Biol. 223:857-871. [DOI] [PubMed] [Google Scholar]
- 49.Litchfield, D. W. 2003. Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem J. 369:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Litchfield, D. W., F. J. Lozeman, M. F. Cicirelli, M. Harrylock, L. H. Ericsson, C. J. Piening, and E. G. Krebs. 1991. Phosphorylation of the β subunit of casein kinase II in human A431 cells. Identification of the autophosphorylation site and a site phosphorylated by p34cdc2. J. Biol. Chem. 266:20380-20389. [PubMed] [Google Scholar]
- 51.Litchfield, D. W., B. Lüscher, F. J. Lozeman, R. N. Eisenman, and E. G. Krebs. 1992. Phosphorylation of casein kinase II by p34cdc2 in vitro and at mitosis. J. Biol. Chem. 267:13943-13951. [PubMed] [Google Scholar]
- 52.McConkey, G. A., and D. F. Bogenhagen. 1988. TFIIIA binds with equal affinity to somatic and major oocyte 5S RNA genes. Genes Dev. 2:205-214. [DOI] [PubMed] [Google Scholar]
- 53.Meggio, F., and L. A. Pinna. 2003. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 17:349-368. [DOI] [PubMed] [Google Scholar]
- 54.Millstein, L., P. Eversole-Cire, J. Blanco, and J. M. Gottesfeld. 1987. Differential transcription of Xenopus oocyte and somatic-type 5S genes in a Xenopus oocyte extract. J. Biol. Chem. 262:17100-17110. [PubMed] [Google Scholar]
- 55.Mulner-Lorillon, O., P. Cormier, J.-C. Labbé, M. Dorée, R. Poulhe, H. Osborne, and R. Bellé. 1990. M-phase-specific cdc2 protein kinase phosphorylates the β subunit of casein kinase II and increases casein kinase II activity. Eur. J. Biochem. 193:529-534. [DOI] [PubMed] [Google Scholar]
- 56.Nieuwkoop, P. D., and J. Faber. 1956. Normal tables of Xenopus laevis Daudin. Elsevier-North Holland, Amsterdam, The Netherlands.
- 57.Nolte, R. T., R. M. Conlin, S. C. Harrison, and R. S. Brown. 1998. Differing roles for zinc fingers in DNA recognition: structure of a six-finger transcription factor IIIA complex. Proc. Natl. Acad. Sci. USA 95:2938-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Panetta, G., M. Buttinelli, A. Flaus, T. J. Richmond, and D. Rhodes. 1998. Differential nucleosome positioning on Xenopus oocyte and somatic 5S RNA genes determines both TFIIIA and H1 binding: a mechanism for selective H1 repression. J. Mol. Biol. 282:683-697. [DOI] [PubMed] [Google Scholar]
- 59.Peck, L. J., L. Millstein, P. Eversole-Cire, J. M. Gottesfeld, and A. Varshavsky. 1987. Transcriptionally inactive oocyte-type 5S RNA genes of Xenopus laevis are complexed with TFIIIA in vitro. Mol. Cell. Biol. 7:3503-3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pelham, H. R. B., and D. D. Brown. 1980. A specific transcription factor that can bind either the 5S RNA gene or 5S RNA. Proc. Natl. Acad. Sci. USA 77:4170-4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pinna, L. A. 2002. Protein kinase CK2: a challenge to canons. J. Cell Sci. 115:3873-3878. [DOI] [PubMed] [Google Scholar]
- 62.Rawlings, S. L., G. D. Matt, and P. W. Huber. 1996. Analysis of the binding of Xenopus transcription factor IIIA to oocyte 5S rRNA and to the 5S rRNA gene. J. Biol. Chem. 271:869-877. [DOI] [PubMed] [Google Scholar]
- 63.Reynolds, W. F. 1993. The tyrosine phosphatase cdc25 selectively inhibits transcription of the Xenopus oocyte-type tRNAtyrC gene. Nucleic Acids Res. 21:4372-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rollins, M. B., S. Del Rio, A. L. Galey, D. R. Setzer, and M. T. Andrews. 1993. Role of TFIIIA zinc fingers in vivo: analysis of single-finger function in developing embryos. Mol. Cell. Biol. 13:4776-4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sakonju, S., D. F. Bogenhagen, and D. D. Brown. 1980. A control region in the center of the 5S RNA gene directs specific initiation of transcription. I. The 5′ border of the region. Cell 19:13-25. [DOI] [PubMed] [Google Scholar]
- 65a.Scherly, D., F. Stutz, N. Lin-Marq, and S. G. Clarkson. 1993. La proteins from Xenopus laevis. cDNA cloning and developmental expression. J. Mol. Biol. 231:196-204. [DOI] [PubMed]
- 66.Schlissel, M. S., and D. D. Brown. 1984. The transcriptional regulation of Xenopus 5S RNA genes in chromatin: the roles of active stable transcription complexes and histone H1. Cell 37:903-913. [DOI] [PubMed] [Google Scholar]
- 67.Sera, T., and A. P. Wolffe. 1998. Role of histone H1 as an architectural determinant of chromatin structure and as a specific repressor of transcription on Xenopus oocyte 5S rRNA genes. Mol. Cell. Biol. 18:3668-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shastry, B. S., B. M. Honda, and R. G. Roeder. 1984. Altered levels of a 5S gene-specific transcription factor (TFIIIA) during oogenesis and embryonic development of Xenopus laevis. J. Biol. Chem. 259:11373-11382. [PubMed] [Google Scholar]
- 69.Theunissen, O., F. Rudt, U. Guddat, H. Mentzel, and T. Pieler. 1992. RNA and DNA binding zinc fingers in Xenopus TFIIIA. Cell 71:679-690. [DOI] [PubMed] [Google Scholar]
- 70.Turner, D. L., and H. Weintraub. 1994. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 8:1434-1447. [DOI] [PubMed] [Google Scholar]
- 71.Ura, K., H. Kurumizaka, S. Dimitrov, G. Almouzni, and A. P. Wolffe. 1997. Histone acetylation: influence on transcription, nucleosome mobility and positioning, and linker histone-dependent transcriptional repression. EMBO J. 16:2096-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vitolo, J. M., C. Thiriet, and J. J. Hayes. 2000. The H3-H4 N-terminal tail domains are the primary mediators of transcription factor IIIA access to 5S DNA within a nucleosome. Mol. Cell. Biol. 20:2167-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wakefield, L., and J. B. Gurdon. 1983. Cytoplasmic regulation of 5S RNA genes in nuclear-transplant embryos. EMBO J. 2:1613-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Westmark, C. J., R. Ghose, and P. W. Huber. 1998. Inhibition of RNA polymerase III transcription by a ribosome-associated kinase activity. Nucleic Acids Res. 26:4758-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Westmark, C. J., R. Ghose, and P. W. Huber. 2002. Phosphorylation of Xenopus transcription factor IIIA by an oocyte protein kinase CK2. Biochem. J. 362:375-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.White, R. J., D. Stott, and P. W. Rigby. 1990. Regulation of RNA polymerase III transcription in response to simian virus 40 transformation. EMBO J. 9:3713-3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wolf, V. J., T. Dang, P. Hartl, and J. M. Gottesfeld. 1994. Role of maturation-promoting factor (p34cdc2-cyclin B) in differential expression of the Xenopus oocyte and somatic-type 5S RNA genes. Mol. Cell. Biol. 14:4704-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wolffe, A. P. 1989. Dominant and specific repression Xenopus oocyte 5S RNA genes and satellite I DNA by histone H1. EMBO J. 8:527-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wolffe, A. P. 1988. Transcription fraction TFIIIC can regulate differential Xenopus 5S RNA gene transcription in vitro. EMBO J. 7:1071-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wormington, W. M., D. F. Bogenhagen, E. Jordan, and D. D. Brown. 1981. A quantitative assay for Xenopus 5S RNA gene transcription in vitro. Cell 24:809-817. [DOI] [PubMed] [Google Scholar]
- 81.Wormington, W. M., and D. D. Brown. 1983. Onset of 5S RNA gene regulation during Xenopus embryogenesis. Dev. Biol. 99:248-257. [DOI] [PubMed] [Google Scholar]
- 82.Wuttke, D. S., M. P. Foster, D. A. Case, J. M. Gottesfeld, and P. E. Wright. 1997. Solution structure of the first three zinc fingers of TFIIIA bound to the cognate DNA sequence: determinants of affinity and sequence specificity. J. Mol. Biol. 273:183-206. [DOI] [PubMed] [Google Scholar]
- 83.Young, D., and D. Carroll. 1983. Regular arrangement of nucleosomes on 5S rRNA genes in Xenopus laevis. Mol. Cell. Biol. 3:720-730. [DOI] [PMC free article] [PubMed] [Google Scholar]