Abstract
It has been only 15 years since studies began on the molecular mechanisms underlying mitochondrial fission and fusion using simple model organisms such as Drosophila, yeast, and C. elegans. Beyond the primary functions of mitochondrial fission and fusion in controlling organelle shape, size and number, it became clear that these dynamic processes are also critical for regulating cell death, mitophagy and organelle distribution. Now, studies suggest that prominent changes occur in mitochondrial dynamics in a broad array of neurodegenerative disease, and there is substantial evidence suggesting a key role in disease pathogenesis, as neurons are among the most energy-consuming cell types and have a highly developed cell shape. Here, we review the recent findings on mitochondrial dynamics in neurodegeneration.
Keywords: mitochondria, fission, fusion, dynamin-related GTPase, neurodegeneration
Mechanisms of Mitochondrial Fission and Fusion
Mitochondria play important roles in many cellular activities, such as energy production, metabolism, aging and cell death [1]. Neurons have a high demand for mitochondrial metabolism and contain many mitochondria throughout the cytoplasm. In neurons and many other cell types, mitochondria are maintained as short tubular structures, which are highly dynamic and move, divide, and fuse [2–4]. The dynamic interactions among mitochondria by fission and fusion establish organelle size, number and shape and allow for the mixing of mitochondrial contents, including proteins, lipids and DNA.
Mitochondrial fission is mediated by an evolutionarily conserved dynamin-related GTPase, Drp1 in mammals and Dnm1p in yeast [2, 5]. Drp1 is a soluble cytosolic protein that assembles into spiral filaments around mitochondrial tubules. Different outer membrane proteins have been proposed as Drp1 receptors, including Mff, Fis1 and the two homologous proteins Mid49 and Mid51 (also called MIEF1) [6–10]. Several different post-translational modifications, including phosphorylation, ubiquitination and sumoylation, of Drp1 regulate its interactions with mitochondria [11, 12]. The Drp1 spiral has been proposed to constrict mitochondrial tubules through conformational changes, driven by GTP hydrolysis, in Drp1 and Dnm1p [13–16]. Interestingly, it has been suggested that tubules of the endoplasmic reticulum wrap around and squeeze mitochondria at the early stage of division, prior to the assembly of Drp1 filaments onto mitochondria [17]. This ER-mediated narrowing of mitochondrial tubules may help Drp1 assemble, as the diameter of Drp1 spirals is smaller than that of mitochondrial tubules. After the completion of mitochondrial fission, Drp1 spirals likely disassemble from mitochondria for future rounds of mitochondrial fission.
Mitochondrial fusion is also mediated by the conserved dynamin-related GTPases mitofusin (Mfn) and optic dominant atrophy 1 (Opa1) [5, 18]. In yeast, the Mfn and Opa1 counterparts are called Fzo1p and Mgm1p, respectively. There are two Mfns, Mfn1 and Mfn2, and these proteins, which are located in the outer membrane, fuse the mitochondrial membranes of adjacent tubules through homotypic and heterotypic interactions [19, 20]. Opa1 is located in the inner mitochondrial membrane and likely interacts with Mfns to form intermembrane protein complexes that couple the fusion of outer membranes to that of the inner membranes [21–23]. Loss of either Mfns or Opa1 leads to a similar mitochondrial fragmentation phenotype, suggesting that both outer and inner membrane fusion processes are affected. Mfn2 and Opa1 are associated with neurodegenerative diseases. Similar to Drp1, these GTPases are also controlled by a variety of mechanisms. Mfns are subjected to ubiquitination and proteosomal degradation [24, 25], while Opa1 is proteolytically regulated by different mitochondrial proteases [26–33].
Many neurodegenerative diseases are associated with alterations in the fission and fusion of mitochondria [34, 35]. In this review, we will first summarize the cellular and physiological functions of mammalian mitochondrial dynamics and their roles in neurodegeneration.
Cellular Functions of Mitochondrial Fission and Fusion
Mitophagy
Mitochondria have multiple quality control mechanisms through degradation, including intramitochondrial proteases, proteosomal degradation, mitochondria-derived vesicles, and mitophagy, which is the autophagy-mediated degradation of mitochondria [36–38]. Parkin, an E3 ubiquitin ligase that is mutated in a large proportion of cases of young onset Parkinson’s disease (PD) [39, 40], is recruited to dysfunctional mitochondria and ubiquitinates mitochondrial proteins for proteosomal degradation and promotes the engulfment of mitochondria by autophagosomes (Fig. 1). During this process, mitochondrial dynamics are shifted toward fission over fusion, partly due to the proteosomal degradation of Mfns [24, 25, 41]. Fission is thought to be important for mitophagy, since inhibition of division slows down mitophagy. Most studies of parkin have involved overexpression of parkin in non-neuronal cells in culture, and mice lacking Parkin are viable with only subtle mitochondrial deficits [42–45]. Therefore, it remains to be determined whether parkin-dependent mitophagy is physiologically relevant in mammalian cells in vivo. Nonetheless, these studies clearly indicate that the regulation of mitochondrial size by fission is important for the clearance of this organelle by mitophagy.
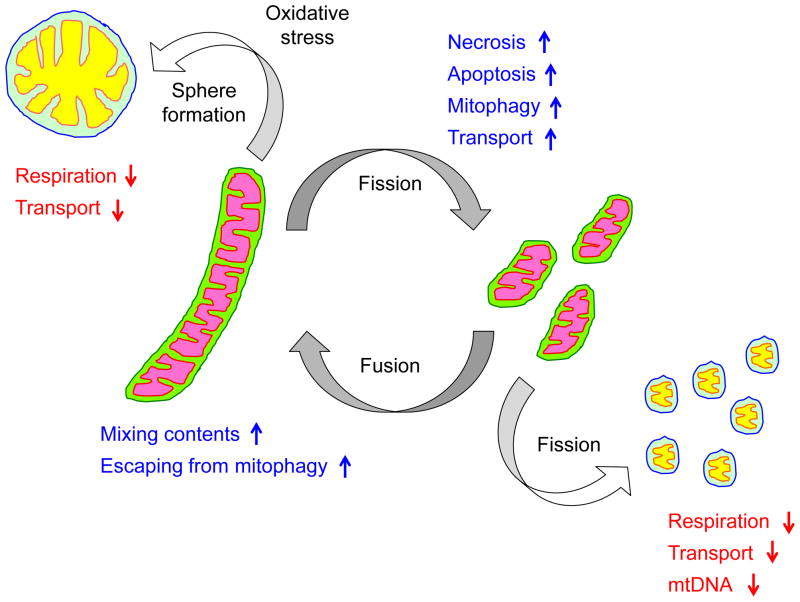
Figure 1.
Cellular and physiological roles of mitochondrial dynamics. Mitochondria divide and fuse, maintaining their short tubular structures. Mitochondrial fission facilitates cell death, mitophagy and organelle transport while fusion allows content mixing and can block mitophagy. Overly fused mitochondria accumulate oxidative damage and further transform their shape into large spheres. Such large round mitochondria are associated with impaired respiration and defective in organelle transport. Excessive fragmentation can generate mitochondria that lack mtDNA, leading to impaired respiration.
Whereas enhanced mitochondrial division facilitates the mitophagy of dysfunctional mitochondria, fission is downregulated during starvation-induced autophagy, thereby resulting in elongated mitochondrial tubules that are protected against degradation (Fig. 1). Upon starvation, Drp1 is phosphorylated at serine637 and dephosphorylated at serine616, leading to the inhibition of mitochondrial fission [46, 47]. Phosphorylation of serine637 by protein kinase A inhibits fission by enhancing the dissociation of Drp1 from mitochondria, likely by separating Drp1 from mitochondria or stimulating spiral disassembly. Phosphorylation of serine616 activates Drp1-mediated mitochondrial fission by an unknown mechanism, but does not stimulate Drp1 GTPase activity. Elongated mitochondria cannot be efficiently engulfed and thus escape degradation, and cells maintain the ability to continuously produce energy from the catabolites generated by autophagic degradation of other cellular components. In cells lacking Opa1 or both Mfn1 and 2, which fail to elongate mitochondria, mitochondria are subjected to degradation upon starvation, and this loss of mitochondria enhances cell death due to energy depletion. Therefore, in response to physiological needs (e.g., promotion or inhibition of mitochondrial degradation), mitochondrial fission and fusion serve as a key regulatory step in determining the fate of mitochondria by simply modulating organelle length and size.
Cell death
Cells die through different mechanisms, such as apoptosis and necrosis, that are interconnected yet distinct in terms of cellular morphology and caspase requirements. During apoptosis and necrosis, changes in mitochondrial morphology have been observed, including the fragmentation of mitochondrial tubules and the remodeling of inner membrane cristae (Fig. 1). Drp1 and Opa1 have been suggested to promote and inhibit, respectively, the release of cytochrome c from mitochondria during apoptosis; however, their roles are controversial and may depend on cell type and physiological cues. The functions of Drp1 and Opa1 in apoptosis have been reviewed elsewhere and are beyond the scope of this discussion [48, 49]. In addition to apoptosis, a recent study has shown that Drp1 also plays a role in necrosis [50]. Although necrosis was traditionally considered a passive cell death mechanism, an increasing number of studies have suggested that necrosis is also programmed and regulated by specific intracellular signaling pathways involving two kinases, RIP1 and RIP3, which form a protein complex called the necrosome, and a mitochondria resident phosphatase, PGAM5 [51]. Upon induction of necrosis, the RIP1/3 complex binds to and phosphorylates PGAM5 to stimulate its phosphatase activity. PGAM5 in turn dephosphorylates Drp1 at serine637 to recruit Drp1 to mitochondria for fission [50]. The loss of Drp1, either by silencing or by treatment with its inhibitor mdivi-1 [52], blocks necrosis. The role of mitochondrial fragmentation in necrosis remains to be determined, but is likely different from that in apoptosis. During apoptosis, mitochondria play an active role in initiating cell death by releasing the proapoptotic factor cytochrome c. In contrast, during necrosis, mitochondrial fragmentation may serve as a means to decrease mitochondrial energy production for efficient cell death.
Transport and distribution
Mitochondria are actively transported throughout the cytosol, mainly along microtubules in mammals and along actin filaments in yeast [4]. In neurons, mitochondrial fission and fusion are important for mitochondrial transport [53]. With regards to fission, inhibiting Drp1 results in enlarged mitochondria that cannot be properly localized to either dendrites or axons, and suppresses synapse formation and function [54, 55]. Although the mechanisms underlying these effects on mitochondrial transport are not well understood, they may result directly from the effects of Drp1 on mitochondrial size (Fig. 1). Indeed, when Drp1 is knocked out before dendrite extension, mitochondrial tubules first become elongated and then, as oxidative damage accumulates in mitochondria due most likely to impaired mitophagy, form large spheres whose diameters are far greater than that of the dendrites [56]. These large, spherical mitochondria may be difficult to transport, as they localize exclusively to the cell body of Drp1-null neurons [56].
Interestingly, Mfns are also important for mitochondrial transport and movement, but perhaps through distinct mechanisms. In Mfn1- or Mfn2-null mouse embryonic fibroblasts (MEFs), mitochondria lose directional movement and appear to move randomly in the cytosol, suggesting that they are detached from microtubules [19]. Supporting this notion, Mfn2 were found to interact with Milton and the calcium-binding GTPase Miro, which form an adapter protein complex connecting the microtubule motor kinesin and mitochondria for anterograde movement [57]. Mfn2-null neurons also show a decreased number of mitochondria within their highly branched dendrites [58].
Respiration
Defects in mitophagy may lead to impairment of respiratory function in mitochondria in Drp1-null neurons and HeLa cells that are depleted for Drp1 likely due to oxidative damage on mitochondrial components; however major targets of oxidative stress have been unidentified [56, 59]. Impaired respiration can lead to further increases in ROS production, therefore it is difficult to pinpoint the primary target. In addition, respiratory defects due to mitochondrial fission deficiency are highly dependent on cell type likely due to basal levels of ROS and contributions of other quality control mechanisms. Indeed, Drp1-null mouse embryonic fibroblasts are normal in terms of activities of electron transport chain complexes and ATP production [60, 61]. Defects in mitochondrial fusion are also associated with impairment of respiration [58]. Excessive mitochondrial division produce many small mitochondrial fragments that lack mitochondrial DNA (mtDNA) and therefore may fail to assemble electron transport chain complexes as several subunits of the electron transport chain complexes are encoded in mtDNA. Although underlying mechanisms are not well understood, loss of mitochondrial fusion leads to decreased number of mtDNA in some cells including mouse embryonic fibroblasts and skeletal muscle cells [62]. However, Opa1-null pancreatic beta cells lose their respiratory function, but contain normal amounts of mtDNA, suggesting the role of mitochondrial fusion in the maintenance of mtDNA may also depend on cell type [63].
In the following section, we will discuss how these functions of mitochondrial dynamics are affected in neurodegenerative diseases.
Neurodegeneration Associated with Altered Mitochondrial Fission and Fusion
Many human diseases are associated with changes in mitochondrial function and morphology. Neurons have an especially high demand for mitochondrial metabolism and are particularly sensitive to decreases in mitochondrial function, which lead to energy deprivation and reactive oxygen species production. By controlling organelle size, mitochondrial fission and fusion also facilitate the efficient microtubule-mediated active transport of mitochondria into the long dendritic and axonal extensions characteristic of highly polarized neurons. Therefore, the diseases described below mainly affect neurons. Abnormalities in mitochondrial dynamics, including the inhibition of mitochondrial fission or fusion and the abnormal activation of mitochondrial fission, have been suggested to occur as early events during the pathogenesis of many diseases.
Postneonatal death caused by a Drp1 mutation in human
A spontaneous, dominant negative mutation in human Drp1 causes neonatal death with impaired brain development and neuronal cell death [64] (Fig. 2). Skin fibroblasts isolated from the patient displayed elongated mitochondria, suggesting a defect in mitochondrial fission. The disease mutation is located in the middle region of the Drp1 protein, and blocks both Drp1 oligomerization and oligomerization-stimulated GTPase activity [65], and also inhibits the recruitment of Drp1 to mitochondria. These findings demonstrate the importance of mitochondrial division in human brain development. The role of mitochondrial fission in development has been further studied using Drp1 knockout mice. Complete loss of Drp1 results in embryonic lethality at E11.5, most likely due to developmental defects in the placenta and heart [60, 61]. Therefore, tissue-specific deletion has been used to elucidate the functions of Drp1 in neurons. Consistent with the case report for the patient described above, the loss of Drp1 leads to defective cell proliferation, viability and synapse formation in neurons during brain development. Furthermore, when Drp1 is deleted in postmitotic cerebellar Purkinje cells after the completion of brain development, mitochondria become elongated due to the lack of organelle division [56, 66]. Due most likely to impaired turnover, these elongated mitochondria accumulate oxidative damage and lose respiratory function, leading to the further transformation of their morphology into large spheres as described above. These mitochondrial defects eventually lead to the degeneration of Drp1-null Purkinje cells. The formation of large mitochondrial spheres and cell death are directly caused by oxidative damage, since both phenotypes are suppressed by treating Drp1-null cells with antioxidants. In contrast to Purkinje cells, Drp1-null MEFs maintain normal respiration and do not show increased cell death [60, 61]. Mitochondria in Drp1-null MEFs are highly interconnected and elongated due to fission defects, but become large spheres only when treated with hydrogen peroxide, suggesting that MEFs have lower amounts of reactive oxygen species than neurons [56]. These studies suggest that mitochondrial fission is a quality control mechanism to protect against oxidative damage in neurons.
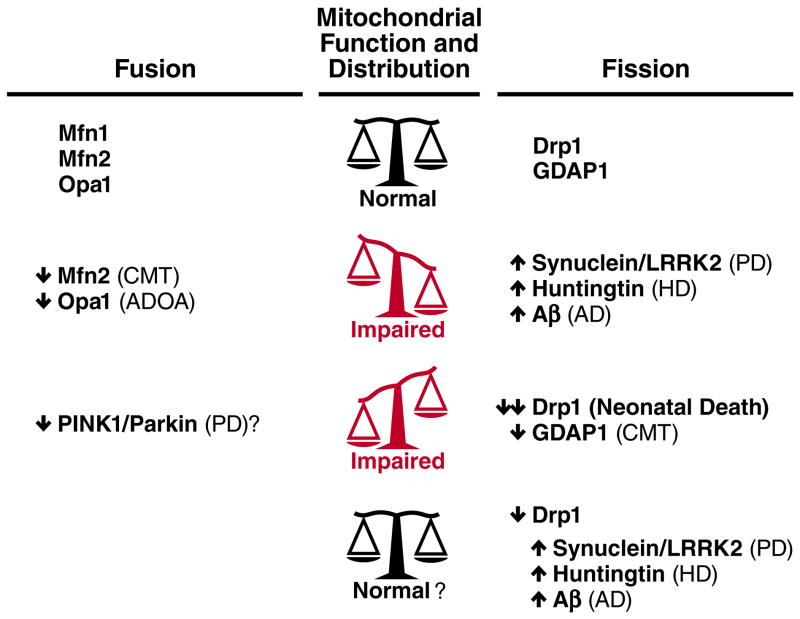
Figure 2.
Disruption of the balance between mitochondrial fusion and fission in neurodegenerative disease. Mitochondrial morphology is normally regulated by the balance between mitochondrial fusion proteins (e.g., Mfn1, Mfn2 and Opa1) and fission proteins (including Drp1 and GDAP1). Disruption of either fusion or fission proteins can result in neurologic disease. Mutation and/or overexpression of a variety of proteins implicated in neurodegenerative diseases including PD, HD and AD, also disrupt mitochondrial dynamics, and normalizing the fusion-fission balance may have therapeutic value. The bottom row shows one strategy in which a normal level of fission is restored by decreasing Drp1, thus compensating for increased fission by disease proteins. It remains unknown if normalizing mitochondrial morphology will restore function.
Neurologic disorders caused by defects in fusion and fission machineries: Autosomal dominant optic atrophy and Charcot-Marie-Tooth neuropathy
Autosomal dominant optic atrophy (ADOA) is a rare degenerative disorder characterized by visual loss due to retinal ganglion cell degeneration and optic nerve atrophy. Based on genetic linkage studies, eight loci have been associated with ADOA, implicating at least three mitochondrial membrane proteins, Opa1, Opa3 and Opa7 [67]. Mutations in the inner membrane protein Opa1 are responsible for disease in the majority of ADOA patients (75%), while mutations in the outer membrane protein Opa3 account for a minor population (1%). Opa1 mediates the fusion of mitochondrial membranes and is also involved in the maintenance of the inner membrane cristae structure, which helps prevent cytochrome c from being released to the cytoplasm during apoptosis (Fig. 2). Most pathological Opa1 mutations cause haploinsufficiency. Consistent with this idea, heterozygous Opa1 knockout mice show the degeneration of optic nerves at advanced ages [68]. Analysis of Opa3 function has just started, but studies suggest that Opa3 controls Drp1-independent mitochondrial fission [69]. The function of Opa7 is currently unknown.
Interestingly, another neurologic disorder, Charcot-Marie-Tooth neuropathy (CMT), can also be caused by defects in mitochondrial dynamics (Fig. 2). This heterogenous disorder, which is characterized by distal muscle weakness and sensory loss, has multiple genetic etiologies, including mutations in Mfn2 [70], a mitochondrial outer membrane protein important for fusion. Although the mechanisms by which loss of Mfn2 produces CMT are unclear, loss of Mfn2 leads to decreases in the amount of mtDNA and increases in its mutations and deletions, and ultimately results in impaired aerobic respiration in mice [62]. Alternatively, the deficit may not be in the intrinsic function of the mitochondria, but rather due to changes in the axonal transport of mitochondria [57]. This could result in the accumulation of damaged mitochondria and/or the mislocalization of the mitochondria in processes, which in turn might produce regional bioenergetics failure and/or regional absence of other key mitochondrial functions. Indeed, mouse Mfn2 is critical for the movement of mitochondria, and Mfn2-null neurons show an impaired distribution of mitochondria.
Mutations in ganglioside-induced differentiation-associated protein 1 (GDAP1) can also cause several variants of CMT[71]. GDAP1 is a mitochondrial outer membrane protein that has been implicated in mitochondrial fission, although the exact role of GDAP1 in organelle division is less understood than that of Drp1 [72, 73]. Recent studies have reported two cases in which CMT patients carry mutations in both Mfn2 and GDAP1 [74, 75]. Since Mfn2 is a fusion protein and GDAP1 a fission protein, it is surprising that, in both cases, symptoms are more severe than those of patients with single mutations. In one case, mitochondrial morphology is normal, but the combination of uncoupled respiration resulting from the Mfn2 mutation and complex I defects resulting from the GDAP1 mutation leads to additive respiration impairments [75]. The apparent normal mitochondrial morphology in these patients is consistent with the restoration of nearly wild-type morphology in yeast mutant cells lacking both fission and fusion, highlighting the need for balance between these opposite activities, which antagonistically regulate mitochondrial shape [76]. However, the additive respiration defects seen in these patients clearly indicate the importance of mitochondrial division and fusion beyond the regulation of mitochondrial shape. In another case in which both Mfn2 and GDAP1 are mutated, mitochondrial enlargement is observed in axons, suggesting that fusion is occurring at relatively higher rates than fission [74].
Alzheimer’s disease
Alzheimer’s disease (AD) is a devastating neurodegenerative disorder that affects roughly 5.4 million Americans and whose prevalence is increasing [77]. Pathologically, the disease is characterized by the extracellular deposition of amyloid plaques, followed by the accumulation of intraneuronal neurofibrillary tangles of hyperphosphorylated tau. This is associated with the loss of synapses and then neuronal death, initially in focal areas including the entorhinal cortex and hippocampus, and ultimately more broadly in the cortex [78]. Clinically, early memory loss progresses to broad cognitive dysfunction [79]. Although the pathogenesis of AD is not well understood, accumulating evidence has implicated mitochondria. Of particular interest to this review are prominent changes in the expression of mitochondrial fission and fusion proteins [80], suggesting an imbalance in the fusion-fission process. Although the mechanisms leading to these changes are unclear, they may represent direct effects of amyloid beta (Aβ), as increased Aβ fragments mitochondria and decreases mitochondrial mass in the neurites of cultured neurons. Interestingly, this process depends on S-nitrosylation-mediated stimulation of Drp1 activity [81] (Fig. 2).
However, this mechanism is controversial, and an alternative hypothesis has also been suggested, in which Drp1 is phosphorylated for recruitment to the surface of mitochondria [82]. Other possible mechanisms for Drp1 involvement in AD are increases in both Drp1 expression levels and its interactions with Aβ and phosphorylated tau in AD patients [83, 84]. However, the physiologic relevance of these changes remains unclear, and other studies have reported normal or decreased levels of Drp1 in AD patients and animal models [80, 82, 85]. Therefore, whether changes in Drp1 levels are directly associated with the pathogenesis of AD remains to be determined.
Parkinson’s disease
Parkinson’s disease (PD) is a common and disabling neurodegenerative disorder, that results in substantial suffering and financial cost to society [86]. It features the degeneration of nigrostriatal DA neurons and accumulation of α-synuclein [87], resulting in characteristic deficits in movement. Although the pathogenesis of PD is unknown, abundant evidence from autopsy studies and dopaminergic neurotoxins indicates a central role for mitochondria [88–91]. Additional evidence for mitochondrial involvement, including an important role for mitochondrial dynamics, has emerged from recent work on proteins whose mutation and/or overexpression produces familial forms of PD, particularly the autosomal recessive PD proteins PINK1 and parkin. Indeed, a number of studies in Drosophila have demonstrated that PINK1 and Parkin function as fission proteins, and that the toxicity of losing either depends on the level of Drp1 [92, 93] (Fig. 2).
However, the mechanisms by which changes in these proteins might affect mitochondrial morphology are unclear, as both Parkin (an E3 ubiquitin ligase) and PINK1 (a serine/threonine kinase) have multiple proposed targets [40, 94], but it is unknown which are physiologically relevant. One mechanism by which Parkin and PINK1 might impact mitochondrial dynamics is through their roles in mitophagy. Indeed, Parkin is recruited to mitochondria in which the membrane potential is decreased in a PINK1-dependent manner [95, 96]. The ubiquitination of mitochondrial proteins stimulates their proteosomal degradation and the formation of autophagosomes for mitophagy. In addition to mitophagy, Parkin and PINK1 might also affect mitochondrial distribution by regulating mitochondrial transport. Indeed, a recent study has shown that PINK1 and Parkin act to arrest mitochondrial motility in axons. When it is exposed on the surface of dysfunctional mitochondria, Pink1 phosphorylates miro, which connects mitochondria to the microtubule motor kinesin [97]. Phosphorylated Miro is then ubiquitinated by Parkin and degraded by proteosomes. The degradation of Miro dissociates kinesin from mitochondria and prevents organelle transport. Therefore, PINK1 and Parkin may inhibit the movement of dysfunctional mitochondria into neurites and mark these organelles for degradation. However, it is important to note that the effects of PINK1 and Parkin on mitochondrial morphology in mammalian systems have been less clear and are controversial, and remain to be validated in vivo.
Interestingly, yet another autosomal recessive PD protein, DJ-1, also regulates mitochondrial morphology, perhaps as a secondary consequence of increased reactive oxygen species (ROS) production. The effects of DJ-1 on mitochondrial fragmentation can be blocked by overexpression of either Parkin or PINK1 [98, 99].
Loss of PINK1, parkin or DJ-1 produces relatively rare forms of PD that show degeneration that is more restricted to dopamine neurons than most cases of idiopathic PD, and can also lack Lewy bodies, raising questions about their relevance to sporadic PD [100]. However, overexpression or mutation of the autosomal dominant PD proteins synuclein and LRRK2 can also affect mitochondrial dynamics [101–105], potentially indicating a broader role for these proteins in idiopathic PD. Indeed, synuclein accumulates in the brains of essentially all patients with sporadic PD, in the form of Lewy bodies and Lewy neurites [106]. With LRRK2, the most common genetic cause of PD [107], mitochondrial fragmentation may occur through effects on Drp1 [105]. In contrast, with α-synuclein, mitochondrial fragmentation appears to occur through a direct interaction between synuclein and mitochondrial membranes. Interestingly, the process can occur independently of Drp1, although the precise effects on fusion versus fission remain unclear [101, 102]. In addition, although mitochondrial fragmentation by synuclein precedes neuronal death from elevated synuclein [102], it is not clear if the fragmentation is actually causal.
Huntington’s disease
Huntington’s disease (HD) is a devastating neurodegenerative disease. The disorder results from an expansion of CAG triplet repeats in the polyglutamine region of the huntingtin protein (Htt), leading to the accumulation of intracellular Htt aggregates. [108]. Neurodegeneration occurs in the striatum during early pathogenesis, but broad areas of the brain are also affected during later stages of disease progression. As with AD and PD, considerable evidence has implicated mitochondrial failure in the pathogenesis of HD [109], and striatal neurons that degenerate in HD are remarkably susceptible to inhibition of complex II [110, 111],
More recently, changes in mitochondrial dynamics have also been found in HD. For example, in HD patients and animal models of the disease, mitochondria are excessively fragmented and show decreased motility and respiration [109]. Interestingly, mutant Htt can associate with the surface of mitochondria [112], although it is unclear if this interaction mediates the effects of Htt on mitochondrial morphology. Indeed, Htt aggregates bind to many proteins, and mitochondria can be damaged as a secondary consequence of overall decreases in cellular health. Recent studies have identified Drp1 as a potential target of mutant Htt. All of these studies have shown that excessive activation of Drp1 occurs in HD, although several distinct models have been proposed to underlie these findings.
For instance, Htt aggregates directly bind to Drp1 in vitro and stimulate its GTPase activity [113] (Fig. 2). In neurons expressing mutant forms of Htt, Drp1 colocalizes with Htt aggregates on mitochondria. Since the GTPase activity of Drp1 is stimulated by its assembly, mutant Htt may facilitate abnormal assembly of Drp1 oligomers on the surface of mitochondria, thereby activating fission. The mitochondrial fragmentation and increased cell death that are induced by mutant Htt can be rescued by introducing a dominant negative form of Drp1 [113, 114], supporting the idea that Drp1 activation is a major target of mutant Htt. In contrast, increased levels of cytoplasmic calcium, rather than direct interactions between Htt and Drp1, could activate Drp1 through dephosphorylation by the calcium-dependent protein phosphatase calcineurin [115]. Although the basal level of Drp1 phosphorylation is low, the calcineurin-mediated dephosphorylation of Drp1 has been shown to promote its association with mitochondria [12]. Htt aggregates may function as a scaffold on the mitochondrial surface that brings different kinases and phosphatases to mitochondria.
Although the above studies observed normal levels of Drp1 in HD models, increased amounts of Drp1 and decreased levels of Mfns and Opa1 are found in HD patients, suggesting that mitochondrial dynamics are shifted toward fission over fusion [116]. Since mRNA amounts of Drp1 and Fis1 are elevated in HD models, mutant Htt may also regulate mitochondrial dynamics at the level of gene transcription. Consistent with the observed changes in Drp1 and Fis1 mRNA levels, Htt has been suggested to control the gene expression of mitochondrial proteins via transcriptional regulation, possibly through PGC-alpha [117].
Concluding remarks
Imbalances between mitochondrial division and fusion have been proposed to cause neurodegenerative diseases. Many studies have shown that acute readjustment of these imbalances has beneficial effects on mitochondrial structure, function and cell survival in different disease models. Since these studies have mainly used in vitro culture systems or non-mammalian models such as Drosophila, it remains to be determined whether normalization of mitochondrial dynamics is beneficial in mammals in vivo. This can be interrogated by a number of approaches, including the inhibition of Drp1 through expressing dominant negative forms, silencing, or treating with its inhibitor mdivi-1, and the overexpression of other fusion and fission components. However, the long-term simultaneous inhibition of mitochondrial fission and fusion may cause harmful effects, as seen in CMT patients with mutations in both Mfn2 and GDAP1. Indeed, the evolution of extensive fission and fusion machinery strongly suggests that a basal level is required, and that simply restoring a “normal” mitochondrial morphology may not be sufficient. Therefore, as potential therapeutic approaches, it would be important not to block fusion or fission completely, but rather to partially suppress these activities. To achieve such balance, the examination of mitochondrial structure and dynamics may provide a guide for the appropriate level of inhibition or activation of fusion and fission components. In addition to restoring the normal balance between fusion and fission, a complementary therapeutic approach would be to target the adverse consequences that result from loss of mitochondrial fission. For instance, since impaired mitochondrial fission or fusion can produce oxidative damage [56, 59, 118] and may also produce regional bioenergetics failure in neuronal processes lacking mitochondria [55], approaches that enhance antioxidant defenses and/or promote bioenergetics function may also prove beneficial. Currently, there are a number of ongoing clinical trials to evaluate the use of antioxidant treatments and to stimulate bioenergetics function for many neurodegenerative diseases, including CMT, AD, PD and HD (www.clincaltrials.gov). Assessing mitochondrial dynamics during these studies would be of great interest and would further our understanding of the pathogenesis of these diseases. Importantly, at present, we are limited by our inability to directly measure mitochondrial morphology in neurodegenerative disease, since the morphology certainly changes before brains are examined. As a result, we are forced to rely on the levels of fusion-fission proteins, without knowing what the actual morphology was. To overcome this problem, it is thus critical that we develop new technologies to monitor mitochondrial morphology and dynamics in human patients.
Acknowledgments
We acknowledge the many researchers who contributed to our understanding of mitochondrial dynamics and diseases, and apologize that we could not cite all of the relevant research due to space restrictions. We thank Kristen F. Swaney and Gary Howard for critically reading the manuscript. This work was supported by a Burroughs-Wellcome Medical Scientist Fund Career Awards (to KN) and NIH grants to K.N. (1KO8NS062954-01A1 and P30NS069496), M.I. (GM084015) and H.S. (GM089853).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kageyama Y, et al. Mitochondrial division: molecular machinery and physiological functions. Current opinion in cell biology. 2011;23:427–434. doi: 10.1016/j.ceb.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Westermann B. Bioenergetic role of mitochondrial fusion and fission. Biochimica et biophysica acta. 2012 doi: 10.1016/j.bbabio.2012.02.033. [DOI] [PubMed] [Google Scholar]
- 4.Frederick RL, Shaw JM. Moving mitochondria: establishing distribution of an essential organelle. Traffic. 2007;8:1668–1675. doi: 10.1111/j.1600-0854.2007.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tamura Y, et al. SnapShot: Mitochondrial dynamics. Cell. 2011;145:1158, e1151. doi: 10.1016/j.cell.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palmer CS, et al. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO reports. 2011;12:565–573. doi: 10.1038/embor.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Otera H, et al. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol. 2010;191:1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao J, et al. Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. The EMBO journal. 2011;30:2762–2778. doi: 10.1038/emboj.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2008;19:2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoon Y, et al. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol. 2003;23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson TJ, et al. Cell signaling and mitochondrial dynamics: Implications for neuronal function and neurodegenerative disease. Neurobiology of disease. 2012 doi: 10.1016/j.nbd.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang CR, Blackstone C. Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1. Ann N Y Acad Sci. 2010;1201:34–39. doi: 10.1111/j.1749-6632.2010.05629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoon Y, et al. Mammalian dynamin-like protein DLP1 tubulates membranes. Mol Biol Cell. 2001;12:2894–2905. doi: 10.1091/mbc.12.9.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ford MG, et al. The crystal structure of dynamin. Nature. 2011;477:561–566. doi: 10.1038/nature10441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mears JA, et al. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nature structural & molecular biology. 2011;18:20–26. doi: 10.1038/nsmb.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukushima NH, et al. The GTPase effector domain sequence of the Dnm1p GTPase regulates self-assembly and controls a rate-limiting step in mitochondrial fission. Mol Biol Cell. 2001;12:2756–2766. doi: 10.1091/mbc.12.9.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman JR, et al. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen H, Chan DC. Physiological functions of mitochondrial fusion. Annals of the New York Academy of Sciences. 2010;1201:21–25. doi: 10.1111/j.1749-6632.2010.05615.x. [DOI] [PubMed] [Google Scholar]
- 19.Chen H, et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koshiba T, et al. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 2004;305:858–862. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]
- 21.Cipolat S, et al. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc Natl Acad Sci U S A. 2004;101:15927–15932. doi: 10.1073/pnas.0407043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olichon A, et al. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J Biol Chem. 2003;278:7743–7746. doi: 10.1074/jbc.C200677200. [DOI] [PubMed] [Google Scholar]
- 23.Song Z, et al. Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol Biol Cell. 2009;20:3525–3532. doi: 10.1091/mbc.E09-03-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka A, et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gegg ME, et al. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Human molecular genetics. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Head B, et al. Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. The Journal of cell biology. 2009;187:959–966. doi: 10.1083/jcb.200906083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ehses S, et al. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J Cell Biol. 2009;187:1023–1036. doi: 10.1083/jcb.200906084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cipolat S, et al. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell. 2006;126:163–175. doi: 10.1016/j.cell.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 29.Ishihara N, et al. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. Embo J. 2006;25:2966–2977. doi: 10.1038/sj.emboj.7601184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duvezin-Caubet S, et al. OPA1 Processing Reconstituted in Yeast Depends on the Subunit Composition of the m-AAA Protease in Mitochondria. Mol Biol Cell. 2007 doi: 10.1091/mbc.E07-02-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Griparic L, et al. Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. J Cell Biol. 2007;178:757–764. doi: 10.1083/jcb.200704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song Z, et al. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J Cell Biol. 2007;178:749–755. doi: 10.1083/jcb.200704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quiros PM, et al. Loss of mitochondrial protease OMA1 alters processing of the GTPase OPA1 and causes obesity and defective thermogenesis in mice. The EMBO journal. 2012;31:2117–2133. doi: 10.1038/emboj.2012.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho DH, et al. Mitochondrial dynamics in cell death and neurodegeneration. Cell Mol Life Sci. 2010;67:3435–3447. doi: 10.1007/s00018-010-0435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knott AB, Bossy-Wetzel E. Impairing the mitochondrial fission and fusion balance: a new mechanism of neurodegeneration. Ann N Y Acad Sci. 2008;1147:283–292. doi: 10.1196/annals.1427.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shutt TE, McBride HM. Staying cool in difficult times: Mitochondrial dynamics, quality control and the stress response. Biochimica et biophysica acta. 2012 doi: 10.1016/j.bbamcr.2012.05.024. [DOI] [PubMed] [Google Scholar]
- 37.Karbowski M, Youle RJ. Regulating mitochondrial outer membrane proteins by ubiquitination and proteasomal degradation. Current opinion in cell biology. 2011;23:476–482. doi: 10.1016/j.ceb.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baker MJ, et al. Quality control of mitochondrial proteostasis. Cold Spring Harb Perspect Biol. 2011:3. doi: 10.1101/cshperspect.a007559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marder KS, et al. Predictors of parkin mutations in early-onset Parkinson disease: the consortium on risk for early-onset Parkinson disease study. Arch Neurol. 2010;67:731–738. doi: 10.1001/archneurol.2010.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dawson TM, Dawson VL. The role of parkin in familial and sporadic Parkinson’s disease. Mov Disord. 2010;25(Suppl 1):S32–39. doi: 10.1002/mds.22798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ziviani E, et al. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Von Coelln R, et al. Loss of locus coeruleus neurons and reduced startle in parkin null mice. Proc Natl Acad Sci U S A. 2004;101:10744–10749. doi: 10.1073/pnas.0401297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palacino JJ, et al. Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J Biol Chem. 2004;279:18614–18622. doi: 10.1074/jbc.M401135200. [DOI] [PubMed] [Google Scholar]
- 44.Goldberg MS, et al. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J Biol Chem. 2003;278:43628–43635. doi: 10.1074/jbc.M308947200. [DOI] [PubMed] [Google Scholar]
- 45.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nature reviews Molecular cell biology. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomes LC, et al. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nature cell biology. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rambold AS, et al. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:10190–10195. doi: 10.1073/pnas.1107402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Landes T, Martinou JC. Mitochondrial outer membrane permeabilization during apoptosis: the role of mitochondrial fission. Biochimica et biophysica acta. 2011;1813:540–545. doi: 10.1016/j.bbamcr.2011.01.021. [DOI] [PubMed] [Google Scholar]
- 49.Sheridan C, Martin SJ. Mitochondrial fission/fusion dynamics and apoptosis. Mitochondrion. 2010;10:640–648. doi: 10.1016/j.mito.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 50.Wang Z, et al. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell. 2012;148:228–243. doi: 10.1016/j.cell.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 51.Christofferson DE, Yuan J. Necroptosis as an alternative form of programmed cell death. Current opinion in cell biology. 2010;22:263–268. doi: 10.1016/j.ceb.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lackner LL, Nunnari J. Small molecule inhibitors of mitochondrial division: tools that translate basic biological research into medicine. Chem Biol. 2010;17:578–583. doi: 10.1016/j.chembiol.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sheng ZH, Cai Q. Mitochondrial transport in neurons: impact on synaptic homeostasis and neurodegeneration. Nature reviews Neuroscience. 2012;13:77–93. doi: 10.1038/nrn3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Z, et al. The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell. 2004;119:873–887. doi: 10.1016/j.cell.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 55.Verstreken P, et al. Synaptic mitochondria are critical for mobilization of reserve pool vesicles at Drosophila neuromuscular junctions. Neuron. 2005;47:365–378. doi: 10.1016/j.neuron.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 56.Kageyama Y, et al. Mitochondrial division ensures the survival of postmitotic neurons by suppressing oxidative damage. The Journal of cell biology. 2012;197:535–551. doi: 10.1083/jcb.201110034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Misko A, et al. Mitofusin 2 is necessary for transport of axonal mitochondria and interacts with the Miro/Milton complex. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2010;30:4232–4240. doi: 10.1523/JNEUROSCI.6248-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen H, et al. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130:548–562. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 59.Parone PA, et al. Preventing mitochondrial fission impairs mitochondrial function and leads to loss of mitochondrial DNA. PLoS One. 2008;3:e3257. doi: 10.1371/journal.pone.0003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ishihara N, et al. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat Cell Biol. 2009;11:958–966. doi: 10.1038/ncb1907. [DOI] [PubMed] [Google Scholar]
- 61.Wakabayashi J, et al. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J Cell Biol. 2009;186:805–816. doi: 10.1083/jcb.200903065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen H, et al. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Z, et al. The dynamin-related GTPase Opa1 is required for glucose-stimulated ATP production in pancreatic beta cells. Molecular biology of the cell. 2011;22:2235–2245. doi: 10.1091/mbc.E10-12-0933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waterham HR, et al. A lethal defect of mitochondrial and peroxisomal fission. The New England journal of medicine. 2007;356:1736–1741. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]
- 65.Chang CR, et al. A lethal de novo mutation in the middle domain of the dynamin-related GTPase Drp1 impairs higher order assembly and mitochondrial division. J Biol Chem. 2010;285:32494–32503. doi: 10.1074/jbc.M110.142430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Z, et al. Mitochondrial division prevents neurodegeneration. Autophagy. 2012:8. doi: 10.4161/auto.21213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lenaers G, et al. Dominant optic atrophy. Orphanet J Rare Dis. 2012;7:46. doi: 10.1186/1750-1172-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Williams PA, et al. Mouse models of dominant optic atrophy: what do they tell us about the pathophysiology of visual loss? Vision Res. 2011;51:229–234. doi: 10.1016/j.visres.2010.08.031. [DOI] [PubMed] [Google Scholar]
- 69.Ryu SW, et al. Optic atrophy 3 as a protein of the mitochondrial outer membrane induces mitochondrial fragmentation. Cellular and molecular life sciences: CMLS. 2010;67:2839–2850. doi: 10.1007/s00018-010-0365-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reilly MM, et al. Charcot-Marie-Tooth disease. Journal of the peripheral nervous system: JPNS. 2011;16:1–14. doi: 10.1111/j.1529-8027.2011.00324.x. [DOI] [PubMed] [Google Scholar]
- 71.Palau F, et al. The role of mitochondrial network dynamics in the pathogenesis of Charcot-Marie-Tooth disease. Adv Exp Med Biol. 2009;652:129–137. doi: 10.1007/978-90-481-2813-6_9. [DOI] [PubMed] [Google Scholar]
- 72.Niemann A, et al. GDAP1 mutations differ in their effects on mitochondrial dynamics and apoptosis depending on the mode of inheritance. Neurobiol Dis. 2009;36:509–520. doi: 10.1016/j.nbd.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 73.Estela A, et al. Charcot-Marie-Tooth-related gene GDAP1 complements cell cycle delay at G2/M phase in Saccharomyces cerevisiae fis1 gene-defective cells. The Journal of biological chemistry. 2011;286:36777–36786. doi: 10.1074/jbc.M111.260042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vital A, et al. A French family with Charcot-Marie-Tooth disease related to simultaneous heterozygous MFN2 and GDAP1 mutations. Neuromuscul Disord. 2012;22:735–741. doi: 10.1016/j.nmd.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 75.Cassereau J, et al. Simultaneous MFN2 and GDAP1 mutations cause major mitochondrial defects in a patient with CMT. Neurology. 2011;76:1524–1526. doi: 10.1212/WNL.0b013e318217e77d. [DOI] [PubMed] [Google Scholar]
- 76.Sesaki H, Jensen RE. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J Cell Biol. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thies W, Bleiler L. 2011 Alzheimer’s disease facts and figures. Alzheimers Dement. 2011;7:208–244. doi: 10.1016/j.jalz.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 78.Huang Y, Mucke L. Alzheimer mechanisms and therapeutic strategies. Cell. 2012;148:1204–1222. doi: 10.1016/j.cell.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cummings JL. Alzheimer’s disease. The New England journal of medicine. 2004;351:56–67. doi: 10.1056/NEJMra040223. [DOI] [PubMed] [Google Scholar]
- 80.Wang X, et al. Impaired balance of mitochondrial fission and fusion in Alzheimer’s disease. J Neurosci. 2009;29:9090–9103. doi: 10.1523/JNEUROSCI.1357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cho DH, et al. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bossy B, et al. S-Nitrosylation of DRP1 does not affect enzymatic activity and is not specific to Alzheimer’s disease. Journal of Alzheimer’s disease: JAD. 2010;20(Suppl 2):S513–526. doi: 10.3233/JAD-2010-100552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Manczak M, Reddy PH. Abnormal interaction between the mitochondrial fission protein Drp1 and hyperphosphorylated tau in Alzheimer’s disease neurons: implications for mitochondrial dysfunction and neuronal damage. Human molecular genetics. 2012;21:2538–2547. doi: 10.1093/hmg/dds072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Manczak M, et al. Impaired mitochondrial dynamics and abnormal interaction of amyloid beta with mitochondrial protein Drp1 in neurons from patients with Alzheimer’s disease: implications for neuronal damage. Human molecular genetics. 2011;20:2495–2509. doi: 10.1093/hmg/ddr139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Trushina E, et al. Defects in mitochondrial dynamics and metabolomic signatures of evolving energetic stress in mouse models of familial Alzheimer’s disease. PLoS One. 2012;7:e32737. doi: 10.1371/journal.pone.0032737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guttman M, et al. Burden of parkinsonism: a population-based study. Mov Disord. 2003;18:313–319. doi: 10.1002/mds.10333. [DOI] [PubMed] [Google Scholar]
- 87.Braak H, et al. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol Aging. 2003;24:197–211. doi: 10.1016/s0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- 88.Langston JW, et al. Pargyline prevents MPTP-induced parkinsonism in primates. Science. 1984;225:1480–1482. doi: 10.1126/science.6332378. [DOI] [PubMed] [Google Scholar]
- 89.Betarbet R, et al. Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nat Neurosci. 2000;3:1301–1306. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- 90.Schapira AHV, et al. Anatomic and disease specificity of NADH CoQ1 reductase (complex I) deficiency in Parkinson’s disease. J Neurochem. 1990;55:2142–2145. doi: 10.1111/j.1471-4159.1990.tb05809.x. [DOI] [PubMed] [Google Scholar]
- 91.Zheng B, et al. PGC-1alpha, a potential therapeutic target for early intervention in Parkinson’s disease. Sci Transl Med. 2010;2:52ra73. doi: 10.1126/scitranslmed.3001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Poole AC, et al. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci U S A. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang Y, et al. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc Natl Acad Sci U S A. 2008;105:7070–7075. doi: 10.1073/pnas.0711845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shin JH, et al. PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson’s disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vives-Bauza C, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Matsuda N, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. The Journal of cell biology. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang X, et al. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147:893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang X, et al. Parkinson’s disease-associated DJ-1 mutations impair mitochondrial dynamics and cause mitochondrial dysfunction. Journal of neurochemistry. 2012;121:830–839. doi: 10.1111/j.1471-4159.2012.07734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Irrcher I, et al. Loss of the Parkinson’s disease-linked gene DJ-1 perturbs mitochondrial dynamics. Human molecular genetics. 2010;19:3734–3746. doi: 10.1093/hmg/ddq288. [DOI] [PubMed] [Google Scholar]
- 100.Ahlskog JE. Parkin and PINK1 parkinsonism may represent nigral mitochondrial cytopathies distinct from Lewy body Parkinson’s disease. Parkinsonism Relat Disord. 2009;15:721–727. doi: 10.1016/j.parkreldis.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kamp F, et al. Inhibition of mitochondrial fusion by alpha-synuclein is rescued by PINK1, Parkin and DJ-1. EMBO J. 2010;29:3571–3589. doi: 10.1038/emboj.2010.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nakamura K, et al. Direct membrane association drives mitochondrial fission by the Parkinson disease-associated protein alpha-synuclein. The Journal of biological chemistry. 2011;286:20710–20726. doi: 10.1074/jbc.M110.213538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xie W, Chung KK. Alpha-synuclein impairs normal dynamics of mitochondria in cell and animal models of Parkinson’s disease. J Neurochem. doi: 10.1111/j.1471-4159.2012.07769.x. [DOI] [PubMed] [Google Scholar]
- 104.Gui YX, et al. Extracellular signal-regulated kinase is involved in alpha-synuclein-induced mitochondrial dynamic disorders by regulating dynamin-like protein 1. Neurobiol Aging. 2012 doi: 10.1016/j.neurobiolaging.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 105.Wang X, et al. LRRK2 regulates mitochondrial dynamics and function through direct interaction with DLP1. Human molecular genetics. 2012;21:1931–1944. doi: 10.1093/hmg/dds003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jellinger KA. Neuropathology of sporadic Parkinson’s disease: evaluation and changes of concepts. Mov Disord. 2012;27:8–30. doi: 10.1002/mds.23795. [DOI] [PubMed] [Google Scholar]
- 107.Clark LN, et al. Frequency of LRRK2 mutations in early- and late-onset Parkinson disease. Neurology. 2006;67:1786–1791. doi: 10.1212/01.wnl.0000244345.49809.36. [DOI] [PubMed] [Google Scholar]
- 108.Zheng Z, Diamond MI. Huntington disease and the huntingtin protein. Prog Mol Biol Transl Sci. 2012;107:189–214. doi: 10.1016/B978-0-12-385883-2.00010-2. [DOI] [PubMed] [Google Scholar]
- 109.Bossy-Wetzel E, et al. Mutant huntingtin and mitochondrial dysfunction. Trends Neurosci. 2008;31:609–616. doi: 10.1016/j.tins.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Massieu L, et al. Neurotoxicity of glutamate uptake inhibition in vivo: correlation with succinate dehydrogenase activity and prevention by energy substrates. Neuroscience. 2001;106:669–677. doi: 10.1016/s0306-4522(01)00323-2. [DOI] [PubMed] [Google Scholar]
- 111.Greene JG, Greenamyre JT. Characterization of the excitotoxic potential of the reversible succinate dehydrogenase inhibitor malonate. J Neurochem. 1995;64:430–436. doi: 10.1046/j.1471-4159.1995.64010430.x. [DOI] [PubMed] [Google Scholar]
- 112.Panov AV, et al. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nature neuroscience. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- 113.Song W, et al. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nature medicine. 2011;17:377–382. doi: 10.1038/nm.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang H, et al. Effects of overexpression of huntingtin proteins on mitochondrial integrity. Hum Mol Genet. 2009;18:737–752. doi: 10.1093/hmg/ddn404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Costa V, et al. Mitochondrial fission and cristae disruption increase the response of cell models of Huntington’s disease to apoptotic stimuli. EMBO molecular medicine. 2010;2:490–503. doi: 10.1002/emmm.201000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shirendeb UP, et al. Mutant huntingtin’s interaction with mitochondrial protein Drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degeneration in Huntington’s disease. Human molecular genetics. 2012;21:406–420. doi: 10.1093/hmg/ddr475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cui L, et al. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 118.Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. Embo J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]