Abstract
Our understanding of kinetoplastid RNA (kRNA) editing has centered on this paradigm: guide RNAs (gRNAs) provide a blueprint for uridine insertion/deletion into mitochondrial mRNAs by the RNA editing core complex (RECC). Yet the characterization of constituent subunits of the mitochondrial RNA-binding complex 1 (MRB1) implies that it too is vital to the editing process. The recently elucidated MRB1 architecture will be instrumental in putting functional data from individual subunits into context. Our model depicts two functions for MRB1: mediating multi-round kRNA editing by coordinating the exchange of multiple gRNAs required by RECC to edit lengthy regions of mRNAs, and then linking kRNA editing with other RNA processing events.
Keywords: Kinetoplastida, trypanosome, RNA editing, protein complexes, RECC, MRB1
Kinetoplastid RNA editing and the mitochondrial RNA binding complex 1
Much has been learned about kRNA editing since its discovery in the mitochondrion of trypanosomes by Benne and colleagues in 1986 [1], such as the role of small guide RNAs (gRNAs; Glossary) in providing information for each uridine (U) insertion or deletion [2], validation of the enzyme cascade model [3], as well as detailed characterization of the RNA editing core complex (RECC; Box 1), the molecular machine that orchestrates the required enzymatic steps [4–6]. As a process shared by causative agents of African sleeping sickness, Chagas disease, leishmaniases and other serious illnesses, kRNA editing is being explored as a possible drug target [7]. Nonetheless, perplexing questions remain about both the mechanism of kRNA editing and about how this process is integrated into the general mitochondrial RNA metabolism.
Box 1. RNA editing core complex (RECC).
The RECC complex is also referred to in the literature as the L-complex [4] or the 20 S editosome [5]. The latter name reflects the size of the editosome in ultracentrifugation studies. The complex contains about 20 proteins that encapsulate the enzymatic activities needed for a single round of U-insertion/deletion editing (Figure 1a and described in main text). In comparison to MRB1, the composition of RECC appears to be static. Nevertheless, recent work has shown that there are three major forms of RECC, each bearing a defining endonuclease [44, 45]. One of them specifically cleaves edited sites (ESs) that require U-insertion while another does so for ESs slated to undergo U-deletion. A third is special in that it cleaves coxII transcripts. The editing of coxII does not need traditional gRNAs since it bears its own within its 3 ′-UTR, thus acting incis [37]. The deletion endonuclease associates with a U-specific exonuclease that trims superfluous Us from the ES. It is still not clear whether these three RECCs represent stable complexes, or if the appropriate endonuclease, and associated proteins, are recruited by a RECC already processing a gRNA:mRNA duplex.
Another complex called the mitochondrial RNA binding (MRB1) complex is also required for editing and rivals RECC in terms of complexity. The current state of knowledge we review here indicates that MRB1 may be the key to many unanswered questions. Several of the proteins that comprise MRB1 bear signature motifs and domains that clearly indicate a role in RNA metabolism; others lack identifiable domains and, furthermore, are unique to the excavate order Kinetoplastea [8–11]. Intense work has revealed a dizzying array of potential functions of these proteins in kRNA editing and metabolism [11–20], making it difficult to pinpoint the function(s) of MRB1. Recently the architecture of MRB1 was reported [16], which has allowed us to better interpret functional analysis of the constituents identified thus far. Here, we present a model of how MRB1 may work in kRNA metabolism by mediating the exchange of gRNAs required by RECC in processing mRNAs that need multi-round kRNA editing, and by linking kRNA editing with other RNA processing events.
Herein, we use the MRB1 nomenclature for some of the constituent subunits as introduced elsewhere [14], which is the ‘MRB’ prefix followed by the last digits of the trypanosome genome database TriTrypDB (http://tritrypdb.org/tritrypdb/) accession number. Some proteins, such as the gRNA associated proteins (GAPs), TbRGG2 and RNA editing helicase 2 (REH2) were named before this nomenclature was introduced.
The basic mechanism of kRNA editing mediated by RECC
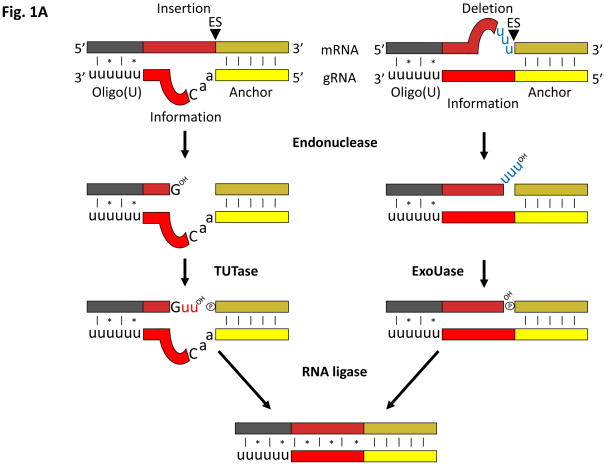
In Trypanosoma brucei, 12 out of 18 protein-coding mitochondrial genes require kRNA editing for their maturation. Editing initiates with the 5′-most portion of the small gRNA, called the anchor domain, hybridizing with a complementary part of the pre-edited (pre)-mRNA, with some additional interactions within the gRNA 3′ oligo(U) tail (Figure 1a) [21]. The editing site (ES) where a U-insertion/deletion occurs on the pre-mRNA is indicated by the first base-pair mismatch between the gRNA:mRNA duplex (Figure 1a). The pre-mRNA is cleaved here, in the first RECC-catalyzed kRNA editing step, to yield 5′ and 3′ fragments that are bridged by the bound gRNA. Depending on the gRNA information domain sequence downstream of the anchor, one or more Us are added or removed from the 5′-fragment. RECC enzymes catalyze both events: a 3′-terminal uridylyltransferase (TUT) adds Us and a U-specific exonuclease deletes them. After the ES has been edited, the pre-mRNA fragments are sealed together by a RECC RNA ligase. A single gRNA carries information for several ESs along a stretch of the pre-mRNA called the editing block; as editing progresses, the mRNA becomes complementary to the gRNA via both normal and non-canonical Watson-Crick base-paring.
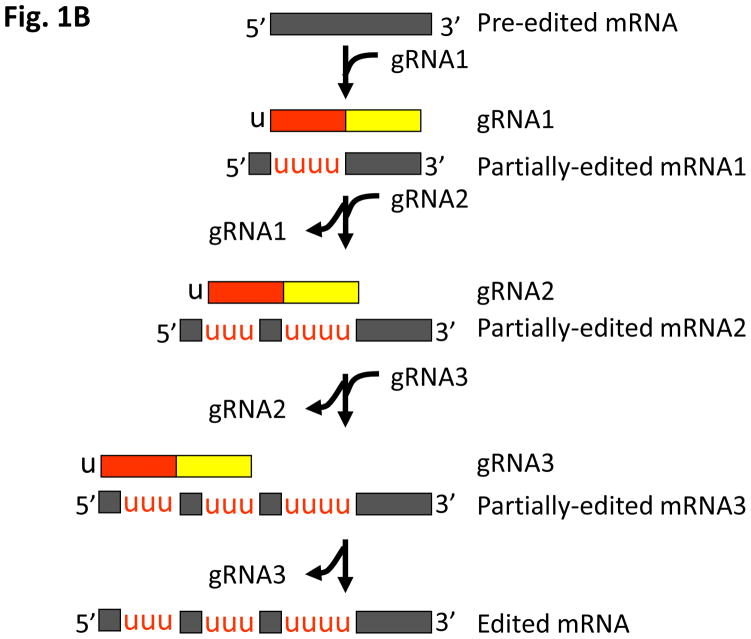
Figure 1. Single-versus multi-round kRNA editing.
(a) A single round of kRNA editing as mediated by one of the appropriate RECCs through an enzyme cascade. The first base pair mismatch in the duplex between gRNA (lower strand; domains in solid colors; yellow = anchor, red = information; black U = 3′-oligo(U) tail) and mRNA (top strand; regions on mRNA are indicated in a different shade of color to that of the corresponding gRNA domains) define the editing site (ES; arrowhead), which is then cleaved on the mRNA by an endonuclease (all enzymatic activities in bold). In U-insertion, Us (in red) are added to the 3′-end of the 5′-mRNA cleavage product by a terminal uridylyltransferase (TUTase) as guided by the gRNA. In U-deletion, a U-specific exonuclease (exoUase) prunes away extra Us (in blue) on the 3′-end of the 5′-mRNA cleavage product. An RNA ligase then seals the two mRNA fragments back together again after the appropriate editing event is completed. (b) Pan-edited mRNAs require multi-rounds of kRNA editing. A cascade of gRNAs is required to decrypt a pan-edited mRNA (black striped strand). The first gRNA (solid colors using same scheme to distinguish domains as in (a); black U signifies 3′-oligo(U) tail) guides RNA editing (signified by inserted red Us) along a block (current editing block underlined in red) defining several editing sites. When the editing events dictated by the gRNA are completed, the gRNA is unwound from the mRNA. A second gRNA binds to a newly edited sequence on the mRNA that can now hybridize its anchor domain. As this process is repeated, editing progresses from the 3′ to 5′ direction until the mRNA is fully edited. The mRNA intermediates between the pre-edited molecule and the fully-edited form are referred to as partially-edited. Abbreviations: OH, 3′-hyroxyl group; P, 5′-phosphate group; *, non-canonical base pairing; |, canonical Watson-Crick base pairing.
Massively processed pan-edited mRNAs require a cascade of gRNAs
Three quarters of the edited mRNAs in T. brucei are pan-edited, an extreme form of kRNA editing. For example, 547 Us are inserted and 41 are deleted over 223 ESs in the cytochrome c oxidase (cox) III transcript, representing more than half of the mature mRNA [22]. Pan-editing occurs in the 3′ to 5′ direction along the pre-mRNA and requires numerous gRNAs, unlike the one or two needed by less-edited transcripts [23]. When editing requires multiple gRNAs (Figure 1b), a portion of the fully processed pre-mRNA editing block generated from the first gRNA sequence serves as the annealing site for the anchor domain of the next gRNA. This process is repeated until the pre-mRNA is fully-edited, containing the decrypted open reading frame (ORF) ready for translation. The pan-editing intermediates, in which the 5′-proximal sequences remain unedited, are referred to as partially-edited transcripts. When provided with an mRNA and cognate gRNA, the core editing activities encapsulated in RECC can be reconstituted in vitro [3]. However, in vitro editing is limited to one ES rather than the entire editing block specified by the gRNA. Consequently, the cascade of gRNA exchange also cannot take place in vitro. Together, these findings suggest that other factors are required for editing in vivo. Furthermore, the mechanisms of gRNA removal from a fully-processed editing block, and recruitment and binding of the next required gRNA are unknown. Because this process involves the unwinding of a tightly bound gRNA:edited mRNA duplex, helicase activity is likely required. The RNA helicase, REH1, appears to act in gRNA removal in some capacity [24], although it does not contribute the majority of the unwinding activity in the mitochondria [13].
Other steps in kRNA metabolism
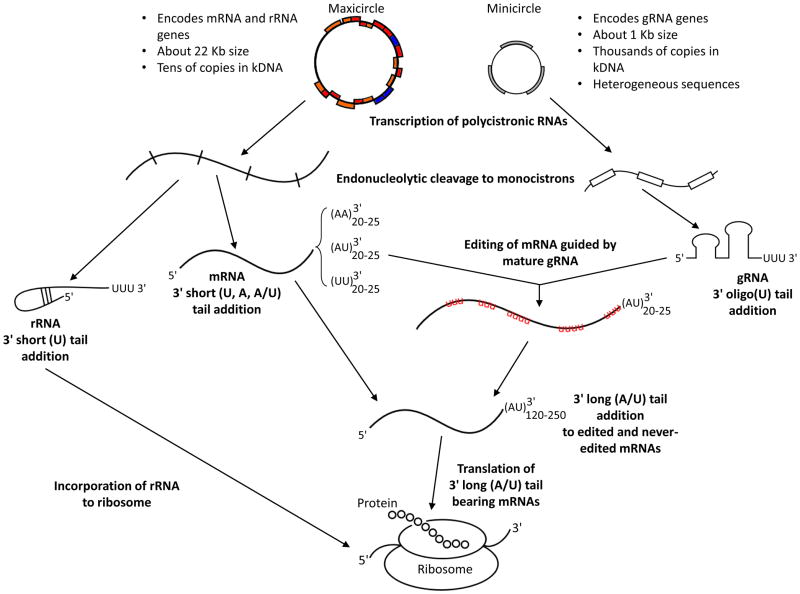
While editing, due to its uniqueness, stands out as a kRNA maturation process, it is but one step from the mitochondrial genome to translation of the encoded proteins (Figure 2). Transcription of T. brucei maxi- and minicircle kinetoplast DNAs (kDNAs), which encode all mRNAs and rRNAs or virtually all gRNAs, respectively, is performed by a single RNA polymerase [12, 25]. Because maxicircle and minicircle transcripts are polycistronic, endonucleolytic cleavage is needed to free individual cistrons. An endonuclease called mitochondrial RNA precursor-processing endonuclease 1(mRPN1) is involved in processing minicircle pre-RNAs into single gRNAs [26]. The genes on polycistronic maxicircle transcripts are very close together or overlapping and are cleaved into cistrons by a hitherto unknown enzyme(s) [27].
Figure 2. Summary of kRNA metabolism.
The maxi- and minicircles of the kDNA are depicted on the top with a summary of their major characteristics adjacent. Genes encoding pan-edited mRNAs are in red, minimally-edited in blue and never-edited in orange. Genes encoding gRNAs are grey on the minicircles. RNA metabolism steps are indicated in bold. Transcription of both yields polycistronic transcripts that are subsequently cleaved to yield monocistrons. The rRNAs obtain short 3′ (U) tails and are incorporated into ribosomes; mRNAs initially get short 3′ tails (as long as 20–25 nt, comprised of U, A or A/U as indicated in subscript next to tails in the figure); gRNAs get short 3′-oligo(U) tails. The mature gRNAs mediate editing of mRNAs. Fully edited and never edited mRNAs are eventually marked with long 3′ (A/U) tails (120–250 nt) for translation. Turnover of mRNAs could occur at any step of this process.
RNA turnover and physical shuttling of RNA between relevant complexes and binding proteins are two other processes occurring in the mitochondria. Both may be influenced by yet another post-transcriptional event: the addition of non-encoded 3′ tails comprised of adenine (A), U or some combination thereof to most mitochondrial RNAs [10, 28–31]. The kinetoplast poly(A) polymerase 1 (KPAP1) attaches the As, while the TUTase RET1 adds the Us into these tails. These structures occur in two different populations according to their lengths. Short 3′ mRNA tails appear to impact transcript stability in a variety of ways [10, 28, 32, 33], while long tail addition, facilitated by two pentatricopeptide (PPR) proteins, likely marks mRNAs as translationally competent [34]. Long A/U tail addition is hypothesized to serve as a bridge between editing and translation [34]. Several of the protein players mentioned in these processes, plus RECC, associate to some degree with the MRB1 complex. However, very little is known about how mRNAs are physically transcribed or transition from processing to editing, degradation and translation.
The architecture of MRB1
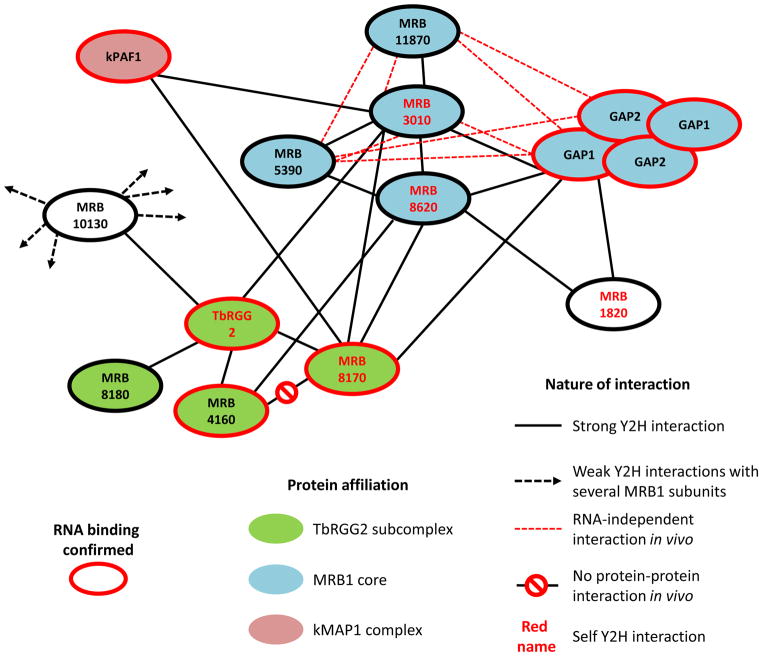
Reports of MRB1 purification from T. brucei and Leishmania tarentolae have described both overlapping and varying subunits [8–11, 13–16]. In the former category are GAPs 1 and 2 (also known as GRBC2 and 1), MRB3010, MRB8620, MRB5390 and MRB11870, which have been reassuringly present in virtually all MRB1 purifications. Others, such as the DExD-box RNA helicase REH2 plus the RNA-binding proteins TbRGG2 and MRB1680, associate with MRB1 in a variable fashion. Many interactions were found to be due to proteins binding to the same RNA. We propose that some of the more tenuous interactions reflect a dynamic composition of MRB1, in which proteins were incorporated into several subcomplexes [16]. This notion that MRB1 is made up of several subcomplexes and/or that some subunits shuttle between those involved in other aspects of RNA metabolism is supported by the observed heterodispersion of individual parts in ultracentrifugation sedimentation studies [8, 9, 12–14, 16].
To address the confusing nature of the MRB1 composition, a comprehensive yeast two-hybrid screen was used to map the interactions between the 31 proteins found in various MRB1 isolations [16]. Surprisingly, only six proteins were responsible for the majority of interactions. Four of these are components of a core identified by multiple RNase-resistant co-purifications (Figure 3). Subunits from this core interact with the TbRGG2 subcomplex, comprised of the eponymous protein and either one of the paralogs MRB8170 or MRB4160; additionally TbRGG2 interacts with MRB8180 and MRB10130, although the temporal association of all of these proteins is unknown [15, 16, 26]. The TbRGG2 subcomplex interacts with the MRB1 core in an RNase-sensitive manner, in contrast to the direct binding exhibited among most of the core subunits. MRB10130, a polypeptide almost entirely comprised of ARM/HEAT repeats that often act in protein-protein interactions, also associates with the core subcomplex and proteins involved in other steps in kRNA metabolism [35].
Figure 3. Architecture of MRB1.
A summary of the pairwise yeast two-hybrid (Y2H) mapped MRB1 subunit interactions plus those determined in vivo as adapted from Ammerman and colleagues [16]. The figure is updated to reflect the mutually exclusive interaction of MRB8170 and MRB4160 with TbRGG2 [15] and the heterotetrameric nature of the GAP1 and GAP2 interaction [11]. A key to the figure is shown at the bottom. The black lines represent ‘strong’ Y2H interactions in at least one direction. The promiscuous ‘weak’ Y2H interactions of the ARM/HEAT domain containing MRB10130 with various MRB1 subunits are also indicated by dashed black lines projecting out.
As mentioned previously, many of the intra-MRB1 interactions are enhanced or even dependent on the presence of RNA. One such protein is MRB6070, which contains zinc fingers, suggesting that it is an RNA binding protein. Although it does not interact with other MRB1 proteins via yeast two-hybrid analysis, it does associate with MRB1 complex purified from cells in an RNA-dependent manner [16]. The RNA-dependent interactions within the MRB1 complex and the observation that several subunits self-interact in the yeast two-hybrid screen and/or seem to form oligomers in vivo makes it difficult to pinpoint the MRB1 subunit stoichiometry. In addition, it remains an open question whether the bona fide protein-protein interactions among the MRB1 subunits reflect constant interactions, as is observed for many RECC constituents, or are dynamic.
MRB1 interaction with other protein complexes involved in kRNA metabolism
In addition to intra-complex interactions, several MRB1 subunits have been found to interact with proteins of other complexes acting in the various kRNA metabolic processes enumerated above. Sub-stoichiometric amounts of RECC have been retained in TbRGG2, REH2, and core MRB1 component purifications in an RNase-sensitive manner [11, 13, 18]. The complex containing KPAP1 and the kinetoplast polyadenylation/uridylation factor 1 (KPAF1) PPR protein associates with MRB1 in a similar fashion [10, 11, 13, 34]. Furthermore, KPAF1 was shown to interact with the MRB1 core and the TbRGG2 subcomplex in the yeast two-hybrid screen (Figure 3) [16]. MRB1 subunit purifications from T. brucei also intermittently co-purify mitochondrial edited mRNA stability factor 1, a protein named according to its apparent function in the organelle [8, 11]. Thus it seems that MRB1 (sub)complexes are also involved with those playing a role in mitochondrial mRNA maturation and stability.
Interestingly, KPAP1 and the PPR protein KPAF1 associate with the mitochondrial ribosomes, the endpoint for all translatable mRNAs, in addition to MRB1. MRB1 proteins pulled down substoichiometric amounts of ribosomal proteins as well [8, 11, 13], although it must be noted that ribosomal proteins are common contaminants in proteomic analyses of protein purifications in general. A more refined study demonstrated a preferential association of the MRB1 core GAPs, along with the TUTase RET1 and RECC, with the large subunit of the ribosome [34]. These observations support a model in which MRB1 physically and functionally interacts with the mitochondrial protein translation machinery.
Since MRB1 contains subunits that have RNA binding activity (e.g., GAPs and REH2; see below), it is perhaps not surprising that the complex associates with other RNA-interacting proteins and complexes. For example, the endonuclease mRPN1 involved in processing gRNAs has been reported to associate with the TbRGG2 subcomplex [26]. Additionally, the mitochondrial-RNA-binding-proteins 1/2 complex, which is an mRNA:gRNA matchmaker in vitro, has a substoichiometric, RNase-sensitive association with MRB1 [13, 15, 36]. Therefore, these associations link MRB1 to the processes of gRNA biogenesis and utilization.
Nuts and bolts: the functional characterization of MRB1 subunits
Functional studies of MRB1 subunits based on RNAi-mediated depletion of individual components reveal multifarious phenotypes. Of the six MRB1 core proteins, GAP1, GAP2, MRB3010, MRB5390, MRB8620, and MRB11870, all but the last two have been analyzed in this way. These proteins proved to be essential for the viability of T. brucei by affecting different aspects of the RNA editing process (Table 1).
Table 1.
Summary of MRB1 subunit RNAi phenotypes
| RNAi Growth inhibition | RNA abundance effect due to RNAi | RNA binding? | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Namea | TriTrpDB Ascension # | PF | BF | Never-edited RNA | Pan-edited RNA | Minimally-edited RNA | gRN A | Proven | Predicted domainb | Complex | Notes | Refs |
| GAP1 (GRBC2) | Tb927.2.3800 | Y | Y | N | Y | Y (not coxII) | Y | Y | MRB1 core | -gRNA bidning -Paralog GAP2 |
[11, 12] | |
| GAP2 (GRBC1) | Tb927.7.2570 | Y | Y | N | Y | Y (not coxII) | Y | Y | MRB1 core | -gRNA binding -Paralog GAP1 |
[11, 12] | |
| MRB3010 | Tb927.5.3010 | Y | Y | N | Y | Y | N | N | MRB1 core | -Role in early step kRNA editing | [14] | |
| MRB5390 | Tb11.02.5390 | Y | ND | N | Y (subset) | Y (subset) | ND | ND | MRB1 core | [17] | ||
| TbRGG2 | Tb927.10.10830 | Y | Y | N | Y | N | N | Y | RGG, RRM | TbRGG2 subcomplex | -3′-5′ progression RNA editing | [17–20] |
| MRB8[15] 170 | Tb927.8.8170 | N | ND | N | Y | N | N | Y | TbRGG2 subcomplex | -paralog MRB4160 | [15] | |
| MRB4160 | Tb927.4.4160 | N | ND | N | N | N | N | Y | TbRGG2 subcomplex | -paralog MRB8170 | [15] | |
| MRB8170/MRB4160c | -- | Y | Y | N | Y | Y | N | -- | -- | -RNAi affects pan-edited more than minimally | [15] | |
| MRB1680 | Tb927.6.1680 | Y | ND | N | Y | N | ND | ND | C2H2 Zn Finger | [17] | ||
| REH2 | Tb927.4.1500 | Y | ND | N | Y | Y (not coxII) | Y | Y | dsRBP, DExD-box | -gRNA binding -RNA unwinding activity |
[12, 13] | |
Aliases given in parentheses
Common abbreviations of motifs.
Describes simultaneous RNAi-silencing of the paralogs MRB8170 and MRB4160.
Abbreviations:Y, yes; N, no; ND, not determined; PF, procyclic form; BF, bloodstream form; gRNA, guide RNA
GAP1 and 2 are paralogs that associate in a heterotetramer, and repression of either results in loss of both proteins [11, 12]. The GAPs bind gRNAs in vitro despite lacking domains known to confer this activity [11]. Consistent with this finding, their repression results in decreased gRNA levels, which subsequently affect editing of mRNAs that require these trans-elements. As such, the minimally edited coxII, whose gRNA acts in cis as it is contained in the 3′-UTR of the mRNA [37], is not affected. Unlike the GAPs, the core protein MRB3010 does not have in vitro RNA binding activity, and its down-regulation in T. brucei did not affect bulk gRNA levels [14]. However, RNAi did result in depletion of both pan- and minimally-edited RNAs, although the latter were affected to a lesser degree. Simultaneously, there was an increase in pre-edited and early partially-edited RNAs, consistent with MRB3010 playing a role at an early step in editing. MRB5390 repression similarly resulted in depletion of edited transcripts, although to a more modest effect [17].
Like the core MRB1 proteins, TbRGG2 is essential for RNA editing [17–20]. Yet unlike the aforementioned knockdowns, TbRGG2 RNAi-silencing only affects pan-edited transcripts, resulting in a buildup of partially-edited RNAs at the expense of the fully edited forms. This observation suggests a role in promoting the 3′ to 5′ progression of editing, a notion supported by the in vitro characterization of its N-terminal G-rich region, with GWG and RG repeats, and C-terminal RRM domain [19, 20]. TbRGG2 has an N-terminal RNA annealing activity, which is crucial for editing, while the C-terminal domain confers a RNA melting activity.
The TbRGG2 subcomplex also contains either MRB8170 or MRB4160 in a mutually exclusive fashion [15, 16]. These two proteins are highly similar paralogs in T. brucei, having arisen from a chromosomal duplication in this clade, being a single copy in other trypanosomatids [15, 38]. Like TbRGG2, both paralogs have in vitro RNA binding activity. Furthermore, their simultaneous RNAi-silencing preferentially results in a decrease of pan-edited RNAs without affecting gRNAs. While this phenotype is ostensibly reminiscent of TbRGG2, there is an important contrast: minimally edited RNAs are also affected (but not when either MRB8170 or MRB4160 are individually silenced), and some pre-edited forms of the pan-edited mRNAs are also decreased [15].
Some MRB1 subunits that are neither part of the core nor TbRGG2 subcomplex have also been characterized by RNAi. The RNA helicase REH2 is a gRNA binding protein, and like the GAPs, its depletion results in a decrease of gRNAs [12, 13]. Furthermore, REH2 associates with the bulk of RNA unwinding activity from mitochondrial lysates, which distinguishes it from REH1. MRB1680, a putative RNA-binding protein with five zinc finger domains, has a RNAi phenotype that resembles that of TbRGG2: a decrease of pan-edited RNAs with minor effects on minimally-edited transcripts [17].
Building models of MRB1 function from the nuts and bolts
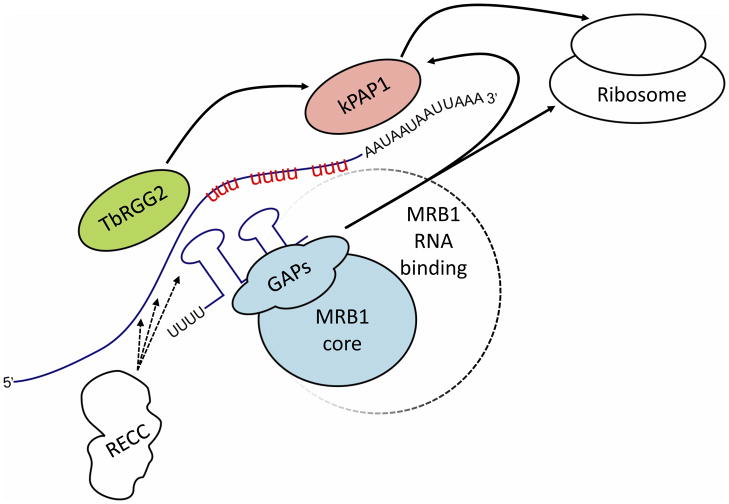
The determined architecture of MRB1 has been instrumental in putting all of the RNAi studies into context. By combining these two datasets, we propose one model describing two functions for MRB1, namely mediating the recruitment and exchange of gRNAs required by RECC in processing pan-edited mRNAs that require multiple rounds of the process, and linking kRNA editing to other kRNA processing events (Figure 4).
Figure 4. Proposed model of MRB1 function.
The assembly of the MRB1 core and TbRGG2 subcomplex(es) plus RECC and the kPAP1 complex onto an mRNA is depicted using the same color scheme as in Figure 3. The GAPs bring gRNAs to the reaction center. The core associates with the transcript via the putative RNA binding proteins of MRB1 (depicted as dashed circle radiating from the MRB1 core). The TbRGG2 subcomplex(es) promotes gRNA:mRNA annealing and/or unwinding of these double-stranded structures. The kPAP1 complex is responsible for 3′-tail addition. The solid black arrows signify verified interactions among complexes. Substrate RNAs are depicted in blue. Red ‘u’ stretches are sequence already edited within the mRNA. Dashed arrows from RECC indicate that it associates with RNAs in cooperation with MRB1. This scheme does not depict the potential dynamic movement of certain proteins among the complexes.
The first function has MRB1 acting in tandem with RECC as dual core processors of pan-edited RNAs. This notion is supported by the predominant MRB1 RNAi phenotype, in which pan-edited RNAs are more severely compromised than their minimally-edited counterparts because they require more gRNAs for editing. While the paucity of data makes predicting exactly what the core MRB1 subcomplex is doing in this scenario challenging, the gRNA-binding GAPs may serve to bring these molecules to or maintain them within the reaction center. In this model, the MRB1 core serves as a scaffold for creating the reaction center, since the core MRB3010 protein seems to halt an early step(s) in editing, possibly linked to RECC via the mRNA undergoing pan-editing. The TbRGG2 subcomplex serves as an engine for the 3′ to 5′ progression by promoting proper gRNA:mRNA duplex unwinding after an editing block for a given gRNA has been completed and/or promoting the formation of the next duplex. The in vitro characterizedRNA binding, annealing, and unwinding activities of TbRGG2 could potentially be employed in such a task, aided by either of the RNA binding MRB8170 and MRB4160 subunits.
The second function of MRB1 is to link kRNA editing to other processing events required for the expression of mitochondrial genes. The available data suggest an entanglement between the protein players in kRNA editing, gRNA processing, and mRNA 3′-tail addition, RNA stability/turnover and translation, as MRB1 subunits are always present in association with these processes [10, 11, 13, 26, 34]. Furthermore, there is evidence for functional coordination between multiple steps in kRNA metabolism. For instance, a short oligo(A) tail stabilizes mRNA only following the editing of the first gRNA-dictated editing block [10, 33]. The addition of a long (A/U) tail occurs after an RNA is completely edited (or nearly so), and this 3′ extension is thought to act as an important factor signaling the transition from editing to translation [10, 32, 34]. It seems reasonable to assume that shuttling between different steps of kDNA gene expression is a necessary process for organellar biogenesis, and MRB1 appears to be a strong candidate for such a role.
Additional components of MRB1 may very well affect RNA editing and/or link it to other metabolic processes but are not specified in our model. REH2 and MRB1680 are putative RNA binding proteins, the depletion of which impacts kRNA editing [13, 17]. Furthermore, it is difficult at this time to proffer whether the identified subcomplexes represent static or dynamic structures, although the described studies seem to indicate the latter. It will be interesting to see how future results will refine this model.
Our data can also be used to exclude some putative MRB1 functions, such as roles in transcription or cleavage of polycistronic maxicircle RNAs into individual cistrons. We presume that if one of these were true, changes in abundance of never-edited mRNAs would be a predominant MRB1 RNAi phenotype, which is not the case. The MRB1 subunits that were examined by RNAi in both T. brucei bloodstream and procyclic in vitro cell cultures exhibited comparable phenotypes, indicating that MRB1 may not play a major role in regulation of editing among the life cycle stages (Table 1). However, not all MRB1 subunits have been analyzed in this way.
MRB1 contributes to the irremediable complexity of RNA editing
Future studies will likely refine our current MRB1 functional model (Box 2); however, even now it is clear that MRB1 is an emerging key player in kRNA editing in conjunction with RECC. The discovery of MRB1 certainly adds to kRNA editing complexity. More than 70 proteins –and counting– are required for editing mitochondrial RNAs and generating gRNAs in T. brucei [39]. This count is in addition to the presumably large number of proteins required for maturation of never-edited mRNAs as well as rRNAs, the latter of which are assembled into mitochondrial ribosomes, requiring more than 100 proteins to construct this machine that ultimately translates the mature mRNAs [40].
Box 2. Outstanding questions.
What are the precise mechanisms by which MRB1 impacts kRNA editing?
What is the spatial and temporal distribution of MRB1 and other complexes within the single reticulated mitochondrion?
How is RNA routed into the appropriate complexes devoted to a specific processing step?
What are the regulatory elements in the whole pathway of kDNA gene expression from transcription via editing, processing and turnover to translation?
Why would so many proteins be needed for the expression of just a handful of gene products in a eukaryotic cell? According to the most plausible adaptive explanations, kRNA editing generates mitochondrial protein diversity via an alternative editing strategy [41], or it provides another level of gene regulation among life stages [42]. Nevertheless, these appealing theories remain unsupported.
An alternative view, albeit one that is also difficult to address experimentally, suggests that the irremediable complexity of kRNA editing has arisen due to constructive neutral evolution [39, 43]. In essence, this idea suggests that the complexity that typifies many biochemical systems can emerge even in the absence of any positive selection pressure. Thus, as a biological process like kRNA editing grows more complex over time, various interacting components become essential without serving any adaptive advantage. These components have become interdependent and the cell is consequently stuck with an intricate clockwork in which each gear is intrinsically essential to its proper running. In the case of kRNA metabolism, MRB1 and RECC, along with other players involved in the processing of the various kRNA species, are each essential players in expression of kDNA-encoded genes.
Acknowledgments
This work was supported by the Grant Agency of the Czech Republic (204/09/1667 and P506/12/3157), the RNPnet FP7 program (289007), and the Praemium Academiae award to J.L., as well as NIH awards RO1 AI061580 to L.K.R., and F32 AI092902 to S.L.Z. J.L. is Fellow of the Canadian Institute for Advanced Research.
Glossary
- Anchor domain
A 5′-proximal part of a gRNA that facilitates the formation of a gRNA:mRNA duplex by hybridizing to a mRNA that is to undergo editing
- ARM/HEAT repeats
repeat motifs initially described from the Drosophila Armadillo protein and four cytoplasmic proteins in mammals and yeast, respectively, which often act in protein-protein interactions
- Editing block
A stretch of sequence on an edited mRNA that contains several ESs, as determined by the information domain of a gRNA
- Editing site (ES)
Cleavage site on a mRNA where either a uridine-insertion or –deletion takes place, as defined by the first base pair mismatch on the mRNA;gRNA duplex
- Fully-edited mRNA
Transcripts in which all the needed editing steps have been completed
- guide RNA (gRNA)
Small RNAs that act in trans to supply sequence information for decrypting edited mRNAs (one gRNA contained in the 3′-UTR of the coxII mRNA acts in cis)
- Information domain
The part of the gRNA that dictates the ESs along an editing block on an edited mRNA; the fully processed editing block is complementary to the anchor domain via normal and non-canonical Watson-Crick base-pairing
- Maxicircle DNA
The approximately 22 Kb component of kDNA that encodes mRNA and rRNA genes, there are tens of copies of these DNAs, and the gene-encoding region is homogenous in sequence
- Minicircle DNA
The approximately 1 Kb component making up the bulk of kDNA encoding gRNA genes; the thousands of copies are heterogeneous in sequence as they represent all the gRNAs needed to decrypt edited mRNAs
- Minimally-edited mRNA
Transcripts that require kRNA editing only in a small portion of their sequence requiring few gRNAs; sometime also called moderately-edited
- Never-edited mRNA
Transcripts that forgo kRNA editing
- Pan-edited mRNA
Transcripts that undergo kRNA editing throughout their sequence and require many gRNAs
- Partially-edited mRNA
Intermediates of pan-edited transcripts undergoing kRNA editing, in which the 5′-proximal sequence remains ‘pre-edited’ while the 3′-proximal sequence is ‘edited’
- Pre-edited (pre-) mRNA
Edited transcripts before they undergo kRNA editing, thus having the same sequence as the kDNA maxicircle cryptogene; also called unedited
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Benne R, et al. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986;46:819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- 2.Blum B, et al. A model for RNA editing in kinetoplastid mitochondria: “guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990;60:189–198. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- 3.Seiwert SD, Stuart K. RNA editing: transfer of genetic information from gRNA to precursor mRNA in vitro. Science. 1994;266:114–117. doi: 10.1126/science.7524149. [DOI] [PubMed] [Google Scholar]
- 4.Simpson L, et al. Mitochondrial proteins and complexes in Leishmania and Trypanosoma involved in U-insertion/deletion RNA editing. RNA. 2004;10:159–170. doi: 10.1261/rna.5170704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stuart KD, et al. Complex management: RNA editing in trypanosomes. Trends Biochem Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Lukeš J, et al. The remarkable mitochondrion of trypanosomes and related flagellates. In: de Souza W, editor. Structures and Organelles in Pathogenic Protists. Springer; 2010. pp. 227–252. [Google Scholar]
- 7.Salavati R, et al. Inhibitors of RNA editing as potential chemotherapeutics against trypanosomatid pathogens. Int J Parasitol Drugs & Drug Res. 2012;2:36–46. doi: 10.1016/j.ijpddr.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hashimi H, et al. TbRGG1, an essential protein involved in kinetoplastid RNA metabolism that is associated with a novel multiprotein complex. RNA. 2008;14:970–980. doi: 10.1261/rna.888808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panigrahi AK, et al. Mitochondrial complexes in Trypanosoma brucei: a novel complex and a unique oxidoreductase complex. Mol Cell Proteomics. 2008;7:534–545. doi: 10.1074/mcp.M700430-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Etheridge RD, et al. 3′ adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. EMBO J. 2008;27:1596–1608. doi: 10.1038/emboj.2008.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weng J, et al. Guide RNA-binding complex from mitochondria of trypanosomatids. Mol Cell. 2008;32:198–209. doi: 10.1016/j.molcel.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hashimi H, et al. Kinetoplastid guide RNA biogenesis is dependent on subunits of the mitochondrial RNA binding complex 1 and mitochondrial RNA polymerase. RNA. 2009;15:588–599. doi: 10.1261/rna.1411809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hernandez A, et al. REH2 RNA helicase in kinetoplastid mitochondria: ribonucleoprotein complexes and essential motifs for unwinding and guide RNA (gRNA) binding. J Biol Chem. 2010;285:1220–1228. doi: 10.1074/jbc.M109.051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ammerman ML, et al. MRB3010 is a core component of the MRB1 complex that facilitates an early step of the kinetoplastid RNA editing process. RNA. 2011;17:865–877. doi: 10.1261/rna.2446311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kafková L, et al. Functional characterization of two paralogs that are novel RNA binding proteins influencing mitochondrial transcripts of Trypanosoma brucei. RNA. 2012;18:1846–1861. doi: 10.1261/rna.033852.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ammerman ML, et al. Architecture of the trypanosome RNA editing accessory complex, MRB1. Nucl Acids Res. 2012;40:5637–5650. doi: 10.1093/nar/gks211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Acestor N, et al. The MRB1 complex functions in kinetoplastid RNA processing. RNA. 2009;15:277–286. doi: 10.1261/rna.1353209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisk JC, et al. TbRGG2, an essential RNA editing accessory factor in two Trypanosoma brucei life cycle stages. J Biol Chem. 2008;283:23016–23025. doi: 10.1074/jbc.M801021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ammerman ML, et al. TbRGG2 facilitates kinetoplastid RNA editing initiation and progression past intrinsic pause sites. RNA. 2010;16:2239–2251. doi: 10.1261/rna.2285510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foda BM, et al. Multifunctional G-rich and RRM-containing domains of TbRGG2 perform separate yet essential functions in trypanosome RNA editing. Eukaryot Cell. 2012;11:1119–1131. doi: 10.1128/EC.00175-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McManus MT, et al. Trypanosoma brucei guide RNA poly(U) tail formation is stabilized by cognate mRNA. Mol Cell Biol. 2000;20:883–891. doi: 10.1128/mcb.20.3.883-891.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feagin JE, et al. Developmentally regulated addition of nucleotides within apocytochrome b transcripts in Trypanosoma brucei. Cell. 1987;49:337–345. doi: 10.1016/0092-8674(87)90286-8. [DOI] [PubMed] [Google Scholar]
- 23.Maslov DA, Simpson L. The polarity of editing within a multiple gRNA-mediated domain is due to formation of anchors for upstream gRNAs by downstream editing. Cell. 1992;70:459–467. doi: 10.1016/0092-8674(92)90170-h. [DOI] [PubMed] [Google Scholar]
- 24.Li F, et al. Trypanosome REH1 is an RNA helicase involved with the 3′–5′ polarity of multiple gRNA-guided uridine insertion/deletion RNA editing. Proc Natl Acad Sci USA. 2011;108:3542–3547. doi: 10.1073/pnas.1014152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grams J, et al. A trypanosome mitochondrial RNA polymerase is required for transcription and replication. J Biol Chem. 2002;277:16952–16959. doi: 10.1074/jbc.M200662200. [DOI] [PubMed] [Google Scholar]
- 26.Madina BR, et al. Guide RNA biogenesis involves a novel RNase III family endoribonuclease in Trypanosoma brucei. RNA. 2011;17:1821–1830. doi: 10.1261/rna.2815911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koslowsky DJ, Yahampath G. Mitochondrial mRNA 3′ cleavage/polyadenylation and RNA editing in Trypanosoma brucei are independent events. Mol Biochem Parasit. 1997;90:81–94. doi: 10.1016/s0166-6851(97)00133-3. [DOI] [PubMed] [Google Scholar]
- 28.Aphasizheva I, Aphasizhev R. RET1-catalyzed uridylylation shapes the mitochondrial transcriptome in Trypanosoma brucei. Mol Cell Biol. 2010;30:1555–1567. doi: 10.1128/MCB.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adler BK, et al. Modification of Trypanosoma brucei mitochondrial rRNA by posttranscriptional 3′ polyuridine tail formation. Mol Cell Biol. 1991;11:5878–5884. doi: 10.1128/mcb.11.12.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blum B, Simpson L. Guide RNAs in kinetoplastid mitochondria have a nonencoded 3′ oligo(U) tail involved in recognition of the preedited region. Cell. 1990;62:391–397. doi: 10.1016/0092-8674(90)90375-o. [DOI] [PubMed] [Google Scholar]
- 31.Ryan CM, Read LK. UTP-dependent turnover of Trypanosoma brucei mitochondrial mRNA requires UTP polymerization and involves the RET1 TUTase. RNA. 2005;11:763–773. doi: 10.1261/rna.7248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zimmer SL, et al. Additive and transcript-specific effects of KPAP1 and TbRND activities on 3′ non-encoded tail characteristics and mRNA stability in Trypanosoma brucei. PLoS One. 2012;7:e37639. doi: 10.1371/journal.pone.0037639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kao CY, Read LK. Opposing effects of polyadenylation on the stability of edited and unedited mitochondrial RNAs in Trypanosoma brucei. Mol Cell Biol. 2005;25:1634–1644. doi: 10.1128/MCB.25.5.1634-1644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aphasizheva I, et al. Pentatricopeptide repeat proteins stimulate mRNA adenylation/uridylation to activate mitochondrial translation in trypanosomes. Mol Cell. 2011;42:106–117. doi: 10.1016/j.molcel.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrade MA, et al. Comparison of ARM and HEAT protein repeats. J Mol Biol. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- 36.Aphasizhev R, et al. A 100-kD complex of two RNA-binding proteins from mitochondria of Leishmania tarentolae catalyzes RNA annealing and interacts with several RNA editing components. RNA. 2003;9:62–76. doi: 10.1261/rna.2134303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Golden DE, Hajduk SL. The 3′-untranslated region of cytochrome oxidase II mRNA functions in RNA editing of African trypanosomes exclusively as a cis guide RNA. RNA. 2005;11:29–37. doi: 10.1261/rna.7170705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson AP. Tandem gene arrays in Trypanosoma brucei: comparative phylogenomic analysis of duplicate sequence variation. BMC Evol Biol. 2007;7:54. doi: 10.1186/1471-2148-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lukeš J, et al. How a neutral evolutionary ratchet can build cellular complexity. IUBMB Life. 2011;63:528–537. doi: 10.1002/iub.489. [DOI] [PubMed] [Google Scholar]
- 40.Zíková A, et al. Trypanosoma brucei mitochondrial ribosomes: affinity purification and component identification by mass spectrometry. Mol Cell Proteomics. 2008;7:1286–1296. doi: 10.1074/mcp.M700490-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ochsenreiter T, et al. Alternative mRNA editing in trypanosomes is extensive and may contribute to mitochondrial protein diversity. PLoS One. 2008;3:e1566. doi: 10.1371/journal.pone.0001566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Speijer D. Is kinetoplastid pan-editing the result of an evolutionary balancing act? IUBMB Life. 2006;58:91–96. doi: 10.1080/15216540600551355. [DOI] [PubMed] [Google Scholar]
- 43.Gray MW, et al. Cell biology. Irremediable complexity? Science. 2010;330:920–921. doi: 10.1126/science.1198594. [DOI] [PubMed] [Google Scholar]
- 44.Carnes J, et al. Endonuclease associations with three distinct editosomes in Trypanosoma brucei. J Biol Chem. 2011;286:19320–19330. doi: 10.1074/jbc.M111.228965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carnes J, et al. RNA editing in Trypanosoma brucei requires three different editosomes. Mol Cell Biol. 2008;28:122–130. doi: 10.1128/MCB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]