Abstract
Neuroglobin (Ngb) has been demonstrated to be a novel neuroprotective protein that protects against hypoxia/ischemia and oxidative stress-induced injury in the nervous system. However, the regulation mechanisms of Ngb gene expression under both normal resting and stress conditions have not been fully elucidated. The cyclic AMP response element binding protein (CREB) is a key transcription factor that regulates a variety of pro-survival genes, but its role in regulating the neuroprotective gene Ngb has not been studied. In this study we investigated the transcriptional regulation of mouse Ngb gene by CREB in mouse neuroblastoma cell line N2a. Our results showed that CREB knockdown decreased Ngb gene expression, and overexpression of the wild-type CREB, but not the mutant CREB, significantly increased Ngb gene expression in N2a cells. Moreover, a cAMP response element (CRE) site located at −854 in the promoter region of mouse Ngb gene was found to be responsible for both basal and CREB-induced Ngb promoter activity. Using chromatin immunopreciptation (ChIP) assays, we found that CREB could bind to the Ngb promoter region spanning from −1016 to −793 that harbors the CRE site. Taken together, our results suggested that transcription factor CREB participates in the transcriptional regulation of mouse Ngb gene.
Keywords: Neuroglobin, CREB, transcriptional regulation, mouse, N2a cells
Introduction
Neuroglobin (Ngb) is an oxygen-binding globin that was first identified in 2000 [2]. As Ngb is predominantly expressed in brain neurons and highly conserved during evolution, it has been suggested that Ngb is a functionally important protein in the nervous system [2, 6, 22]. Accumulating experimental findings have demonstrated that Ngb is a novel endogenous neuroprotectant in response to a variety of insults including cerebral ischemia and neurodegenerative diseases[7]. Increased Ngb expression confers resistance of neurons against hypoxic/ischemic [10, 21]and oxidative injuries[3, 12]. Ngb also protects neurons from beta-amyloid-induced neurotoxicity and attenuates phenotype in transgenic mice of Alzheimer disease [11], indicating that Ngb is protective in neurodegenerative diseases as well. Thus, Ngb can be a novel target for endogenous neuroprotection against stroke and related neuroglogical disorders [7]. However, the Ngb gene regulation mechanisms remain to be fully understood in order to better develop the Ngb up-regulation therapy approach. Previous reports from our lab and others have shown that Ngb gene can be regulated by transcription factors SP1 and SP3 [23], Egr 1, NFkB and HIF1α [8, 13]. Interestingly, the involvements of multiple transcriptional regulators and the delicate responses of Ngb gene expression to various physiological conditions strongly suggested that there might be other transcriptional factors that participate in Ngb gene regulation [4].
As a key transcription factor in the nervous system, CREB plays critical roles in neuronal growth, survival, and synaptic function [15, 17], thus it may be another candidate serving as a transcriptional regulator of Ngb gene expression. Previous studies indicated that CREB exerts neuroprotective function in response to harmful or stressful stimuli by regulating the expression of a variety of anti-apoptotic proteins, such as BDNF [18], Bcl2 [5, 16], MnSOD [1] and Mcl-1 [20]. Since Ngb is a neuron survival and protection molecule, and considering CREB function in maintaining basal and stress-induced pro-survival genes expression, in this study we tested our hypothesis that CREB may also mediate transcriptional regulation of Ngb gene.
Material and Methods
Reagents
Cell culture supplies including DMEM (Dulbecco’s modified Eagle’s medium), geneticin, penicillin, streptomycin and LipofectamineTM 2000 were purchased from Invitrogen. FBS (Fetal bovine serum), L-glutamine, glutamic acid, 0.05% trypsin-EDTA were purchased from Gibco. The chicken anti-Ngb primary antibody was purchased from BioVendor. Antibodies against CREB and phospho-CREB at ser133 (p-CREB) were purchased form Cell Signaling Technology. All siRNAs (small interfering RNAs) were obtained form Santa Cruz Biotechnology. pCMV-HA vector and CREB Dominant-Negative Vector Set were purchase form Clontech Laboratories. pGL3-Basic vector, Dual-Luciferase® Reporter Assay System were purchased from Promega.
Reporter plasmid construction
Mouse Ngb gene promoter constructs including P-2033 (−2027/+6), P-1539 (−1533/+6), P-1004 (−998/+6), P-886 (−880/+6), P-554 (−549/+6), P-341 (−335/+6) and P-554 (Egr1M) were cloned as described before [13]. Contructs with mutations to the CRE1 and CRE2 sites that were derived form the P-2033 (−2027/+6) construct were generated by overlapping extension PCR as described previously [14]. In brief, in the first round, two PCR were performed in parallel using the P-2033 (−2027/+6) construct as template: one with a wild-type forward primer P-2033F (5′-GGGGTACCGTCCACTACTCCCATGAATTC-3′) and a reverse mutated primer CRE1delR (5′-CATGGAGTTTGCCATCGCACGACCTACTGA-3′) or CRE2delR (5′-TCTGTTGGAGCCCCCAGACAGTAACTAGTA-3′), the other with a forward mutated primer CRE1delF (5′-GCGATGGCAAACTCCATGAATGTGTACGCA-3′) or CRE2delF (5′-TCTGGGGGCTCCAACAGACCTGAGATTGGA-3′) and a wild-type reverse primer P-2033R (5′ -CCAAGCTTCTCCATGCTCTCTCTCCCAG-3′). In the second round, equal molar mixture of the two PCR products was used as template, P-2033F and P-2033R were used as primers. The final PCR products were cloned into pGL3-Basic vector with a KpnI/HindIII site. The sequences of all constructs were confirmed by direct sequencing.
Cell culture and transfection
The mouse N2a (Neuro2a) neuroblastoma cell culture was performed as described before [13]. In brief, N2a cells were maintained in DMEM supplemented with 10% FBS, 0.3 mM L-glutamine and 50 U/ml penicillin/streptomycin. For transfection, N2a cells with 70% confluence were transiently transfected with DNA constructs or siRNAs using LipofectamineTM 2000 following the manufacturer’s protocol.
Luciferase reporter assay
Luciferase reporter assay was performed as described before [13]. Briefly, N2a cells were transiently transfected with DNA constructs and/or siRNAs as well as plasmid pRL-TK encoding Renilla luciferase which was used as a control for transfection efficiency for 24 hours. The cells were then lysed in 1X passive lysis buffer and the lysates were collected and assayed for luciciferase activity using the Dual-Luciferase® Reporter Assay System on VeritasTM Microplate Luminometer (Turner BioSystems). Activity was defined as Firefly/Renilla ratio. Each experiment was performed at least three times.
Western blot analysis
Western blotting was performed as described previously [13]. In brief, protein samples from N2a cells were diluted with 4× E-PAGE™ loading buffer and 10× NuPAGE® reducing agent (Invitrogen) and heated at 70°C for 10 min. Then the protein sample was separated by 4%-12% SDS-PAGE gel (Invitrogen) and transferred onto nitrocellulose membrane using the buffer-less, iBlot® dry blotting system (Invitrogen). The membrane was soaked with 5% nonfat dried milk in 10 mM PBS buffer (PH 7.2) for 1 hour at room temperature to block non-specific binding. Immunoblots were then performed at 4°C by incubation with primary antibodies followed by washing three times with TBS/0.1% Tween for removing excess primary antibody. After that, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies for 1 hour at 37°C. The membranes were then washed with TBS/0.1% Tween for another three times. The blots were developed using enhanced chemiluminescence (ECL, Amersham Pharmacia Biotech) according to the manufacturer’s protocol and then exposed to film (X-Omat; Eastman Kodak Co.). The intensity of immunoreactive bands was then quantified by densitometry using ImageJ software.
ChIP (Chromatin Immunoprecipitation) assay
The ChIP assay was performed using an EZ ChIPTM Chromatin Immunoprecipitation kit (Upstate Biotechnology) according to the manufacturer’s protocol as described before [13]. Briefly, approximately 1×106 N2a cells were cross-linked at 37°C for 10 min in 1% formaldehyde. The cells were then collected for sonication in SDS lysis buffer to shear the chromatin with an average size of approximately 300 bp. Then the supernatant was immunoprecipitated by adding anti-CREB or a control rabbit polyclonal anti-IgG antibody, and the mixture was placed on a rotator at 4°C for overnight. DNA-protein cross-linkers were then reversed by heating at 65°C for 4 hours. The DNA products were further amplified by PCR. The mouse Ngb promoter region containing CRE1 and CRE2 sites which spanned from −1016 to −793 upstream of the starting codon ATG was amplified using primers: chCRE2Fw (5′-CCAGCACCTCGTTTCTTAGTTTT -3′) and chCRE2Rs (5′-GCAGAGGCCAAAACATAGACATC-3′). The mouse Ngb promoter region contained CRE3 site which spanned from −2055 to −1843 upstream of ATG was amplified using primers: chCRE3Fw (5′-GGTTCTGGTTCTTCCACTTAGGTC-3′) and chCRE3Rs (5′-TCCTAATGACTTCAGAAGACCCC-3′). Each of the experiments was repeated three times.
Statistical Analysis
All data values are expressed as means ± SD. Statistical significance (P<0.05) were determined by a One-way ANOVA followed by Tukey-Kramer tests.
Results
1. CREB is involved in maintaining mouse Ngb gene expression
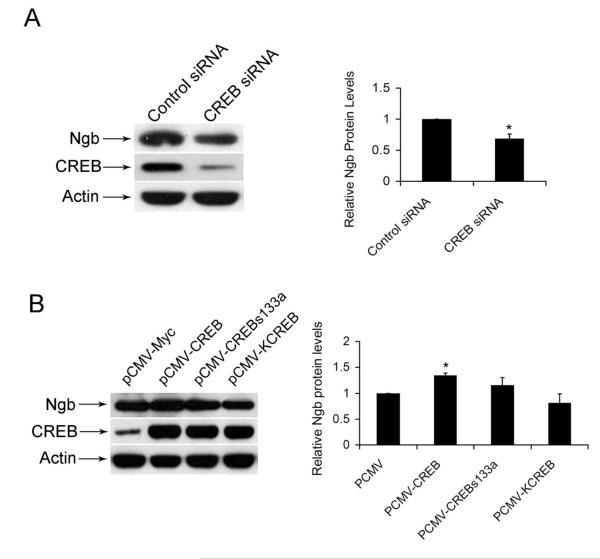
To explore the potential role of CREB in the regulation of Ngb gene expression, pecific siRNA against CREB and control siRNA were transfected into N2a cells, and the endogenous CREB and Ngb protein levels were examined. Our data showed that knock-down of CREB significantly decreased Ngb protein levels (Figure. 1A). To further examine the role of CREB in regulation of Ngb gene expression, N2a cells were transiently transfected with wild-type CREB overexpression vector and dominant mutant CREB vector. Our results showed that CREB overexpression significantly enhanced Ngb protein levels. However, the dominant mutant CREB vector including KCREB and CREBs133a did not significantly increase Ngb protein levels (Figure. 1B).
Figure 1. Effect of transcription factor CREB in mouse Ngb protein levels.
(A). Representative western showed effect of specific mouse CREB siRNA on mouse Ngb protein levels, β-actin served as equal loading controls. (Mean ± SD). n=3, *P<0.05 versus cells transfected with control siRNA. (B). Representative western showed effect of CREB and its mutant on mouse Ngb protein levels, β-actin served as equal loading controls. (Mean ± SD). n=3, *P<0.05 versus cells transfected with control siRNA.
2. CREB is a positive regulator of mouse Ngb gene promoter
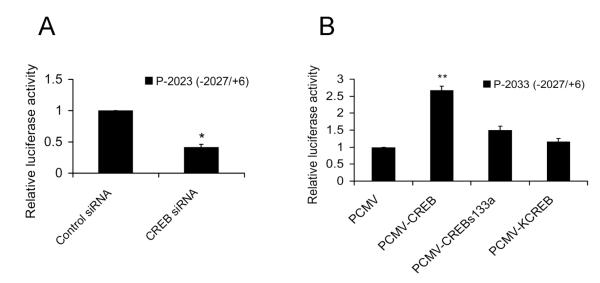
To investigate whether CREB is a transcriptional activator of Ngb gene expression, we examined the effects of CREB on the mouse Ngb promoter activity by luciferase reporter assay. Our results showed that knock-down of CREB by specific CREB siRNA significantly reduced the Ngb promoter activity (Figure 2A), and over-expression of wild-type CREB, but not mutant CREB, significantly increased the Ngb promoter activity of P-2033 (−2027/+6) (Figure 2B). These results indicate that CREB is a positive regulator of mouse Ngb gene promoter.
Figure 2. Effect of transcription factor CREB in mouse Ngb promoter activity.
(A). The luciferase activity (Mean ± SD) was determined at 24 h after transfection. n=4, *P<0.05 versus control cells transfected with control siRNA. (B). The luciferase activity (Mean ± SD) was determined at 24 h after transfection. n=4, ** P<0.01 versus cells transfected with PCMV-Myc.
3. CRE2 site mediates CREB-induced upregulation of mouse Ngb gene promoter
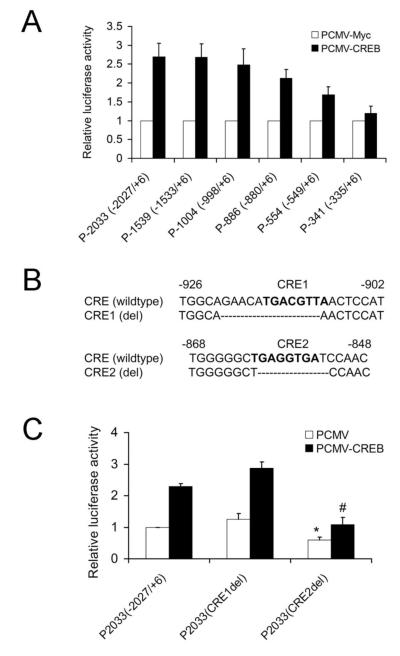
To define the CREB-responsive region in the mouse Ngb gene promoter, we co-transfected the 5′ serial deleted mouse Ngb promoters and CREB expression vectors into N2a cells. Our results showed that deletion of the sequence from −2027 to −998 bp did not affect the transactivation of the mouse Ngb promoter by CREB. However, progressive 5′-deletion from −880 to −335 bp gradually decreased Ngb promoter activity in response to CREB overexpression, suggesting that the region spanning −880 to −335 probably contains CREB-response elements (Figure 3A). Therefore we went on to further identify the CREB binding motifs in the mouse Ngb promoter using the bioinformatic TFBS-prediction tool Jaspar (http://jaspar.genereg.net/). After analysis of the sequence spanning from −998 to −335 bp, we found two potential CREB-binding sites located on −909 and −854, which were designated CRE1 and CRE2 sites respectively (Figure 3B). To investigate the role of these two putative cAMP-response elements (CRE) in CREB-mediated transactivation of the mouse Ngb promoter, we deleted these two CREs from P-2033 (−2027/+6) reporter plasmid, and then co-transfected CREB expression vector and P-2033 (−2027/+6) or one of the CRE deletion plasmids into N2a cells. We found the Ngb promoter activity in P-2033 (−2027/+6) and CRE1 (del) reporter plasmid can be stimulated by CREB, whereas mutation of CRE2 site significantly attenuated CREB-induced Ngb promoter activity (Figure 3C). These results indicated that the CRE2 site is responsible for CREB-induced Ngb promoter activity.
Figure 3. Functional analysis of CRE sites within mouse Ngb promoter.
(A). The luciferase activity (Mean ± SD) was determined at 24 h after transfection. n=4. (B). Deletion mutation of the putative CRE1 and CRE2 sites. Bold letters indicate putative CRE1 and CRE2 sites. (C). Relative luciferase activities of P2033 (−2027/+6), P2033 (CREdel1) and P2033 (CRE2del) in N2a cells. *P<0.05 versus N2a cells co-transfected with PCMV-Myc and P2033 (−2027/+6). # P<0.05 versus N2a cells co-transfected with PCMV-CREB and P2033 (−2027/+6).
4. CREB binds to mouse Ngb promoter in vivo
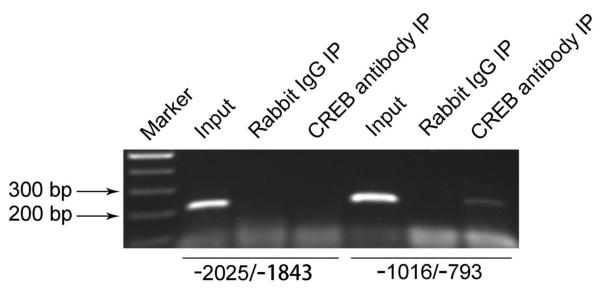
To further confirm the function of CREB in regulating Ngb gene expression, we investigated whether CREB is capable of binding to mouse Ngb promoter region by ChIP assay in N2a cells. Our results indicated that the chromosomal DNA immunoprecipitated by CREB antibody can be amplified by PCR using Ngb promoter-specific primers covering the CRE1 and CRE2 sites from −1016 to −793 bp, but not the primers covering the CRE3 site from −2055 to −1843 bp (Figure 4). These results indicated that CREB specifically binds to the mouse Ngb promoter region covering CRE2 site.
Figure 4. Interactions of CREB with mouse Ngb promoter in N2a cells.
Chromatin immunoprecipitation was performed in N2a cells. Precipitated DNA was immunoprecipitated with anti-CREB, then amplified by PCR using primers specific for mouse Ngb promoter. Rabbit IgG served as a negative control.
Discussion
Experimental results of this study clearly demonstrated that CREB is also an important transcription regulator for basal Ngb gene expression. The transcriptional activation of CREB is critically dependent on phosphorylation of Ser133 by multiple kinases in response to a wide variety of external stimuli [15], while the mutant CREBs133a protein cannot be phosphorylated at Ser133, thus was not able to transactivate the target genes. The mutant KCREB protein contains mutations within its DNA-binding domain that form an inactive dimer with a wild-type CREB, thus blocking its ability to bind CRE of target genes [19]. In this study we also found that in comparison to KCREB, CREBs133a also enhanced Ngb protein levels, which implied that CREB may transactivate Ngb gene in a phospho-Ser133-independent manner.
We also found that the CRE2 site located at −854bp in the promoter of mouse Ngb gene was responsible for both basal and CREB-induced Ngb promoter activity. Furthermore, CREB was found to specifically bind in vivo to the mouse Ngb promoter region that covers the CRE2 site. We have previously reported a large similarity of the mouse Ngb with human Ngb promoter after bioinformatics analysis [13], which identified several putative CRE sites in the promoter of human Ngb gene, thus it is possible that CREB also mediates transcriptional regulation of human Ngb gene. This notion needs to be investigated in future study.
Identification of CREB as a transcriptional factor in Ngb gene regulation is fundamentally and translationally significant. CREB has been well demonstrated as a transcriptional regulator to mediate adaptive responses of neurons to stress stimuli by upregulating the expression of neuron survival and anti-apoptotic proteins, such as BDNF [18], Bcl2 [5, 16], MnSOD [1]. Our findings indicate that by maintaining and upregulating Ngb expression, CREB might promote neuronal survival under pathological conditions. However the regulatory roles of CREB for Ngb in response to neuronal insults and the underlying molecular mechanisms and neuroprotective consequences remain to be elucidated in further investigations. Additionally, we have recently documented that transcription factors NFκB (p65), Sp1 and HIF-1α are involved in Ngb upregulation under hypoxic condition [13]. As CREB has been previously reported to cross-talk with other transcription factors such as NFκB (p65) for transactivating target gene [9], it is interesting that CREB may interact with these transcription factors to regulate Ngb gene in response to some specific stress stimuli. This issue remains to be further defined.
In summary, our findings suggest that transcription factor CREB is a positive regulator of mouse Ngb gene expression in neuronal cells. Further investigation in elucidating the molecular regulation mechanisms of Ngb gene and the role of CREB would lead to a better development of Ngb-regulation based interventions for stroke and other related neurological disorders.
Highlights.
CREB knockdown decreased Ngb gene expression in N2a cells.
Overexpression of wild type CREB, but not mutant CREB, increased Ngb expression.
A cAMP response element site is located at −854 in mouse Ngb promoter region.
CREB can bind to Ngb promoter region in vivo tested by CHIP.
Acknowledgments
This work was supported in part by NIH grant R01-NS049476 (to X.W.) and postdoctoral fellowship (12POST9720007) from American Heart Association (to Z.Y.). We appreciate Dr. Eng H. Lo for his very helpful discussion.
Abbreviations
- Ngb
Neuroglobin
- CREB
the cyclic AMP responsive element binding protein
- CRE
the cAMP-response elements
- ChIP
chromatin immunopreciptation
- N2a
Neuro 2a
- siRNA
small interfering RNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflict of interest.
References
- [1].Bedogni B, Pani G, Colavitti R, Riccio A, Borrello S, Murphy M, Smith R, Eboli ML, Galeotti T. Redox regulation of cAMP-responsive element-binding protein and induction of manganous superoxide dismutase in nerve growth factor-dependent cell survival. J Biol Chem. 2003;278:16510–16519. doi: 10.1074/jbc.M301089200. [DOI] [PubMed] [Google Scholar]
- [2].Burmester T, Weich B, Reinhardt S, Hankeln T. A vertebrate globin expressed in the brain. Nature. 2000;407:520–523. doi: 10.1038/35035093. [DOI] [PubMed] [Google Scholar]
- [3].Duong TT, Witting PK, Antao ST, Parry SN, Kennerson M, Lai B, Vogt S, Lay PA, Harris HH. Multiple protective activities of neuroglobin in cultured neuronal cells exposed to hypoxia re-oxygenation injury. J Neurochem. 2009;108:1143–1154. doi: 10.1111/j.1471-4159.2008.05846.x. [DOI] [PubMed] [Google Scholar]
- [4].Fordel E, Geuens E, Dewilde S, De Coen W, Moens L. Hypoxia/ischemia and the regulation of neuroglobin and cytoglobin expression. IUBMB Life. 2004;56:681–687. doi: 10.1080/15216540500037406. [DOI] [PubMed] [Google Scholar]
- [5].Freeland K, Boxer LM, Latchman DS. The cyclic AMP response element in the Bcl-2 promoter confers inducibility by hypoxia in neuronal cells. Brain Res Mol Brain Res. 2001;92:98–106. doi: 10.1016/s0169-328x(01)00158-9. [DOI] [PubMed] [Google Scholar]
- [6].Fuchs C, Heib V, Kiger L, Haberkamp M, Roesner A, Schmidt M, Hamdane D, Marden MC, Hankeln T, Burmester T. Zebrafish reveals different and conserved features of vertebrate neuroglobin gene structure, expression pattern, and ligand binding. J Biol Chem. 2004;279:24116–24122. doi: 10.1074/jbc.M402011200. [DOI] [PubMed] [Google Scholar]
- [7].Greenberg DA, Jin K, Khan AA. Neuroglobin: an endogenous neuroprotectant. Curr Opin Pharmacol. 2008;8:20–24. doi: 10.1016/j.coph.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Haines B, Demaria M, Mao X, Xie L, Campisi J, Jin K, Greenberg DA. Hypoxia-inducible factor-1 and neuroglobin expression. Neurosci Lett. 2012;514:137–140. doi: 10.1016/j.neulet.2012.01.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kaszubska W, Hooft van Huijsduijnen R, Ghersa P, DeRaemy-Schenk AM, Chen BP, Hai T, DeLamarter JF, Whelan J. Cyclic AMP-independent ATF family members interact with NF-kappa B and function in the activation of the E-selectin promoter in response to cytokines. Mol Cell Biol. 1993;13:7180–7190. doi: 10.1128/mcb.13.11.7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Khan AA, Mao XO, Banwait S, DerMardirossian CM, Bokoch GM, Jin K, Greenberg DA. Regulation of hypoxic neuronal death signaling by neuroglobin. FASEB J. 2008;22:1737–1747. doi: 10.1096/fj.07-100784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Khan AA, Mao XO, Banwait S, Jin K, Greenberg DA. Neuroglobin attenuates beta-amyloid neurotoxicity in vitro and transgenic Alzheimer phenotype in vivo. Proc Natl Acad Sci U S A. 2007;104:19114–19119. doi: 10.1073/pnas.0706167104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Li RC, Morris MW, Lee SK, Pouranfar F, Wang Y, Gozal D. Neuroglobin protects PC12 cells against oxidative stress. Brain Res. 2008;1190:159–166. doi: 10.1016/j.brainres.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Liu N, Yu Z, Xiang S, Zhao S, Tjarnlund-Wolf A, Xing C, Zhang J, Wang X. Transcriptional regulation mechanisms of hypoxia-induced neuroglobin gene expression. Biochem J. 2012;443:153–164. doi: 10.1042/BJ20111856. [DOI] [PubMed] [Google Scholar]
- [14].Liu R, Zhou A, Ren D, He A, Hu X, Zhang W, Yang L, Liu M, Li H, Zhou J, Xiang S, Zhang J. Transcription factor specificity protein 1 (SP1) and activating protein 2alpha (AP-2alpha) regulate expression of human KCTD10 gene by binding to proximal region of promoter. FEBS J. 2009;276:1114–1124. doi: 10.1111/j.1742-4658.2008.06855.x. [DOI] [PubMed] [Google Scholar]
- [15].Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- [16].Meller R, Minami M, Cameron JA, Impey S, Chen D, Lan JQ, Henshall DC, Simon RP. CREB-mediated Bcl-2 protein expression after ischemic preconditioning. J Cereb Blood Flow Metab. 2005;25:234–246. doi: 10.1038/sj.jcbfm.9600024. [DOI] [PubMed] [Google Scholar]
- [17].Sakamoto K, Karelina K, Obrietan K. CREB: a multifaceted regulator of neuronal plasticity and protection. J Neurochem. 2011;116:1–9. doi: 10.1111/j.1471-4159.2010.07080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Tao X, Finkbeiner S, Arnold DB, Shaywitz AJ, Greenberg ME. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron. 1998;20:709–726. doi: 10.1016/s0896-6273(00)81010-7. [DOI] [PubMed] [Google Scholar]
- [19].Walton KM, Rehfuss RP, Chrivia JC, Lochner JE, Goodman RH. A dominant repressor of cyclic adenosine 3′,5′-monophosphate (cAMP)-regulated enhancer-binding protein activity inhibits the cAMP-mediated induction of the somatostatin promoter in vivo. Mol Endocrinol. 1992;6:647–655. doi: 10.1210/mend.6.4.1350057. [DOI] [PubMed] [Google Scholar]
- [20].Wang JM, Chao JR, Chen W, Kuo ML, Yen JJ, Yang-Yen HF. The antiapoptotic gene mcl-1 is up-regulated by the phosphatidylinositol 3-kinase/Akt signaling pathway through a transcription factor complex containing CREB. Mol Cell Biol. 1999;19:6195–6206. doi: 10.1128/mcb.19.9.6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang X, Liu J, Zhu H, Tejima E, Tsuji K, Murata Y, Atochin DN, Huang PL, Zhang C, Lo EH. Effects of neuroglobin overexpression on acute brain injury and long-term outcomes after focal cerebral ischemia. Stroke. 2008;39:1869–1874. doi: 10.1161/STROKEAHA.107.506022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhang C, Wang C, Deng M, Li L, Wang H, Fan M, Xu W, Meng F, Qian L, He F. Full-length cDNA cloning of human neuroglobin and tissue expression of rat neuroglobin. Biochem Biophys Res Commun. 2002;290:1411–1419. doi: 10.1006/bbrc.2002.6360. [DOI] [PubMed] [Google Scholar]
- [23].Zhang W, Tian Z, Sha S, Cheng LY, Philipsen S, Tan-Un KC. Functional and sequence analysis of human neuroglobin gene promoter region. Biochim Biophys Acta. 2011;1809:236–244. doi: 10.1016/j.bbagrm.2011.02.003. [DOI] [PubMed] [Google Scholar]