Abstract
Understanding the spatial clustering of Plasmodium falciparum populations can assist efforts to contain drug-resistant parasites and maintain the efficacy of future drugs. We sequenced single nucleotide polymorphisms (SNPs) in the dihydropteroate synthase gene (dhps) associated with sulfadoxine resistance and 5 microsatellite loci flanking dhps in order to investigate the genetic backgrounds, genetic relatedness, and geographic clustering of falciparum parasites in the Democratic Republic of the Congo (DRC). Resistant haplotypes were clustered into subpopulations: one in the northeast DRC, and the other in the balance of the DRC. Network and clonal lineage analyses of the flanking microsatellites indicate that geographically-distinct mutant dhps haplotypes derive from separate lineages. The DRC is therefore a watershed for haplotypes associated with sulfadoxine resistance. Given the importance of central Africa as a corridor for the spread of antimalarial resistance, the identification of the mechanisms of this transit can inform future policies to contain drug-resistant parasite strains.
Antimalarial drug resistance continually threatens to undermine global efforts to contain and control malaria. Molecular analyses have demonstrated that parasite mutations conferring resistance to chloroquine1 and to pyrimethamine2 originated in Southeast Asia and spread first to East Africa and thence across the African continent. Consequently, these mutations are highly prevalent across sub-Saharan Africa and the clinical drug resistance they confer bedevils malaria control efforts. A better understanding of the geographic spread of drug-resistant malaria may enhance efforts to contain nascent resistance to newer antimalarials3.
In contrast to mutations which confer resistance to chloroquine and pyrimethamine, those which confer resistance to sulfadoxine are not yet fixed in Africa4. This resistance is mediated by the stepwise accumulation of single nucleotide polymorphisms (SNPs) in the dihyropteroate synthase (dhps) gene which encodes the drug’s target5; the most common SNPs encode single amino acid changes at codons 436, 437, 540, and 581 of dhps. The ancestral wildtype haplotype is denoted SAKA to indicate wildtype amino acids at these 4 codons; the S436A mutation is not clearly associated with reduced drug susceptibility and is considered an alternate wildtype (haplotype AAKA). Prior analyses of these dhps mutations in sub-Saharan Africa have demonstrated that resistant dhps haplotypes have multiple origins and are geographically restricted to finite regions4,6. Specifically, resistant parasites in East Africa typically harbor haplotypes with the A437G and K540E mutations (SGEA), sometimes with the addition of the A581G mutation (SGEG); those in West Africa more typically display haplotypes with the single A437G mutation (SGKA) or coupled with the alternate wildtype codon 436 (AGKA)4,7,8.
Sulfadoxine, when co-formulated with pyrimethamine (SP), is no longer a recommended antimalarial therapy in sub-Saharan Africa, but it remains an important tool for malaria prevention owing to its use as intermittent preventive therapies during pregnancy (IPTp), in infancy (IPTi), and in childhood (IPTc)9,10,11. Additionally, it is employed in some countries as a partner drug for artemisinin-based therapies. Because resistance to pyrimethamine is more prevalent than against sulfadoxine, these interventions may be undermined by the ongoing accumulation of parasites bearing SNPs in dhps. These SNPs confer reduced susceptibility to sufladoxine in vitro that is broadly proportional to the number of amino acid changes in dhps12; clinical studies of therapeutic efficacy in children from across Africa suggest differential patterns of sulfadoxine susceptibility in vivo, with treatment failure closely linked to parasites bearing the A437G mutation in Congo-Brazzaville13, the K540E mutation in Burkina Faso14 and Uganda15, both the A437G and K540E mutations in the Democratic Republic of the Congo (DRC)16, and to the triple-mutant parasites bearing the A437G, K540E, and A581G mutations in Tanzania17. These clinical data underscore the importance of molecular surveillance as a surrogate tool to monitor evolving trends in clinical efficacy.
The DRC suffers one of the greatest burdens of falciparum malaria, but the epidemiology of parasite populations across this vast nation is poorly described. We recently described the epidemiology and estimated the consequences of P. falciparum for children18 and pregnant women19 in the DRC using specimens collected from a 2007 nationally-representative cross-sectional survey of DRC adults. Herein, we use a random sample of these P. falciparum parasites to describe the prevalence of dhps mutations across the DRC, explore the origins of resistance-mediating haplotypes, and investigate the geographic clustering of dhps haplotypes. We hypothesized that parasites harboring resistant haplotypes would be spatially restricted, with single-mutant (SGKA, AGKA) haplotypes predominating in the West and double (SGEA) and triple-mutant (SGEG) haplotypes in the East, and that these resistant haplotypes would manifest distinct genetic lineages.
Results
Prevalence and distribution of dhps haplotypes
From 179 randomly-selected P. falciparum parasitemias, we obtained pure (i.e. non-mixed) dhps haplotypes from 151 parasites: 53 (35.1%) were SAKA, 30 (19.9%) were AAKA, 2 (1.3%) were CAKA, 41 (27.2%) were SGKA, 3 (2%) were AGKA, 17 (11.3%) were SGEA, and 5 (3.3%) were SGEG. There were no mutations at codon 613 of dhps. Of these 151 parasites, the geographic coordinates were missing for 15 (6 SAKA, 4 AAKA, 3 SGKA, 1 SGEA, and 1 SGEG), allowing us to geo-locate 136 pure dhps haplotypes across the DRC (Figure 1). Inspection of the geographic distribution of haplotypes revealed that the single-mutant SGKA and AGKA haplotypes were largely clustered in the west and the double- and triple-mutant haplotypes SGEA and SGEG were more prevalent in the east. This pattern is consistent with prior analyses in regions bordering the east and west of the DRC4.
Figure 1. Distributions of wildtype (A), single-mutant (B), and double- and triple-mutant (C) dhps haplotypes across the DRC.
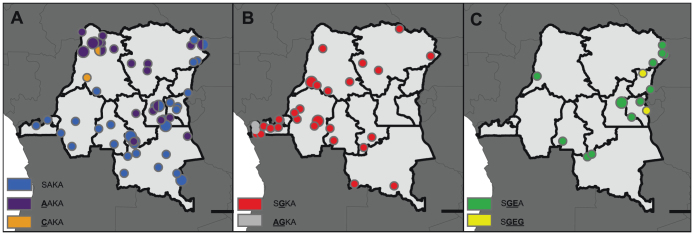
Size of circles is proportional to the number of samples in that location. Haplotypes are indicated by the amino acids at codons 436, 437, 540, and 581 of DHPS; mutant amino acids are underlined and bolded.
Geographic clustering of parasite populations
We investigated this visually-apparent geographic clustering of parasites using an ecological clustering algorithm that measures phylogenetic diversity between microbial communities and assigns confidence to the resolved clusters using jackknifing20. This algorithm clustered the overall parasite population into two communities with >99.9% confidence: an eastern (comprised of North and South Kivu, Orientale, and Maniema provinces) and a western subpopulation (comprised of Bandundu, Equateur, Katanga, Bas-Congo, Kinshasa, Kasai-Occidental, and Kasai-Oriental provinces) (Figure 2). Predicted splits between provinces within these eastern and western subpopulations were not significant. We repeated the clustering algorithm using only microsatellite profiles harbored by parasites with wildtype dhps haplotypes. In this repeat analysis only Maniema was partitioned from the other provinces with >99.9% confidence. This finding may reflect a reduction in statistical power owing to the presence of fewer wildtype dhps parasites in North and South Kivu and Orientale, or that geographic clustering is largely driven by the distributions of mutant dhps haplotypes.
Figure 2. Clustering of provincial falciparum parasite populations.
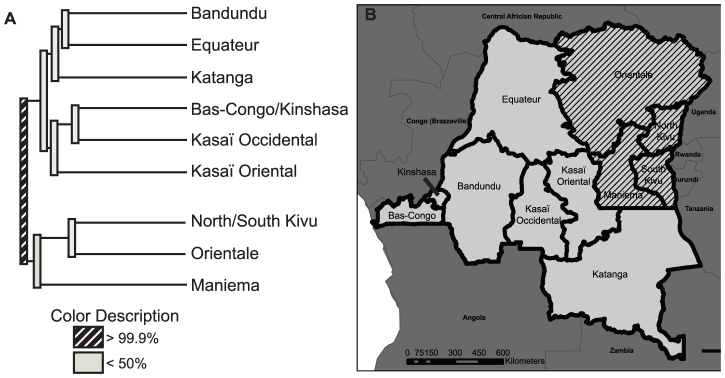
(A) Predicted clustering of microsatellite profiles between DRC provinces into two distinct parasite subpopulations from western DRC (Bandundu, Equateur, Katanga, Bas-Congo/Kinshasa, Kasaï Occidental, Kasaï Oriental) and eastern DRC (North/South Kivu, Orientale, Maniema). Computed by UniFrac with jackknifing and 1000 replicates20. (B) Predicted provincial subpopulations in the DRC. The parasites in the hatched provinces were partitioned by the analysis in (A) to be distinct from those of the un-hatched provinces.
This east-west division of parasites was supported by calculation of nearest-neighbor statistics (Snn), which quantify the likelihood that a closely-related parasite microsatellite profile will be found within the same DRC province21; Snn values close to 1 indicate substantial genetic differentiation between populations while values near 0.5 indicate that the two populations likely belong to a single, panmictic population. Between provinces that were grouped by the Unifrac analysis above into eastern and western subpopulations, several pairwise comparisons returned Snn values close to 1 that were significant despite correction for multiple comparisons by the Bonferroni method (Maniema vs. Bas-Congo/Kinshasa, N/S Kivu vs. Bas-Congo-Kinshasa, Maniema vs. Katanga, and Orientale vs. Katanga) (Table 1). This is consistent with segmentation into different parasite populations. Other pairwise comparisons were non-significant to this stringent level, but mean Snn values were significantly different for pairwise comparisons between eastern and western provinces (0.728) than for those between eastern provinces (0.696) or those between western provinces (0.577; p-value < 0.001 for comparisons of means by ANOVA).
Table 1. Pairwise Hudson’s nearest neighbor (Snn) analyses of parasite populations by DRC province.
| Bas-Congo/Kinshasa | Bandundu | Equateur | Kasai-Occidental | Katanga | Kasai-Oriental | Orientale | Maniema | N./S. Kivu | |
|---|---|---|---|---|---|---|---|---|---|
| Bas-Congo/Kinshasa | 0.59417 | 0.59201 | 0.61019 | 0.63696 | 0.69097 | 0.61067 | 0.9315 | 0.87987 | |
| Bandundu | 0.1800 | 0.46556 | 0.30556 | 0.5098 | 0.56739 | 0.69767 | 0.63167 | 0.65521 | |
| Equateur | 0.1260 | 0.9180 | 0.59928 | 0.61228 | 0.59928 | 0.56332 | 0.70914 | 0.68651 | |
| Kasai-Occidental | 0.3820 | 0.8350 | 0.7460 | 0.47778 | 0.67908 | 0.77366 | 0.76597 | 0.64428 | |
| Katanga | 0.0710 | 0.4110 | 0.1680 | 0.6490 | 0.70833 | 0.78204 | 0.87154 | 0.71491 | |
| Kasai-Oriental | 0.0070 | 0.4360 | 0.1140 | 0.2180 | 0.0250 | 0.7022 | 0.76597 | 0.64428 | |
| Orientale | 0.1460 | 0.1890 | 0.1910 | 0.1360 | 0.0010 | 0.0040 | 0.65208 | 0.67641 | |
| Maniema | <0.0001 | 0.1150 | 0.0090 | 0.0240 | 0.0010 | 0.0030 | 0.0580 | 0.73507 | |
| N./S. Kivu | <0.0001 | 0.0640 | 0.0460 | 0.1080 | 0.0230 | 0.0890 | 0.1210 | 0.0090 |
Global Snn 0.233046 (p = < 0.0001).
Snn values are above the diagonal, and p-values are below. Snn values that are significant at a level of p < 0.0014 (adjusted for multiple comparisons by the Bonferroni method) are bold and italicized. Genetic distances were calculated by the Cavalli-Sforza method, and Snn calculated with 1000 permutations21.
Divergence of dhps haplotypes
Consistent with previous studies in African P. falciparum22, we observed high degrees of expected heterozygosity (He) at microsatellite loci: the overall mean He was 0.564 (standard error [SE] 0.06), and individual loci ranged from 0.404 (for locus −0.13; SE 0.135) to 0.723 (for locus 0.03; SE 0.132) when calculated for the major allele at each locus. These He estimates were not substantially different from those calculated by inputting all alleles that were obtained at mixed loci (data not shown). After partitioning into the 7 unique dhps haplotypes described above, there was evidence of mild selective pressure upon mutant haplotypes: mean He (SE) for SAKA was 0.801 (0.062), for AAKA was 0.765 (0.071), for SGKA was 0.480 (0.081), for AGKA was 0 (0), SGEA was 0.508 (0.110), and for SGEG was 0.393 (0.167) (Figure 3).
Figure 3. Reductions in mean heterozygosity by dhps haplotype.
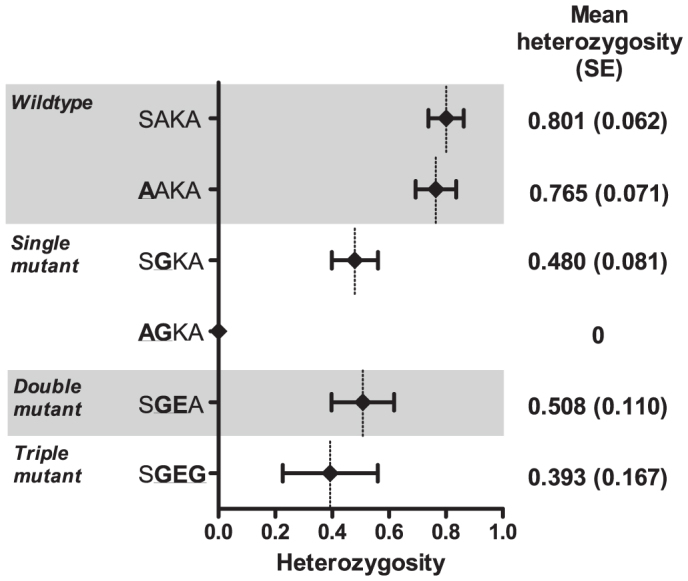
Haplotypes are indicated by the amino acids at codons 436, 437, 540, and 581 of DHPS; mutant amino acids are underlined and bolded. Diamonds are point estimates of the mean, and bars represent standard error. Calculated with GenAlEx v6.5 39.
We first investigated the divergence of mutant and wildtype haplotypes by comparing genetic and geographic distances of the 136 parasites for which we had both dhps haplotypes and geographic coordinates. Overall, there was minimal correlation between geographic and genetic distance by Mantel testing (Rxy = 0.098, p = 0.010) (Figure 4A). However, this relationship was modified by dhps haplotypes: after stratification into wildtype (SAKA, AAKA, and CAKA) and mutant (SGKA, AGKA, SGEA, SGEG) parasites, there was no significant correlation between wildtype parasites (Rxy = 0.046, p = 0.213) but significant correlation for the mutant parasites (Rxy = 0.277, p < 0.001). These results suggest that while wildtype dhps haplotypes are genetically and geographically intermixed, mutant dhps haplotypes are isolated by distance across the DRC.
Figure 4. Correlations of genetic and geographic distance for all parasites (A), wildtype parasites (SAKA, AAKA, CAKA) only (B), and mutant parasites (SGKA, AGKA, SGEA, SGEG) only (C) in the DRC.
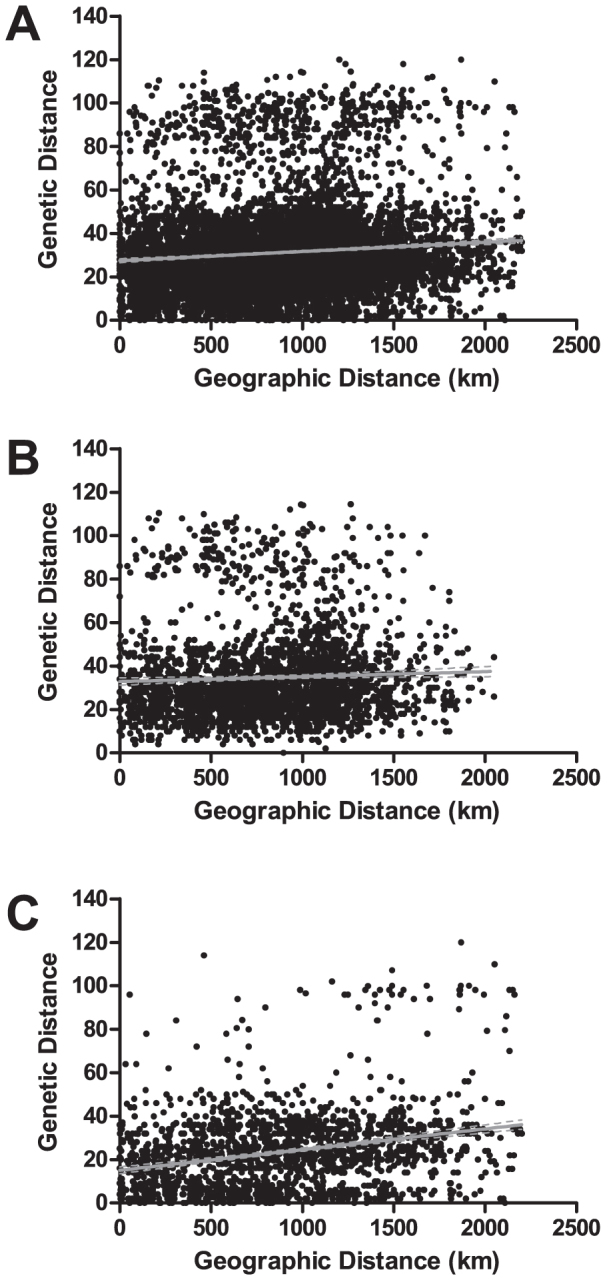
Black dots represent pairwise comparisons, gray line is the best-fit regression line, and dotted gray line is the 95% confidence interval of best-fit line. Mantel values were 0.098 (p = 0.010) for all parasites, 0.046 (p = 0.213) for wildtype parasites only, and 0.277 (p < 0.001) for mutant parasites only. Pearson product-moment correlation coefficient (PMCC) values were 0.09828 (p < 0.0001) for all parasites, 0.04580 (p = 0.0158) for wildtype parasites only, and 0.2771 (p < 0.0001) for mutant parasites only. Genetic and geographic distances and Mantel tests computed using 999 permutations with GenAlex v6.5, and PMCC computed with GraphPad Prizm.
We further explored pairwise divergence between dhps haplotypes using both Nei’s unbiased distance and ΦPT, the latter estimated using AMOVA (Table 2). Nei’s distance measures divergence between populations accounting for both mutation and drift23, while AMOVA estimates the degree of differentiation into subpopulations and is an analog of Wright’s F-statistics24. As expected, the ancestral wild-type SAKA and alternate wild-type AAKA were closely related by both measures, with distance of 0.174 and ΦPT of 0.032 (p = 0.29). Additionally, the double- and triple-mutants SGEA and SGEG were closely related to each other with distance of 0.081 and ΦPT of 0 (p = 0.310). Surprisingly, the single-mutant SGKA (prevalent in Western DRC) was divergent from all other major haplotypes, with distance values over 0.540 and pairwise ΦPT values of 0.133 (p = 0.010) with SAKA, 0.362 (p = 0.010) with SGEA, and 0.380 (p = 0.020) with SGEG (prevalent in Eastern DRC). These data suggested that the SGKA mutant did not arise from the sampled SAKA wildtype parasites and, furthermore, did not give rise to the more resistant SGEA and SGEG haplotypes that we sampled in the DRC.
Table 2. Pairwise Nei’s genetic distance (above diagonal) and ΦPT values (below diagonal) between the major dhps haplotypes in the DRC*.
| SAKA | SGKA | SGEA | SGEG | AAKA | AGKA | |
|---|---|---|---|---|---|---|
| SAKA | 0.540 | 0.765 | 0.735 | 0.174 | 0.879 | |
| SGKA | 0.133 | 0.746 | 0.978 | 0.371 | 0.133 | |
| SGEA | 0.037 | 0.362 | 0.081 | 0.351 | 1.249 | |
| SGEG | 0 | 0.380 | 0 | 0.509 | 1.220 | |
| AAKA | 0 | 0.124 | 0.009 | 0 | 0.666 | |
| AGKA | 0.032 | 0 | 0.217 | 0.588 | 0.008 |
Calculated using GenAlEx v6.5 using 149 specimens with pure (i.e. non-mixed) dhps haplotypes, using 999 permutations over full population and 99 permutations for pairwise comparisons39.
*ΦPT values with a p < 0.05 are indicated in italics and bold. Haplotypes are indicated by the amino acids at codons 436, 437, 540, and 581 of DHPS; mutant amino acids are underlined and bolded.
Origins of dhps haplotypes
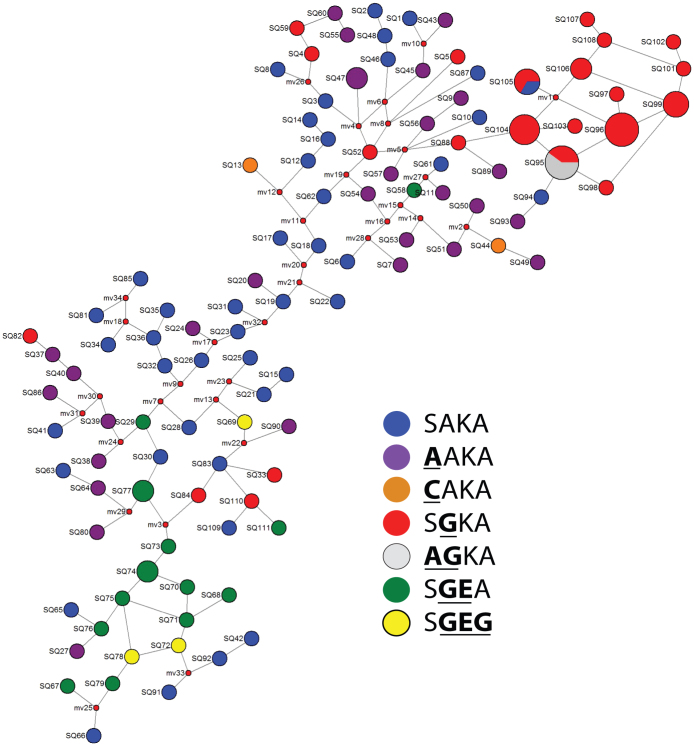
In order to further investigate the origins of the dhps haplotypes in the DRC, we partitioned the 129 specimens with full microsatellite profiles into 111 unique haplotypes. Inspection of a median-joining network built from these 111 haplotypes demonstrated clear segmentation of the resistance lineages in the DRC (Figure 5). Specifically, on a background of unclustered SAKA and AAKA haplotypes (blue and purple circles in Figure 5, respectively), we note clustering of the majority of SGKA and AGKA haplotypes (red and grey circles in Figure 5, respectively) clearly distinct from the major cluster of SGEA and SGEG haplotypes (green and yellow circles in Figure 5, respectively). This segmentation of the single-mutants (SGKA and AGKA) from the double- and triple-mutants (SGEA and SGEG) haplotypes suggested that the dhps haplotypes arose on different genetic backgrounds.
Figure 5. Median-joining network of dhps haplotypes in the DRC.
“SQ” numbers designate microsatellite profile, circles are proportional to the number of parasites bearing that microsatellite profile, and dhps haplotypes are indicated by the color key and are indicated by the amino acids at codons 437, 540, and 581 of DHPS; mutant amino acids are underlined and bolded. Small red nodes are hypothetical median vectors created by the program to connect sampled haplotypes into a parsimonious network. Distances between nodes are arbitrary. Constructed in NETWORK v4.6.10 40,41.
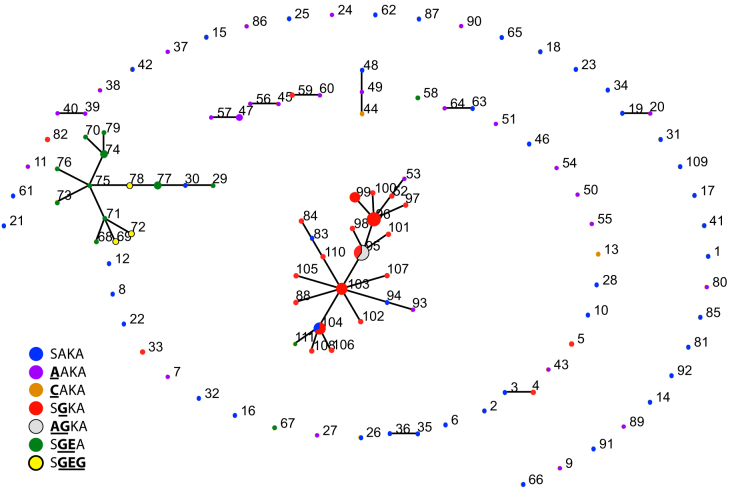
We analyzed this observation further with eBURST, which utilizes multi-locus sequence tags to predict founding genotypes and clusters of descendants25. This algorithm, which is agnostic to dhps haplotype, identified two major clonal clusters against a background of unrelated haplotypes: one cluster contained 15 of 18 SGEA and SGEG haplotypes, and the second cluster contained 33 of 38 of the SGKA and AGKA haplotypes (Figure 6). Thus the genetic origins of the single-mutants SGKA and AGKA were distinct from those of the double- and triple-mutants SGEA and SGEG. Taken together, our spatial and genetic analyses indicated that, among the parasites sampled in our study, haplotypes associated with sulfadoxine resistance are genetically and geographically clustered across the DRC.
Figure 6. Hypothesized lineages of dhps haplotypes in the DRC.
Numbers designate microsatellite profile, nodes are proportional to the number of parasites bearing that profile, and dhps haplotypes are indicated by the color key and are indicated by the amino acids at codons 437, 540, and 581 of DHPS. Nodes with more than one color indicate that the microsatellite profile was borne by more than one type of dhps haplotype. Connected haplotypes are clonal complexes that are hypothesized by the algorithm to have descended from the same founder haplotype. Haplotypes that are connected vary in their microsatellite profile at one locus; unconnected haplotypes are those that do not share 4 loci with any other haplotype. Calculated with eBURST v3 25.
Discussion
In this cross-sectional study of P. falciparum haplotypes in the DRC, parasite populations were geographically clustered into two subpopulations along provincial boundaries running from north-central to southeast DRC. Furthermore, these subpopulations manifested dhps mutant haplotypes with independent lineages, with single-mutant SGKA and AGKA haplotypes in the west largely unrelated to the double- and triple-mutant SGEA and SGEG haplotypes in the east. These twin findings suggest a significant barrier to parasite population flow between eastern and western DRC. Because this corridor across central Africa has been critical to past spread of drug-resistant haplotypes, the identification of these barriers to gene flow may assist in future efforts to control and contain parasite drug resistance.
Central Africa is a known watershed for sulfadoxine-resistant falciparum parasites: in a prior analysis of the origins of dhps haplotypes in sub-Saharan Africa, mutant haplotype prevalences were sharply divided between eastern and western Africa, but few parasites from the DRC were included4. We report a relatively high prevalence of wildtype haplotypes, with 55% (83/151) of parasitemias harboring either the SAKA or AAKA haplotype. These data indicate that resistance to sulfadoxine is still evolving in the DRC and there is minimal selective pressure applied by the relatively infrequent use of sulfadoxine-pyrimethamine for childhood fever18 or IPTp26. Nevertheless, as expected based upon prior studies, there were marked differences in the distributions of mutant dhps haplotypes: single-mutant SGKA and AGKA predominated in the west while double- and triple-mutant SGEA and SGEG haplotypes clustered in the east. These distributions mirror the prevailing mutant haplotypes in neighboring countries: Congo-Brazzaville13 and the Central African Republic27 in the west and Uganda28 and Rwanda29 in the east. We do note, however, a cluster of double-mutant SGEA haplotypes near Mbuji-Mayi in south-central DRC, which may represent the incursion of more highly-resistant parasites into western DRC. The region around Mbuji-Mayi was a major destination for refugees from eastern DRC during the protracted civil war in the DRC30, possibly providing a corridor for parasite transit across geographic and political boundaries.
Microsatellite genotyping of our parasites revealed a high degree of heterozygosity both within and between parasitemias. These findings are consistent with prior studies of African P. falciparum22. Reductions in mean heterozygosities across microsatellites linked to dhps were consistent with selective pressure on haplotypes bearing the A437G, K540E, and A581G mutations, and greatest upon those with all three mutations. These haplotype He values are similar to those from other African studies4,31,32, though our study allows a fuller characterization owing to the DRC’s greater scope of haplotypes and our number of microsatellites analyzed. Interestingly, the degrees of selection upon the mutant SGKA (mean He 0.480), SGEA (mean He 0.508), and SGEG (mean He 0.393) haplotypes were similar to those reported from other regions in which these mutant haplotypes are nearly fixed in the parasite populations6,32,33, despite the DRC continuing to harbor a relatively high prevalence of wildtype haplotypes (55%).
Based upon genetic distance (Figure 4, Table 2), network (Figure 5), and clonal lineage analyses (Figure 6), the lineages of these sulfadoxine-resistant haplotypes appear to be distinct: These analyses largely grouped the SGKA haplotype microsatellite profiles into a cluster distinct from those of the SGEA and SGEG haplotypes. This is supported by prior studies4, though we employed alternate analytic tools to characterize lineages of fine-scale microsatellite profiles in order to more specifically describe these relationships. Because the mutations in dhps are believed to accumulate in a stepwise manner5, we would expect single-, double-, and triple-mutant dhps haplotypes to cluster together in these analyses, but we sampled few SGEA and SGEG haplotypes that were related to the single-mutant SGKA parasites, and few single-mutant SGKA parasites that were closely related to the SGEA and SGEG haplotypes. In western DRC where the SGKA mutant haplotypes predominate, SP use may not be frequent enough to produce sufficient selective pressure to favor the emergence of a related double-mutant: In Congo-Brazzaville to the West, the dhps SGKA haplotype, when combined with the triple-mutant dhfr which is common in the DRC34,35, confers significant clinical resistance to SP as evidenced by clinical treatment failure13. More puzzling is the absence of an abundant ancestral single-mutant SGKA related to the double- and triple-mutant SGEA and SGEG haplotypes. Though several phenomena may account for this, one explanation is that these parasites were imported from neighboring East African countries such as Uganda and Rwanda, where these haplotypes are prevalent28,29. This hypothesis is supported by their increased preponderance in eastern DRC; though one might expect that we would have also detected imported parasites bearing an ancestral SGKA haplotype, the K540E mutation was near fixation in these neighboring countries by the early 2000s28, thus eliminating this ancestral haplotype from their parasite populations. We note the presence of the double-mutant SGEA haplotype in parasitemias from Kasaï-Occidental, Kasaï-Oriental, and Equateur provinces that are far from the eastern borders of the DRC; their clustering with eastern mutant haplotypes indicates that this lineage has migrated across the DRC. Identifying the route and mechanisms of this migration will be critical to controlling and containing drug-resistant parasite populations.
Similar to this genetic divergence of drug-resistant haplotypes, the parasite populations overall in the DRC were geographically divided into one cluster in the West and one in the Northeast (Figure 2B), and pairwise comparisons of parasite genotypes between provinces supported this split (Table 1). These findings suggest that parasite populations are circumscribed within the DRC, despite the absence of obvious barriers to gene flow owing to geography or ecology. Differentiation into divergent populations is dependent upon complex relationships between malaria endemicity, human population density and migration, vector prevalence and efficiency, and malaria treatment and control practices. Future studies to better characterize the mediators of this phenomenon will be critical to understanding the impact of parasite subpopulations on malaria control and elimination.
Our study is subject to several limitations. Multiplicities of infection in our asymptomatic adults were high as evidenced by the return of multiple alleles for many microsatellite loci, from which we selected the dominant allele. Though we may have underestimated minor variants in these mixed parasitemias, this approach has been applied in other studies22 and our analyses were limited to those parasitemias which were pure dhps haplotypes. Additionally, we had no clinical data on the survey recipients who harbored the parasitemias we studied, and thus we cannot gauge the potential impact of personal or aggregate antimalarial use upon the distribution of dhps haplotypes. Nevertheless, this may not have a major impact given the uniformly low use of SP across the DRC: the proportion of febrile children under 5 who received SP ranged from 1.1% (in Kinshasa) to 8.2% (in Bandundu). Finally, we did not have contemporary parasites from neighboring countries with which to compare our dhps lineages. Therefore, future studies will be needed to assess the relationships between our DRC haplotypes and those of East and West Africa.
In our sampling of falciparum parasites across the DRC, we report a full range of dhps haplotypes as well as geographically- and genetically-distinct sulfadoxine-resistant parasites. Our data suggest that more resistant parasite lineages were imported into the DRC from east Africa, and that, despite the overall divergence of parasites into eastern and western subpopulations, these parasites are spreading into western DRC. Given 1) the importance of sulfadoxine to preventive measures in children and pregnant women, 2) the appearance of more highly-resistant dhps haplotypes in East Africa, and 3) the importance of the DRC as a corridor for parasite flow across sub-Saharan Africa, continued molecular surveillance in central Africa is necessary both to characterize the evolving pattern of sulfadoxine-resistant haplotypes and to identify factors governing the spread of drug-resistant parasites.
Methods
Ethics statement
All survey respondents provided verbal informed consent for the collection of blood spots in one of the five main languages spoken in the DRC. Consent was verbally acquired owing to the need for immediate de-identification of all data. Consent procedures, survey administration, and blood sample collection were approved by the Ethics Committees of Macro International and the School of Public Health of the University of Kinshasa, and testing of malaria parasites was approved by the Institutional Review Board of the University of North Carolina.
Specimen collection
The methods of the 2007 Demographic and Health Survey (DHS) in the DRC have been described elsewhere18. Briefly, adults aged over 15 years were selected from 9000 randomly selected households within 300 randomly-selected clusters identified across the DRC based upon the 2006 DRC national census. Survey respondents contributed a finger-prick blood sample which was stored on filter paper as a dried blood spot (DBS), from which three 0.3-inch discs were punched into plastic 96-well plates, with genomic DNA (gDNA) extracted using the invitrogen PureLink 96 kit (invitrogen, Carlsbad, CA, USA). Real-time PCR detection of P. falciparum was performed as previously described18. The DHS was conducted in February and March for clusters in and near Kinshasa, and between May and August for the balance of the DRC.
The original molecular survey of malaria in the DRC identified 2,435 P. falciparum mono-parasitemias in 8,838 adult respondents18. A random subset of 867 specimens that were chosen irrespective of demographic or geographic characteristics yielded 229 P. falciparum parasitemias, of which 50 failed to return dhps genotype data. The resulting 179 parasitemias constitute the unit of analysis for this study. These patients harboring parasitemias did not differ significantly from non-selected parasitemic patients with regard to province or demographics (data not shown).
Genotyping procedures
Specimens were genotyped for SNPs at codons 436, 437, 540, 581, and 613 of dhps by amplification and sequencing as previously described6. Parasites with multiple peaks at any locus in the manually-inspected eletropherogram were scored as mixed at that locus. All parasites were assayed for allele length at five microsatellite loci flanking the dhps gene: −2.9 and −0.13 kB upstream, and 0.03, 0.5, and 9 kB downstream of dhps. Microsatellite loci were amplified as previously described6, PCR products were sized on a 310 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA), and allele lengths were scored using GeneMapper (v4.1, Applied Biosystems). In specimens with multiple peaks, additional allele lengths were recorded if peak heights were > 20% of the major peak. All specimens were amplified and sized in parallel with gDNA from P. falciparum isolate 3d7; the known length of the 5 microsatellites in the 3d7 genome was used to correct allele lengths in the experimental specimens in order to account for inter-run variability in fragment sizing.
Data analyses
For specimens for which geographical coordinates were available, we first mapped dhps haplotypes across the DRC using ArcGIS (v10, ESRI, Redlands, CA, USA). We classified SAKA, AAKA, CAKA haplotypes as wildtype and other haplotypes as mutant consistent with prior studies4, because of the unclear association of mutations at codon 436 with sulfadoxine susceptibility. Prior to mapping, all DHS cluster coordinates are randomly offset by 0 – 2 km (in urban areas) or 0 – 5 km (in rural areas) in order to maintain privacy.
Based upon the microsatellite profiles, we investigated the geographic structure of haplotypes by DRC political province. Pairwise genetic distances between parasites were calculated using the Cavalli-Sforza and Edwards model as implemented in Populations v1.2.2336. The genetic distance matrix created in Populations was used as input for FastME to construct neighbor-joining trees with balanced branch-length estimation37,38. The FastME output in Newick format was used in Fast UniFrac to assess for geographic clustering. Fast UniFrac compares communities of microbes and clusters them with confidence estimates derived from permutation20. 1000 permutations were used to determine significance. For these analyses, we first input microsatellite profiles of all dhps haplotypes, and then repeated them using only parasites bearing wildtype haplotypes as defined above.
To corroborate the conclusions of these geographic phylogenies, we also tested the overall parasite populations for genetic differentiation into subpopulations by calculating pairwise Hudson’s nearest-neighbor statistics (Snn) between DRC provinces21. Snn measures how frequently the nearest neighbors of closely linked genetic marker data are located within the same geographic locality and employs permutation in order to estimate p-values of these measures. Typically, genetically distant populations have values near 1; panmictic populations have values close to 0.5. Populations were defined by DRC province, though we combined Bas-Congo with Kinshasa and North with South Kivu; the resulting 9 geographic categories produced 36 pairwise comparisons, and for this analysis a p-value < 0.0014 (0.05 / 36) was considered significant after correction using the Bonferroni method for multiple comparisons. Means of pairwise provincial Snn values between subpopulations that were identified by UniFrac were compared by analysis of variance (ANOVA) using Stata/IC v10 (Stata Corp, College Station, TX, USA).
We calculated the number of alleles and expected heterozygosity (He) per microsatellite locus in the overall dataset using GenAlEx 6.539. In order to assess the degree of selection on different dhps haplotypes, we calculated He across all five loci for each dhps haplotype. Based upon the microsatellite fragment lengths, we calculated pairwise Nei’s genetic distances between dhps haplotypes in order to estimate genetic relatedness23. We assessed isolation-by-distance using pairwise comparisons of genetic and geographic distance and computing Mantel tests using 999 permutations in GenAlex. We first included all parasites for which dhps haplotype and geographic coordinates were available, then repeated testing among only wildtype parasites and among only mutant parasites. In order to further assess the divergence of wildtype and mutant dhps haplotypes, we calculated ΦPT via an Analysis of Molecular Variance (AMOVA) in GenAlEx using 999 permutations over the full population and 99 pairwise permutations24,39.
In order to further explore the origins of the prevailing dhps haplotypes in the DRC, we first partitioned into unique haplotypes the microsatellite profiles from each specimen that was successfully genotyped at all five loci. Next, we calculated a median-joining network of these unique microsatellite haplotypes using NETWORK v4.6.1040,41; for network construction, we assigned weights to each locus in inverse proportion to the locus’s He, as calculated above. The unique microsatellite haplotypes were then analyzed using eBURST v3, which groups multi-locus sequence types into clonal clusters that are hypothesized to have derived from a common ancestor25. Analyses with NETWORK and eBURST were performed in an unsupervised fashion with regard to dhps haplotype; these haplotypes were added post-hoc to the median-joining network and hypothesized clonal clusters.
Author Contributions
Conceived and designed experiments: SMT, ALA, ME, MTA, VU, AKT, SRM. Performed the experiments: SMT, ALA, MJ, MTA. Analyzed the data: SMT, ALA, CMP, MTA, JJJ, SRM. Contributed reagents/materials/analysis tools: KM, CMP, JJJ, MJ, ME, MTA, VU, AKT. Wrote the paper: SMT, ALA, JJJ, SRM.
Acknowledgments
We thank Dr Augustin Okenge (Programme National de Lutte Contre le SIDA, Kinshasa, DRC), Dr Jeremie Mwonga (Laboratoire National de Reference SIDA et IST, Kinshasa, DRC), and Ann Way, Mohamed Ayad, and Martin Vaessen (all of MeasureDHS, Calverton, MD) for help in obtaining access to the dried blood spots. Stephanie Carrier, Hannah Hoffman, and Jaymin Patel (all of the University of North Carolina, Chapel Hill) assisted in the laboratory. This work would not have been possible without the work of scores of survey administrators. Ultimately, we are indebted to the DHS respondents for their participation. This work was supported by a Gillings Innovation Laboratory award from the UNC Gillings School of Global Public Health (to SRM), a Summer Undergraduate Research Fellowship from the Office for Undergraduate Research at the University of North Carolina at Chapel Hill (to ALA), and the CDC Antimicrobial Resistance Network for the laboratory work performed at CDC (to VU). Dr Taylor is supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number K08AI100924, and Mr Parobek was supported in part by NIH training grant T32GM0088719. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Wootton J. C. et al. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature 418, 320–323 (2002). [DOI] [PubMed] [Google Scholar]
- Roper C. et al. Intercontinental spread of pyrimethamine-resistant malaria. Science 305, 1124 (2004). [DOI] [PubMed] [Google Scholar]
- WHO. World Malaria Report 2011 (World Health Organization, 2012). [Google Scholar]
- Pearce R. J. et al. Multiple origins and regional dispersal of resistant dhps in African Plasmodium falciparum malaria. PLoS Med 6, e1000055 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley C. H. et al. Pyrimethamine-sulfadoxine resistance in Plasmodium falciparum: what next? Trends Parasitol 17, 582–588 (2001). [DOI] [PubMed] [Google Scholar]
- Vinayak S. et al. Origin and evolution of sulfadoxine resistant Plasmodium falciparum. PLoS Pathog 6, e1000830 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita T. et al. Limited geographical origin and global spread of sulfadoxine-resistant dhps alleles in Plasmodium falciparum populations. J Infect Dis 204, 1980–1988 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridaran S. et al. Anti-folate drug resistance in Africa: meta-analysis of reported dihydrofolate reductase (dhfr) and dihydropteroate synthase (dhps) mutant genotype frequencies in African Plasmodium falciparum parasite populations. Malaria J 9, 247 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Kuile F. O., van Eijk A. M. & Filler S. J. Effect of sulfadoxine-pyrimethamine resistance on the efficacy of intermittent preventive therapy for malaria control during pregnancy: a systematic review. JAMA 297, 2603–2616 (2007). [DOI] [PubMed] [Google Scholar]
- Aponte J. J. et al. Efficacy and safety of intermittent preventive treatment with sulfadoxine-pyrimethamine for malaria in African infants: a pooled analysis of six randomised, placebo-controlled trials. Lancet 374, 1533–1542 (2009). [DOI] [PubMed] [Google Scholar]
- Dicko A. et al. Intermittent preventive treatment of malaria provides substantial protection against malaria in children already protected by an insecticide-treated bednet in Mali: a randomised, double-blind, placebo-controlled trial. PLoS Med 8, e1000407 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triglia T., Wang P., Sims P. F., Hyde J. E. & Cowman A. F. Allelic exchange at the endogenous genomic locus in Plasmodium falciparum proves the role of dihydropteroate synthase in sulfadoxine-resistant malaria. EMBO J 17, 3807–3815 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndounga M. et al. Therapeutic efficacy of sulfadoxine-pyrimethamine and the prevalence of molecular markers of resistance in under 5-year olds in Brazzaville, Congo. Trop Med Internat Health 12, 1164–1171 (2007). [DOI] [PubMed] [Google Scholar]
- Dokomajilar C. et al. Roles of specific Plasmodium falciparum mutations in resistance to amodiaquine and sulfadoxine-pyrimethamine in Burkina Faso. Amer J Trop Med Hyg 75, 162–165 (2006). [PubMed] [Google Scholar]
- Dorsey G. et al. Principal role of dihydropteroate synthase mutations in mediating resistance to sulfadoxine-pyrimethamine in single-drug and combination therapy of uncomplicated malaria in Uganda. Amer J Trop Med Hyg 71, 758–763 (2004). [PubMed] [Google Scholar]
- Alker A. P. et al. dhfr and dhps genotype and sulfadoxine-pyrimethamine treatment failure in children with falciparum malaria in the Democratic Republic of Congo. Trop Med Internat Health 13, 1384–1391 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesase S. et al. High resistance of Plasmodium falciparum to sulphadoxine/pyrimethamine in northern Tanzania and the emergence of dhps resistance mutation at codon 581. PloS One 4, e4569 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S. M. et al. Molecular malaria epidemiology: mapping and burden estimates for the Democratic Republic of the Congo, 2007. PloS One 6, e16420 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S. M. et al. Quantification of the burden and consequences of pregnancy-associated malaria in the Democratic Republic of the Congo. J Infect Dis 204, 1762–1771 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M., Lozupone C. & Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 4, 17–27, 7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R. A new statistic for detecting genetic differentiation. Genetics 155, 2011–2014 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson T. J. et al. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol 17, 1467–1482 (2000). [DOI] [PubMed] [Google Scholar]
- Nei M. & Roychoudhury A. K. Sampling variances of heterozygosity and genetic distance. Genetics 76, 379–390 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L., Smouse P. E. & Quattro J. M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479–491 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil E. J., Li B. C., Aanensen D. M., Hanage W. P. & Spratt B. G. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol 186, 1518–1530 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eijk A. M. et al. Coverage of malaria protection in pregnant women in sub-Saharan Africa: a synthesis and analysis of national survey data. Lancet Infect Dis 11, 190–207 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard D., Djalle D., Yapou F., Manirakiza A. & Talarmin A. Frequency distribution of antimalarial drug-resistant alleles among isolates of Plasmodium falciparum in Bangui, Central African Republic. Amer J Trop Med Hyg 74, 205–210 (2006). [PubMed] [Google Scholar]
- Francis D. et al. Geographic differences in antimalarial drug efficacy in Uganda are explained by differences in endemicity and not by known molecular markers of drug resistance. J Infect Dis 193, 978–986 (2006). [DOI] [PubMed] [Google Scholar]
- Karema C. et al. Molecular correlates of high-level antifolate resistance in Rwandan children with Plasmodium falciparum malaria. Antimicrob Agent Chemother 54, 477–483 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns J. Dancing in the Glory of Monsters: The Collapse of the Congo and the Great War of Africa. (PublicAffairs, 2011). [Google Scholar]
- McCollum A. M., Basco L. K., Tahar R., Udhayakumar V. & Escalante A. A. Hitchhiking and selective sweeps of Plasmodium falciparum sulfadoxine and pyrimethamine resistance alleles in a population from central Africa. Antimicrob Agent Chemother 52, 4089–4097 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M. T. et al. Selective sweeps and genetic lineages of Plasmodium falciparum drug -resistant alleles in Ghana. J Infect Dis 203, 220–227 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M. T. et al. Tracking origins and spread of sulfadoxine-resistant Plasmodium falciparum dhps alleles in Thailand. Antimicrob Agent Chemother 55, 155–164 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohuet S. et al. Short report: molecular markers associated with Plasmodium falciparum resistance to sulfadoxine-pyrimethamine in the Democratic Republic of Congo. Amer J Trop Med Hyg 75, 152–154 (2006). [PubMed] [Google Scholar]
- Swarthout T. D. et al. Artesunate + amodiaquine and artesunate + sulphadoxine-pyrimethamine for treatment of uncomplicated malaria in Democratic Republic of Congo: a clinical trial with determination of sulphadoxine and pyrimethamine-resistant haplotypes. Trop Med Internat Health 11, 1503–1511 (2006). [DOI] [PubMed] [Google Scholar]
- Populations v. 1.2.30. http://bioinformatics.org/~tryphon/populations/ (Accessed November 6, 2012) (1999).
- Desper R. & Gascuel O. Fast and accurate phylogeny reconstruction algorithms based on the minimum-evolution principle. J Comput Biol 9, 687–705 (2002). [DOI] [PubMed] [Google Scholar]
- Saitou N. & Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4, 406–425 (1987). [DOI] [PubMed] [Google Scholar]
- Peakall R. & Smouse P. E. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research--an update. Bioinformatics 28, 2537–2539 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluxus Engineering, <http://www.fluxus-engineering.com/sharenet.htm> (Accessed November 6, 2012)
- Bandelt H. J., Forster P. & Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16, 37–48 (1999). [DOI] [PubMed] [Google Scholar]