Abstract
Xeroderma pigmentosum (XP) is a human disorder which is characterized by hypersensitivity to sunlight and elevated incidence of skin cancer. The disease is caused by mutations in genes that encode components of the nucleotide excision repair pathway. The gene product of XP complementation group G (XPG) is a structure-specific endonuclease which makes an incision 3′ to DNA photoproducts and other helix-distorting DNA adducts. In addition, the XPG protein has been implicated in transcription and repair of oxidative DNA damage. Moreover, XPG is capable of cleaving R loops in vitro, a potential intermediate during immunoglobulin heavy-chain class switch recombination. Due to its multiple functions, complete elimination of XPG in mice results in severe postnatal growth defects and premature death. To understand the contribution of the XPG nuclease activity to its function in vivo, we introduced a point mutation into the mouse XPG gene which inactivates the nuclease catalytic site but leaves the remainder of the protein intact. The XPG nuclease-deficient animals develop normally and exhibit no obvious defect in class switch recombination. However, the mutant mice are hypersensitive to UV irradiation. This phenotype suggests that the nuclease activity of XPG is required only for nucleotide excision repair and that other regions of the protein perform independent functions.
Xeroderma pigmentosum (XP) is a human genetic disorder which is characterized by hypersensitivity to sunlight and elevated incidence of skin cancer (2, 5). The disease is caused by mutations in genes encoding components of nucleotide excision repair, which is responsible for removing UV-induced DNA damage as well as bulky base modifications by carcinogenic chemicals. The patients can be divided into seven genetic complementation groups: XPA through -G. The genes that are mutated in each complementation group have been cloned and characterized. These proteins assemble into a repair complex over the DNA lesion and catalyze the excision of the DNA adduct as a 24- to 32-base oligonucleotide (23, 32).
XPG is a structure-specific endonuclease that makes the incision 3′ to the DNA adduct during nucleotide excision repair (15, 22). Besides hypersensitivity to sunlight, patients in the XPG group frequently exhibit complex abnormalities associated with Cockayne syndrome (CS) such as neurological disorders and developmental defects (7, 12, 21). The complexity could be explained by the multiple functions of the XPG protein. Besides acting as the excision nuclease in nucleotide excision repair, XPG also stimulates base excision repair of oxidative DNA damage (3, 10). In addition, the yeast homologue of XPG, Rad2, has been shown to facilitate efficient transcription by RNA polymerase II (13). By analogy, XPG may play a similar role in mammals. Consistent with this possibility, XPG was found to copurify with TFIIH during fractionation of nuclear extracts (17), and this association was further confirmed in immunoprecipitation experiments (1, 9). Since TFIIH is a dual-function transcription/repair factor (26), its interaction with XPG could play a role in transcription as well as nucleotide excision repair.
Patients with large truncations in the XPG protein frequently have features of combined XP-CS, while missense mutations generally give rise to XP only (7, 12, 20, 21). The likely explanation is that large deletions of the XPG protein affect multiple functions while point mutation may eliminate only the nucleotide excision repair function. Similar to the complex abnormalities of XPG patients, complete inactivation of the XPG gene in mice leads to severe developmental defects (8). The mutant mice are runted and die within 3 weeks after birth. Histological examination of the mutant animals revealed abnormalities in multiple organs. By contrast, mice deficient in XPA or XPC exhibit only hypersensitivity to UV irradiation but show no developmental defects (6, 19, 24). Thus, the complex phenotype of XPG knockout mice cannot be attributed to deficiency in nucleotide excision repair. Instead, the developmental defect reflects the involvement of the XPG protein in additional housekeeping functions.
Among the functions of XPG, the best characterized is the nuclease activity. The XPG protein shows sequence homology to a family of structure-specific nucleases, which include RNase H, FEN1, Rad2, and eubacterial DNA polymerases (18). Based on the crystal structure of RNase H, the active site for hydrolysis involves several conserved acidic residues which chelate two catalytic magnesium ions (18). These acidic residues are also conserved in XPG and could potentially serve similar functions. Consistent with this prediction, mutations in these conserved acidic residues completely inactivate the nuclease activity of XPG proteins in vitro (4, 29). On the other hand, the nuclease-deficient XPG protein is capable of stimulating the base excision repair of oxidized bases in vitro (10). Moreover, nuclease-deficient Rad2, the yeast homologue of XPG, is fully competent in promoting transcription (13).
To address the role of the nuclease activity of mammalian XPG in vivo, we introduced a missense mutation, E791A, into the mouse XPG gene. This mutation completely abolishes the nuclease activity of XPG in vitro (4, 29). We found that mice homozygous for this mutation develop normally but show a spectrum of UV-induced lesions characteristic of XP patients.
MATERIALS AND METHOD
Introduction of the E791A mutation into the mouse genome
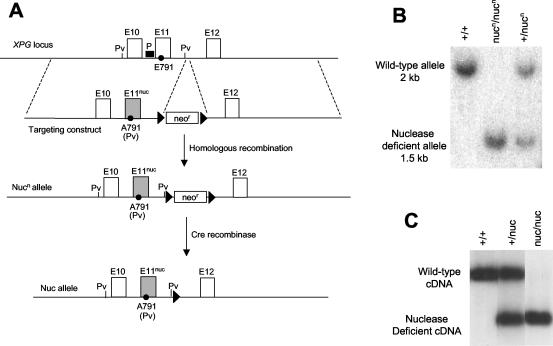
The E791A mutation was introduced into the endogenous XPG gene through gene targeting. To construct the targeting construct, a 4.5-kb SalI/XhoI fragment was used as the 5′ homology arm, while a 5-kb SalI/SpeI fragment served as the 3′ homology arm. E791 is located in exon 11 of the XPG gene (14), which is contained in the 5′ homology arm. The codon for E791 (GAG) was mutated into A791 (GCT). The mutation also creates a novel PvuII site for easy detection of the mutation. These fragments were cloned into the pLN-TK vector to generate the targeting construct. The construct was transfected into TC1 embryonic stem (ES) cells, and stable integrants were selected with G418 and ganciclovir. Correctly targeted clones were identified by diagnostic Southern analysis and were used to generate mutant mice. The mice were genotyped by both Southern analysis and PCR. For Southern analysis, the DNA was digested with PvuII. The digested DNA was electrophoresed on an agarose gel and was detected in Southern blotting with a 200-bp EcoRI/PstI fragment as the probe (Fig. 1A).
FIG. 1.
Generation of XPG nuclease-deficient mice. (A) The diagram describes the strategy to introduce the E791A mutation into the mouse XPG gene. The genomic organization around E791 (exons 10 to 12) is depicted. The shaded exon 11 (E11nuc) represents the mutated version of exon 11, which contains the E791A mutation. The black triangle represents the loxP site. P indicates the probe for Southern analysis. Pv represents the PvuII restriction enzyme site, which was used for the Southern analysis presented in panel B. (B) Southern analysis of mouse tail DNA. The DNA was digested with PvuII, which distinguishes the wild-type and the nuclease-deficient allele. The DNA was hybridized with the probe shown in panel A. (C) RT-PCR analysis of XPG expression in MEFs. The cDNAs were digested with PvuII, which distinguishes between the wild-type and mutant XPG message. The digestion product was hybridized with a probe, which extends from the 5′ end of the cDNA to the E791A mutation.
Analysis of XPG expression by reverse transcriptase-mediated PCR (RT-PCR)
RNA was isolated from mouse embryonic fibroblasts (MEFs) derived from 13.5-day-old embryos, and cDNA was synthesized by reverse transcription. XPG cDNA was amplified with the following primers: 5′-GGC TGG AGC AGG AGG AGC ATG CTG A-3′ and 5′-GCG TCT AGA TTA ATG ATG GTG GTG ATG GTG GGT TTT CTT TTT CCT TCT CTT CAT A-3′. The 5′ ends of the two primers correspond to positions 1,255 and 3,509 bp from the start of the XPG open reading frame, respectively. The PCR product was digested with PvuII to distinguish between the wild-type and mutant cDNAs. The digested cDNA was electrophoresed on an agarose gel and was detected with a probe which contains the region from the 5′ end of the cDNA to the mutation-associated PvuII site.
Class switching assays
For class switching, splenocytes were resuspended at a concentration of 3 × 106 cells/ml (approximately 1 × 106 B cells/ml) in RPMI medium supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 U of penicillin-streptomycin/ml, 100 μM β-mercaptoethanol, 20 μg of lipopolysaccharide (LPS)/ml, and 20 μg of dextran sulfate/ml or 25 ng of recombinant murine interlukin-4/ml. LPS plus dextran sulfate induces class switching to immunoglobulin G2b (IgG2b) and IgG3, while LPS plus interleukin-4 promotes class switching to IgG1 and IgE. Low levels of switching to IgA is also induced by LPS. The culture supernatant was collected 6 days after stimulation, and the concentration of antibodies was determined by enzyme-linked immunosorbent assay (ELISA).
UV sensitivity assays
MEFs were isolated from day 13.5 embryos and were grown in Dulbecco's modified Eagle medium supplemented with 15% fetal bovine serum, 2 mM glutamine, 100 U of penicillin-streptomycin/ml, and 100 μM β-mercaptoethanol. For UV irradiation experiments, 5 × 105 cells were plated per well in a six-well plate. The cells were grown for 2 days to confluency. The cells were irradiated with UVC under 3 ml of medium. The use of medium during irradiation was necessary to attenuate the high emission level of the UV lamp, which kills even wild-type cells in 2 s. Taking into consideration the UV absorbance of the medium, the cells were exposed to UV at a dose rate of 0.25 W/m2 as measured at a wavelength of 254 nm. After irradiation, the cells were trypsinized, and 5 × 105 cells were plated into a new well of a six-well plate. The cells were grown for 2 days and were trypsinized. The number of viable cells was counted in the presence of trypan blue.
Mice that are 8 weeks old were used for irradiation experiments. The back skin of the mice was shaved. The mice were irradiated with a set of four FS-20 UVB lamps, which were filtered by a Kodacel membrane to eliminate residual UVC. The mice received a dose of 2,000 J/m2 for each irradiation. The irradiation was carried out three times a week for a maximum of 15 weeks. For histological examination, dissected tissues were fixed in Boins solution. Sectioned tissues were stained with hematoxylin and eosin.
RESULTS AND DISCUSSION
Introducing the E791A mutation into the XPG gene
Based on mutagenesis experiments in vitro, three acidic residues (D77, E791, and D812) are critical for the nuclease activity of XPG (4, 29). To create a mouse strain with a nuclease-deficient XPG protein, we introduced the E791A mutation into the mouse genome. To achieve this, we replaced exon 11 of the XPG gene, which contains the codon for E791 (14), with a mutated version (E791A) in mouse ES cells using gene targeting technology (Fig. 1A). The mutation creates a novel PvuII restriction enzyme site, which was used for detecting the mutation. Correctly targeted clones were identified by Southern analysis and were used to generate mutant animals (Fig. 1B). We refer to the mutated nuclease-deficient XPG allele as nucn. The superscript “n” indicates the presence of the neomycin resistance (Neor) gene in the intron between exons 11 and 12 to distinguish it from the allele in which the Neor gene was deleted as described below.
Heterozygous animals (+/nucn) developed normally. Interbreeding between heterozygous animals produced progeny at the expected Mendelian ratios (89 mice; +/+, 23%; +/nucn, 52%; nucn/nucn, 25%). The homozygous mutant mice (nucn/nucn) show no obvious abnormalities and are fertile. Since the Neor gene potentially could affect the expression of XPG, the selection marker was deleted through flanking loxP sites by Cre-mediated recombination. To achieve this deletion, we bred heterozygous animals with E2A-cre transgenic mice, which express the Cre recombinase in germ cells (11). Resulting animals in which the Neor gene was deleted in the germ line were used for further breeding to obtain homozygous mutant mice (referred to as nuc/nuc), which are indistinguishable from the nucn/nucn mice.
Since XPG null mice have severe developmental defects, the apparently normal development of the XPG nuclease-deficient mice suggests that the mutant protein is properly expressed. As we do not have an antibody for the murine XPG protein to confirm this point by Western analysis, we used RT-PCR to determine the expression level of the mutant XPG message in MEFs. The E791A mutation creates a novel PvuII site, which can be used to distinguish between the wild-type and mutant XPG cDNAs. By this method, we found that the mutant XPG message is present at levels equal to the wild-type message in heterozygous MEF cells (Fig. 1C).
CSR is normal in XPG nuclease-deficient mice
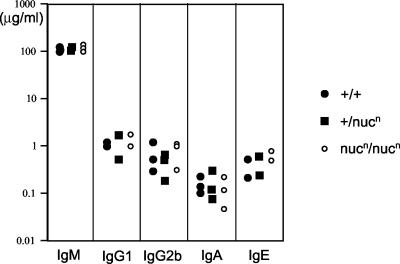
XPG is capable of cleaving R-loop structures in vitro (27). R-loop structures have been implicated as an intermediate in class switch recombination (CSR), which is responsible for generating the various immunoglobulin heavy-chain (IgH) isotypes by B cells (25, 33). The ability to cleave R-loop structures raised the possibility that XPG may be involved in this recombination process (27). To evaluate this possibility in vivo, we isolated splenocytes from XPG nuclease-deficient mice (for this experiment, nucn/nucn mice were used). The splenocytes were induced to undergo CSR to the various IgH isotypes by polyclonal activation via incubation in the presence of bacterial LPS with or without the interleukin-4 cytokine. The homozygous mutant splenocytes proliferated normally after activation. The concentrations of the various IgH isotypes in the culture supernatant were measured by ELISA. We found that the levels of the different IgH isotypes produced by the nuclease-deficient splenocytes was comparable to those of wild-type and heterozygous mutant cells (Fig. 2). While we might miss a subtle effect on CSR by this type of assay, we can conclude that the nuclease activity of XPG is not required for CSR.
FIG. 2.
Normal IgH class switching in XPG-deficient splenocytes. Shown is ELISA analysis of antibody concentrations in the supernatant of in vitro-stimulated splenocytes. The y axis indicates the concentration of antibodies. The x axis indicates the IgH isotype.
It is possible that a redundant nuclease could compensate for the loss of XPG. In this regard, the structure-specific nuclease XPF-ERCC1 also is capable of cleaving R-loop structures in vitro (27). To address the possibility that XPF-ERCC1 is redundant to XPG in CSR, we bred XPG nuclease-deficient animals with mice deficient in XPF (28). Mice doubly deficient in XPG and XPF appear morphologically identical to mice deficient in XPF or ERCC1, which exhibit severe developmental defects and die approximately 3 weeks after birth (16, 28, 30). Based on preliminary analysis, CSR appears grossly normal in the double-mutant splenocytes (data not shown). Similarly, no significant reduction in CSR efficiency was observed in XPF-deficient (28) or ERCC1-deficient (31) mice. We conclude from these results that neither the XPF nor the XPG nuclease activity is required for CSR. However, based on these findings, we cannot exclude the possibility that XPF-ERCC1 and XPG may actually function in CSR but that their role in this process can be replaced by the activity of other factors or DNA repair pathways.
XPG nuclease-deficient cells are hypersensitive to UV irradiation in vitro
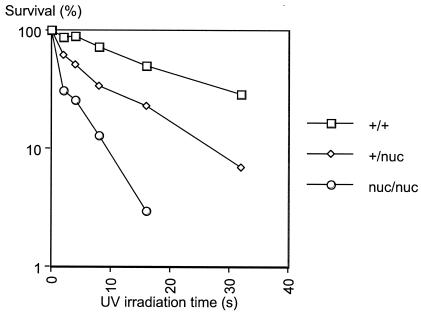
XPG is an integral part of the nucleotide excision repair pathway and makes incisions 3′ to the DNA adduct. Therefore, it is expected that deficiencies in the nuclease activity of XPG should render cells hypersensitive to UV irradiation. To confirm this possibility, we isolated MEFs from 13.5-day-old embryos (for this and the following in vivo experiments, nuc/nuc mice were used). MEFs from the mutant embryos showed no growth defects (data not shown), again in contrast to MEFs derived from XPG null embryos, which senesce prematurely (8). However, the homozygous mutant MEFs were hypersensitive to UVC irradiation as compared to wild-type cells (Fig. 3). Moreover, the heterozygous mutant cells showed intermediate sensitivity to UVC. This phenotype might be explained by the dominant negative nature of the mutation. Since the point mutation specifically affects the catalytic step, the mutant protein could compete with the wild-type protein for the DNA substrate and/or interacting proteins. In previous transfection experiments, overexpression of the E791A mutant XPG protein in cell lines rendered wild-type cells more sensitive to UV irradiation (4). These results confirm the important role of the nuclease activity of XPG in nucleotide excision repair.
FIG. 3.
XPG nuclease-deficient MEFs are hypersensitive to UV irradiation. Shown is the survival curve of UVC-irradiated MEFs. The y axis indicates the percent surviving irradiated cells relative to unirradiated control populations. The x axis indicates the irradiation time.
XPG nuclease-deficient mice are hypersensitive to UV irradiation in vivo
To evaluate the role of the XPG nuclease in repairing UV damage in vivo, we exposed a group of 16 wild-type (+/+), 18 heterozygous (+/nuc), and 16 homozygous (nuc/nuc) mice to UVB irradiation (2,000 J/m2) three times a week. After four or five irradiation treatments, a fraction of the homozygous mutant mice (4 of 16) developed a “sunburn” response consisting of redness and blistering. The sunburn response was also observed in 1 of 18 heterozygous mutant animals. No visible changes were observed in the wild-type animals. After 9 to 12 irradiation treatments, 15 of the 16 homozygous animals showed obvious changes in the ear and eye. One homozygous animal developed obvious eye and ear lesions after 21 irradiation treatments. The ears became smaller and wrinkled, and the eyes appeared opacified. The back skin of the homozygous mutant animals did not show visible changes except for the early acute sunburn response. Some heterozygous animals (5 of 18) showed mild ear and eye changes as well at later stages of the UVB treatment (17 to 22 irradiation treatments). None of the wild-type animals exhibited any obvious changes during the entire irradiation period. In spite of the obvious photodamage, none of the homozygous mutant animals developed any visible tumors after up to 45 irradiation treatments (15 weeks).
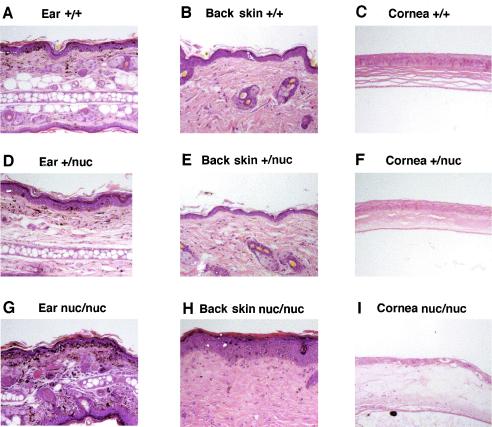
To examine the photodamage in detail, we performed histological analysis with tissue sections of the ear, eye, and back skin of the animals irradiated for 10 weeks (Fig. 4, representative data). There were minimal changes in the ear, cornea, and back skin of irradiated wild-type and heterozygous mice (Fig. 4A to F). The high dose of irradiation did cause low levels of inflammatory cell infiltration, pigment incontinence, and epidermal thickening on the side of the ear receiving more irradiation. Consistent with mild UV-induced lesions in heterozygous mice, stromal sclerosis was observable in the cornea of the +/nuc mice (Fig. 4F). By contrast, tissue sections from the irradiated nuc/nuc mice exhibited more extensive photodamage (Fig. 4G to I). The ear showed increased pigment incontinence, extensive inflammatory infiltration, epidermal thickening, and scale formation in ear skin with disruption of the ear architecture, which correlates with the wrinkled appearance of the ears and is consistent with the healing of repeated severe inflammatory insults. The back skin of nuc/nuc mice showed extensive epidermal thickening and hyperkeratosis, with dermal sclerosis and inflammation. In the eye section, complete blistering of the cornea was observed.
FIG. 4.
XPG nuclease-deficient mice are hypersensitive to UV irradiation. Shown are tissue sections stained with hematoxylin and eosin. The tissues were dissected from mice that were exposed to UVB irradiation for 10 weeks. Tissue identity and genotype are indicated for each panel.
The phenotype of the XPG nuclease-deficient mice suggests that the nuclease activity of XPG is required only for nucleotide excision repair. Since XPG null mice show severe developmental defects (8), XPG must perform functions independent of its nuclease activity, which may include transcription and repair of oxidative DNA damage. Similar to XPA and XPC knockout mice (6, 19, 24) the XPG nuclease-deficient animal is hypersensitive to UV irradiation. One important difference is that UV irradiation did not cause tumor development in XPG nuclease-deficient mice. The reason for this difference is not clear at present. It could potentially be attributed to differences in the genetic background of the animals or to the irradiation conditions. Alternatively, mutations in distinct components of the nucleotide excision repair machinery could have different consequences in tumorigenesis. Definitive resolution of this issue requires side-by-side comparison of the XPG nuclease-deficient mice and mice deficient in XPA or XPC.
Acknowledgments
We thank Laurie Davidson and Dan Foy for mouse work.
This work was supported by National Institutes of Health (NIH) grant A13154 (to F.W.A.) and NIH training grant A107512 (to M.T.). D.A.J. was supported by NIH grant P30AR042689. F.W.A. is an Investigator and R.S. is an Associate of the Howard Hughes Medical Institute.
REFERENCE
- 1.Araujo, S. J., E. A. Nigg, and R. D. Wood. 2001. Strong functional interactions of TFIIH with XPC and XPG in human DNA nucleotide excision repair, without a preassembled repairosome. Mol. Cell. Biol. 21:2281-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berneburg, M., and A. R. Lehmann. 2001. Xeroderma pigmentosum and related disorders: defects in DNA repair and transcription. Adv. Genet. 43:71-102. [DOI] [PubMed] [Google Scholar]
- 3.Bessho, T. 1999. Nucleotide excision repair 3′ endonuclease XPG stimulates the activity of base excision repair enzyme thymine glycol DNA glycosylase. Nucleic Acids Res. 27:979-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Constantinou, A., D. Gunz, E. Evans, P. Lalle, P. A. Bates, R. D. Wood., and S. G. Clarkson. 1999. Conserved residues of human XPG protein important for nuclease activity and function in nucleotide excision repair. J. Biol. Chem. 274:5637-5648. [DOI] [PubMed] [Google Scholar]
- 5.de Boer, J., and J. H. Hoeijmakers. 2000. Nucleotide excision repair and human syndromes. Carcinogenesis 21:453-460. [DOI] [PubMed] [Google Scholar]
- 6.de Vries, A., C. T. van Oostrom, F. M. Hofhuis, P. M. Dortant, R. J. Berg, F. R. de Gruijl, P. W. Wester, C. F. van Kreijl, P. J. Capel, H. van Steeg, and S. J. Verbeek. 1995. Increased susceptibility to ultraviolet-B and carcinogens of mice lacking the DNA excision repair gene XPA. Nature 377:169-173. [DOI] [PubMed] [Google Scholar]
- 7.Emmert, S., H. Slor, D. B. Busch, S. Batko, R. B. Albert, D. Coleman, S. G. Khan, B. Abu-Libdeh, J. J. DiGiovanna, B. B. Cunningham, M. M. Lee, J. Crollick, H. Inui, T. Ueda, M. Hedayati, L. Grossman, T. Shahlavi, J. E. Cleaver, and K. H. Kraemer. 2002. Relationship of neurologic degeneration to genotype in three xeroderma pigmentosum group G patients. J. Investig. Dermatol. 118:972-982. [DOI] [PubMed] [Google Scholar]
- 8.Harada, Y. N., N. Shiomi, M. Koike, M. Ikawa, M. Okabe, S. Hirota, Y. Kitamura, M. Kitagawa, T. Matsunaga, O. Nikaido, and T. Shiomi. 1999. Postnatal growth failure, short life span, and early onset of cellular senescence and subsequent immortalization in mice lacking the xeroderma pigmentosum group G gene. Mol. Cell. Biol. 19:2366-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iyer, N., M. S. Reagan, K. J. Wu, B. Canagarajah, and E. C. Friedberg. 1996. Interactions involving the human RNA polymerase II transcription/nucleotide excision repair complex TFIIH, the nucleotide excision repair protein XPG, and Cockayne syndrome group B (CSB) protein. Biochemistry 35:2157-2167. [DOI] [PubMed] [Google Scholar]
- 10.Klungland, A., M. Hoss, D. Gunz, A. Constantinou, S. G. Clarkson, P. W. Doetsch, P. H. Bolton, R. D. Wood, and T. Lindahl. 1999. Base excision repair of oxidative DNA damage activated by XPG protein. Mol. Cell 3:33-42. [DOI] [PubMed] [Google Scholar]
- 11.Lakso, M., J. G. Pichel, J. R. Gorman, B. Sauer, Y. Okamoto, E. Lee, F. W. Alt, and H. Westphal. 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. USA 93:5860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lalle, P., T. Nouspikel, A. Constantinou, F. Thorel, and S. G. Clarkson. 2002. The founding members of xeroderma pigmentosum group G produce XPG protein with severely impaired endonuclease activity. J. Investig. Dermatol. 118:344-351. [DOI] [PubMed] [Google Scholar]
- 13.Lee, S. K., S. L. Yu, L. Prakash, and S. Prakash. 2002. Requirement of yeast RAD2, a homolog of human XPG gene, for efficient RNA polymerase II transcription. Implications for Cockayne syndrome. Cell 109:823-834. [DOI] [PubMed] [Google Scholar]
- 14.Ludwig, D. L., J. S. Mudgett, M. S. Park, A. V. Perez-Castro, and M. A. MacInnes. 1996. Molecular cloning and structural analysis of the functional mouse genomic XPG gene. Mamm. Genome 7:644-649. [DOI] [PubMed] [Google Scholar]
- 15.Matsunaga, T., D. Mu, C. H. Park, J. T. Reardon, and A. Sancar. 1995. Human DNA repair excision nuclease. Analysis of the roles of the subunits involved in dual incisions by using anti-XPG and anti-ERCC1 antibodies. J. Biol. Chem. 270:20862-20869. [DOI] [PubMed] [Google Scholar]
- 16.McWhir, J., J. Selfridge, D. J. Harrison, S. Squires, and D. W. Melton. 1993. Mice with DNA repair gene (ERCC-1) deficiency have elevated levels of p53, liver nuclear abnormalities and die before weaning. Nat. Genet. 5:217-224. [DOI] [PubMed] [Google Scholar]
- 17.Mu, D., C. H. Park, T. Matsunaga, D. S. Hsu, J. T. Reardon, and A. Sancar. 1995. Reconstitution of human DNA repair excision nuclease in a highly defined system. J. Biol. Chem. 270:2415-2418. [DOI] [PubMed] [Google Scholar]
- 18.Mueser, T. C., N. G. Nossal, and C. C. Hyde. 1996. Structure of bacteriophage T4 RNase H, a 5′ to 3′ RNA-DNA and DNA-DNA exonuclease with sequence similarity to the RAD2 family of eukaryotic proteins. Cell 85:1101-1112. [DOI] [PubMed] [Google Scholar]
- 19.Nakane, H., S. Takeuchi, S. Yuba, M. Saijo, Y. Nakatsu, H. Murai, Y. Nakatsuru, T. Ishikawa, S. Hirota, Y. Kitamura, Y. Kato, Y. Tsunoda, H. Miyauchi, T. Horio, T. Tokunaga, T. Matsunaga, O. Nikaido, Y. Nishimune, Y. Okada, and K. Tanaka. 1995. High incidence of ultraviolet-B- or chemical-carcinogen-induced skin tumours in mice lacking the xeroderma pigmentosum group A gene. Nature 377:165-168. [DOI] [PubMed] [Google Scholar]
- 20.Nouspikel, T., and S. G. Clarkson. 1994. Mutations that disable the DNA repair gene XPG in a xeroderma pigmentosum group G patient. Hum. Mol. Genet. 3:963-967. [DOI] [PubMed] [Google Scholar]
- 21.Nouspikel, T., P. Lalle, S. A. Leadon, P. K. Cooper, and S. G. Clarkson. 1997. A common mutational pattern in Cockayne syndrome patients from xeroderma pigmentosum group G: implications for a second XPG function. Proc. Natl. Acad. Sci. USA 94:3116-3121. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.O'Donovan, A., A. A. Davies, J. G. Moggs, S. C. West, and R. D. Wood. 1994. XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature 371:432-435. [DOI] [PubMed] [Google Scholar]
- 23.Petit, C., and A. Sancar. 1999. Nucleotide excision repair: from E. coli to man. Biochimie 81:15-25. [DOI] [PubMed] [Google Scholar]
- 24.Sands, A. T., A. Abuin, A. Sanchez, C. J. Conti, and A. Bradley. 1995. High susceptibility to ultraviolet-induced carcinogenesis in mice lacking XPC. Nature 377:162-165. [DOI] [PubMed] [Google Scholar]
- 25.Shinkura, R., M. Tian, M. Smith, K. Chua, Y. Fujiwara, and F. W. Alt. 2003. The influence of transcriptional orientation on endogenous switch region function. Nat. Immunol. 4:435-441. [DOI] [PubMed] [Google Scholar]
- 26.Svejstrup, J. Q., P. Vichi, and J. M. Egly. 1996. The multiple roles of transcription/repair factor TFIIH. Trends Biochem. Sci. 21:346-350. [PubMed] [Google Scholar]
- 27.Tian, M., and F. W. Alt. 2000. Transcription-induced cleavage of immunoglobulin switch regions by nucleotide excision repair nucleases in vitro. J. Biol. Chem. 275:24163-24172. [DOI] [PubMed] [Google Scholar]
- 28.Tian, M., R. Shinkura, N. Shinkura, and F. W. Alt. 2004. Growth retardation, early death, and DNA repair defects in mice deficient for the nucleotide excision repair enzyme XPF. Mol. Cell. Biol. 24:1200-1205. [DOI] [PMC free article] [PubMed]
- 29.Wakasugi, M., J. T. Reardon, and A. Sancar. 1997. The non-catalytic function of XPG protein during dual incision in human nucleotide excision repair. J. Biol. Chem. 272:16030-16034. [DOI] [PubMed] [Google Scholar]
- 30.Weeda, G., I. Donker, J. de Wit, H. Morreau, R. Janssens, C. J. Vissers, A. Nigg, H. van Steeg, D. Bootsma, and J. H. Hoeijmakers. 1997. Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr. Biol. 7:427-439. [DOI] [PubMed] [Google Scholar]
- 31.Winter, A. G., K. Samuel, K. T. Hsia, and D. W. Melton. 2003. The repair and recombination enzyme ERCC1 is not required for immunoglobulin class switching. DNA Repair 2:561-569. [DOI] [PubMed] [Google Scholar]
- 32.Wood, R. D., S. J. Araujo, R. R. Ariza, D. P. Batty, M. Biggerstaff, E. Evans, P. H. Gaillard, D. Gunz, B. Koberle, I. Kuraoka, J. G. Moggs, J. K. Sandall, and M. K. Shivji. 2000. DNA damage recognition and nucleotide excision repair in mammalian cells. Cold Spring Harbor Symp. Quant. Biol. 65:173-182. [DOI] [PubMed] [Google Scholar]
- 33.Yu, K., F. Chedin, C. L. Hsieh, T. E. Wilson, and M. R. Lieber. 2003. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 4:442-451. [DOI] [PubMed] [Google Scholar]