Abstract
A chromosome fragmentation assay was used to measure the efficiency and genetic control of break-induced replication (BIR) in Saccharomyces cerevisiae. Formation of a chromosome fragment by de novo telomere generation at one end of the linear vector and recombination-dependent replication of 100 kb of chromosomal sequences at the other end of the vector occurred at high frequency in wild-type strains. RAD51 was required for more than 95% of BIR events involving a single-end invasion and was essential when two BIR events were required for generation of a chromosome fragment. The similar genetic requirements for BIR and gene conversion suggest a common strand invasion intermediate in these two recombinational repair processes. Mutation of RAD50 or RAD59 conferred no significant defect in BIR in either RAD51 or rad51 strains. RAD52 was shown to be essential for BIR at unique chromosomal sequences, although rare recombination events were detected between the subtelomeric Y′ repeats.
DNA double-strand breaks (DSBs) are potentially lethal lesions that can occur spontaneously during normal cellular metabolism or by treatment of cells with DNA-damaging agents (38). DSBs also function as initiators of regulated recombination processes, such as mating type switching in Saccharomyces cerevisiae, meiotic recombination, and the rearrangement of immunoglobulin and T-cell receptor genes (8, 43, 48, 51). DSBs generated by endonucleases or ionizing radiation produce two free DNA ends that can be repaired by homologous recombination by utilizing a sister chromatid or homologous chromosome as a template or else by end joining independent of extensive sequence homology.
In S. cerevisiae, genes of the RAD52 epistasis group (RAD50, RAD51, RAD52, RAD54, RAD55, RAD57, RAD59, MRE11/RAD58, XRS2, and RDH54/TID1) are required for the repair of DSBs by homologous recombination (54). RAD52 is required for virtually all homology-dependent recombination. However, the requirement for the other genes is not so absolute and can vary depending on the configuration of the template sequences. For instance, RAD51, RAD54, RAD55, and RAD57 are required for repair of DSBs by gene conversion (54) but not for the RAD52-dependent single-strand annealing pathway or for the RAD52-dependent amplification of TG1-3 repeats observed in telomerase-deficient strains (21, 27, 29, 56).
In contrast to DSBs made by endonucleases or irradiation, breaks produced by replication fork collapse leave DNA molecules with only a single end whose repair is obligatorily by homologous recombination. Studies of E. coli have shown an essential role for the RecA and RecBCD proteins in the restoration of chromosomal replication following replication fork collapse (24, 37, 45). RecBCD prepares a 3′ end for loading of RecA to promote joint molecule formation between the intact and broken sister arms (24). The D-loop formed by RecA is recognized by PriA protein, which then functions in the assembly of the primosome for lagging-strand DNA synthesis and at the 3′ end of the invading strand to initiate leading-strand synthesis (63). Similarly, late replication of bacteriophage T4 requires UvsX-dependent strand invasion (26).
The mechanisms for repair of two-ended DSBs are well established in eukaryotes, particularly in yeast. However, contrary to the situation with prokaryotes, the genetic control of one-ended invasion events in repair of collapsed replication forks has not been well established in higher systems. Strong circumstantial evidence implicating an essential role for RAD51 comes from investigation with cultured chicken cells conditionally expressing RAD51. Here, accumulation of unrepaired chromosome breaks during S phase is concomitant with depletion of RAD51 (28, 47, 59). These findings suggest that RAD51 is essential for maintenance or restart of stalled replication forks and/or for the repair of collapsed replication forks. Evidence implicating RAD51 as an important player in one-ended repair events also comes from observations in yeast. RAD51-dependent recombination between subtelomeric Y′ elements leads to survivors in the absence of telomerase (27, 29, 56), a process thought to occur by a one-ended invasion resulting in replication to the end of the chromosome (13). On the other hand, Malkova et al., also studying yeast, reported repair of an HO-endonuclease break on the right arm of chromosome III by recombination-dependent replication in the absence of RAD51 function (33). These recombination events were interpreted as resulting from one-ended break-induced repair leading to duplication of distal sequences all the way to the end of the chromosome. A plasmid-based break-induced replication (BIR) assay has also been described, but these events occurred at low efficiency due to the requirement for nonhomologous end joining to complete the repair event (25).
To learn more about the genetic control of one-ended invasion events, and to attempt to resolve the paradoxical role of RAD51, we planned a new experimental system for studying break-induced replication. The design was based on the chromosome fragmentation vector system of Hieter et al. in which a linearized vector transformed into yeast cells undergoes two independent recombination-dependent replication events to generate a stable chromosome fragment (40). The vector used for transformation was modified to include a TG1-3 tract to provide a site for de novo telomere addition at one end of the linear vector, and the other end consists of a unique chromosomal region for strand invasion of homologous chromosomal sequences. The advantages of this system compared with HO-induced break systems is that repair cannot occur by gene conversion. We report here that most of the observed repair events occur by a RAD51-dependent pathway (including RAD54, RAD55, and RAD57) but with no requirement for RAD50 or RAD59. These results support the emerging view that the essential role of RAD51 in vertebrates is in recombination-dependent restoration of collapsed replication forks.
MATERIALS AND METHOD
Media, growth conditions, and genetic methods
Standard genetic methods were followed. YPD (1% yeast extract, 2% peptone, 2% dextrose) and synthetic complete (SC) medium lacking the appropriate amino acid or nucleic acid base were prepared as described previously (1). Transformations were performed by the lithium acetate method (19). Yeast cells were grown at 30°C, unless otherwise stated.
Yeast strains and plasmids
S. cerevisiae strains used in this study are RAD5 derivatives of W303-1A and W303-1B (57), unless otherwise noted, and are listed in Table 1. Strains containing the rad52::TRP1, rad51::HIS3, and rad59::LEU2 alleles have been described previously (5). The strains containing multiple rad mutations were made by crossing strains from the laboratory collection, dissection of tetrads, and screening of the haploid segregants for those with the desired genotype. Chromosome fragmentation vectors (CFV) CFV/D8B-Y′, CFV/D8B-tg, CFV/MRC1-tg, CFV/PCA1-tg, CFV/YBR235-tg, and CFV/Y′-tg are derivatives of pYCF2/D8B, which has been described previously (40). To construct CFV/D8B-Y′, a SnaBI oligonucleotide linker was cloned into the BglII site in pYCF2/D8B, resulting in a unique SnaBI site between the D8B region and the Y′ subtelomeric repeat. To construct CFV/D8B-tg, oligonucleotides containing a HindIII half site, two Rap1 binding sites, a SnaBI site, and a BglII half site,5′-AGCTTTGTGTGGTGTGTGGGTGTGTGTGGGTGTGTGGGTGTGTGGGTACGTAA and 5′-GATCTTACGTACCCACACACCCACACACCCACACACACCCACACACCACACAA, were annealed and then cloned into the HindIII and BglII sites of pYCF2/D8B, resulting in a unique SnaBI site between the D8B region and the Rap1 binding sites and deleting the Y′ region. To construct CFV/MRC1-tg, CFV/PCA1-tg, and CFV/YBR235-tg, an approximately 2-kb region of each open reading frame was amplified by PCR, subcloned into pGEM-T (Promega), and then cloned into the BglII and EcoNI sites of CFV/D8B-tg, replacing the D8B sequences with another unique chromosomal locus. To construct CFV/Y′-tg, oligonucleotides containing a BglII half site, a SnaBI site, two Rap1 binding sites, and an EcoNI half site, 5′-GATCTTACGTACCCACACACCCACACACCCACACACACCCACACACCACACACCTAA and 5′-ATTAGGTGTGTGGTGTGTGGGTGTGTGTGGGTGTGTGGGTGTGTGGGTACGTAA, were annealed and then cloned into the BglII and EcoNI sites of pYCF2/D8B, resulting in a unique SnaBI site between the Y′ region and the GT tract and deleting the D8B region.
TABLE 1.
Yeast strains
| Strain no. | Genotypea | Source or reference |
|---|---|---|
| W1588-4C | MATa | R. Rothstein |
| W1588-4A | MATα | R. Rothstein |
| HKY604-17A | MATα rad50::hisG | H. Klein |
| HKY604-17C | MATarad50::hisG | H. Klein |
| HKY596-2B | MATα rad54::LEU2 | H. Klein |
| HKY597-2C | MATα rad55::LEU2 | H. Klein |
| HKY598-8B | MATα rad57::LEU2 | H. Klein |
| LSY841 | MATarad51::HIS3 rad59::LEU2 met17-sna ADE2 | 6 |
| LSY949 | MATarad52::TRP1 | L. Langston |
| LSY950 | MATα rad52::TRP1 | L. Langston |
| LSY1253-5D | MATα rad51::HIS3 lys2 | This study |
| LSY1253-7A | MATarad51::HIS3 lys2 | This study |
| LSY1253-7B | MATα rad59::LEU2 lys2 | This study |
| LSY1253-20D | MATarad59::LEU2 met17-sna | This study |
| LSY1253-18A | MATarad51::HIS3 rad59::LEU2 | This study |
| LSY1253-16A | MATα rad51::HIS3 rad59::LEU2 lys2 | This study |
| LSY1254-53A | MATarad50::hisG rad51::HIS3 | This study |
| LSY1254-35A | MATarad50::hisG rad51::HIS3 lys2 | This study |
| LSY1254-2C | MATarad50::hisG rad59::LEU2 lys2 | This study |
| LSY1254-5D | MATα rad50::hisG rad59::LEU2 lys2 met17-sna | This study |
| LSY1254-42A | MATarad50::hisG rad51::HIS3 rad59::LEU2 met17-sna | This study |
| LSY1254-53C | MATarad50::hisG rad51::HIS3 rad59::LEU2 | This study |
| LSY1040 | MATa/MATα rad51::LEU2/rad51::HIS3 | This study |
| LSY1307 | MATa/MATα | This study |
All strains are derivatives of W303 (leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his 3-11,15 RAD5); only mating type and differences from this genotype are given.
Determination of frequency of stable Ura+ transformants
One microgram of the chromosome fragmentation vector, digested with SnaBI, was used to transform competent yeast cells, selecting for Ura+ transformants. For CFV/D8B-Y′, CFV/D8B-tg, CFV/MRC1-tg, CFV/PCA1-tg, and CFV/YBR235-tg, two classes of transformants were expected: circular plasmids, due to end joining or contamination of the linear vector DNA with undigested plasmids, and linear chromosome fragments. The mitotic stability of the Ura+ transformants was determined to distinguish between these two classes. For ade2-1 strains, Ura+ transformants were struck onto nonselective YPD medium and were analyzed for a red sectoring phenotype. The ade2-1 red-colony-color phenotype is partially suppressed by SUP11, which is contained on the vectors. Transformants that showed a nonsectoring phenotype (white) were grown on nonselective solid medium and then replica plated to SC-URA to score for Ura− segregants. Transformants that failed to show red/white sectoring but showed Ura− segregants after nonselective growth were generally due to petite formation or a secondary mutation that eliminated the red-colony-color phenotype. For diploids and ADE2 strains, Ura+ transformants were struck onto nonselective YPD medium, replica plated to SC-URA, and scored for Ura+/Ura− phenotype. The frequency of stable Ura+ transformants presented in the tables is the number of mitotically stable Ura+ transformants per microgram of linearized DNA transformed divided by the number of Ura+ transformants per microgram of circular plasmid DNA transformed. The mean BIR frequencies (with standard deviations) presented in the tables are from at least three independent transformations of each strain. Statistical analyses were performed by using the Student's t test.
For CFV/Y′-tg, genomic DNA was purified from Ura+ transformants and was digested with EagI. DNA fragments were separated on 0.8% agarose gels, transferred to nylon membranes (Amersham Hybond-N+), and hybridized with a PCR fragment generated by amplification of CEN4 sequences adjacent to the Rap1 binding sites in CFV/Y′-tg. Transformants in which the vector recircularized contained a vector-length EagI-linearized fragment. Transformants containing a linear chromosome, formed by Y′ recombination and de novo telomere addition, were identified by an approximately 2-kb fragment containing CEN4 and the non-Y′ telomere. The frequency of BIR is the number of Ura+ transformants that have created the recombinant linear DNA molecule per microgram of linearized DNA transformed divided by the number of Ura+ transformants per microgram of circular plasmid DNA transformed.
Visualization of long (>100 kb) chromosome fragments
Agarose plugs of intact chromosomal DNA were prepared from stable Ura+ transformants as described by Schwartz and Cantor (44a). Chromosomes were separated by electrophoresis through 1% pulsed-field gel electrophoresis (PFGE)-certified agarose (Bio-Rad) at 14°C in 0.5× Tris-borate-EDTA, using a CHEF-DR II Pulsed Field Electrophoresis system (Bio-Rad). Gels were stained with SYBR gold (Molecular Probes), and the DNA was transferred to nylon membranes and hybridized with a PCR product generated by amplification of the URA3 or YBR235 open reading frame. Transformants in which the CFV were repaired by BIR contained an approximately 110-kb stable chromosome fragment.
Visualization of 30-kb chromosome fragments
Genomic DNA was purified from stable Ura+ transformants derived from CFV/MRC1-tg or CFV/PCA1-tg. Undigested DNA was separated on 0.8% gels, transferred to nylon membranes, and hybridized with a PCR fragment generated by amplification of pBR322 sequences present in the original chromosome fragmentation vector. Transformants in which the vector was repaired by BIR contained an approximately 30-kb fragment.
Visualization of vector recircularization events
Genomic DNA was purified from Ura+ transformants classified as mitotically unstable for the SUP11 and URA3 markers. Undigested DNA was separated on 0.8% gels, transferred to nylon membranes, and hybridized with a PCR fragment generated by amplification of pBR322 sequences present in the original chromosome fragmentation vector. Transformants that arose by a plasmid rejoin event or by contamination with uncut plasmid DNA exhibited two bands, corresponding to supercoiled or relaxed circular forms of the plasmid. The relaxed circular form of the plasmid migrated more slowly than bulk genomic DNA under the electrophoresis conditions used. In some cases the plasmids were shorter than the original CFV, consistent with imprecise end joining.
RESULT
Experimental system
To study the role of the RAD52 group genes in BIR we developed a chromosome fragmentation assay based on the pioneering system described by Hieter and colleagues (40, 61) (Fig. 1A). The Hieter system utilized a chromosome fragmentation vector (CFV) containing unique chromosomal sequences to target recombination, the URA3 selectable marker, and part of the Y′ subtelomeric repeat. When the vector was linearized in vitro and used to transform yeast, those investigators observed that recombination occurred between sequences present at the two ends of the CFV and the corresponding chromosomal regions leading to duplication of sequences to the telomeres, thereby forming a stable chromosome fragment (CF). Ura+ transformants were also found to arise by direct joining of the two ends of the CFV to form a self-replicating plasmid.
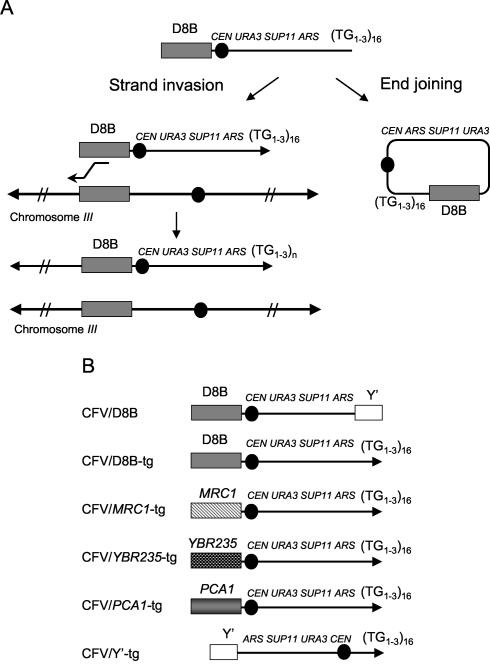
FIG. 1.
(A) Schematic representation of the chromosome fragmentation assay. The linear CFV undergoes de novo telomere addition to the tg tract at one end of the vector, and the other end invades the endogenous chromosomal locus duplicating sequences from the region of homology between the vector and native chromosome to the telomere. (B) Maps of the CFVs used for chromosome fragmentation. All contain URA3, SUP11, CEN4, and an ARS element. D8B refers to a 5-kb BglII fragment located 100 kb from the left telomere of chromosome III.
We modified the pCF2/D8B vector described by Morrow et al. (40) in two ways. The first was to change the site of linearization so that blunt ends would be formed in order to minimize repair by end joining. The second, and more important, was to replace the Y′ sequence with a 42-nucleotide tract of TG1-3 repeats containing two Rap1 binding sites designed to promote de novo telomere formation (31, 32). Thus, CFs generated from the modified CFV (CFV/D8B-tg) would undergo strand invasion at only one end, rather than the two ends required to form a stable CF according to the original vector design, and telomere addition at the other end of the vector.
The two potential classes of transformants derived from CFV/D8B-tg, i.e., 110-kb CFs and end-joined plasmids, can be distinguished by their mitotic stability. Centromere-containing plasmids are lost at a rate of about 1%/cell/generation, whereas large artificial chromosomes are about 100-fold more stable (17, 42). The SUP11 marker present on the CFV suppresses the ade2-1 mutation present in the yeast strain background used leading to white colonies, whereas cells lacking the CFV form red colonies (17). Transformants that showed very low sectoring following nonselective growth were scored as containing stable CFs and transformants showing high red or white sectoring were scored as plasmid end-joined events. These two phenotypic classes were confirmed by a second genetic screen for mitotic stability of the URA3 marker and by physical analysis (Fig. 2).
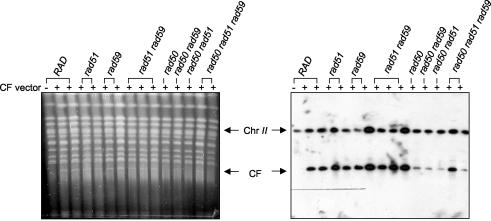
FIG. 2.
Identification of chromosome fragments by PFGE. Plugs were prepared from stable Ura+ transformants derived from CFV/YBR235-tg. The panel on the left shows the gel stained with SYBR gold, the panel on the right is a Southern blot of the same gel probed with a PCR product derived from YBR235 to indicate chromosome II and the CF.
RAD51, RAD52, RAD54, RAD55, and RAD57 are required for BIR
Transformation of recombination-proficient cells with linearized CFV/D8B-tg yielded 5.7 × 104 Ura+ transformants/μg of DNA, only fivefold less than that observed using the uncut plasmid (Table 2). Of these Ura+ transformants, more than 95% were stable for the URA3 and SUP11 markers and were shown to contain the expected 110-kb CF, in addition to a normal-length copy of chromosome III, by PFGE gel analysis (data not shown). Thus, de novo telomere addition and BIR are highly efficient processes in wild-type cells.
TABLE 2.
Effect of rad52 group mutations on the frequency of BIRa
| Relevant genotype | Frequency of stable Ura+ (10−2) with CFV/D8B-tg | Frequency relative to wild type |
|---|---|---|
| RAD | 20 ± 10 | 100 |
| rad52 | <0.0027 | <0.014 |
| rad51 | 0.14 ± 0.05 | 0.70 |
| rad54 | 0.20 ± 0.1 | 1.0 |
| rad55 | 0.47 ± 0.3 | 2.4 |
| rad57 | 0.52 ± 0.2 | 2.6 |
| rad59 | 12 ± 7.7 | 60 |
| rad51 rad59 | 0.07 ± 0.03 | 0.35 |
| rad50 | 11 ± 0.4 | 55 |
| rad50 rad51 | 0.47 ± 0.29 | 2.4 |
| rad50 rad59 | 3.5 ± 1.6 | 18 |
| rad50 rad51 rad59 | 0.34 ± 0.16 | 1.7 |
| RAD/RAD | 17 ± 5.3 | 100 |
| rad51/rad51 | 0.17 ± 0.04 | 1 |
The frequency of BIR is the number of stable Ura+ transformants per microgram with cut DNA divided by the number of transformants per microgram with uncut DNA transformed.
To determine the frequency of BIR, the number of stable Ura+ transformants derived from the linearized CFV was normalized to the transformation efficiency using uncut replicating plasmid for wild-type and each of the rad52 group mutant strains (Table 2). Consistent with previous studies showing an essential role for RAD52 in homologous recombination, no stable Ura+ transformants were recovered from the rad52 strain. There was a 140-fold decrease in the BIR frequency in the rad51 strain (P < 0.0001), and further analysis by PFGE revealed that only 50% of the stable Ura+ transformants contained CFs (data not shown). The transformants with a stable Ura+ phenotype lacking CFs are probably due to nonhomologous integration of the linear fragment or conversion or reversion of the chromosomal ura3-1 marker. The BIR frequency was also significantly reduced in rad54, rad55, and rad57 mutants (Table 2). Because rad55 and rad57 mutants exhibit more severe recombination and repair defects at low temperature, the transformations were repeated with cells grown at 18°C. By using cells grown at low temperature, the frequency of BIR was reduced more than 500-fold for rad51, rad54, rad55, and rad57 mutants compared with that of the wild type, suggesting that the residual RAD51-independent BIR events are temperature dependent (data not shown).
The BIR studies by Malkova et al. (33, 34) and Signon et al. (46) utilized diploid yeast strains. Because mating type heterozygosity is known to regulate several DNA repair pathways and to suppress the defect of some recombinational repair mutants (39), the transformation experiments were repeated using diploid strains. The efficiency of BIR was the same in wild-type haploid and diploid strains, and the same reduction in BIR was observed in rad51 diploids as was observed in haploids (Table 2). Therefore, the requirement for RAD51 in BIR is not specific for haploid strains.
To determine whether the low level of BIR seen in rad51 strains using CFV/D8B-tg was a general phenomenon or specific for this sequence, three other CFVs were generated (Fig. 1B). These all contain the TG1-3 tract for de novo telomere formation on one end of the linearized vector and a unique sequence for strand invasion at the other. The sequences inserted included a 2-kb fragment located 20 kb from the telomere of the left arm of chromosome III (MRC1), a 1.5-kb fragment located 125 kb from the telomere of the right arm of chromosome II (YBR235w) and a 1.5-kb fragment located 20 kb from the telomere of the left arm of chromosome II (PCA1). The frequency of BIR observed in the wild-type strain for the vectors containing chromosome II sequences was slightly decreased when compared with the vectors containing chromosome III sequences (Table 3). This could be due to the shorter region of homology present in the chromosome II-containing vectors, or to some feature of chromosome III that is more permissive for recombination-dependent replication. For all of these vectors, at least a 33-fold decrease in BIR frequency was observed in the rad51 strain, and physical analysis confirmed the presence of the expected size CF in stable Ura+ transformants (Fig. 2). The decreased intensity of the CF band by PFGE in rad50 strains is most likely due to the lower stability of the CF in this strain background.
TABLE 3.
Effect of the rad50, rad51, and rad59 mutations on the frequency of BIRa at other loci
| Relevant genotype | Frequency of BIR at:
|
|||
|---|---|---|---|---|
| CFV/D8B-tg | CFV/MRC1-tg | CFV/YBR235-tg | CFV/PCA1-tg | |
| RAD | 20 ± 10 | 39 ± 19 | 6.3 ± 2.4 | 12.8 ± 9.4 |
| rad51 | 0.14 ± 0.05 | 0.95 ± 0.1 | 0.19 ± 0.05 | 0.36 ± 0.2 |
| rad50 | 11 ± 0.4 | 76 ± 19 | 8.9 ± 6.1 | 31 ± 23 |
| rad59 | 12 ± 7.7 | 27 ± 15 | 5.0 ± 0.7 | 17 ± 5.3 |
The frequency of BIR for each vector is the number of stable Ura+ transformants per microgram with cut DNA divided by the number of transformants per microgram with uncut DNA transformed (10−2).
BIR is independent of RAD50 and RAD59
Previous studies suggested the RAD51-independent pathway for BIR requires the RAD50 and RAD59 genes (46). To test the requirement for these factors in RAD51-dependent and RAD51-independent BIR, rad50, rad59, rad51 rad50, rad51 rad59, rad50 rad59, and rad51 rad50 rad59 mutants were transformed with the CFV/D8B-tg vector. There was no significant decrease in the number of stable Ura+ transformants in the rad50 and rad59 strains compared to that of the wild type, and the frequency of BIR was much higher than that observed in rad51 strains (Table 2). The frequency of BIR in the rad51 rad59 strain was significantly lower than that observed for the wild-type strain (P < 0.01), but there was no difference from that of the rad51 strains. The significant increase in the frequency of BIR in the rad50 rad51 strain, compared to that of the rad51 strain (P = 0.013), could be due to increased stability of the linearized CFV in the absence of the RAD50-controlled nuclease activity (22). BIR was reduced in the rad50 rad59 strain compared to that of the wild type, but this reduction was not significant compared to that of the rad50 or rad59 single mutants. The rad50 and rad59 mutants also showed no significant reduction in BIR using the chromosome II-containing CFVs (Table 3) and all of the stable Ura+ transformants contained the expected size CF (Fig. 2).
RAD51 is essential for two independent BIR events
The results presented above show that RAD51 is important, but not absolutely essential, for strand invasion from one end of a linear chromosome. To test the requirement for RAD51 in the repair of two chromosome ends by BIR, the original D8B CFV containing Y′ sequences but modified to include a SnaBI site was used to transform wild-type, rad50, rad51, rad52, and rad59 strains. Stable Ura+ transformants derived from this vector occur by strand invasion of one end at the D8B region of chromosome III, and invasion of one of the multiple Y′ sequences present in the subtelomeric regions of most yeast chromosomes by the other end. The wild-type strain exhibited high-frequency BIR, but the rad51 and rad52 strains showed a more than 1,000-fold decrease in the number of stable Ura+ transformants compared to that of the wild-type strain (Table 4). Of five independent transformations of the rad51 strain, only one yielded stable Ura+ transformants, and of the three colonies produced, none contained a CF by PFGE analysis. Southern blot analysis of these transformants failed to detect vector sequences, suggesting they probably arose by conversion or reversion of the ura3-1 marker. Similarly, transformation of most of the double- and triple-mutant strains that contained the rad51 mutation failed to yield stable Ura+ transformants (Table 4). As before, the differences between the wild-type strain and rad50 or rad59 mutants were not significant.
TABLE 4.
Effect of rad52 group mutations on the frequency of two BIRa events
| Relevant genotype | Frequency of BIR (10−2) | Frequency relative to wild type |
|---|---|---|
| RAD | 17 ± 15 | 100 |
| rad52 | <0.004 | <0.02 |
| rad51 | <0.009 | <0.05 |
| rad59 | 7.8 ± 5.2 | 46 |
| rad51 rad59 | <0.008 | <0.05 |
| rad50 | 13 ± 11 | 76 |
| rad50 rad51 | 0.018 ± 0.03 | 0.1 |
| rad50 rad59 | 5.6 ± 3 | 33 |
| rad50 rad51 rad59 | <0.005 | <0.03 |
The frequency of BIR is the number of stable Ura+ transformants per microgram with cut DNA divided by the number of transformants per microgram with uncut DNA transformed and corrected for the number containing CFs.
Y′ recombination is reduced in rad51 mutants
The failure to recover CFs from CFV/D8B-Y′ could be due to an essential requirement for RAD51 in strand invasion of Y′ sequences. RAD51 is known to be required for the formation of survivors in telomerase-defective cells by promoting amplification of Y′ sequences (27, 56). To test this hypothesis, a CFV was constructed containing both Y′ sequences and TG1-3 sequences (Fig. 1B). Upon linearization with SnaBI and transformation of yeast, linear CFs are generated by de novo telomere addition to the TG1-3 repeats and strand invasion of the Y′ sequences at endogenous Y′ elements. The efficiency of BIR using the CFV/Y′-tg was very high, with almost the same number of transformants obtained from cut as with uncut DNA in the wild-type, rad50, and rad59 strains (Table 5). This high frequency is probably due to the increased number of donor sequences for recombination. The number of Ura+ transformants obtained from the linearized CFV was reduced about 200-fold in the rad52 strain and about 20-fold in the rad51 strains compared to that for transformation with uncut DNA. Because short linear chromosomes are much less stable than long CFs, mitotic stability could not be used to distinguish between BIR and end-joining events among the Ura+ transformants (12, 42). Instead, 14 to 17 Ura+ transformants from each of three independent transformations of each strain (45 to 50 total for each strain) were analyzed by Southern blotting. Genomic DNA was digested with EagI, which generates two fragments from CF-containing transformants, one of which is predicted to be approximately 2 kb and to be diffuse due to the heterogeneity of the telomere tract. Precise end joining is expected to generate a single fragment of 12 kb. More than 90% of the transformants analyzed from the wild-type, rad50, and rad59 strains contained the expected linear chromosome. As expected, the telomere-containing fragment from the rad50 transformants was shorter than observed in the wild type due to the defect in telomere maintenance conferred by the rad50 mutation (23). About 15% of the Ura+ transformants derived from rad51 strains contained end-joined plasmids, and the rest contained linear plasmids, indicative of Y′ BIR. Surprisingly, two of the transformants generated in the rad52 strain contained linear plasmids. Previous studies have also shown rare recombinational healing of chromosome ends after telomere loss in rad52 strains (35). Only strains containing rad52 or rad51 mutations showed a significant reduction in BIR compared to that of the wild-type strain (P < 0.01) and, as observed for CFV/D8B-tg, the rad50 rad51 strain showed higher BIR than the rad51 strain (P = 0.03). These results demonstrate Y′ BIR is reduced about 25-fold by the rad51 mutation and suggest that the failure to detect CFs in rad51 mutants using the D8B-Y′ CFV is due to a failure to complete two BIR events rather than a failure to initiate BIR from Y′ sequences.
TABLE 5.
Effect of the rad52 group mutations on Y′ BIRa
| Relevant genotype | BIR frequency (10−2) | Frequency relative to wild type |
|---|---|---|
| RAD | 63 ± 17 | 100 |
| rad52 | 0.015 ± 0.015 | 0.024 |
| rad51 | 2.7 ± 1.2 | 4.3 |
| rad59 | 70 ± 25 | 111 |
| rad51 rad59 | 2.1 ± 0.5 | 3.3 |
| rad50 | 93 ± 27 | 148 |
| rad50 rad51 | 5.4 ± 0.75 | 8.6 |
| rad50 rad59 | 158 ± 74 | 251 |
| rad50 rad51 rad59 | 2.6 ± 1.4 | 4.1 |
BIR frequencies were determined from the number of Ura+ transformants containing linear chromosome fragments (see Materials and Methods).
DISCUSSION
We used a chromosome fragmentation assay to measure the efficiency and genetic control of recombination-dependent replication (BIR) in yeast. Three important conclusions can be drawn from our analysis. First, BIR occurs with high efficiency in wild-type cells, even when two independent strand invasion-replication events are required to yield a stable chromosome fragment. Second, in contrast to results of previous studies, we showed that RAD51 is required for more than 97% of BIR events involving a single-end invasion and is essential when CF formation demands more than one BIR event. Third, and also in contrast to results of previous studies, RAD50 and RAD59 are not required for interchromosomal BIR in RAD51 or rad51 strains. These conclusions are discussed in more detail below.
Rad51 is required for single-ended strand invasion
Previous studies have shown that the major pathway to repair plasmid and chromosomal DSBs is RAD51-dependent gene conversion (6, 33). In the absence of RAD51, repair of an HO-endonuclease-induced DSB at the MAT locus in diploids can occur by strand invasion and duplication of sequences distal to the break site (33). This inefficient RAD51-independent repair pathway was found to require a cis-acting element, termed the BIR facilitator, located about 30 kb from the site of the DSB (34). In the absence of the BIR facilitator, induction of a DSB in the rad51 strains resulted primarily in chromosome loss. It is not clear if the BIR facilitator is only found on the right arm of chromosome III, or if similar sequences are dispersed throughout the genome. In another study, repair of an HO-induced DSB on chromosome VII was reduced more than 20-fold in rad51 and rad52 mutants, but some RAD51-independent ionizing-radiation induced BIR events were detected (15). Here we show a 33- to 140-fold decrease in BIR in rad51 mutants. The level of RAD51-independent recombination was about the same for all of the vectors used, with some variability in the efficiency of repair in wild-type strains. When formation of a CF required two independent strand invasion events, a severe defect was observed in the rad51 mutant, resulting in no detectable CFs. The rare stable Ura+ transformants generated were most likely due to conversion of the endogenous ura3-1 locus by the linear transforming DNA. Previous studies have shown only a fivefold reduction in gene targeting in rad51 strains relative to the wild type (44).
Studies in chicken DT-40 cells indicate an essential role for RAD51 during S-phase progression (47). Based on the results presented here, we suggest that RAD51 is required to repair chromosomal DSBs generated by replication fork collapse by a one-ended strand invasion process. Although repair of collapsed replication fork requires recombination between sister chromatids and the system described here measures strand invasion between homologous chromosomes, we believe that this represents a valid assay for one-ended strand invasion. The Escherichia coli in vitro assay system for replication fork restart relies on strand invasion between homologous plasmids, not sister chromatids, and uses the same factors known from genetic studies to be required for repair of collapsed replication forks (63). In eukaryotes with large genomes replication fork collapse is expected to occur multiple times, and thus complete genome replication would be dependent on RAD51. In organisms with smaller genomes, such as S. cerevisiae, replication fork collapse might be predicted to occur less than once per cell cycle, and therefore RAD51 would not be essential. However, RAD51 is essential for vegetative yeast growth when replication is perturbed (53, 58).
RAD50 and RAD59 are not required for interchromosomal BIR
RAD51-independent BIR of an HO-induced DSB at the MAT locus on chromosome III requires RAD50 and RAD59 and is dependent on the presence of a cis-acting element termed the BIR facilitator (34, 46). We found no significant role for RAD50 in either RAD51-dependent or RAD51-independent BIR by using the chromosome fragmentation assay. Instead, RAD51-independent BIR occurred at higher frequency in rad50 mutants. Previous studies have shown a reduced rate of processing DSBs in rad50 mutants, raising the possibility that the linear CFV is more stable in this strain (22). If the linear CFV persisted longer in rad50 strains, this would allow more time for rare RAD51-independent strand invasion to occur. The requirement for RAD50 in the BIR assay described by Signon et al. (46) could be an indirect effect of reduced resection of the HO-induced break. Because RAD51-independent BIR of the HO cut site at the chromosomal MAT locus requires strand invasion at a site 30 kb upstream of the HO cut site, it is possible that the delayed resection in rad50 mutants prevents invasion from occurring at the BIR facilitator. Alternatively, the BIR facilitator could represent a preferred sequence for RAD50-dependent strand invasion. Previous studies identified a RAD50- and RAD59-dependent pathway for telomere maintenance in the absence of telomerase and RAD51 (10). This pathway involves recombination between the telomere repeat sequences and could be considered a pathway of short repeat recombination. Consistent with this hypothesis, Ira et al. identified a RAD51-independent, RAD50-dependent pathway for intrachromosomal recombination between short repeats (18). Similarly, the requirement for RAD59 in RAD51-independent BIR mediated through the facilitator sequence and for telomere maintenance in rad51 tlc1 mutants could be explained by a requirement for this gene in short repeat recombination (10, 18, 49).
MRE11 and RAD50 are essential for mouse early embryonic development and for viability of vertebrate cell lines (30, 62). Studies using an MRE11 conditional allele suggest an essential role for MRE11 in S phase, and Mre11 has been found to colocalize with PCNA at replication forks in dividing cells (36, 64). Antibody depletion of Mre11 from Xenopus oocyte extracts results in incomplete replication of DNA added to the extract and accumulation of DSBs during DNA synthesis (11). These results suggest that Mre11 is required during replication to prevent replication fork collapse or for the repair of collapsed forks. Mre11 forms a stable complex with Rad50 and Xrs2 (NBS1 in vertebrates) (9, 60), and this complex is thought to function specifically in sister chromatid DSB repair but not in recombination between ectopic repeats or interchromosomal recombination (3, 7, 14, 16, 20). The lack of a requirement for RAD50 in the chromosome fragmentation assay is most likely because CF formation requires interchromosomal strand invasion rather than recombination between sister chromatids.
RAD52 is essential for BIR
Studies with S. cerevisiae indicate an essential role for RAD52 in most homologous recombination events (54). In some mitotic recombination assays, significant levels of RAD51-independent recombination are observed, and these all require RAD52. Rad52 catalyzes annealing between complementary single-stranded DNA and promotes Rad51-dependent strand invasion by targeting Rad51 to replication protein A-coated single-stranded DNA (41, 50, 52). In the absence of Rad51, it is assumed that Rad52 can promote some type of strand invasion process, and this occurs with greatest efficiency in the presence of Rad59 (4, 5). The Rad52-promoted strand invasion could possibly occur by annealing between a single-stranded region and transiently unwound donor duplex DNA.
CF formation is more efficient than gene targeting
We cannot eliminate the possibility that some of the chromosome fragments derived from CFVs are the result of reciprocal exchange between the end(s) of the linear CFV and chromosomal sequences. Because the efficiency of BIR using CFV/D8B-Y′ and CFV/D8B-tg is about 50-fold higher than that typically observed for gene targeting (44, 55), it seems unlikely that crossing over at the ends of the fragment is the primary mechanism for CF formation. To recover CFs in haploid strains, reciprocal exchange would have to occur in G2 and be followed by segregation of the CF and the intact sister to the same daughter cell and, thus, would be expected to occur at even lower efficiency than conventional gene targeting. Furthermore, a reciprocal exchange between the linear CFV and a sister chromatid would generate a broken chromosome in addition to the CF. The broken chromosome would then be expected to engage in another recombination event, repeating a cycle of futile crossing over. To eliminate the possibility of G2 crossovers, Morrow et al. showed that CFs could also be generated from a CFV containing two different sequences from the D8B region of chromosome III at the ends on the linear fragment, thus requiring two independent BIR events into the same chromosomal sequence (or one invasion of chromosomal sequences followed by intrachromosomal BIR) (40). Generation of stable Ura+ transformants from the Iso-CFV cannot occur by a simple G2 crossover mechanism and is also dependent on RAD51 (unpublished data).
In summary, we describe a simple genetic assay to determine the frequency of one-ended strand invasion followed by extensive replication in yeast. This process is efficient in wild-type cells and is dependent on the same genes that are required for DSB-induced gene conversion (RAD51, RAD52, RAD54, RAD55, and RAD57), suggesting a common strand invasion intermediate. The frequency of plasmid gap repair (a gene conversion process) and BIR following transformation with linear vectors is quite similar and is reduced by the same amount in the rad mutants, suggesting that one-ended strand invasion is the limiting step and is catalyzed by the same proteins (Table 2) (6). The key difference between gene conversion and BIR is that the tract of DNA synthesis accompanying gene conversion is short, whereas several hundred kilobase pairs can be duplicated during BIR. At this time we do not know which replication factors are required in these two systems, but the expectation is that BIR should require a more processive replication fork. Interchromosomal BIR is potentially detrimental because it could result in loss of heterozygosity or formation of nonreciprocal translocations if it were to occur between ectopic repeats. BIR does not normally occur when breaks present with two ends, suggesting that the second end regulates the extent of DNA synthesis or acts as a barrier to BIR (2).
Acknowledgments
We thank P. Hieter for providing pCF2 vectors and W. K. Holloman and members of the Symington laboratory for stimulating discussions and critical reading of the manuscript. We thank A. Lustig for advice on design and cloning of the oligonucleotides used for de novo telomere addition.
The research described in this article was supported by Public Health Service grant GM41784 from the National Institutes of Health.
REFERENCE
- 1.Adams, A., D. E. Gottschling, C. A. Kaiser, and T. Stearns. 1998. Methods in yeast genetics, a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Aguilera, A. 2001. Double-strand break repair: are Rad51/RecA-DNA joints barriers to DNA replication? Trends Genet. 17:318-321. [DOI] [PubMed] [Google Scholar]
- 3.Alani, E., R. Padmore, and N. Kleckner. 1990. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 61:419-436. [DOI] [PubMed] [Google Scholar]
- 4.Bai, Y., A. P. Davis, and L. S. Symington. 1999. A novel allele of RAD52 that causes severe DNA repair and recombination deficiencies only in the absence of RAD51 or RAD59. Genetics 153:1117-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai, Y., and L. S. Symington. 1996. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 10:2025-2037. [DOI] [PubMed] [Google Scholar]
- 6.Bartsch, S., L. E. Kang, and L. S. Symington. 2000. RAD51 is required for the repair of plasmid double-stranded DNA gaps from either plasmid or chromosomal templates. Mol. Cell. Biol. 20:1194-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bressan, D. A., B. K. Baxter, and J. H. Petrini. 1999. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7681-7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao, L., E. Alani, and N. Kleckner. 1990. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61:1089-1101. [DOI] [PubMed] [Google Scholar]
- 9.Carney, J. P., R. S. Maser, H. Olivares, E. M. Davis, M. Le Beau, J. R. Yates III, L. Hays, W. F. Morgan, and J. H. Petrini. 1998. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93:477-486. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Q., A. Ijpma, and C. W. Greider. 2001. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 21:1819-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costanzo, V., K. Robertson, M. Bibikova, E. Kim, D. Grieco, M. Gottesman, D. Carroll, and J. Gautier. 2001. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol. Cell. 8:137-147. [DOI] [PubMed] [Google Scholar]
- 12.Dani, G. M., and V. A. Zakian. 1983. Mitotic and meiotic stability of linear plasmids in yeast. Proc. Natl. Acad. Sci. USA 80:3406-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunn, B., P. Szauter, M. L. Pardue, and J. W. Szostak. 1984. Transfer of yeast telomeres to linear plasmids by recombination. Cell 39:191-201. [DOI] [PubMed] [Google Scholar]
- 14.Freedman, J. A., and S. Jinks-Robertson. 2002. Genetic requirements for spontaneous and transcription-stimulated mitotic recombination in Saccharomyces cerevisiae. Genetics 162:15-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galgoczy, D. J., and D. P. Toczyski. 2001. Checkpoint adaptation precedes spontaneous and damage-induced genomic instability in yeast. Mol. Cell. Biol. 21:1710-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez-Barrera, S., F. Cortes-Ledesma, R. E. Wellinger, and A. Aguilera. 2003. Equal sister chromatid exchange is a major mechanism of double-strand break repair in yeast. Mol. Cell. 11:1661-1671. [DOI] [PubMed] [Google Scholar]
- 17.Hieter, P., C. Mann, M. Snyder, and R. W. Davis. 1985. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell 40:381-392. [DOI] [PubMed] [Google Scholar]
- 18.Ira, G., and J. E. Haber. 2002. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol. Cell. Biol. 22:6384-6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivanov, E. L., V. G. Korolev, and F. Fabre. 1992. XRS2, a DNA repair gene of Saccharomyces cerevisiae, is needed for meiotic recombination. Genetics 132:651-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivanov, E. L., N. Sugawara, J. Fishman-Lobell, and J. E. Haber. 1996. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics 142:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanov, E. L., N. Sugawara, C. I. White, F. Fabre, and J. E. Haber. 1994. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:3414-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kironmai, K. M., and K. Muniyappa. 1997. Alteration of telomeric sequences and senescence caused by mutations in RAD50 of Saccharomyces cerevisiae. Genes Cells 2:443-455. [DOI] [PubMed] [Google Scholar]
- 24.Kowalczykowski, S. C. 2000. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25:156-165. [DOI] [PubMed] [Google Scholar]
- 25.Kraus, E., W. Y. Leung, and J. E. Haber. 2001. Break-induced replication: a review and an example in budding yeast. Proc. Natl. Acad. Sci. USA 98:8255-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kreuzer, K. N. 2000. Recombination-dependent DNA replication in phage T4. Trends Biochem. Sci. 25:165-173. [DOI] [PubMed] [Google Scholar]
- 27.Le, S., J. K. Moore, J. E. Haber, and C. W. Greider. 1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152:143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim, D. S., and P. Hasty. 1996. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol. 16:7133-7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lundblad, V., and E. H. Blackburn. 1993. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell 73:347-360. [DOI] [PubMed] [Google Scholar]
- 30.Luo, G., M. S. Yao, C. F. Bender, M. Mills, A. R. Bladl, A. Bradley, and J. H. Petrini. 1999. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc. Natl. Acad. Sci. USA 96:7376-7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lustig, A. J. 1992. Hoogsteen G-G base pairing is dispensable for telomere healing in yeast. Nucleic Acids Res. 20:3021-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lustig, A. J., S. Kurtz, and D. Shore. 1990. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science 250:549-553. [DOI] [PubMed] [Google Scholar]
- 33.Malkova, A., E. L. Ivanov, and J. E. Haber. 1996. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl. Acad. Sci. USA 93:7131-7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malkova, A., L. Signon, C. B. Schaefer, M. L. Naylor, J. F. Theis, C. S. Newlon, and J. E. Haber. 2001. RAD51-independent break-induced replication to repair a broken chromosome depends on a distant enhancer site. Genes Dev. 15:1055-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mangahas, J. L., M. K. Alexander, L. L. Sandell, and V. A. Zakian. 2001. Repair of chromosome ends after telomere loss in Saccharomyces. Mol. Biol. Cell. 12:4078-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maser, R. S., O. K. Mirzoeva, J. Wells, H. Olivares, B. R. Williams, R. A. Zinkel, P. J. Farnham, and J. H. Petrini. 2001. Mre11 complex and DNA replication: linkage to E2F and sites of DNA synthesis. Mol. Cell. Biol. 21:6006-6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michel, B. 2000. Replication fork arrest and DNA recombination. Trends Biochem. Sci. 25:173-178. [DOI] [PubMed] [Google Scholar]
- 38.Michel, B., S. D. Ehrlich, and M. Uzest. 1997. DNA double-strand breaks caused by replication arrest. EMBO J. 16:430-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morgan, E. A., N. Shah, and L. S. Symington. 2002. The requirement for ATP hydrolysis by Saccharomyces cerevisiae Rad51 is bypassed by mating-type heterozygosity or RAD54 in high copy numbers. Mol. Cell. Biol. 22:6336-6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morrow, D. M., C. Connelly, and P. Hieter. 1997. “Break copy” duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics 147:371-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mortensen, U. H., C. Bendixen, I. Sunjevaric, and R. Rothstein. 1996. DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl. Acad. Sci. USA 93:10729-10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murray, A. W., and J. W. Szostak. 1983. Construction of artificial chromosomes in yeast. Nature 305:189-193. [DOI] [PubMed] [Google Scholar]
- 43.Roth, D. B., P. B. Nakajima, J. P. Menetski, M. J. Bosma, and M. Gellert. 1992. V(D)J recombination in mouse thymocytes: double-strand breaks near T cell receptor delta rearrangement signals. Cell 69:41-53. [DOI] [PubMed] [Google Scholar]
- 44.Schiestl, R. H., J. Zhu, and T. D. Petes. 1994. Effect of mutations in genes affecting homologous recombination on restriction enzyme-mediated and illegitimate recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:4493-4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44a.Schwartz, D. C., and C. R. Cantor. 1984. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell 37:67-75. [DOI] [PubMed] [Google Scholar]
- 45.Seigneur, M., V. Bidnenko, S. D. Ehrlich, and B. Michel. 1998. RuvAB acts at arrested replication forks. Cell 95:419-430. [DOI] [PubMed] [Google Scholar]
- 46.Signon, L., A. Malkova, M. L. Naylor, H. Klein, and J. E. Haber. 2001. Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol. Cell. Biol. 21:2048-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sonoda, E., M. S. Sasaki, J. M. Buerstedde, O. Bezzubova, A. Shinohara, H. Ogawa, M. Takata, Y. Yamaguchi-Iwai, and S. Takeda. 1998. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 17:598-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strathern, J. N., A. J. Klar, J. B. Hicks, J. A. Abraham, J. M. Ivy, K. A. Nasmyth, and C. McGill. 1982. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell 31:183-192. [DOI] [PubMed] [Google Scholar]
- 49.Sugawara, N., G. Ira, and J. E. Haber. 2000. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell. Biol. 20:5300-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sugiyama, T., and S. C. Kowalczykowski. 2002. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem. 277:31663-31672. [DOI] [PubMed] [Google Scholar]
- 51.Sun, H., D. Treco, N. P. Schultes, and J. W. Szostak. 1989. Double-strand breaks at an initiation site for meiotic gene conversion. Nature 338:87-90. [DOI] [PubMed] [Google Scholar]
- 52.Sung, P. 1997. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 272:28194-28197. [DOI] [PubMed] [Google Scholar]
- 53.Symington, L. S. 1998. Homologous recombination is required for the viability of rad27 mutants. Nucleic Acids Res. 26:5589-5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Symington, L. S. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66:630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Symington, L. S., L. E. Kang, and S. Moreau. 2000. Alteration of gene conversion tract length and associated crossing over during plasmid gap repair in nuclease-deficient strains of Saccharomyces cerevisiae. Nucleic Acids Res. 28:4649-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teng, S. C., J. Chang, B. McCowan, and V. A. Zakian. 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell. 6:947-952. [DOI] [PubMed] [Google Scholar]
- 57.Thomas, B. J., and R. Rothstein. 1989. The genetic control of direct-repeat recombination in Saccharomyces: the effect of rad52 and rad1 on mitotic recombination at GAL10, a transcriptionally regulated gene. Genetics 123:725-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tishkoff, D. X., N. Filosi, G. M. Gaida, and R. D. Kolodner. 1997. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell 88:253-263. [DOI] [PubMed] [Google Scholar]
- 59.Tsuzuki, T., Y. Fujii, K. Sakumi, Y. Tominaga, K. Nakao, M. Sekiguchi, A. Matsushiro, Y. Yoshimura, and T. Morita. 1996. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl. Acad. Sci. USA 93:6236-6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Usui, T., T. Ohta, H. Oshiumi, J. Tomizawa, H. Ogawa, and T. Ogawa. 1998. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95:705-716. [DOI] [PubMed] [Google Scholar]
- 61.Vollrath, D., R. W. Davis, C. Connelly, and P. Hieter. 1988. Physical mapping of large DNA by chromosome fragmentation. Proc. Natl. Acad. Sci. USA 85:6027-6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiao, Y., and D. T. Weaver. 1997. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 25:2985-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu, L., and K. J. Marians. 2003. PriA mediates DNA replication pathway choice at recombination intermediates. Mol. Cell. 11:817-826. [DOI] [PubMed] [Google Scholar]
- 64.Yamaguchi-Iwai, Y., E. Sonoda, M. S. Sasaki, C. Morrison, T. Haraguchi, Y. Hiraoka, Y. M. Yamashita, T. Yagi, M. Takata, C. Price, N. Kakazu, and S. Takeda. 1999. Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J. 18:6619-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]