Abstract
Single-molecule fluorescence spectroscopy offers real-time, nanometer-resolution information. Over the past two decades, this emerging single-molecule technique has been rapidly adopted to investigate the structural dynamics and biological functions of proteins. Despite this remarkable achievement, single-molecule fluorescence techniques must be extended to macromolecular protein complexes that are physiologically more relevant for functional studies. In this review, we present recent major breakthroughs for investigating protein complexes within cell extracts using single-molecule fluorescence. We outline the challenges, future prospects and potential applications of these new single-molecule fluorescence techniques in biological and clinical research.
Keywords: single-molecule fluorescence, macromolecular complex, cell lysate, single-molecule immunoprecipitation, single-molecule pull-down, protein
Single-molecule protein studies
Single-molecule techniques have become potent tools for the discovery of novel protein mechanisms by allowing high spatiotemporal resolution. Sub-nanometer resolution, the ultimate scale of biological systems, has been reached with single-molecule fluorescence microscopy[1] and spectroscopy[2]; single-molecule force and torque spectroscopy[3, 4]; atomic force microscopy[5] and spectroscopy[6]; and nanopores[7]. Measurements can be carried out on the biologically relevant time scales of microseconds to minutes.
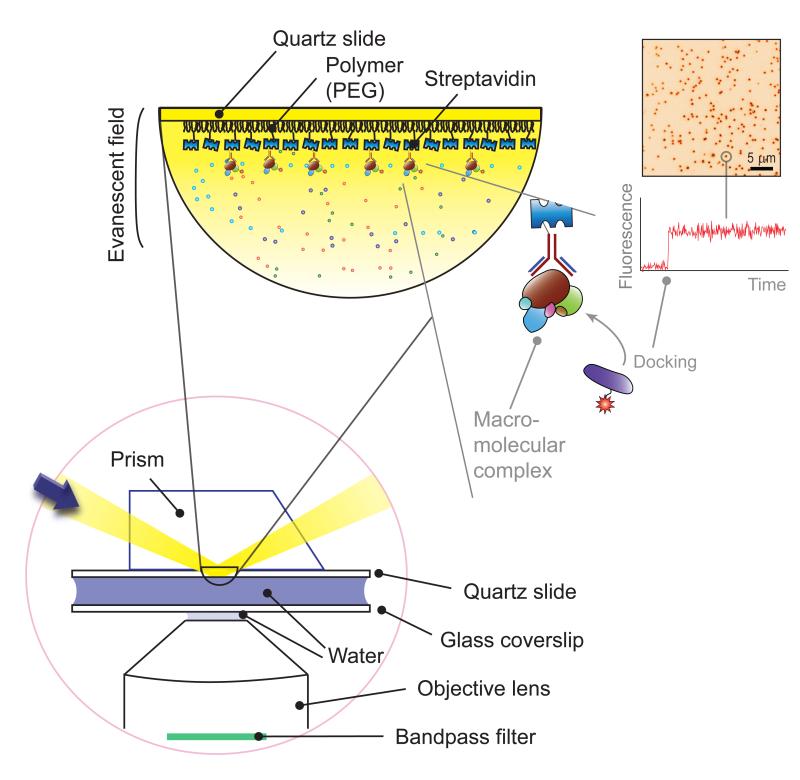
Among these techniques, single-molecule fluorescence spectroscopy has enabled researchers to unveil the action mechanism of a protein by imaging protein activity in real time. Benefitting from advances in general microscopy[1], detection devices[8], and fluorophore physics[9, 10], single-molecule fluorescence spectroscopy has reached sub-nanometer spatial resolution[11] and microsecond temporal resolution[12, 13]. Single-molecule fluorescence imaging is carried out primarily with total internal reflection, confocal, and zero-mode waveguide microscopy[2]. Among these, total internal reflection fluorescence microscopy has been employed by several research teams in the development of novel techniques described in this review[14-17]. With total internal reflection microscopy (Figure 1), a localized fluorescence spot on a CCD (charge-coupled device) screen represents the interaction of a fluorescent molecule with a surface-immobilized molecule. The time of this docking event and the ensuing time trajectory deliver valuable kinetic information on biochemical processes such as protein complex assemblies, protein-protein interactions, protein-nucleic acid interactions, and protein-lipid interactions.
Figure 1. Total internal reflection microscopy.
(Bottom left) In total internal reflection microscopy, a sheer layer of ~100 nm on a glass surface is excited via total internal reflection at the interface of water and glass (here, quartz). Background signals from the solution are thereby effectively minimized, which is essential for harvesting a finite number of photons from single fluorophores. As the excitation is confined to the glass surface, the molecules of interest are immobilized on a surface for long-term observation. (Top left) To immobilize molecules of interest, biotin-streptavidin-biotin conjugation is used. Here, streptavidin is bound to a biotinylated polymer (PEG, poly-ethylene glycol) surface, the streptavidin proteins bind to biotinylated antibodies, and the macromolecular protein complexes are bound to the antibodies. (Top right) In this immobilization scheme, the docking of a fluorescent partner molecule leads to a sudden increase in fluorescence signals over a localized spot, as shown in a representative CCD image and fluorescence time trace. Adapted from Yeom et al. [16] with permission.
Despite the success of single-molecule fluorescence techniques over the past two decades, technical hurdles remain for the techniques to be applied more widely in biological disciplines. With single-molecule fluorescence spectroscopy, one can observe single isolated recombinant proteins interacting with their sole partner molecules; however, virtually no cellular processes occur through this type of idealized interaction. Rather, cellular processes result from proteins constantly interacting with each other and forming large protein complexes[18]. Although this shortcoming calls for more physiologically relevant single-molecule studies, there is little knowledge of how to deal with inherently dynamic macromolecular complexes at the single-molecule level.
Several research groups have recently made progress in creating more physiologically relevant single-molecule studies by introducing two traditional biochemical methods, cell extract isolation and immunoprecipitation (IP), to single-molecule fluorescence spectroscopy (Table 1). Using cell extracts, Gelles and colleagues visualized spliceosome assembly[14, 19], and van Oijen and colleagues reconstituted a eukaryotic replisome on a single-molecule surface[15, 20]. Using protein immunoprecipitates, Joo and colleagues have studied the regulation process of microRNA biogenesis [16], and Ha and colleagues have demonstrated single-molecule complex analysis[17, 21]. These new approaches have a key advantage over conventional in vitro single-molecule techniques: the proteins continuously interact with other cellular proteins, spontaneously form a macromolecular complex, and dissociate during observation. We anticipate that these new approaches will pave the way for single-molecule studies of intrinsically complex protein systems. Here, we review the novel technical achievements made in these recent single-molecule fluorescence studies.
Table 1.
Single-Molecule Protein Complex Studies
| Labeling-Based Selective Observation | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| Interactions | Systems | Cell extracts | Labeled objects | Labeling by | Substrates | Refs |
| Spliceosome | Yeast | - | - | RNA | 26, 27 | |
| Protein- | Spliceosome | Yeast | NTC, snRNAs (U1, U2, U5) | DHFR-, SNAP-mediated | RNA | 14 |
| Nucleic Acid | Replisome | Xenopus egg § | DNA duplication products | Fluorescent DIG antibody | lambda DNA | 15, 42 |
| Replisome | Xenopus egg § | DNA clamp (PCNA) | PCNA antibody ++ | lambda DNA | 20 | |
| Replisome | Xenopus egg § | Fen1 | mKikGR | lambda DNA | 45 | |
| DNA polymerase * | E. coli | Polymerases (PolBI, DinB) | FGE-mediated | DNA | 35 | |
| Immobilization-Based Selective Observation | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| Interactions | Systems | Cell extracts | Immobilized objects | Immobilization by | Substrates/Partners | Refs |
| Protein- | Poly(U) polymerase complex | Human §§ | FLAG-mCherry-tagged TUT4 | RFP antibody + | RNA | 16 |
| Nucleic Acid | Helicase * | E. coli | His-tagged PcrA | His antibody + | DNA | 17 |
|
| ||||||
| Replisome | Human | T7-tagged ORCA | T7 antibody + | YFP-Orc1, Cdt1, Geminin | 49 | |
| Protein kinase | Human | FLAG-mCherry-tagged PKA | FLAG antibody + | YFP-PKA | 17 | |
| Protein kinase holoenzyme | Human | PKA | PKA antibody ++ | AKAP | 17 | |
| Protein- | Protein kinase holoenzyme | Human | FLAG-tagged SKIP | FLAG antibody + | YFP-PKA | 48 |
| Protein | Peptide-loading complex | Human | TAP1 | TAP1 antibody+ | TAP2, YFP-Tapasin | 50 |
| Membrane protein complex | Human | YFP-tagged β2AR | YFP antibody + | - | 17 | |
| Mitochondrial protein | Human | YFP-tagged MAVS | YFP antibody + | - | 17 | |
| mTOR signaling complex | Human | FLAG-tagged mTOR | FLAG antibody + | HA-Raptor +++ | 17 | |
The system of interest was not a protein complex.
fractionated
immunoprecipitated
biotinylated
immobilized via biotinylated secondary antibody
probed with fluorescent secondary antibody
AKAP: A-kinase anchoring protein
β2AR: β2-adrenergic receptor
DHFR: dihydrofolate reductase
DIG: Digoxigenin
Fen1: flap endonuclease 1
FGE: formylglycine-generating enzyme
NTC: Prp19-complex
PCNA: proliferating cell nuclear antigen
RFP: red fluorescent protein
SKIP: sphingosine kinase interacting protein
SNAP: a variant of O6-alkylguanine-DNA-alkyltransferase
snRNA: small nuclear RNA
TAP: transporter associated with antigen processing
TUT4: terminal uridylyl transferase 4
YFP: yellow fluorescent protein
Single-molecule observations using cell extracts
Unlike purified recombinant proteins, crude cell extracts provide more biologically relevant environment in which molecules of interest can interact not only with their partners but also with other cellular components. It was anticipated to combine this approach with single molecule techniques and observe the assembly and function of macromolecular complexes in real time. However, technical hurdles related to the specificity and efficiency of labeling molecules of interest within crude cell extracts had been reported[22-24]. In order to overcome this limitation, several groups recently developed original approaches. Hoskins et al. used protein complexes specifically tagged with fluorophores[14, 19], and Yardimci et al. immunostained reaction products using specific fluorescent antibodies[15, 20].
Real-time observation of spliceosome assembly
Despite the relatively small number of genes in the human genome, we have extraordinary transcriptomic and proteomic diversity due to alternative splicing via the spliceosome. This highly complex and dynamic macromolecular machine is involved in intron excision from a nascent eukaryotic transcript. The spliceosome is a mega-Dalton complex consisting of five small nuclear RNAs and more than 100 cofactor proteins[25]. This large number of components makes reconstituting the spliceosome with recombinant proteins unfeasible. Due to this limitation, the use of cell extracts represents a promising alternative that has been successfully adopted to perform in vitro splicing of labeled pre-mRNA at the single-molecule level[26, 27]. Although it was anticipated that the kinetics of the assembly and action of the spliceosome would be discovered soon after the development and use of single-molecule observations, the dye labeling of spliceosome subunits has only recently been accomplished[14] due to the difficulty of labeling within heterogeneous cell extracts.
A variety of chemical strategies are used for protein labeling[22-24]. These approaches can be reliably practiced with purified proteins, but many of these schemes suffer from an undesirably low quality of conjugation when they are performed intracellularly or within crude cell extracts. Two exceptions are enzyme-mediated and protein-directed conjugation technologies (Box 1), which deliver remarkable specificity. These technologies are commercially available; therefore, they are also easily accessible. Hoskins et al. adopted protein-directed conjugation methods based on the DHFR (dihydrofolate reductase) protein and the SNAP tag (a variant of O6-alkylguanine-DNA-alkyltransferase). The DHFR protein forms a non-covalent, tight complex with a trimethoprim-tagged fluorophore. The SNAP tag forms a covalent complex with a benzylguanine-tagged fluorophore. As the DHFR-trimethoprim and SNAP-benzylguanine formations are highly selective, fluorophore labeling can be reliably practiced within crude cell extracts.
Box 1. Labeling proteins with organic fluorophores within cell extracts.
In general, organic fluorophores are favored over fluorescent proteins in single-molecule measurements because their superior photostability (i.e., a large number of total photons emitted) and minimal photoblinking (i.e., a uniform number of photons emitted per time bin) enable reliable real-time observation over a biologically relevant time scale (milliseconds to minutes). In addition, organic fluorophores are so small that their perturbation of the 3-dimensional structure and functionality of a protein is minimal.
Labeling proteins within cell extracts with organic fluorophores involves the practical issues of achieving high selectivity and high conjugation efficiency within a heterogeneous environment. Two types of technology stand out for their high performance. The first is a series of ‘protein-directed’ labeling techniques including DHFR[28], SNAP-tag[29], CLIP-tag[29], and Halo-tag[30]-based conjugations. The conjugations between these proteins and their substrates exhibit high selectivity and high affinity, even within cells and cell extracts[24]. The second technology is a series of ‘enzyme-mediated’ labeling strategies such as biotin ligase-mediated[31], phosphopantetheine transferase-mediated[32], lipoic acid ligase-mediated[33], and formylglycine-generating enzyme-mediated[34, 35] approaches. These techniques involve the recognition of specific amino-acid motifs of cellular proteins that are then modified with remarkably high specificity. Detailed descriptions of the protein-directed and protein-mediated labeling strategies can be found in other reviews[22-24].
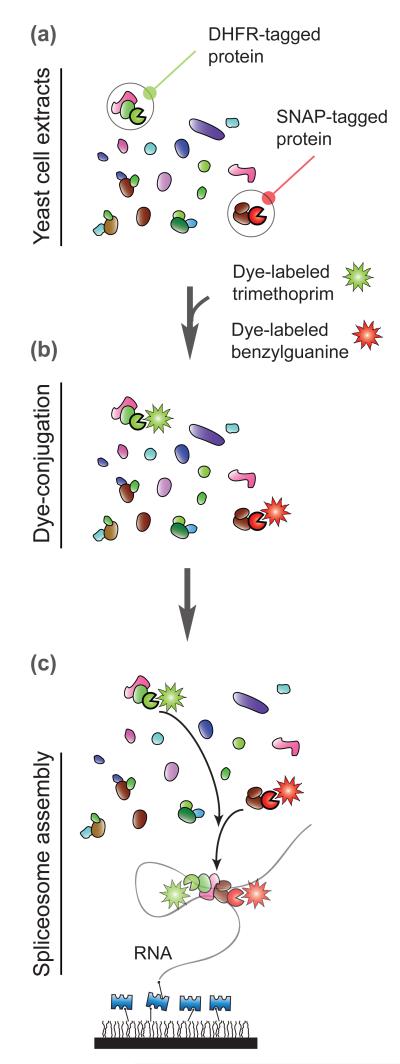
Using these methods, Hoskins et al. labeled small nuclear ribonucleoprotein particles and the multiprotein Prp19-complex with fluorophores in different colors (Figure 2). Equipped with multicolor single-molecule fluorescence spectroscopy, they determined the order of spliceosome assembly. Surprisingly, they found that spliceosome assembly occurs efficiently without any kinetic barriers and that the assembly process is dynamic and reversible at any step. In the future, concurrent observation of this assembly process and splicing activity should yield further insight. Further experimental details can be found in a recent review article[25].
Figure 2. Spliceosome assembly.
(a) Hoskins et al. prepared yeast whole extracts containing DHFR (dihydrofolate reductase) and SNAP (a variant of O6-alkylguanine-DNA-alkyltransferase tagged spliceosome components[14]. The incomplete circles in green and red represents the DHFR and SNAP tag, respectively. (b) The DHFR- and SNAP-tagged proteins were mixed with dye-labeled trimethoprim and benzylguanine within cell extracts. (c) These cell extracts with dye-conjugated proteins were introduced to a surface where RNA splicing substrates were immobilized. A glass surface is coated with polymer and layered with Streptavidin as shown in Figure 1.
Immunostaining of a eukaryotic replisome system
When single-molecule techniques were first applied to biological systems, bacterial and viral DNA polymerases were favored as model systems because their replication assays are relatively simple. Whereas these systems have been progressively studied[36], eukaryotic replication systems have received less attention at the molecular level due to the complexity of the many cofactors involved (Box 2). For the first single-molecule observation of a eukaryotic replisome, Yardimci et al.[15] employed Xenopus egg extracts that were naturally enriched with replication protein complexes (Figure 3) [37].
Box 2. Technical challenges in performing a physiologically relevant single-molecule measurement.
Challenges arising from low concentrations
A single-molecule assay performed with subnanomolar or lower concentrations might fail to reproduce observations made with bulk assays[43]. For example, a biochemical study completed prior to the work of Yardimci et al. [15] showed that eukaryotic DNA duplication occurred only in the presence of a high concentration of DNA, as it was unexpectedly discovered that DNA is a cofactor of its own duplication[44]. Therefore, it was essential for Yardimci et al. to perform their single-molecule experiment with extra DNA strands included in the observation chamber so that surface-immobilized DNA molecules that were separated by distances greater than their contour lengths (16 μm) would be still triggered for replication. Care should be taken when designing any other single-molecule experiments that involve cofactors that are weakly associated with a macromolecular complex.
Challenges arising from high concentrations
A large fraction of cellular reactions occurs through weak molecular interactions. While biochemical studies of such transient interactions require a protein concentration as high as micromolar, it is known that a high concentration of fluorescent molecules in solution leads to severe background signals hampering single-molecule detection. Loveland et al developed a novel technique that overcomes this barrier utilizing photoactivable fluorescent proteins[45]. They tagged a protein of interest, Fen1 (flap endonuclease 1), with mKikGR photoactivable fluorescent protein. They supplemented a micromolar concentration of this tagged protein to Xenopus cell extracts and probed the activity of Fen1 during DNA replication. The high concentration of Fen1 would have led to severe background signals if all the mKikKR had been fluorescent. But, since they selectively photoactivated Fen1-mKikKR within a thin volume near a surface using total internal reflection excitation, they could image DNA-bound Fen1-mKikKR with single-molecule sensitivity.
Challenges arising from ectopic expression
To investigate a biological system under physiologically relevant conditions, the gene of interest must be weakly expressed without perturbing the cellular system. This provides protein stability and minimizes any undesirable effects such as misfolding, denaturation, aggregation, degradation, and improper stoichiometry between an expressed protein and its endogenous partner proteins. Yeom et al have shown that weak expression is crucial to obtaining a sufficient number of functional protein complexes[16] and have pursued a method of expressing the gene of interest ectopically at a concentration comparable to that of endogenous proteins. Hoskins et al. adopted another approach for expressing their protein of interest by modifying the yeast genome using homologous recombination[14].
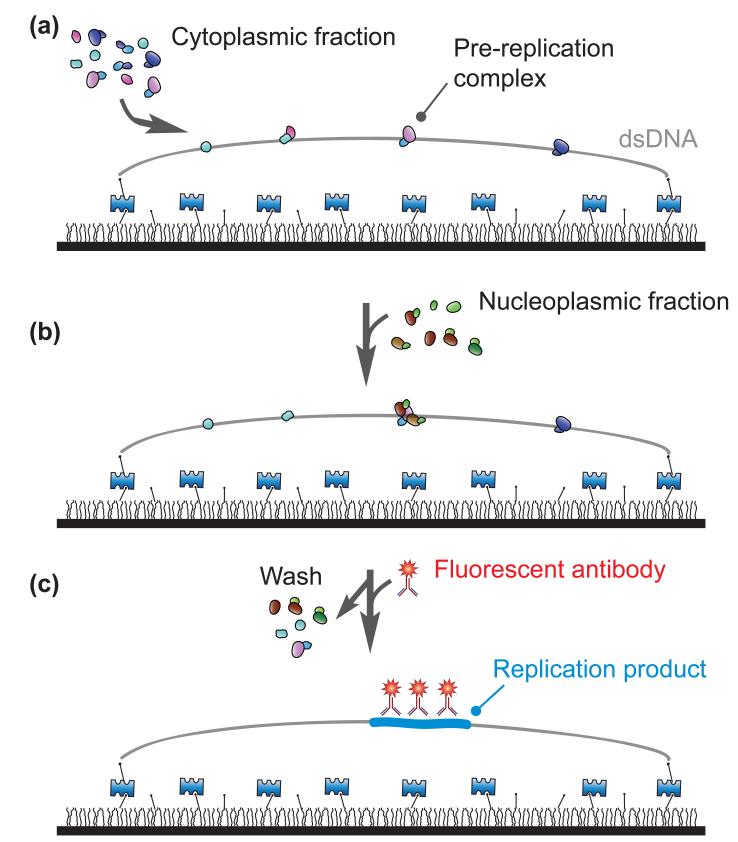
Figure 3. Eukaryotic DNA replication.
(a) Yardimci et al. immobilized bacteriophage λ DNA molecules (double-stranded DNA) on a surface[15]. To emulate the initiation of DNA replication at the G1 phase, they incubated the tethered DNA strands with the cytoplasmic fractions of egg extracts. (b) Next, they replaced the environment of the pre-replication complex with the nucleoplasmic fractions to emulate the G1-S transition. (c) Then, to visualize the distribution of the nascent duplication products, which was tagged with Digoxigenin, they washed their samples to remove cell extracts and introduced fluorescent antibodies against Digoxigenin. A glass surface is coated with polymer and layered with Streptavidin as shown in Figure 1.
The researchers first immobilized long double-stranded DNA molecules on a glass surface in an end-specific manner. This immobilization scheme allowed them to change the local environment around the DNA strands, and the researchers then triggered replication by introducing Xenopus egg extracts into an observation chamber. Included in the cell extracts were chemically modified nucleotides that were incorporated during DNA replication. The replication products were visualized when fluorescent antibodies that were specific to the artificial chemical groups (Digoxigenin) were introduced into the chamber. Despite the specificity limitations of any antibody, this step-by-step procedure effectively prevented non-specific tagging of cell extract components. A similar approach has been used to immunostain replication proteins[20]. Further experimental details can be found elsewhere[20].
The duplication of a eukaryotic genome involves DNA replication from multiple origins. At each origin, DNA synthesis progresses in a bidirectional manner coordinated by two sister replisomes[38]. The spatial and temporal relationships between eukaryotic sister replisomes remain unclear, and experimental data are controversial. Using the single-molecule assay described above, Yardimci et al. examined whether eukaryotic sister replisomes function independently or as a dimeric complex at a replication bubble[38, 39]. Specifically, using a long double-stranded DNA molecule stretched by laminar flow[40, 41] (Figure 3), they revealed that the conformation of a DNA strand does not influence the activity of a replication complex and concluded that eukaryotic systems do not use a coupled mechanism in vitro. The same group utilized this single-molecule replisome assay to investigate the DNA helicase (Cdc45, MCM2-7, and GINS) of the replisome and revealed that this helicase translocates along a single-stranded DNA with a 3′ to 5′ directionality [42].
In summary, by introducing the cell extract isolation method to the single-molecule fluorescence spectroscopy, the two teams have answered the need for single-molecule techniques that use more biologically relevant conditions. This breakthrough has been achieved by adopting recently developed dye-conjugation methods and designing a single-molecule immunostaining assay. Several caveats should be carefully considered in investigating other protein complexes (see Boxes 1 and 2).
Single-molecule observations using immunoprecipitates
In a cellular environment, nearly all proteins interact with other cellular factors to perform biological functions. Understanding the spatiotemporal relationships between a protein and its partners within a macromolecular complex is crucial to elucidating the function of the protein. Co-IP coupled with Western blotting or mass spectroscopy is conventionally used to determine interactions between proteins within macromolecular complexes. However, these approaches are static and cannot provide reliable stoichiometric or dynamic information. To deal with this limitation, two teams have developed a promising approach based on single-molecule IP[16, 17]. In this approach, streptavidin is layered via biotin-streptavidin binding on a polymer-coated glass surface, and biotinylated antibodies are conjugated to the streptavidin layer (Figure 4a). When cell extracts are introduced onto this surface, target proteins are immobilized, which completes the single-molecule IP. When protein substrates or partner proteins with fluorescent tags are flowed into an observation chamber, fluorescent signals are detected from the surface via total internal reflection microscopy (Figure 1), and these signals report on interactions between the fluorescent molecules and the immobilized proteins.
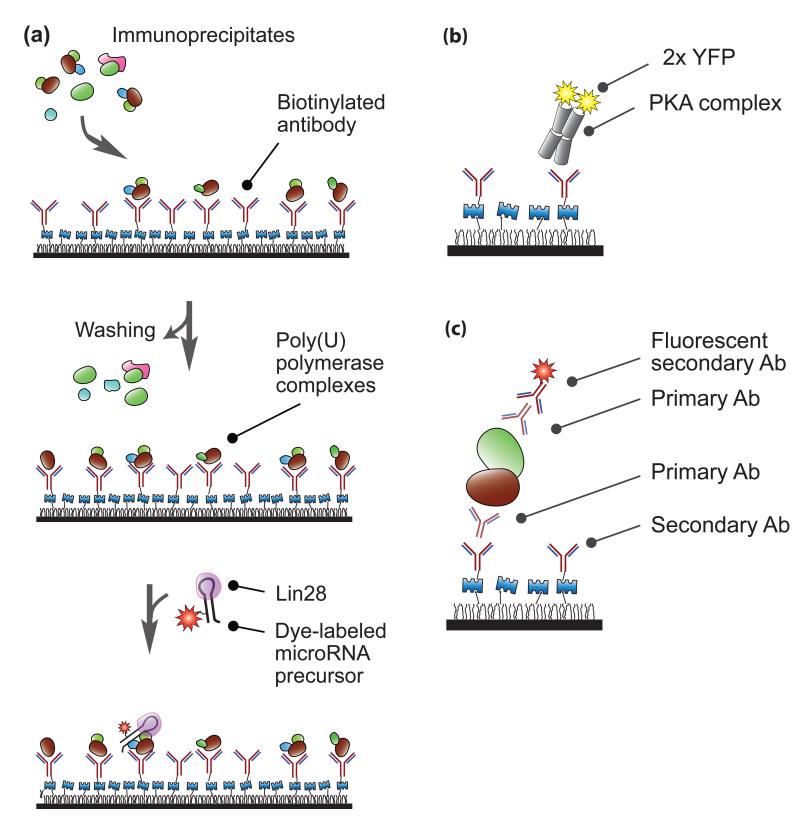
Figure 4. Single-molecule immunoprecipitation.
(a) MicroRNA modification. Using surface-bound antibodies, Yeom et al. immobilized poly(U) polymerase immunoprecipitates obtained from human cell extracts[16]. After washing to remove unbound immunoprecipitates, they introduced fluorescently tagged Lin28-bound precursor microRNA. By recording fluorescent signals from the labeled RNA substrates, they visualized interactions between the RNA strands and the protein complexes in real time. (b-c) Single-molecule complex analysis. (b) Jain et al. pulled down a protein kinase A complex using surface-immobilized antibodies[17]. The stoichiometry of the complex was analyzed based on the number of YFP (yellow fluorescent protein) photobleaching steps. (c) Endogenous protein complexes were pulled down and visualized via the combination of primary antibodies and secondary antibodies. “Ab” stands for antibody. A quartz slide is coated with polymer and layered with Streptavidin as shown in Figure 1.
Single-molecule enzymology with immunoprecipitated complexes
In IP, cellular proteins are naturally pulled down with their cofactors. Therefore, single-molecule IP provides an ideal platform for studying macromolecular protein complexes at the molecular level. The system investigated by Yeom et al. was a human poly(U) polymerase complex, which is involved in the regulation of microRNA biogenesis. As its cellular function has only recently been identified[46, 47], this protein complex has not yet been well characterized and thus it was not feasible to reconstitute the protein complex by assembling its individual components. Therefore, Yeom et al. carried out single-molecule IP using human cell extracts and reconstituted this protein complex on a single-molecule surface (Figure 4a). They then introduced a microRNA substrate labeled with a dye and bound by its cofactor (Lin28) onto this surface. The uridylation of the microRNA substrate was visualized in real time[16]. By recording weak interactions between the polymerase complex and the RNA substrate, which could not have been detected from a bulk measurement, they discovered that the cofactor protein Lin28 is a processivity factor that enhances the binding affinity between the poly(U) polymerase and its microRNA substrate.
The development of the single-molecule IP technique has revealed that the reconstitution of functional protein complexes on a single-molecule surface requires careful consideration at each step of immunoprecipitate preparation. Attention must be paid to the possibility of antibody cross-reactivity toward unwanted protein species, even if the antibody is of high quality. This non-specific binding of unwanted protein species to an antibody can be reduced by harsh washing during single-molecule IP as demonstrated by Jain et al in pulling down PcrA helicase[17]. However, harsh conditions will induce the dissociation of protein cofactors from protein complexes, making single-molecule IP unsuitable for protein complex studies. Alternatively, problems with antibody specificity can be overcome by preparing immunoprecipitates at higher purities using a tandem purification scheme in which two rounds of immunoprecipitation are carried out using two orthogonal antibodies[16] or by labeling proteins of interest with fluorophores within cell extracts[14, 35] (Box 1). Another important consideration is whether overexpressing proteins might affect their function (Box 2).
Single-molecule complex analysis
Single-molecule IP is a novel tool used to analyze the composition of single protein complexes through a form of Western blotting. In a recent work by Jain et al.[17, 21], proof-of-concept of single-molecule complex analysis was achieved with various fluorescent proteins and antibody probes (Figure 4b).
Jain et al. first demonstrated single-molecule co-IP using a tetrameric complex of protein kinase A (Figure 4b). This complex consists of two regulatory and two catalytic subunits. The researchers tagged the catalytic subunits with YFP (yellow fluorescent protein). When they immunoprecipitated the regulatory subunits on the imaging surface, they could detect YFP from co-immunoprecipitated catalytic subunits. The stoichiometry of the catalytic subunits could be confirmed by counting the number of YFP molecules per complex. The researchers have also reported proof-of-principle studies using systems such as mTOR (mammalian target of rapamycin) protein complexes and membrane receptor proteins[17]. The same group has extended its research to stoichiometry analyses of AKAPs (A-kinase anchoring proteins)[48], ORCA (origin-recognition complex-associated) protein complexes[49], and peptide loading complexes[50].
Jain et al. also demonstrated the analysis of endogenous proteins obtained from mouse brain and heart tissues (Figure 4c). Although it is often demanding to analyze endogenous proteins due to their low abundance, this difficulty can be overcome by the high sensitivity of single-molecule complex analysis. This cutting-edge approach might lead to clinical applications. For example, establishing the precise diagnosis of a patient requires a relatively large amount of human tissue, but this is often not feasible for certain diseases such as neurological disorders, in which the amount of tissue that can be removed by a clinician is limited. Single-molecule complex analysis can overcome this challenge by allowing the analysis of biological samples of limited abundance and obtaining quantitative information.
In summary, the two teams have developed new IP techniques to reconstitute macromolecular complexes at the molecular level. They have used these techniques to determine the function and stoichiometry of a protein complex. It is anticipated that these new approaches will become a universal tool to provide reliable stoichimetric and dynamic information about the spatiotemporal relationships between a protein and its partners within a macromolecular complex.
Concluding remarks
New single-molecule approaches[14-17] will enable biologists to explore protein complexes at the nanoscale and will provide opportunities for tackling challenges that require more physiologically relevant systems than purified proteins.
Among a wide range of potential applications, these approaches are particularly useful for intrinsically heterogeneous systems where multiple proteins form complexes with varying stoichiometries. In the case of a protein involved in multiple functionally distinct complexes, we will be able to examine the function of each complex on a single-molecule surface without segregating individual complexes. An interesting example would be mTOR, a central regulator of cell signaling that has two functionally distinct complexes, mTORC1 and mTORC2[51]. The new approaches will also be valuable in investigating posttranslational modifications. These subjects have not been explored in conventional single-molecule studies because of their dynamic temporal and spatial profiles within the cell.
The new techniques discussed herein can be applied to studies of other protein activities such as protein-lipid[52-54] and receptor-ligand interactions. It will be particularly useful when observing transient interactions which are often difficult to detect using conventional biochemical tools. These techniques can be further expanded to study protein complexes from cellular organelles such as lysosomes, endosomes, exocytic vesicles, and mitochondria, as well as studies of the organelles themselves. Moreover, proteins, protein complexes, and vesicles secreted into biological fluids[55, 56] can be the subjects of future research.
Single-molecule macromolecular complex approaches will also become an innovative platform for analyzing complex compositions. Using a fluorescent antibody that targets a component of a complex, we will be able to measure the stoichiometry of a protein complex. These approaches also have the potential to identify unknown cofactors of a protein complex. When combined with single-molecule ELISA (enzyme-linked immunosorbent assay)[57], single-molecule sequencing technologies[58-60], single-molecule multi-color FRET (fluorescence resonance energy transfer)[61-63], and screening tools, the new techniques discussed in this review will acquire versatility and be used for practical analysis in the near future, complementing mass spectroscopy techniques.
These new techniques might also be useful in clinical applications, especially in the discovery of new drugs. Although high-throughput screening using libraries of compounds is a proven way to identify novel entities[64], these highly artificial and non-physiological assay systems have low success rates. Moreover, this process is time-consuming and sample-intensive. With their high sensitivity, single-molecule techniques offer an effective alternative as they require only a small amount of sample and could reduce development time. As most eukaryotic proteins form macromolecular complexes, single-molecule macromolecular complex techniques could revolutionize the field of screening by providing biologically relevant structural and functional data that will allow for the design and discovery of new drugs to inhibit the assembly and function of macromolecular complexes involved in diseases.
Molecular biologists will soon take advantage of these new techniques when investigating the structural and functional properties of macromolecular complexes. Despite this prospect, there are eminent limitations. These new approaches cannot reconstitute all the physiological parameters of a cell due to the dilution of biomolecules and the loss of cellular compartmentalization during sample preparation. Ideally, it would be pertinent to observe the biomolecules interacting with other cellular components inside living cells. Recently, several groups developed single-molecule fluorescence techniques to study macromolecular complexes within living cells. Using a total internal reflection microscope, Ulbrich et al. determined the subunit stoichiometry of membrane-bound proteins in Xenopus laevis oocytes by counting the number of GFP molecules[65], and Madl et al. determined the stoichiometry of Orai1 channel proteins in mammalian cells by carrying out brightness analysis of GFP signals[66]. Using slimfield fluorescence microscopy, Reyes-Lamothe et al. investigated the replisome stoichiometry and architecture in a bacteria[67]. Other examples of quantitative stoichiometry analysis are found in a review by Coffman and Wu[68]. More recently, Ries et al investigated the dynamics of intracellular structures at the nanometer scale in neurons and yeasts using single-molecule superresolution microscopy and high-affinity fluorescent nanobodies[69]. Coupling these in vivo single-molecule imaging methods with the cell extract isolation- and IP-based approaches will be a promising strategy towards macromolecular complex investigation for their complementary advantages and disadvantages.
Outstanding questions
When studying macromolecular protein complexes at the single-molecule level, what strategy should we use to overcome the complication arising from the heterogeneity of the complexes?
What probing scheme should we use to achieve the most accurate determination of the composition of a protein complex at the single-molecule level?
Can we integrate these new approaches into existing biochemical analysis tools? Can we use these integrated tools for identifying novel cofactors and discovering new drugs?
Can we combine these new approaches with other advanced single-molecule tools for deeper nanoscopic insights?
Can we develop these techniques further so that we can study macromolecular complexes even more physiologically relevant conditions that overcome the limitation from the dilution of cell extracts and the loss of the cellular compartments?
Acknowledgements
C. J. was supported by Starting Grants (ERC-StG-2012-309509) through the European Research Council. M. F. was supported by Fondation pour la Recherche Medicale. V. N. was supported by the Research Center Program (EM1202) of IBS (Institute for Basic Science); and the National Honor Scientist Program (20100020415) through the National Research Foundation of Korea (NRF). We appreciate helpful discussions with Jeff Gelles and Antoine van Oijen. We also appreciate critical comments from Elio Abbondanzieri, Timothy Blosser, Stanley Chandradoss, Jetty van Ginkel, Anna Haagsma, Inha Heo, Sungchul Hohng, Kaushik Ragunathan, and Kyu-Hyeon Yeom. We apologize for being unable to cite other research papers and reviews due to space limitations.
References
- 1.Toomre D, Bewersdorf J. A new wave of cellular imaging. Annu Rev Cell Dev Biol. 2010;26:285–314. doi: 10.1146/annurev-cellbio-100109-104048. [DOI] [PubMed] [Google Scholar]
- 2.Joo C, et al. Advances in single-molecule fluorescence methods for molecular biology. Annu Rev Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- 3.Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koster DA, et al. Cellular strategies for regulating DNA supercoiling: a single-molecule perspective. Cell. 2010;142:519–530. doi: 10.1016/j.cell.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katan AJ, Dekker C. High-speed AFM reveals the dynamics of single biomolecules at the nanometer scale. Cell. 2011;147:979–982. doi: 10.1016/j.cell.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 6.Noy A. Force spectroscopy 101: how to design, perform, and analyze an AFM-based single molecule force spectroscopy experiment. Curr Opin Chem Biol. 2011;15:710–718. doi: 10.1016/j.cbpa.2011.07.020. [DOI] [PubMed] [Google Scholar]
- 7.Dekker C. Solid-state nanopores. Nat Nanotechnol. 2007;2:209–215. doi: 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]
- 8.Rasnik I, et al. Electronic cameras for low-light microscopy. Methods Cell Biol. 2007;81:219–249. doi: 10.1016/S0091-679X(06)81012-5. [DOI] [PubMed] [Google Scholar]
- 9.Rasnik I, et al. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- 10.Altman RB, et al. Enhanced photostability of cyanine fluorophores across the visible spectrum. Nat Methods. 2012;9:428–429. doi: 10.1038/nmeth.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pertsinidis A, et al. Subnanometre single-molecule localization, registration and distance measurements. Nature. 2010;466:647–651. doi: 10.1038/nature09163. [DOI] [PubMed] [Google Scholar]
- 12.Chung HS, et al. Single-molecule fluorescence experiments determine protein folding transition path times. Science. 2012;335:981–984. doi: 10.1126/science.1215768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campos LA, et al. A photoprotection strategy for microsecond-resolution single-molecule fluorescence spectroscopy. Nat Methods. 2011;8:143–146. doi: 10.1038/nmeth.1553. [DOI] [PubMed] [Google Scholar]
- 14.Hoskins AA, et al. Ordered and dynamic assembly of single spliceosomes. Science. 2011;331:1289–1295. doi: 10.1126/science.1198830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yardimci H, et al. Uncoupling of sister replisomes during eukaryotic DNA replication. Mol Cell. 2010;40:834–840. doi: 10.1016/j.molcel.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeom KH, et al. Single-molecule approach to immunoprecipitated protein complexes: insights into miRNA uridylation. EMBO Rep. 2011;12:690–696. doi: 10.1038/embor.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jain A, et al. Probing cellular protein complexes using single-molecule pull-down. Nature. 2011;473:484–488. doi: 10.1038/nature10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker M. Proteomics: The interaction map. Nature. 2012;484:271–275. doi: 10.1038/484271a. [DOI] [PubMed] [Google Scholar]
- 19.Hoskins AA, et al. New insights into the spliceosome by single molecule fluorescence microscopy. Curr Opin Chem Biol. 2011;15:864–870. doi: 10.1016/j.cbpa.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yardimci H, et al. Single-molecule analysis of DNA replication in Xenopus egg extracts. Methods. 2012 doi: 10.1016/j.ymeth.2012.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain A, et al. Single-molecule pull-down for studying protein interactions. Nat Protoc. 2012;7:445–452. doi: 10.1038/nprot.2011.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wombacher R, Cornish VW. Chemical tags: applications in live cell fluorescence imaging. J Biophotonics. 2011;4:391–402. doi: 10.1002/jbio.201100018. [DOI] [PubMed] [Google Scholar]
- 23.Stephanopoulos N, Francis MB. Choosing an effective protein bioconjugation strategy. Nat Chem Biol. 2011;7:876–884. doi: 10.1038/nchembio.720. [DOI] [PubMed] [Google Scholar]
- 24.Henriques R, et al. PALM and STORM: unlocking live-cell super-resolution. Biopolymers. 2011;95:322–331. doi: 10.1002/bip.21586. [DOI] [PubMed] [Google Scholar]
- 25.Hoskins AA, Moore MJ. The spliceosome: a flexible, reversible macromolecular machine. Trends Biochem Sci. 2012;37:179–188. doi: 10.1016/j.tibs.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crawford DJ, et al. Visualizing the splicing of single pre-mRNA molecules in whole cell extract. RNA. 2008;14:170–179. doi: 10.1261/rna.794808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abelson J, et al. Conformational dynamics of single pre-mRNA molecules during in vitro splicing. Nat Struct Mol Biol. 2010;17:504–512. doi: 10.1038/nsmb.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller LW, et al. In vivo protein labeling with trimethoprim conjugates: a flexible chemical tag. Nat Methods. 2005;2:255–257. doi: 10.1038/nmeth749. [DOI] [PubMed] [Google Scholar]
- 29.Gautier A, et al. An engineered protein tag for multiprotein labeling in living cells. Chem Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 30.Los GV, et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 31.Howarth M, Ting AY. Imaging proteins in live mammalian cells with biotin ligase and monovalent streptavidin. Nat Protoc. 2008;3:534–545. doi: 10.1038/nprot.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin J, et al. Site-specific protein labeling by Sfp phosphopantetheinyl transferase. Nat Protoc. 2006;1:280–285. doi: 10.1038/nprot.2006.43. [DOI] [PubMed] [Google Scholar]
- 33.Uttamapinant C, et al. A fluorophore ligase for site-specific protein labeling inside living cells. Proc Natl Acad Sci U S A. 2010;107:10914–10919. doi: 10.1073/pnas.0914067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rabuka D, et al. Site-specific chemical protein conjugation using genetically encoded aldehyde tags. Nat Protoc. 2012;7:1052–1067. doi: 10.1038/nprot.2012.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi X, et al. Quantitative fluorescence labeling of aldehyde-tagged proteins for single-molecule imaging. Nat Methods. 2012 doi: 10.1038/nmeth.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Oijen AM, Loparo JJ. Single-molecule studies of the replisome. Annu Rev Biophys. 2010;39:429–448. doi: 10.1146/annurev.biophys.093008.131327. [DOI] [PubMed] [Google Scholar]
- 37.Almouzni G, Wolffe AP. Nuclear assembly, structure, and function: the use of Xenopus in vitro systems. Experimental cell research. 1993;205:1–15. doi: 10.1006/excr.1993.1051. [DOI] [PubMed] [Google Scholar]
- 38.Botchan M, Berger J. DNA replication: making two forks from one prereplication complex. Mol Cell. 2010;40:860–861. doi: 10.1016/j.molcel.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ligasova A, et al. Organization of human replicon: singles or zipping couples? J Struct Biol. 2009;165:204–213. doi: 10.1016/j.jsb.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim S, et al. Multiplexed single-molecule assay for enzymatic activity on flow-stretched DNA. Nat Methods. 2007;4:397–399. doi: 10.1038/nmeth1037. [DOI] [PubMed] [Google Scholar]
- 41.Gorman J, Greene EC. Visualizing one-dimensional diffusion of proteins along DNA. Nat Struct Mol Biol. 2008;15:768–774. doi: 10.1038/nsmb.1441. [DOI] [PubMed] [Google Scholar]
- 42.Fu YV, et al. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell. 2011;146:931–941. doi: 10.1016/j.cell.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Oijen AM. Cutting the forest to see a single tree? Nat Chem Biol. 2008;4:440–443. doi: 10.1038/nchembio0808-440. [DOI] [PubMed] [Google Scholar]
- 44.Lebofsky R, et al. DNA is a co-factor for its own replication in Xenopus egg extracts. Nucleic Acids Res. 2010;39:545–555. doi: 10.1093/nar/gkq739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loveland AB, et al. A general approach to break the concentration barrier in single-molecule imaging. Nat Methods. 2012;9:987–992. doi: 10.1038/nmeth.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heo I, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 47.Hagan JP, et al. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Means CK, et al. An entirely specific type I A-kinase anchoring protein that can sequester two molecules of protein kinase A at mitochondria. Proc Natl Acad Sci U S A. 2011;108:E1227–1235. doi: 10.1073/pnas.1107182108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen Z, et al. Dynamic association of ORCA with pre-RC components regulates DNA replication initiation. Molecular and cellular biology. 2012 doi: 10.1128/MCB.00362-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Panter MS, et al. Dynamics of Major Histocompatibility Complex Class I Association with the Human Peptide-loading Complex. J Biol Chem. 2012;287:31172–31184. doi: 10.1074/jbc.M112.387704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saci A, et al. Rac1 regulates the activity of mTORC1 and mTORC2 and controls cellular size. Mol Cell. 2011;42:50–61. doi: 10.1016/j.molcel.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Diao J, et al. A single vesicle-vesicle fusion assay for in vitro studies of SNAREs and accessory proteins. Nat Protoc. 2012;7:921–934. doi: 10.1038/nprot.2012.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karatekin E, Rothman JE. Fusion of single proteoliposomes with planar, cushioned bilayers in microfluidic flow cells. Nat Protoc. 2012;7:903–920. doi: 10.1038/nprot.2012.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Christensen SM, Stamou DG. Sensing-applications of surface-based single vesicle arrays. Sensors (Basel) 2010;10:11352–11368. doi: 10.3390/s101211352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vlassov AV, et al. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 57.Rissin DM, et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotechnol. 2010;28:595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harris TD, et al. Single-molecule DNA sequencing of a viral genome. Science. 2008;320:106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 59.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 60.Clarke J, et al. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol. 2009;4:265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 61.Lee J, et al. Single-molecule four-color FRET. Angew Chem Int Ed Engl. 2010;49:9922–9925. doi: 10.1002/anie.201005402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gambin Y, Deniz AA. Multicolor single-molecule FRET to explore protein folding and binding. Mol Biosyst. 2010;6:1540–1547. doi: 10.1039/c003024d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stein IH, et al. Single-molecule four-color FRET visualizes energy-transfer paths on DNA origami. J Am Chem Soc. 2011;133:4193–4195. doi: 10.1021/ja1105464. [DOI] [PubMed] [Google Scholar]
- 64.Macarron R, et al. Impact of high-throughput screening in biomedical research. Nat Rev Drug Discov. 2011;10:188–195. doi: 10.1038/nrd3368. [DOI] [PubMed] [Google Scholar]
- 65.Ulbrich MH, Isacoff EY. Subunit counting in membrane-bound proteins. Nat Methods. 2007;4:319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Madl J, et al. Resting state Orai1 diffuses as homotetramer in the plasma membrane of live mammalian cells. J Biol Chem. 2010;285:41135–41142. doi: 10.1074/jbc.M110.177881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reyes-Lamothe R, et al. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science. 2010;328:498–501. doi: 10.1126/science.1185757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Coffman VC, Wu JQ. Counting protein molecules using quantitative fluorescence microscopy. Trends Biochem Sci. 2012 doi: 10.1016/j.tibs.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ries J, et al. A simple, versatile method for GFP-based super-resolution microscopy via nanobodies. Nat Methods. 2012;9:582–584. doi: 10.1038/nmeth.1991. [DOI] [PubMed] [Google Scholar]