Abstract
Two DNA-based methods were compared for the ability to assign serotype to 139 isolates of Salmonella enterica ssp. I. Intergenic sequence ribotyping (ISR) evaluated single nucleotide polymorphisms occurring in a 5S ribosomal gene region and flanking sequences bordering the gene dkgB. A DNA microarray hybridization method that assessed the presence and the absence of sets of genes was the second method. Serotype was assigned for 128 (92.1%) of submissions by the two DNA methods. ISR detected mixtures of serotypes within single colonies and it cost substantially less than Kauffmann–White serotyping and DNA microarray hybridization. Decreasing the cost of serotyping S. enterica while maintaining reliability may encourage routine testing and research.
Keywords: epidemiology, food safety, single nucleotide polymorphism
Introduction
Serotyping of Salmonella enterica ssp. I is the basis of national and international surveillance and communications, it facilitates determining associations between the pathogen and sources, and it gives some guidance in regards to preventing transmission (P. Fields, pers. commun.) (Foodnet, 2011). The historical method used to serotype S. enterica is the antibody-based Kauffman–White (KW) scheme (Kauffman & Edwards, 1952; Brenner et al., 2000). Positive results generate an antigenic formula based on structural details of the H-antigen of flagella and the O-antigen of lipopolysaccharide (H- and O-antigens, respectively) (Bopp et al., 1999; Popoff & Le Minor, 2001).
A major advantage of DNA analysis is that it is not impacted by variable expression of cell-surface antigens as are antibody-based agglutination assays like the KW scheme. Major obstacles to genome typing of S. enterica becoming broadly available include expense, the need for highly specialized equipment, and in some cases, sophisticated bioinformatics (Wattiau et al., 2011). A DNA-based method for assigning serotype to S. enterica at comparatively low cost and with readily accessible laboratory equipment commonly used for culturing and conducting the polymerase chain reaction (PCR) would be beneficial. A discrete region within S. enterica ssp. I was previously shown to differentiate closely related serotypes (Morales et al., 2006). The region of interest spans from the end of a 23S ribosomal gene, across a 5S gene and includes the last base pair preceding a tRNA aspU ribosomal gene neighboring dkgB (previously yafB) (Fig. 1). We wanted to know if dkgB-linked intergenic sequence ribotyping (ISR) would assign serotype similarly to an AOAC International certified DNA microarray hybridization method (DNAhyb) (Check & Trace by Checkpoints, Certificate 121001) (Malorny et al., 2003; Wattiau et al., 2008; Madajczak & Szych, 2010). The set of isolates examined were previously assigned a KW serotype and this historical information is included.
Fig. 1.
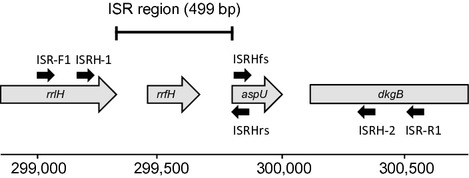
Description of the ISR region within the genome of Salmonella enterica serovar Enteritidis strain P125109 (GenBank AM933172). The nucleotide sequence of each ISR is serotype specific and size ranges from 257 to 530 bp. Sequence is required to assign serotype to a submission. An ISR region includes sequence from the end of the rrlH gene (rRNA-23S ribosomal gene) and the start of the aspU (tRNA-Asp) gene that is adjacent to the dkgB gene.
Materials and methods
Salmonella enterica submissions included for analysis
The investigators providing the submissions listed in Table 1 reported serotype. In this laboratory, cultures were streaked on brilliant green (BG) agar (Acumedia; Neogen Corporation, Lansing, MI) and incubated for 24–48 h at 37 °C to obtain well-separated large colonies. One colony was then transferred to brain heart infusion (BHI) broth (Acumedia) and incubated for 16 h at 37 °C with shaking. For submissions that later appeared to have mixed cultures and for those with disagreement between methods, agglutination reactions for single colonies using commercially available absorbed antisera (Difco, BD, Franklin Lakes, NJ) were carried out, and in some cases, isolates were submitted for serotyping (Silliker, South Holland, IL). Thus, single colonies were processed by both ISR and DNAhyb (Check & Trace, Check Points, Wageningen, the Netherlands). In cases of disagreement between methods, a maximum of 10 well-isolated colonies were selected from agar plates and then transferred to BHI broth for individual analysis.
Table 1.
Serotype of Salmonella enterica ssp. I as determined by the KW scheme, DNAhyb, and dkgB-linked ISR
| Accession Number | KW scheme | DNAhyb | ISR | ISR size | Supplier | Source | Locality | Category |
|---|---|---|---|---|---|---|---|---|
| (a) Submissions with agreement between DNA-based genomic methods DNAhyb and ISR, and with no conflict to serotype as reported using the KW scheme [115] | ||||||||
| 21027 | Enteritidis | Enteritidis 2994.G | Enteritidis | 499 | ESQRU, ARS, USDA | Mouse spleen | Northeast US | TP |
| 21046 | Enteritidis | Enteritidis 2994.G | Enteritidis | 499 | ESQRU, ARS, USDA | Mouse spleen | Northeast US | TP |
| 22079 | Enteritidis | Enteritidis 2994.G | Enteritidis | 499 | UC | Creek | California | TP |
| 23023 | Pullorum | Gallinarum Pullorum 2978.H | Pullorum | 361 | ESQRU, ARS, USDA | Unknown | Unknown | TP |
| 24018 | Newport | Newport 12427 | Newport | 498 | NVSL | Unknown | Unknown | TP |
| 25001 | Agona | Agona 7205 | Agona | 498 | SGSC | Unknown | Peru | TP |
| 25006 | Choleraesuis | Choleraesuis or Paratyphi C 13545 | Cholerasuis | 499 | SGSC | Unknown | Thailand | TP |
| 25012 | Dublin | Dublin (probability 99.92%) 2488 | Dublin | 499 | SGSC | Cattle | Idaho | TP |
| 25013 | Dublin | Dublin (probability 99.92%) 2488 | Dublin | 499 | SGSC | Bovine | France | TP |
| 25021 | Gallinarum | Gallinarum Gallinarum 2978 | Gallinarum | 498 | SGSC | Human | Connecticut | TP |
| 25026 | Infantis | Infantis 9381 | Infantis_2 | 500 | SGSC | Human | North Carolina | TN |
| 25030 | Montevideo | Montevideo 6690 | Montevideo_1 | 362 | SGSC | Human | Georgia | TP |
| 25031 | Montevideo | Montevideo 6702 | Montevideo_2 | 361 | SGSC | Human | Florida | TP |
| 25042 | Paratyphi A | Paratyphi A 14413 | Paratyphi A | 498 | SGSC | Unknown | Unknown | TP |
| 25049 | Paratyphi C | Choleraesuis or Paratyphi C 13545 | Paratyphi C | 395 | SGSC | Human | France | TP |
| 25052 | Pullorum | Gallinarum Pullorum 2978.H | Pullorum | 361 | SGSC | Unknown | Germany | TP |
| 25063 | Typhi | Typhi 7241 | Typhi | 267 | SGSC | Unknown | Dakar | TP |
| 25064 | Typhi | Typhi 7241 | Typhi | 267 | SGSC | Unknown | Dakar | TP |
| 26022 | Cerro | Cerro 4237 | Cerro | 361 | EMSFL, ARS, USDA | Fecal (dairy cow) | Unknown | TP |
| 26023 | Cerro | Cerro 4237 | Cerro | 361 | EMSFL, ARS, USDA | Lung (dairy cow) | Unknown | TP |
| 26024 | Cerro | Cerro 4237 | Cerro | 361 | EMSFL, ARS, USDA | Fecal (dairy cow) | Unknown | TP |
| 26028 | Oranienburg | Oranienburg 6717 r | Oranienburg | 365 | EMSFL, ARS, USDA | Fecal (dairy cow) | Unknown | TP |
| 26029 | Oranienburg | Oranienburg 6717 | Oranienburg | 365 | EMSFL, ARS, USDA | Fecal (dairy cow) | Unknown | TP |
| 26030 | Typhimurium 5- | Typhimurium 10909 | Typhimurium | 498 | EMSFL, ARS, USDA | Fecal (dairy cow) | Unknown | TP |
| 26039 | Montevideo | Montevideo 6702 | Montevideo_3 | 362 | EMSFL, ARS, USDA | Milk | Unknown | TN |
| 26050 | Agona | Agona 7205 | Agona | 498 | EMSFL, ARS, USDA | Fecal (dairy cow) | Unknown | TP |
| 26080 | Agona | Agona 7205 | Agona | 498 | EMSFL, ARS, USDA | Fecal (dairy cow) | Unknown | TP |
| 29047 | Typhimurium | Typhimurium 10909 | Typhimurium | 498 | ESQRU, ARS, USDA | Unknown | Unknown | TP |
| 29054 | Typhimurium | Typhimurium 10909 | Typhimurium | 498 | ESQRU, ARS, USDA | Unknown | Unknown | TP |
| 29056 | Typhimurium | Typhimurium 10909 | Typhimurium | 498 | ESQRU, ARS, USDA | Unknown | Unknown | TP |
| 29096 | Schwarzengrund | Schwarzen. or Grumpensis 14909.B | Schwarzengrund | 257 | PPSPR, ARS, USDA | Scalder tank water | Georgia | TP |
| 99113 | Pullorum | Gallinarum Pullorum 2978.H | Pullorum | 361 | CFIA | Chicken House | Unknown | TP |
| 99117 | Gallinarum | Gallinarum Gallinarum 2978 | Gallinarum | 498 | CFIA | Chicken House | Unknown | TP |
| 99163 | Typhimurium 5- | Typhimurium 10909 | Typhimurium | 498 | USDA, ARS, TX | Pigeon | Unknown | TP |
| 99164 | Typhimurium 5- | Typhimurium 10909 | Typhimurium | 498 | ESQRU, ARS, USDA | Unknown | Unknown | TP |
| 99172 | Typhimurium 5- | Typhimurium 10909 | Typhimurium | 498 | USDA, ARS | Pigeon | Unknown | TP |
| 100304.05 | Kentucky | Kentucky 10299 | Kentucky | 492 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100304.07 | Kentucky | Kentucky 10299 | Kentucky | 492 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100304.08 | Kentucky | Kentucky 10299 | Kentucky | 492 | PPSPR, ARS, USDA | Scalder tank foam | Georgia | TP |
| 100304.19 | 1,4,[5],12:i:- | 1,4,[5],12:i:- 2717 | 1,4,[5],12:i:- | 498 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100304.32 | Typhimurium 5- | Typhimurium 10909 | Typhimurium | 498 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100304.43 | Heidelberg | Heidelberg 15835 | Heidelberg | 498 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100304.48 | Heidelberg | Heidelberg 15835 | Heidelberg | 498 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100304.52 | Heidelberg | Heidelberg 15835 | Heidelberg | 498 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100304.57 | Typhimurium 5- | Typhimurium 10909 | Typhimurium | 498 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100304.63 | Typhimurium 5- | Typhimurium 10909 | Typhimurium | 498 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100304.69 | Thompson | Thompson 14415 | Thompson | 259 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100304.74 | Senftenberg | Senftenberg 2156 | Senftenberg | 362 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100304.75 | Senftenberg | Schwarzen. or Grumpensis 14909.B | Schwarzengrund | 257 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100304.76 | Senftenberg | Schwarzen. or Grumpensis 14909.B | Schwarzengrund | 257 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100304.78 | Thompson | Thompson 14415 | Thompson | 259 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100304.79 | Thompson | Thompson 14415 | Thompson | 259 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100616.101 | Kentucky | Kentucky 10299 | Kentucky | 492 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100616.87 | 1,4,[5],12:i:- | 1,4,[5],12:i:- 2717 | 1,4,[5],12:i:- | 498 | PPSPR, ARS, USDA | Scalder tank foam | Georgia | TP |
| 100616.89 | Typhimurium | Typhimurium 10909 | Typhimurium | 498 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100616.9 | Typhimurium 5- | Typhimurium 10909 | Typhimurium | 498 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100616.91 | Typhimurium | Typhimurium 10909 | Typhimurium | 498 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100709.01 | Kentucky | Kentucky 10299 | Kentucky | 492 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100709.02 | Kentucky | Kentucky 10299 | Kentucky | 492 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100709.03 | Kentucky | Kentucky 10299 | Kentucky | 492 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100709.04 | Kentucky | Kentucky 10299 | Kentucky | 492 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100709.05 | Senftenberg | Senftenberg 2156 | Senftenberg | 362 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100709.09 | Heidelberg | Heidelberg 15835 | Heidelberg | 498 | PPSPR, ARS, USDA | Scalder tank foam | Georgia | TP |
| 100721.01-2 | 1,4,[5],12:i:- | 1,4,[5],12:i:- 2717 | 1,4,[5],12:i:- | 498 | PPSPR, ARS, USDA | Scalder tank water | Georgia | TP |
| 100721.02 | Infantis | Infantis 9381 | Infantis_1 | 500 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100721.05 | Typhimurium | Typhimurium 10909 | Typhimurium | 498 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100723.09 | Enteritidis | Enteritidis 2994.G | Enteritidis | 499 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100304.10 | 1,4,[5],12:i:- | 1,4,[5],12:i:- 2717 | 1,4,[5],12:i:- | 498 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100304.50 | Heidelberg | Heidelberg 15835 | Heidelberg | 498 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100709.10 | Thompson | Thompson 14415 | Thompson | 259 | PPSPR, ARS, USDA | Scalder dip tank foam | Georgia | TP |
| 100723.10 | Enteritidis | Enteritidis 2994.G | Enteritidis | 499 | PPSPR, ARS, USDA | Scalder dip tank foam | Georgia | TP |
| 100304.58-2 | Kentucky | Kentucky 10299 | Kentucky | 492 | PPSPR, ARS, USDA | Scalder tank foam | Georgia | TP |
| 100304.62-2 | Typhimurium 5- | Typhimurium 10909 | Typhimurium | 498 | PPSPR, ARS, USDA | Scalder dip tank foam | Georgia | TP |
| 100616.86-2 | Typhimurium | Typhimurium 10909 | Typhimurium | 498 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100723.01-2 | Kentucky | Kentucky 10299 | Kentucky | 492 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100723.14-2 | Schwarzengrund | Schwarzen. or Grumpensis 14909.B | Schwarzengrund | 257 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100723.02-2 | Heidelberg | Heidelberg 15835 | Heidelberg | 498 | PPSPR, ARS, USDA | Scalder tank foam | Georgia | TP |
| 25010 | Derby | Derby 50 | Derby | 498 | SGSC | Swine | Minnesota | TP |
| 25032 | Muenchen | Muenchen 11942 | Muenchen | 404 | SGSC | Unknown | Unknown | TP |
| 25039 | Panama | Panama 14909 | Panama | 362 | SGSC | Unknown | Italy | TP |
| 26063 | Mbandaka | Mbandaka 11813.C | Mbandaka | 499 | EMSFL, ARS, USDA | Fecal (dairy cow) | Unknown | TP |
| 26066 | Mbandaka | Mbandaka 11813.C | Mbandaka | 499 | EMSFL, ARS, USDA | Fecal (dairy cow) | Unknown | TP |
| 100616.100 | Rough | Infantis 9381 | Infantis_1 | 500 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100616.102 | Rough | Enteritidis 2866.G | Enteritidis | 499 | PPSPR, ARS, USDA | Scalder tank water | Georgia | TP |
| 100616.84 | Rough | Typhimurium 10909 | Typhimurium | 498 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100616.97 | Rough | Typhimurium 10909 | Typhimurium | 498 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100616.99 | Rough | Typhimurium 10909 | Typhimurium | 498 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100709.06 | Rough | Senftenberg 2156 | Senftenberg | 362 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100709.07 | Rough | 1,4,[5],12:i:- 2717 | 1,4,[5],12:i:- | 498 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100709.12 | Rough | Infantis 9381 | Infantis_1 | 500 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100723.04 | Rough | Kentucky 10299 | Kentucky | 492 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100723.08 | Auto agglutinator | Heidelberg 15835 | Heidelberg | 498 | PPSPR, ARS, USDA | Scalder tank foam | Georgia | TP |
| 100723.12 | Rough | Senftenberg 2156 r | Senftenberg | 362 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100723.15 | Rough | Schwarzen. or Grumpensis 14909.B | Schwarzengrund | 257 | PPSPR, ARS, USDA | Scalder tank water | Georgia | TP |
| 25046 | Salmonella (B) | Abony 5325 | Abony | 498 | SGSC | Water | United Kingdom | TP |
| 25050 | Salmonella (C1) | Oranienburg 6717 r | Oranienburg | 361 | SGSC | Human | France | TP |
| 29063 | Salmonella (C1) | Newport 13444 | Newport_1 | 499 | ESQRU, ARS, USDA | Unknown | Unknown | TP |
| 29065 | Salmonella (C1) | Oranienburg 6717 r | Oranienburg | 365 | ESQRU, ARS, USDA | Unknown | Unknown | TP |
| 29066 | Salmonella (B) | Typhimurium 10909 | Typhimurium | 498 | ESQRU, ARS, USDA | Unknown | Unknown | TP |
| 29067 | Salmonella (D) | Enteritidis 2994.G | Enteritidis | 499 | ESQRU, ARS, USDA | Unknown | Unknown | TP |
| 25004 | No O or H antigen | Anatum 15087 | Anatum | 499 | SGSC | Swine | Minnesota | TP |
| 25037 | Newport | Newport 13519 | Newport_2 | 395 | SGSC | Human | Mexico | TP |
| 25038 | Newport | Newport 14539 | Newport_3 | 498 | SGSC | Snake | Massachusetts | TP |
| 101116-10* | Fresno (D2) | Genovar 5216 | UN0019 (D1) | 258 | Breeder farm | Chickens | Alabama or Tennessee | TP |
| 101116-12* | Fresno (D2) | Genovar 5216 | UN0019 (D1) | 258 | Breeder farm | Chickens | Alabama or Tennessee | TP |
| 25005 | Choleraesuis | Genovar 14861 | UN0009 (C1) | 361 | SGSC | Unknown | Switzerland | TP |
| 25007 | Choleraesuis | Genovar 11439 N | UN0010 (C1) | 498 | SGSC | Unknown | Australia | TP |
| 25017 | Enteritidis | Genovar 6660 | UN0002 (D1) | 360 | SGSC | Unknown | Brazil | TP |
| 25019* | Enteritidis | Genovar 2370 | UN0003 (-) | 498 | SGSC | Unknown | Switzerland | TP |
| 25009 | Derby | Genovar 6176 | UN0022 (B) | 498 | SGSC | Avian | Oklahoma | TP |
| 25014 | Dublin | Genovar 5324 | UN0012 (D1) | 498 | SGSC | Unknown | Thailand | TP |
| 25027 | Infantis | Genovar 14892 | UN0023 (C1) | 499 | SGSC | Unknown | Senegal | TP |
| 25034 | Muenchen | Genovar 13646 | UN0036 (C2C3) | 258 | SGSC | Human | France | TP |
| 99167 | Typhimurium 5- | Typhimurium 10909 | Typhimurium 5- | 395 | USDA, ARS | Pigeon | Unknown | TN |
| 99168 | Typhimurium 5- | Typhimurium 10909 | Typhimurium 5- | 395 | USDA, ARS | Pigeon | Unknown | TN |
| (b) Submissions with agreement between DNAhyb and ISR, but disagreeing with serotype as reported using the KW scheme [13] | ||||||||
| 25040 | Panama (D1) | Javiana 12917 | Javiana (D1) | 367 | SGSC | Human | North Carolina | TP |
| 25041 | Panama (D1) | Javiana 12917 | Javiana (D1) | 367 | SGSC | Human | North Carolina | TP |
| 25035 | Muenchen (C2) | Manhattan 11706 | Manhattan (C2) | 396 | SGSC | Human | North Carolina | TP |
| 25051-1†‡ | Pullorum (D1) | Oranienburg 6717 r | Oranienburg (-) | 361 | SGSC | Unknown | Unknown | TP |
| 26026 | Mbandaka (C1) | Tennessee 2108 | Tennessee (C1) | 258 | EMSFL, ARS, USDA | Fecal (dairy cow) | Unknown | TP |
| 100304.58-1†‡ | Kentucky (C2C3) | Typhimurium 10909 | Typhimurium (B) | 498 | PPSPR, ARS, USDA | Scalder tank foam | Georgia | TP |
| 100304.62-1†‡ | Typhimurium 5- (B) | Schwarzen. or Grumpensis 14909.B | Schwarzengrund (B) | 257 | PPSPR, ARS, USDA | Scalder dip tank foam | Georgia | TP |
| 100616.86-1†‡ | Typhimurium (B) | Infantis 9381 | Infantis_1 (C1) | 500 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100616.98-1† | Typhimurium 5- (B) | Kentucky 10299 | Kentucky (C2C3) | 492 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TP |
| 100721.01-1†‡ | 1,4,[5],12:i:- (B) | Kentucky 10299 | Kentucky (C2C3) | 492 | PPSPR, ARS, USDA | Scalder tank water | Georgia | TP |
| 100304.53 | Senftenberg (B) | Typhimurium 10909 | Typhimurium (B) | 498 | PPSPR, ARS, USDA | Scalder tank water | Georgia | TP |
| 25011* | Derby (B) | Genovar 5160 | UN0025 (C2C3) | 498 | SGSC | Turkey | Pennsylvania | TP |
| 25048* | Paratyphi C (C1) | Genovar 14375 | UN0024 (B) | 499 | SGSC | Unknown | France | TP |
| (c) Submissions with disagreement between DNAhyb and ISR, but with DNAhyb in agreement with serotype as reported by the KW scheme [11] | ||||||||
| 25036 | Newport | Newport 13444 | UN0034 | 498 | SGSC | Human | North Carolina | FP |
| 25043-2 | Paratyphi B (B) | Paratyphi B (possibly Java) 13383 | UN0015 | 498 | SGSC | Unknown | Unknown | FP |
| 25044 | Paratyphi B (B) | Paratyphi B (possibly Java) 13383 | UN0011 | 499 | SGSC | Food | Middle East | FP |
| 25051-2 | Pullorum (D1) | Gallinarum Pullorum 2978.H | UN0008 | 530 | SGSC | Unknown | Unknown | FP |
| 25057 | Schwarzengrund | Schwarzen. or Grumpensis 14909.B | UN0006 | 365 | SGSC | Unknown | Scotland | FP |
| 26078 | Kentucky | Kentucky (10791) 6 | UN0028 | 499 | EMSFL, ARS, USDA | Fecal (dairy cow) | Unknown | FP |
| 101116-14 | Javiana | Javiana 12917 | UN0007 | 361 | Breeder farm | Chickens | Alabama or Tennessee | FP |
| 25043-1†‡ | Paratyphi B (B) | Paratyphi B (possibly Java) 13383 | Javiana (D1) | 367 | SGSC | Unknown | Unknown | TN |
| 100723.01-1†‡ | Heidelberg (B) | Heidelberg 15835 | Kentucky (C2C3) | 498 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TN |
| 100723.02-1†‡ | Heidelberg (B) | Heidelberg 15835 | Kentucky (C2C3) | 492 | PPSPR, ARS, USDA | Scalder tank foam | Georgia | TN |
| 100723.14-1†‡ | Kentucky (C2C3) | Kentucky 10299 | Schwarzengrund (B) | 492 | PPSPR, ARS, USDA | Carcass rinse | Georgia | TN |
O-antigen immunoreactivity group of KW scheme did not match ISR results, but O-antigens D1 and D2 are cross-reactive.
Submissions with hyphenation with numerical extension (-1) were classified as potentially containing a mixture of at least two serotypes as evidenced by forward/reverse ISR sequences that did not match when first evaluated. If a second serotype was isolated, it has extension -2 and is listed elsewhere in the table.
Mixtures of serotypes were confirmed by processing at least 10 CFU.
Determination of ISR
The locations where primers hybridize the reference genome in S. enterica ssp. I Enteritidis strain P125109 are shown in Fig. 1. Forward (ISR-F1) and reverse (ISR-R1) primers incorporate the rRNA-23S ribosomal ribose nucleic acid (RNA) region neighboring dkgB (previously known as yafB). Reference genomes and primers used in these analyses are listed in Tables 2 and 3. Primers ISR-F1 and ISR-R1 replaced previously published primers ISRH-1 and ISRH-2 (Morales et al., 2006).
Table 2.
Reference ISR sequences available in public databases for Salmonella enterica ssp. I
| Serotype designation | ISR size (bp) | Refseq | GenBank accession |
|---|---|---|---|
| (a) Genome sequences used to obtain ISRs for Salmonella enterica ssp. I at The National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) | |||
| 4,[5],12:i:- str. CVM23701 | 498 | NZ_ABAO00000000 | ABAO00000000 |
| Agona str. SL483 | 498 | NZ_ABEK00000000 | ABEK00000000 |
| Choleraesuis str. SC-B67 | 499 | NC_006905 | AE017220 |
| Dublin str. CT_02021853 | 499 | NZ_ABAP00000000 | ABAP00000000 |
| Heidelberg str. SL476 | 498 | NC_011083 | CP001120 |
| Heidelberg str. SL486 | 498 | NZ_ABEL00000000 | ABEL00000000 |
| Javiana str. GA_MM04042433 | 367 | NZ_ABEH00000000 | ABEH00000000 |
| Kentucky str. CDC 191 | 492 | NZ_ABEI00000000 | ABEI00000000 |
| Kentucky str. CVM29188 | 492 | NZ_ABAK00000000 | ABAK00000000 |
| Newport str. SL254 | 492 | NC_011080 | CP000604 |
| Newport str. SL317 | 492 | NZ_ABEW00000000 | ABEW00000000 |
| Paratyphi A str. ATCC 9150 | 498 | NC_006511 | CP000026 |
| Paratyphi B SPB7 | 498 | NC_010102.1 | CP000886 |
| Paratyphi C RKS4594 | 395 | NC_012125.1 | CP000857 |
| Saintpaul str. SARA23 | 498 | NZ_ABAM00000000 | ABAM00000000 |
| Saintpaul str. SARA29 | 498 | NZ_ABAN00000000 | ABAN00000000 |
| Schwarzengrund str. CVM19633 | 267 | NC_011094 | CP001127 |
| Schwarzengrund str. SL480 | 267 | NZ_ABEJ00000000 | ABEJ00000000 |
| Typhi Ty2 | 267 | NC_004631 | AE014613 |
| Typhi str. CT18 | 267 | NC_003198 | AL513382 |
| Typhimurium LT2 | 498 | NC_003197 | AE006468 |
| Virchow str. SL491 | 499 | Not yet completed | na |
| (b) Genome sequences used to obtain ISRs for Salmonella enterica ssp. I available at The Sanger Institute (http://www.sanger.ac.uk/Projects/Salmonella/) | |||
| Enteritidis PT4 NCTC 13349 | 499 | NC_011294.1 | AM933172.1 |
| Gallinarum 287/91 NCTC 13346 | 498 | NC_011274.1 | AM933173.1 |
| Hadar | 498 | NA* | NA |
| Infantis | 500 | NA | NA |
| Typhimurium DT104 NCTC 13348 | 498 | NA | NA |
| Typhimurium DT2 | 498 | NA | NA |
| Typhimurium SL1344 NCTC 13347 | 498 | NA | NA |
| Typhimurium D23580 | 498 | NA | NA |
| (c) ISR sequences for Salmonella enterica ssp. I submitted to the National Center for Biotechnology Information† | |||
| Schwarzengrund_2 | 257 | In process | In process |
| Cerro | 361 | In process | JN105120 |
| Infantis_1 | 500 | In process | JN105121 |
| Oranienburg | 365 | In process | JN105122 |
| Pullorum | 361 | In process | JN105123 |
| Senftenberg | 362 | In process | JN105124 |
| Thompson | 259 | In process | JN105125 |
| Tennessee | 258 | BankIt1458394 SEQ_018 | JN092310 |
| Mbandaka | 499 | BankIt1458394 SEQ_030 | JN092322 |
| Montevideo_1 | 362 | BankIt1458394 SEQ_031 | JN092323 |
| Montevideo_2 | 361 | BankIt1458394 SEQ_032 | JN092324 |
| Montevideo_3 | 362 | BankIt1458394 SEQ_033 | JN092325 |
| Manhatten | 258 | BankIt1458394 SEQ_036 | JN092328 |
| UN0001 | 404 | BankIt1458394 SEQ_001 | JN092293 |
| UN0002 | 360 | BankIt1458394 SEQ_002 | JN092294 |
| UN0003 | 498 | BankIt1458394 SEQ_003 | JN092295 |
| UN0004 | 362 | BankIt1458394 SEQ_004 | JN092296 |
| UN0005 | 361 | BankIt1458394 SEQ_005 | JN092297 |
| UN0006 | 365 | BankIt1458394 SEQ_006 | JN092298 |
| UN0007 | 361 | BankIt1458394 SEQ_007 | JN092299 |
| UN0008 | 530 | BankIt1458394 SEQ_008 | JN092300 |
| UN0009 | 361 | BankIt1458394 SEQ_009 | JN092301 |
| UN0010 | 498 | BankIt1458394 SEQ_010 | JN092302 |
| UN0011 | 499 | BankIt1458394 SEQ_011 | JN092303 |
| UN0012 | 498 | BankIt1458394 SEQ_012 | JN092304 |
| UN0013 | 498 | BankIt1458394 SEQ_013 | JN092305 |
| UN0014 | 395 | BankIt1458394 SEQ_014 | JN092306 |
| UN0015 | 489 | BankIt1458394 SEQ_015 | JN092307 |
| UN0016 | 396 | BankIt1458394 SEQ_016 | JN092308 |
| UN0017 | 395 | BankIt1458394 SEQ_017 | JN092309 |
| UN0019 | 258 | BankIt1458394 SEQ_019 | JN092311 |
| UN0021 | 499 | BankIt1458394 SEQ_021 | JN092313 |
| UN0022 | 498 | BankIt1458394 SEQ_022 | JN092314 |
| UN0023 | 499 | BankIt1458394 SEQ_023 | JN092315 |
| UN0024 | 499 | BankIt1458394 SEQ_024 | JN092316 |
| UN0025 | 498 | BankIt1458394 SEQ_025 | JN092317 |
| UN0026 | 498 | BankIt1458394 SEQ_026 | JN092318 |
| UN0027 | 499 | BankIt1458394 SEQ_027 | JN092319 |
| UN0028 | 499 | BankIt1458394 SEQ_028 | JN092320 |
| UN0034 | 498 | BankIt1458394 SEQ_034 | JN092326 |
| UN0035 | 498 | BankIt1458394 SEQ_035 | JN092327 |
Not available due to incomplete annotation.
Serotype names replace the unique accession number following multiple confirmations and agreement between methods.
Table 3.
Primers used to correlate genotype to serotype of Salmonella enterica by ISR
| Primer name | Orientation | Primer sequence (5′–3′) | Reference | Amplicon size (bp)* |
|---|---|---|---|---|
| ISR-F1 | Forward | GCCAATGGCACTGCCCGGTA | This study | 14641 |
| ISR-R1 | Reverse | TACCGTGCGCTTTCGCCCAG | ||
| ISRH-1 | Forward | GATGCGTTGAGCTAACCGGTACTA | Morales et al. (2006) | Does not apply |
| ISRH-2 | Reverse | ATTCTTCGACAGACACGGCATCAC | ||
| ISRHfs | Forward | GTGGAGCGGTAGTTCAGTTGGTTA | Morales et al. (2006) | Does not apply |
| ISRHrs | Reverse | TAACCAACTGAACTACCGCTCCAC |
Amplicon size in S. Typhimurium str. LT2 genome (GenBank AE006468).
For primer amplification, DNA was extracted from 1 mL of pelleted cells using the PureLink Genomic DNA Mini Kit (Invitrogen Life Technologies, Grand Island, NY). One microliter of DNA was added to 2× Gene Amp Fast PCR Master Mix (Applied Biosystems, Foster City, CA) and 200 nM forward (ISR-F1) and reverse (ISR-R1) primers in a final volume of 30 μL. The PCR was performed on a Veriti 96 well Fast Thermal cycler (Applied Biosystems) as follows: 95 °C for 10 s, 35 cycles at 94 °C, 40 s at 64 °C, and 10 s at 72 °C. After confirmation of the predicted amplicon of approximately 1400 bp by gel electrophoresis, PCR products were purified using the QIAquick PCR purification kit (Qiagen, Valencia, CA). DNA concentrations were measured (NanoDrop, Wilmington, DE) prior to submitting PCR products for Sanger sequencing (Retrogen Inc., San Diego, CA) on an Applied Biosystems, Incorporated (ABI) Prism® 3730 DNA Analyzer using primers ISRHfs and ISRHfr.
Analysis and naming of ISR sequences
ISR sequences were aligned using SeqMan Pro of the Lasergene 8 software (DNASTAR, Madison, WI). Parameters were set to 100% minimum match percentage and a match size of at least 50 bp. Primers were designed to assure linkage to dkgB, which is required to assure that the correct region is under investigation. Serotype names were assigned to an ISR sequence when a 100% match was made to a reference sequence or when DNAhyb and KW serotyping agreed. Otherwise, ISR sequences are identified as ‘UN’ followed by four-digit numbers. The initial (5′ ATGTTTTGGCG 3′) and final (5′ CGGTGGAGCGG 3′) eleven nucleotides should be similar for ISRs, with the exception that the first nucleotide in the ISR sequence can sometimes be a cytosine rather than an adenine nucleotide.
DNAhyb assay
The DNAhyb protocol was performed as directed (Check & Trace) on single Salmonella colonies grown for 24–48 h on BG agar at 37 °C. Large colonies are recommended to reach the recommended DNA concentration. Images of products were obtained on a single-channel ATR03 reader and processed by the Salmonella Check-Points software which indicates a serotype name, or alternatively, a genovar number. Images of spot patterns were discussed with the manufacturer in unusual situations, such as finding genetic variants of Salmonella Kentucky. In this case, three isolates were submitted to the source of the kit for independent verification that new variants were being identified and that all positive and negative controls worked.
Results
ISR and DNAhyb assign serotype to S. enterica similarly
Details from analysis of S. enterica by ISR and DNAhyb are shown in Table 1. Of the 139 submissions, 115 (82.7%) had substantial agreement between DNAhyb and ISR, as well as the reported KW serotype (Table 1a). Some genetic variation was noted in ISRs in this grouping, but serotype association was maintained in comparison with both DNAhyb and the KW scheme and thus these were counted as agreements. Further analysis of ISR variation showed that 15 named serotypes had at least three independent submissions. Of this group, 10 (66.7%) had uniform ISRs with no variation. Five (5) serotypes had multiple ISRs, namely Salmonella Infantis (two variants), Salmonella Typhimurium (two variants), S. Kentucky (two variants), Salmonella Newport (four variants), and Salmonella Montevideo (three variants). For the purpose of determining specificity and sensitivity in this study, results with disagreement about how much variation is accounted for by DNAhyb vs. ISR were counted as true negatives (TN), because the ISR method produced information somewhat different, but not necessarily in disagreement, to DNAhyb. In summary, 128 of the 139 isolates (92.1%) had agreement between DNAhyb and ISR (Table 1b + 1a). Detail about ISR variation within a single named serotype will be discussed in following text.
For 10 submissions, the forward/reverse (F/R) sequences did not match. Individually processing multiple colonies for nine of these submissions in this category recovered a second serotype. The frequency with which a second serotype accounted for disagreement between genomic methods and KW serotype suggests that minority subpopulations are common. In addition, techniques appear to vary in their ability to detect multiple serotypes. Seven other submissions (25011, 25048, 25040, 25041, 25035, 26026, and 100304.53) had KW serotypes with O-antigens that did not match what was received for analysis but there was no disagreement in F/R sequences. In these cases, misinterpretation of the second cell-surface molecule flagella could have contributed to misinterpretation of serotype. Alternatively, these submissions could have also been mixed when initially examined for KW serotype and undergone separation of serotypes prior to analysis during the current study. For submissions 101116-10 and 101116-12, which were classified as Salmonella Fresno and ISR UN0019, O-antigen D2 epitopes would be expected to cross-react with factor 9 antisera used to detect D1 epitopes (Curd et al., 1998).
Table 1c shows paired samples with disagreement between DNAhyb and ISR. Four of these submissions yielded mixed serotypes, namely 25043-1, 100723.01-1, 100723.02-1, and 100723-14.1. They are included in Table 1c to reflect the incidence with which disagreement was encountered. However, disagreements between DNAhyb and ISR and KW were resolved for these four isolates. The other seven submissions in Table 1c had a unique (UN) ISR sequence that was identified by DNAhyb as a genovar with an available reference sequence. However, slight differences in the ISR suggested genetic variants could be present as observed for the first group in Table 1a. These submissions were grouped in Table 1c because the serovar they might be associated with could not be further verified at this time. Acquisition of more isolates is needed to resolve the relationship between KW serotype, DNAhyb and ISR sequence in these cases.
ISR appears more sensitive than DNAhyb for the detection of genetic variation within serotype
ISR appears to give a more sensitive assessment of serotype than DNAhyb. To explain further, some specific examples are cited. These are as follows:
ISR sometimes detected two types of S. Typhimurium, namely Typhimurium and Typhimurium 5-, whereas DNAhyb did not (Table 1, accession numbers 100304.57, 100304.74, 100304.32, 100304.62-2, 100616.9, 99163, 99164 and 99172). Thus, DNAhyb is not currently capable of detecting the 5- variant. ISR indicated that UN0014 was linked to the Typhimurium 5- KW serotype, but this correlation was not observed for all examples (see 100304.62-2). Thus, variation in expression of cell-surface epitopes may account for the 5- variant in addition to genetic variation as observed by ISR.
Only one publicly available reference sequence out of 27 disagreed with assignment of serotyping by the three methods. The ISRs for the reference sequences of S. Schwarzengrund NC_011094 and NZ_ABEJ00000000 were 267 bp and no SNP differences were present. The strains of S. Schwarzengrund analyzed here had ISRs of 257 bp regardless of method used to assign serotype. Further analysis is required to explain this discrepancy, which could be due to strain variation or some discrepancy with annotation in the reference strain.
Five submissions of S. Newport had different ISR sequences and four DNAhyb patterns despite having a single serotype assigned by the KW scheme. Alignment of the sequence of five ISRs from submissions that serotyped as S. Newport showed that ISR UN0017 had a deletion within the intergenic region between the end of the 23S and 5S rRNA genes (Fig. 2, Top).
O-antigen group D submissions had an even more complex ISR outcome than did S. Newport (Fig. 2, Bottom). Alignment showed that, in comparison with S. Enteritidis, ISR UN0002 had a somewhat similar deletion as that seen occurring within Salmonella Pullorum in the intergenic region between the 5S rRNA and tRNA aspU. In this same region, ISR UN0008 has an insertion that was 100% similar to a region from S. Newport found in a different ribosomal region. The insert accounted for the exceptional length of the ISR. Specifically, there was a 146 bp insert in ISR UN0008 that was the same as base pairs 4165975-4166120 in the genome of S. Newport strain SL254 (NC_011080). Other serotypes that had ISR variants within a serotype included S. Kentucky, S. Montevideo, and S. Infantis.
Fig. 2.
Alignment of ISR sequences to evaluate variation within serotype. (Top) Alignment of ISRs from Salmonella enterica serovar Newport. The shortest ISR shown is UN0017, which had a deletion occurring before the 5S ribosomal gene. The 5S ribosomal gene of Salmonella Enteritidis was included for reference purposes. (Bottom) Alignment of ISRs from the O-antigen group D serotypes of S. enterica. The submission with ISR UN0002 was S. Enteritidis by the KW scheme and Genovar 6660 by DNAhyb. ISR UN0008 was Salmonella Pullorum by the KW scheme and Genovar Gallinarum Pullorum 2978H by DNAhyb. Alignment of ISR sequences from Group D serotypes indicated that UN0002 is more like S. Pullorum. ISR UN0008 is more like S. Gallinarum or S. Enteritidis, except that it has a 146 bp insertion also found in S. Newport.
ISR and DNAhyb are limited to assignment of serotype to S. enterica
The limit of detection of ISR was found by analysis of S. Enteritidis submissions 22079, 21027, and 21046, which were included to as control strains because they were previously characterized by whole-genome analysis (Guard et al., 2011). Strains 21027 and 21046 are clonally related and are within the same phage type lineage 13a/8, whereas 22079 is phage type 4. Despite belonging to the same phage type, strains 21027 and 21046 are phenotypically distinct and are known to have 16 genes with altered open reading frames as well as other SNPs. All three subpopulations of S. Enteritidis had the same KW serotype, DNAhyb genovar, and ISR. Thus, the two DNA methods were equivalent in regards to determining serotype only.
Determination of sensitivity and specificity of ISR in comparison with DNAhyb
Table 1 includes the category of each ISR for the purposes of determining sensitivity and specificity in comparison with DNAhyb. True positives (TP) were defined as submissions assigned a serotype by ISR in complete agreement with DNAhyb, TN indicated ISR was different for the serotype assigned by DNAhyb due to the presence of genetic variation (including mixtures of serotypes), false positives (FP) were assigned a serotype by ISR but not by DNAhyb, and false negatives (FN) should have returned an ISR sequence but did not. As all submissions were S. enterica ssp. I and produced an ISR and a DNAhyb genovar, the FN value is 0. Calculating sensitivity from the values 124/124 + 0 (TP/TP + FN) suggests unity (similar performance) of the two methods. Calculating specificity from the values 8/7 + 8 = 0.53 (TN/FP + TN) suggests that DNAhyb is more specific, or in other words, it detects less genetic variation than does ISR. Removing submissions with mixtures did not change the finding that ISR appears to give more specific information than DNAhyb. Given that detection of new serotypes is a continuous process for S. enterica ssp. I, application of ISR has the potential to expand knowledge about diversity of serotypes. In these analyses, we used S. enterica serovar Enteritidis to provide a crucial control that shows ISR does not provide fine-scale differentiation achieved with whole-genome sequencing.
Discussion
A limit of detection of ISR is that it targets a single region of the bacterial chromosome. Homologous recombination and other genomic events that mobilize DNA could generate a hybrid strain with potential to alter the correlation between an ISR region and the rest of the chromosome (Porwollik & McClelland, 2003). Methods that target multiple regions around the bacterial chromosome, such as DNAhyb and whole-genome sequencing, will thus still be required for critical stages of analysis. The primary use proposed for ISR is to facilitate routine and inexpensive serotyping of S. enterica. The method has been applied to processing DNA samples from South America in cooperation with the United States, and further development of software that incorporates a validated database will streamline analysis for users (Pulido-Landinez et al., 2012). SNP analysis by ISR complements methods such as DNAhyb that evaluate the whole genome, and each genome method can be used to check the quality of results from the other.
Disagreement between the KW scheme and genotyping by either DNA method could be attributed to at least four causes with a biological or molecular explanation.
Flagella H-antigen immunoreactivity may contribute disproportionately to interpretive differences between investigators;
A genetic variant may have a unique ISR or DNAhyb genovar that, in consensus with previous knowledge, is a genetic variant of an existing serotype;
Mixtures of serotypes could be present within cultures, which can be detected by some methods but not others.
The most troublesome group was new variants with undefined relationships to named serotypes. ISR UN0002 (ISR 360 bp) and UN0008 (ISR 530 bp) were associated with submissions serotyped by the KW scheme as S. Enteritidis and S. Pullorum, respectively, despite their unique ISR sequences. Classifying them by the KW scheme as S. Enteritidis or S. Pullorum could have unintended consequences, because the biological impact of these strains on susceptible hosts is not known. For example, S. Pullorum on-farm can initiate depopulation of chickens in order to protect poultry health (http://www.aphis.usda.gov/animal_health/animal_dis_spec/poultry/), whereas S. Enteritidis in people and foods can initiate control measures to protect human health (http://www.fda.gov/Food/FoodSafety/Product-SpecificInformation/EggSafety/EggSafetyActionPlan/ucm170746.htm). No information is available on the comparative virulence properties of UN0002 (ISR 360 bp) and UN0008 (530 bp) to S. Pullorum (ISR 361 bp) or S. Enteritidis (ISR 499 bp). Further research using biological assays is needed to characterize the virulence of strains identified by ISR as being potentially new strains of concern to either human or animal health.
Assay costs were from $10 to $12 per sample for ISR, $35 to $185 for KW serotyping and $50 for the method of DNAhyb used here. The point of comparison begins when a colony is identified on agar that is suspected of being Salmonella. The low cost and simplicity of conducting ISR make it a method that supports public health laboratories and food producers with in-house laboratories in their efforts to monitor S. enterica. Other efficiencies such as submission of DNA to centralized facilities and applying robotics for sample preparation may lower the cost of conducting ISR further. If a simple method for serotyping S. enterica is available, farm management and plant processors may test samples and monitor environments more frequently. The ability of ISR to detect a mixture of serotypes, its independence of cell-surface epitopes, cost, and simple software requirements are relative strengths. We suggest that it will a useful addition for assigning serotype at minimal cost rather than being another typing method with no clear advantage (Achtman, 1996; Achtman et al., 2012).
Acknowledgments
Authors have no conflict of interest, financial or otherwise. Use of commercially available products by the United States Department of Agriculture (USDA) does not constitute endorsement. This manuscript was primarily supported by USDA-ARS Project Number 6612-32000-007-00 “Genetic Analysis of Poultry-Associated Salmonella enterica (abbr).” Submissions were collected in association with USDA-ARS Project Numbers 1265-32000-078-00, 6612-41420-017-00 and 6612-32420-001-00.
References
- Achtman M. A surfeit of YATMs? J Clin Microbiol. 1996;34:1. doi: 10.1128/jcm.34.7.1870-1870.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achtman M, Wain J, Weill FX, et al. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 2012;8:e1002776. doi: 10.1371/journal.ppat.1002776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bopp C, Brenner FW, Wells J, Strokbine N. EscherichiaShigella, and Salmonella. In: Murray P, Baron E, Pfaller M, Tenover F, Yolken R, editors. Manual of Clinical Microbiology. Washington, DC: ASM Press; 1999. pp. 459–474. [Google Scholar]
- Brenner FW, Villar RG, Angulo FJ, Tauxe R, Swaminathan B. Salmonella nomenclature. J Clin Microbiol. 2000;38:2465–2467. doi: 10.1128/jcm.38.7.2465-2467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curd H, Liu D, Reeves PR. Relationships among the O-antigen gene clusters of Salmonella enterica groups B, D1, D2, and D3. J Bacteriol. 1998;180:1002–1007. doi: 10.1128/jb.180.4.1002-1007.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foodnet. Vital signs: incidence and trends of infection with pathogens transmitted commonly through food–foodborne diseases active surveillance network, 10 U.S. sites, 1996–2010. MMWR Morb Mortal Wkly Rep. 2011;60:749–755. [PubMed] [Google Scholar]
- Guard J, Morales CA, Fedorka-Cray P, Gast RK. Single nucleotide polymorphisms that differentiate two subpopulations of Salmonella Enteritidis within phage type. BMC Res Notes. 2011;4:369. doi: 10.1186/1756-0500-4-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman F, Edwards P. A simplification of the Kauffmann-White schema. Am J Clin Path. 1952;22:692–697. doi: 10.1093/ajcp/22.7_ts.692. [DOI] [PubMed] [Google Scholar]
- Madajczak G, Szych J. Evaluation of PremiTest Salmonella kit for identification of not-typable by conventional methods Salmonella. Med Dosw Mikrobiol. 2010;62:29–36. [PubMed] [Google Scholar]
- Malorny B, Guerra B, Zeltz P, Rissler K, Helmuth R. Typing of Salmonella by DNA-microarrays. Berl Munch Tierarztl Wochenschr. 2003;116:482–486. [PubMed] [Google Scholar]
- Morales CA, Gast R, Guard-Bouldin J. Linkage of avian and reproductive tract tropism with sequence divergence adjacent to the 5S ribosomal subunit rrfH of Salmonella enterica. FEMS Microbiol Lett. 2006;264:48–58. doi: 10.1111/j.1574-6968.2006.00432.x. [DOI] [PubMed] [Google Scholar]
- Popoff M, Le Minor L. Antigenic Formulas of the Salmonella Serovars. Institut Pasteur, Paris: World Health Organization Collaborating Centre for Reference and Research on Salmonella; 2001. 8th revision. [Google Scholar]
- Porwollik S, McClelland M. Lateral gene transfer in Salmonella. Microbes Infect. 2003;5:977–989. doi: 10.1016/s1286-4579(03)00186-2. [DOI] [PubMed] [Google Scholar]
- Pulido-Landinez M, Laviniki V, Sanchez-Ingunza R, Guard J, Pinheiro do Nascimento V. Use of FTA cards for the transport of DNA samples of Salmonella spp. from poultry products from southern Brazil. Acta Sci Vet. 2012;40:1073. [Google Scholar]
- Wattiau P, Van Hessche M, Schlicker C, Vander Veken H, Imberechts H. Comparison of classical serotyping and PremiTest assay for routine identification of common Salmonella enterica serovars. J Clin Microbiol. 2008;46:4037–4040. doi: 10.1128/JCM.01405-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattiau P, Boland C, Bertrand S. Methodologies for Salmonella enterica subsp. enterica subtyping: gold standards and alternatives. Appl Environ Microbiol. 2011;77:7877–7885. doi: 10.1128/AEM.05527-11. [DOI] [PMC free article] [PubMed] [Google Scholar]



