Abstract
G-quadruplexes are higher-order nucleic acid structures that can form in G-rich telomeres and promoter regions of oncogenes. Telomeric quadruplex stabilization by small molecules can lead to telomere uncapping, followed by DNA damage response and senescence, as well as chromosomal fusions leading to deregulation of mitosis, followed by apoptosis and downregulation of oncogene expression. We report here on investigations into the mechanism of action of tetra-substituted naphthalene diimide ligands on the basis of cell biologic data together with a National Cancer Institute COMPARE study. We conclude that four principal mechanisms of action are implicated for these compounds: 1) telomere uncapping with subsequent DNA damage response and senescence; 2) inhibition of transcription/translation of oncogenes; 3) genomic instability through telomeric DNA end fusions, resulting in mitotic catastrophe and apoptosis; and 4) induction of chromosomal instability by telomere aggregate formation.
Introduction
G-rich nucleotide sequences comprising repetitive short G-tracts, such as in telomeric DNA, diverse gene promoter regions, and the 5′-untranslated regions of many mRNAs, may fold into G-quadruplex higher-order nonduplex structures (Burge et al., 2006). These can be involved in telomere maintenance or DNA damage signaling and may act as molecular switches to regulate transcription and translation (Lipps and Rhodes, 2009; Bochman et al. 2012).
G-quadruplex stabilization by small molecules has diverse cellular effects, depending on the target nucleic acid. It can activate a DNA damage response in cancer cells, indicated by DNA double-strand break foci with phosphorylated histone γ-H2AX and chromosomal fusions in metaphase (Gomez et al., 2003; Burger et al., 2005; Pennarun et al., 2005; Tahara et al., 2006; Salvati et al., 2007; Leonetti et al., 2008; Rodriguez et al., 2008). This activates cell-cycle checkpoints, growth arrest, senescence, and apoptosis (Tauchi et al., 2003; Pennarun et al., 2005; Qi et al., 2006). The DNA damage response is a result of telomeric protein uncapping, notably the end-protection POT1 protein (Zaug et al., 2005). Small-molecule quadruplex-binding ligands inhibit POT1 binding to the telomeric overhang (Gomez et al., 2006a,b; Gunaratnam et al., 2007) and may dissociate other proteins from the telomere, such as TRF2 (Karlseder et al., 1999; Tahara et al., 2006; Gunaratnam et al., 2007; Zhou et al., 2009). The ligand telomestatin displaces POT1 in both normal and cancer cells, but it displaces TRF2 only in the latter, resulting in selective DNA damage response activation (Gomez et al., 2006a, b; Tahara et al., 2006). TRF2 represses Ataxia telangiectasia mutated (ATM) kinase, which is activated on dissociation from the complex (Denchi et al., 2007; Pennarun et al., 2008; Rizzo et al., 2009). Chromatin immunoprecipitation sequencing analysis of the effects of a quadruplex ligand on a cancer cell genome (Rodriguez et al., 2012) has shown that damage is not restricted to telomeres but also occurs at other gene loci.
Checkpoint signaling on telomere damage can proceed in a p53-dependent or p16INK4a/Rb dependent manner (Smogorzewska and de Lange, 2002; Feldser and Greider, 2007). Upregulation of senescence-associated cyclin-dependent kinase inhibitors p21 and p16INK4a produces chromosomal fusions in prostate cancer cells treated with the compound BRACO-19 (9-[4,N,N-dimethylamino)phenylamine]-3.6-box)3-[urrp;pdom-propionamio)acridine x 3HCl) (Incles et al., 2004). Treatment of leukemia cells with telomestatin leads to phosphorylation of Chk2, upregulation of p21, senescence, and cell-cycle arrest in G1 phase (Tauchi et al., 2003). Other G-quadruplex ligands, such as TMPyP4, cause G2/M phase arrest in MCF7 cells at subcytotoxic concentrations (Izbicka et al., 1999). Multiple effects on cell-cycle progression, such as activation of S-phase checkpoint and increase of M-phase, can also occur (Pennarun et al., 2005), as well as S- and G2/M phase arrest (Leonetti et al., 2004; Huang et al., 2008).
These cell-cycle effects may depend on cell type, individual mutations in the genes involved in checkpoint signaling in particular cancer cell lines, the nature and concentration of ligand, as well as its binding mode and selectivity for particular G-quadruplexes. Most ligands can affect telomere stability, with the formation of chromosomal end fusions, anaphase bridges (Capra et al., 2010), and translocations resulting in mitotic catastrophe (Tauchi et al., 2003; Pennarun et al., 2005; Shimizu et al., 2005). Nevertheless, not only telomeric end-to-end fusions but also the reorganization of telomeres in three-dimensional (3D) interphase nuclei (e.g., the formation of so-called telomeric aggregates) can result in chromosomal instability (Chuang et al., 2004; Louis et al., 2005), an effect that possibly also plays a role in G-quadruplex effects on the genome. Additionally, G-quadrplex stabilization may hinder cell-cycle progression directly: DNA needs to be unwound for DNA synthesis in S-phase; accordingly, stabilization may stop S-phase progression (Paeschke et al., 2008).
Several G-quadruplex ligands have been shown to decrease oncogene expression (reviewed by Qin and Hurley, 2008; Balasubramanian et al., 2011). In theory, oncogenes associated with all six hallmarks of cancer (Hanahan and Weinberg, 2011) can be targeted with G-quadruplex ligands. G-quadruplex ligands can inhibit c-kit expression and activity in a c-kit-expressing gastrointestinal stromal tumor cell line (Gunaratnam et al., 2009; Bejugam et al., 2010; McLuckie et al., 2011). The G-quadruplex ligand quarfloxin decreases RNA polymerase I transcription by inhibiting nucleolin binding to ribosomal DNA G-quadruplexes in the nucleoli (Drygin et al., 2011). A study of quarfloxin using the COMPARE computer program did not show any correlation to known compounds, suggesting a unique mechanism of action.
We previously reported on a family of tetra-substituted naphthalene diimide (ND) ligands, which show elevated G-quadruplex DNA stabilization and potent antiproliferative activity in a panel of cancer cell lines (Cuenca et al., 2008; Gunaratnam et al., 2009; Hampel et al., 2010; Gunaratnam et al., 2011; Collie et al., 2012). We report here on studies of the cellular mechanism of action of three selected ND compounds from this family (BMSG-SH-3–5; Fig. 1). Compound BMSG-SH-3 (N,N’-bis(3-(4-methylpiperazin-1-yl)propylamino)-2,6-bis(3-(4-methylpiperazin-1-yl)pentylamino)-1,2,5,8-naphthalenetetracarboxylic acid diimide) is the particular focus of this study since previous xenograft studies have demonstrated in vivo antitumor activity against the MIA-Pa-Ca-2 pancreatic cancer (Gunaratnam et al., 2011). It is shown here that this compound induces DNA damage and senescence, may inhibit protein synthesis, and may initiate genomic instability, which has an antimitotic effect. A number of other ND compounds with G-quadruplex affinity have been described (see, e.g., Nadai et al., 2011; Doria et al., 2012a,b; Wang et al., 2012), with telomerase activity and telomere targeting ability in some instances.
Fig. 1.
Structures of BMSG-SH-3–5.
Materials and Methods
FRET Melting Temperature.
The oligonucleotide sequences were purchased from Eurogentec, Southampton, UK. From a 50-μM nucleotide stock solution, a 400-nM solution in FRET buffer (50 mM sodium cacodylate, pH 7.4) was prepared. The nucleotide was annealed by heating the sample up to 90°C for 10 min and allowing it to cool down to room temperature within 4 h. A 10-mM stock solution of BMSG-SH-1 in deionized water was diluted to double of the required concentrations with FRET buffer. Each well of RT-PCR 96-well plates (MJ Research, Waltham, MA) was loaded with 50 μl of oligonucleotide solution and 50 μl of drug solution. Drug concentrations of 0.1, 0.2, 0.5, 1, 2, 5, and 10 μM were used and repeated three times. Measurements were conducted on a DNA Opticon Engine (MJ Research) with excitation at 450–495 nm and detection at 515–545 nm. The fluorescence was read at intervals of 0.5°C, over the range 30–100°C. Before each reading, the temperature was held constant for 30 s. The raw data were processed using Origin (Version 7.0, OriginLab Corp., Northampton, MA). The graphs were smoothed using a 10-point running average and normalized. The melting temperature was obtained by determining the maximum of the first derivative of the smoothed melting curve. The value of ΔTm is the difference in melting temperatures between that for the quadruplex in the presence of the drug at a certain concentration and that for the native quadruplex.
Tissue Culture.
The human carcinoma cell lines MCF7, A549, MIA-Pa-Ca-2 (European Collection of Cell Cultures), and human pancreatic adenoma carcinoma (HPAC; American Type Culture Collection) were maintained in monolayer culture in 75 cm2 flasks (TPP, Trasadingen, Switzerland) under a humidified 5% CO2 atmosphere at 37°C. Incubations were also carried out under these conditions unless specified otherwise. For the cell lines MCF7 and A549, Dulbecco’s modified Eagle’s medium (DMEM) was used (GIBCO 21969, Invitrogen, UK) supplemented with L-glutamine (2 mM, GIBCO 25030, Invitrogen), essential amino acids (1%, GIBCO 11140, Invitrogen), fetal calf serum (10%, S1810, Biosera, UK), and hydrocortisone (0.5 μg/ml, Acros Organics, UK). For MIA-Pa-Ca-2, Dulbecco’s MEM, supplemented with L-glutamine (2 mM) and fetal calf serum (10%) was used. For HPAC, D-MEM/F12 (1:1) (21331, Invitrogen) supplemented with L-glutamine (2 mM), insulin (0.002 mg/ml, GIBCO 12585-014, Invitrogen), transferrin (0.005 mg/ml, 11108-016, Invitrogen), hydrocortisone (40 ng/ml), epidermal growth factor (10 ng/ml, 53003-018, Invitrogen), and fetal calf serum (10%) was used. To passage the cells, they were washed with phosphate-buffered saline (PBS; GIBCO 14040; Invitrogen), treated with trypsine (GIBCO 25300; Invitrogen), and reseeded into fresh medium, resulting in an initial cell density of approximately 1 × 104 cells/ml of medium. Cells were counted using a Neubauer hemocytometer (Assistant, Germany) by microscopy or a MacsQuant flow cytometer (Miltenyi Biotech, Germany) on a suspension of cells obtained by washing with PBS, trypsinization, centrifugation at 8°C at 8000 rpm for 3 min, and resuspension in fresh medium.
Cell Uptake.
Cover glasses (16-mm diameter, thickness #1, FB58700, Fisher, UK) were cleaned in HCl (2 M) overnight, rinsed with deionized water, and sterilized in ethanol (70% v/w) overnight, dried, and coated with polylysine solution (100 μg/ml, P4832, Sigma Aldrich, UK) under sterile conditions. In a six-well plate (Nunc A/S), 0.5 ml of indicator-free medium (GIBCO 31053, Invitrogen) containing 2 × 105 cells was placed on top of one dry cover glass per well. After incubation overnight, leading to enhancement of the cells to the cover glasses, the suitable amount of a 1-mM stock solution of the drug was added, and the cells were incubated for 30 min. The media were then removed, and the cells were washed with PBS and fixed with fresh formaldehyde solution (2% in PBS) for 10 min at room temperature. After washing with PBS (5 × 2 ml), the cover glasses were placed face down on microscopy slides (631-0114, VWR International) using mounting medium (6 μl, Vectashield; Vector Laboratories, CA). The edges of the cover glasses were sealed with nail varnish. Visualization was performed with a LSM 710 META confocal microscope (Zeiss, Germany) with a plan-apochromat 40×/1.3 oil DIC M27 oil submersion lens and a 410- to 533-nm filter. Cell morphology was visualized by light microscopy, and the drug was visualized by its fluorescence in the red light spectrum.
Long-Term Growth Inhibition.
After counting, 3*104 cells were seeded in 10 ml of medium containing the compound to be tested in a 75-cm2 flask and incubated for 7 days. The cells were then harvested and counted, and 3*104 of the cells were reseeded; the process was repeated for another week. Population doublings were calculated using the following formula:
where N0 is the number of cells at the time of seeding (3*104), Nf is the number of cells at the time of counting, and pd is the number of population doublings.
Senescence Staining.
Staining for senescence-associated β-galactosidase was carried out following the protocol from the supplier (Cell Signaling Technology, Inc., Beverly, MA); 1 × 105 cells obtained from the long-term studies were seeded in 35-mm six-well plates (Nunc A/S) in 2 ml medium and incubated overnight. The medium was then removed, the cells washed, fixed with formaldehyde (2%) and glutaraldehyde (0.2%) in PBS for 15 min, and washed twice more. Then 1 ml of a staining solution [citric acid/sodium phosphate (40 mM), pH 6.0, NaCl (0.15 M), MgCl2 (2 mM), potassium ferrocyanide (5 mM), potassium ferricyanide (5 mM), X-gal (1 mg/ml, 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside] was added to each well. The cells were incubated overnight and examined by light microscopy (magnification 200×) for the characteristic senescence-associated blue stain.
Flow Cytometry and Cell-Cycle Analysis.
Flow cytometry experiments were carried out on a MACS Quant flow cytometer (Miltenyi Biotech, Germany) with specific MACS Quantify software. After incubation with the compound, cells were harvested by trypsinization and centrifuged (4 min at 8000 rpm). Eventual floating cells in the media were also collected. Cells were washed with PBS and fixed with ice-cold ethanol (70%) while mixing and stored at 4°C. The ethanol was removed by centrifugation (8 min at 12,000 rpm) and the cells washed with PBS twice. The pellet was incubated with ribonuclease A solution (50 μl, 100 μg/ml, 15 min at room temperature, Sigma Aldrich), and propidium iodide (PI) solution (450 μl of a 50 μg/ml solution in PBS, Sigma Aldrich) was added to the mixture. During flow cytometric analysis, at least 10,000 events were collected. Events on the PI channel were collected on a linear scale, and in a dot plot displaying PI area versus PI height, doublets, clumps, and subG1 cells were gated out.
γ-H2AX Staining.
After incubation with the compound, cells were harvested by trypsinization and centrifuged (4 min at 8000 rpm). Eventual floating cells in the media were also collected. The cells were washed with PBS and fixed with paraformaldehyde (2% in PBS) for 15 min at room temperature while mixing. Ice-cold PBS (1 ml) was added, and the solvent removed by centrifugation (8 min at 12,000 rpm). The cells were permeabilized with triton x (0.1% in PBS, Sigma Aldrich, 1 ml) for 30 min at room temperature and washed twice with blocking buffer (3% BSA (Bovine serum albumin), Sigma Aldrich, and 0.05% triton x in PBS). Phospho histone H2AX (Ser139)(20E3) rabbit monoclonal antibody alexa fluor 488 conjugate (5 μl, #9719, Cell Signaling, USA) was added, and the cells were incubated for 30 min at room temperature. The cells were washed once with PBS and analyzed by flow cytometry after resuspension in PBS (200 μL). At least 10,000 events were collected.
Fluorescence in Situ Hybridization (FISH) and 3D Microscopy.
To investigate the telomere organization in 3D interphase cells, experiments were performed on the HaCaT-myc cell line cultured in DMEM with 10% FCS (Fetal Calf Serum), and 1% penicillin/streptomycin and grown on coverslips. The cells were either treated with the ligand or, for control purposes, incubated only with solvent HCl. Additionally, an extra set of treated and untreated cells were exposed to 50 J/m2 ultraviolet (UV)-C light. In all cases, the cells were fixed 48 h post treatment.
To visualize the telomeres in the cells, 3D FISH, applying a telomere-specific, CY3-labeled PNA probe, was performed. Therefore, the slides were fixed with 3.7% formaldehyde for 10 min and then washed once in PBS/ 0.1% glycine followed by 12 min PBS/0.2% triton × 100 and twice for 5 min in PBS. Afterward, the slides were dehydrated with 70%, 85%, and 100% ethanol and air-dried. For hybridization, a CY3-labeled telomeric PNA probe (DAKO) was applied to the slides and codenatured for 3 min at 80°C. Hybridization took place for 2 hours at 30°C. The slides were then washed twice for 15 min in 70% formamide/10 mM Tris base, 1 min in PBS, and 5 min in 0.1 × SSC (Saline-Sodium Citrate) at 55°C. The slides were subsequently washed twice for 5 min in PBS/0.05% tween 20 and mounted with 20 μl Vectashield containing 500 ng/ml DAPI.
After FISH, the slides were examined with an Axioplan 2 microscope (Zeiss, Germany), equipped with appropriate filter sets, an HRM CCD camera (Zeiss), and an automated z-stage with a 63 × oil objective lens. For each region of interest, 100 pictures were made for each fluorescent filter, with 0.2-μm distance in Z between the pictures. For each experiment, at least 100 cells were examined. Deconvolution was performed using the nearest neighbor algorithm and visually examined in 3D for telomere organization.
All telomeres within each cell were inspected for the formation of double, triple, or more than triple telomere associations or the formation of telomeric aggregates. In cases of telomeric aggregate formation, the number of telomeres involved in this abnormal formation cannot be determined and clearly disturbs the even and random distribution of telomeres through the 3D interphase nucleus.
Results
Quadruplex Binding.
FRET melting temperature assays have shown that compounds BMSG-SH-3–5 selectively stabilize particular G-quadruplex structures (Table 1). They have high affinity for the telomeric G-quadruplex sequence F21T, with ΔTm values between 23.8 and 31.7°C, and low affinity for the T-loop duplex DNA sequence. Additionally, the compounds stabilize other G-quadruplex–forming sequences, such as those found in the promoter regions of the oncogenes c-kit and hif-1α and in the 5′-UTR of the cell survival gene bcl-2. Affinity for the c-kit1 sequence is low, with ΔTm values from 1.5 to 4.5°C and modest for the c-kit2 sequence with ΔTm values from 7.7° to 15.1°C. Compound BMSG-SH-3 shows low stabilization of the bcl-2 sequence, whereas BMSG-SH-4 and -5 display high stabilization with ΔTm values above 20°C. They also show moderate affinity for the hif-1α sequence, with ΔTm values of 5.9 K and 4.9°C, respectively.
TABLE 1.
FRET melting temperature experiments with BMSG-SH-3–5 on various nucleotide sequences
| Compound or Nucleotide | F21T (G4 DNA, K+) | c-kit1 (G4 DNA, K+) | c-kit2 (G4 DNA, K+) | bcl-2 (G4 RNA, Na+) | hif-1α (G4 DNA, K+) | T-Loop (Duplex DNA, K+) |
|---|---|---|---|---|---|---|
| BMSG-SH-3 | 23.8 | 1.5 | 7.7 | 3.8 | n. a. | 0.2 |
| BMSG-SH-4 | 33.0 | 4.5 | 8.4 | 22.5 | 5.9 | 3.8 |
| BMSG-SH-5 | 31.7 | 1.6 | 15.1 | 21.0 | 4.9 | 3.8 |
FRET, fluorescence resonance energy transfer.
Cellular Effects.
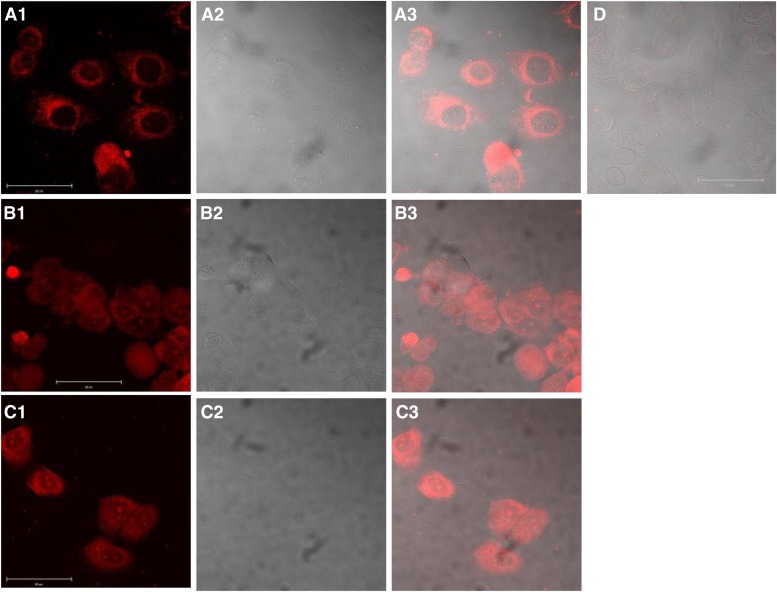
Because of their fluorescent properties, compounds BMSG-SH-3–5 were readily visualized in MCF7 cells (Fig. 2), albeit with considerable variations in the concentration required. BMSG-SH-3 was visualized in the cytoplasm only after incubation at high concentration, 50 μM, whereas BMSG-SH-4 and -5 accumulated in the cytoplasm and in nucleoli after incubation at a lower concentration, 500 nM, for 30 min; increased incubation times gave similar results. BMSG-SH-3 displayed significantly less intense fluorescence than BMSG-SH-4 and -5 (i.e., excitation 600 nm, emission 650–675 nm, see Supplementary Fig. S1), which, however, increased at low pH.
Fig. 2.
Cell uptake study of BMSG-SH-3–5 in the breast cancer cell line MCF7. (A1) BMSG-SH-3, 50 μM, 30 min, fluorescence image. (A2) Transmitted light image. (A3) Merge of images A1 and A2. (B1) BMSG-SH-4, 500 nM, 30 min, fluorescence image. (B2) Transmitted light image. (B3) Merge of images B1 and B2. (C1) BMSG-SH-5, 500 nM, 30 min, fluorescence image. (C2) Transmitted light image. (C3) Merge of images C1 and C2. (D) Untreated sample, merge of fluorescence and transmitted light image. Scale: 50 μm.
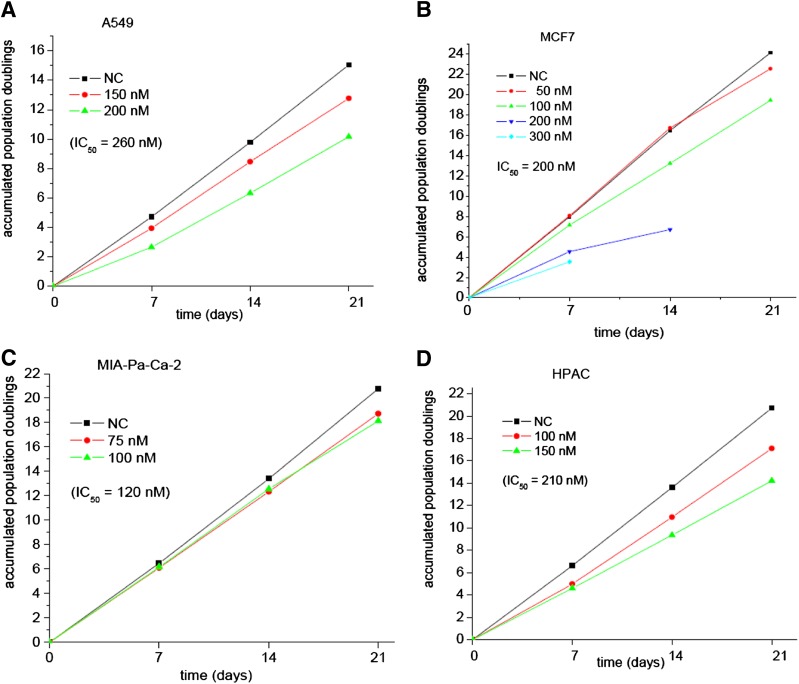
BMSG-SH-3 decreased growth of the A549, MCF7, MIA-Pa-Ca-2, and HPAC cancer cell lines in long-term studies and at various subcytotoxic concentrations (Fig. 3). Growth of A549, MCF7, and HPAC decreased to 88%–50% after incubation with BMSG-SH-3 for 1 week, whereas a decrease to 90% in the growth of MIA-Pa-Ca-2 cells was observed after 2 weeks. Cell growth of MCF7, MIA-Pa-Ca-2, and HPAC decreased further, to up to 40% in subsequent weeks.
Fig. 3.
Long-term growth inhibition of cancer cell lines in the presence of BMSG-SH-3. (A) A549 cells. (B) MCF7 cells. (C) MIA-Pa-Ca-2 cells. (D) HPAC cells.
A549 cells incubated with BMSG-SH-3 at a 200-nM concentration for 2 weeks showed senescence, in terms of flattened, enlarged morphology and positive β-galactosidase activity (with associated senescence staining at pH 6) (Fig. 4).
Fig. 4.
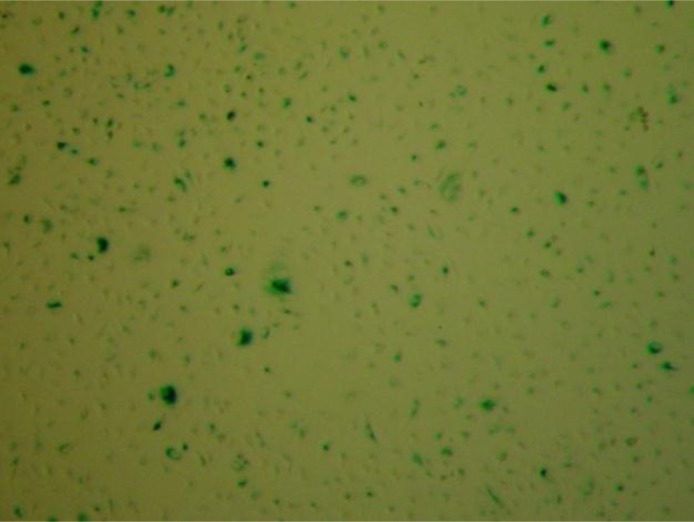
Senescence staining of A549 cells incubated with BMSG-SH-3 (200 nM) for 14 days.
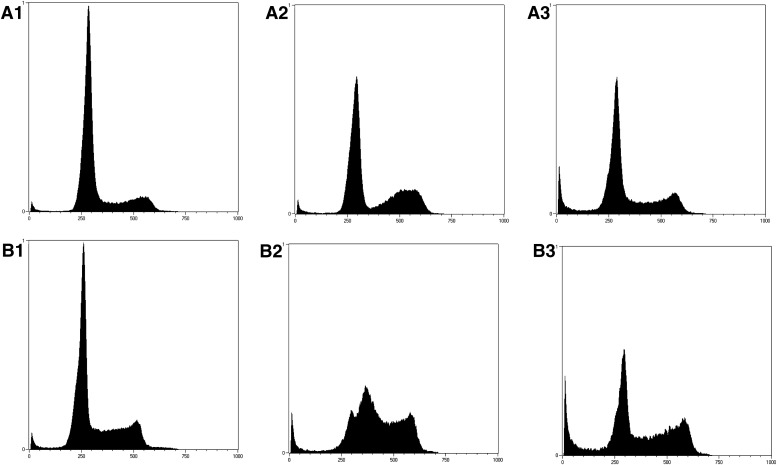
The effects of BMSG-SH-3 on cell-cycle behavior of A549 and MIA-Pa-Ca-2 cells were investigated by PI staining and flow cytometric analysis. At subcytotoxic concentrations, no significant effects were found (results not shown); however, at 1-μM concentration, BMSG-SH-3 produced G2/M phase arrest and apoptosis, manifest as an increased sub-G1 population (Fig. 5, A3 and B3). The reference compound etoposide is known to cause DNA damage leading to mitotic catastrophe (Tanaka et al., 2007), which is shown as G2/M phase arrest after incubation at 30-μM concentration (Fig. 5, A2 and B2).
Fig. 5.
Cell-cycle analysis of cancer cell lines treated with BMSG-SH-3 and etoposide, which is known to cause DNA damage and mitotic catastrophe. (A1) A549 cells, untreated. (A2) A549 cells treated with etoposide (30 μM) for 14 h. (A3) A549 cells treated with BMSG-SH-3 (1 μM) for 14 h. (B1) MIA-Pa-Ca-2 cells, untreated. (B2) MIA-Pa-Ca-2 cells treated with etoposide (30 μM) for 14 h. (B3) MIA-Pa-Ca-2 cells treated with BMSG-SH-3 (1 μM) for 14 h.
Effects at the Genome Level.
Since genomic and genetic changes give rise to genomic instability, we investigated the effects of BMSG-SH-3 at the genome level. Here, we focused on two parameters: the induction of double-strand breaks (DNA-damage; γ-H2AX foci) as well as telomeric aberrations in terms of telomere length and telomere organization.
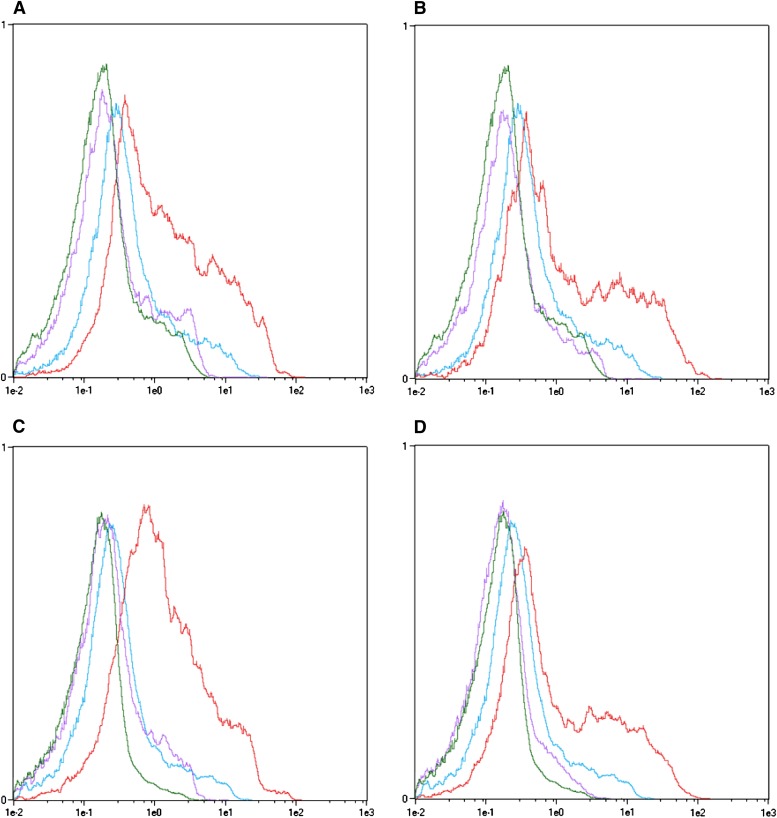
DNA damage in A549 and MIA-Pa-Ca-2 cells caused by BMSG-SH-3 was assessed using flow cytometry (see, for example, Huang and Darzynkiewicz, 2006), to detect γ-H2AX (Fig. 6). After 14 h of incubation with BMSG-SH-3 at 1-μM concentration, a significant level of γ-H2AX was detected (Fig. 6, B and D). The reference compound etoposide, a known DNA-damaging agent (Tanaka et al., 2007), produced a large amount of γ-H2AX after incubation at a higher concentration of 30 μM (Fig. 6, A and C).
Fig. 6.
DNA damage in the form of H2AX phosphorylation caused by BMSG-SH-3 in cancer cells. Color code: Green—untreated, unstained. Blue—untreated, anti γ-H2AX stained. Purple— treated, unstained. Red—treated, anti γ-H2AX stained. (A) A549 cells treated with etoposide, 30 μM, 14 h. (B) A549 cells treated with BMSG-SH-3, 1 μM, 14 h. (C) MIA-Pa-Ca-2 cells treated with etoposide, 30 μM, 14 h. (D) MIA-Pa-Ca-2 cells treated with BMSG-SH-3, 1 μM, 14 h.
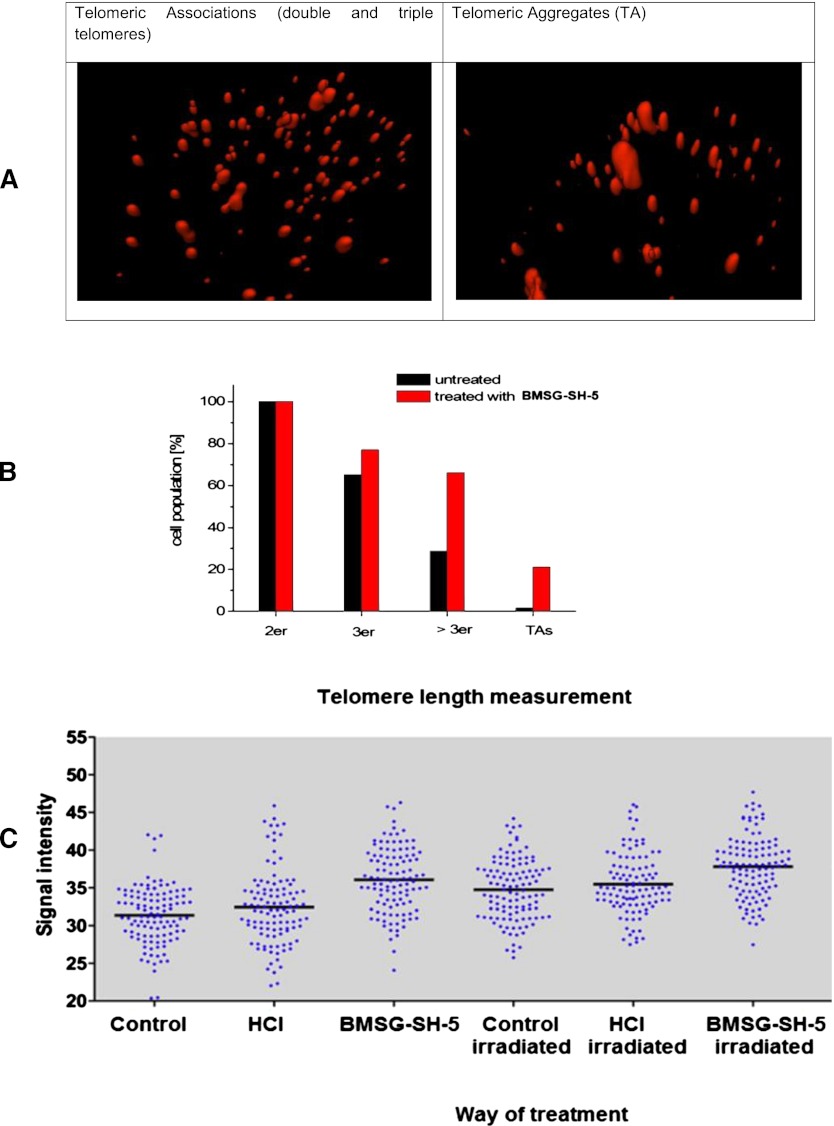
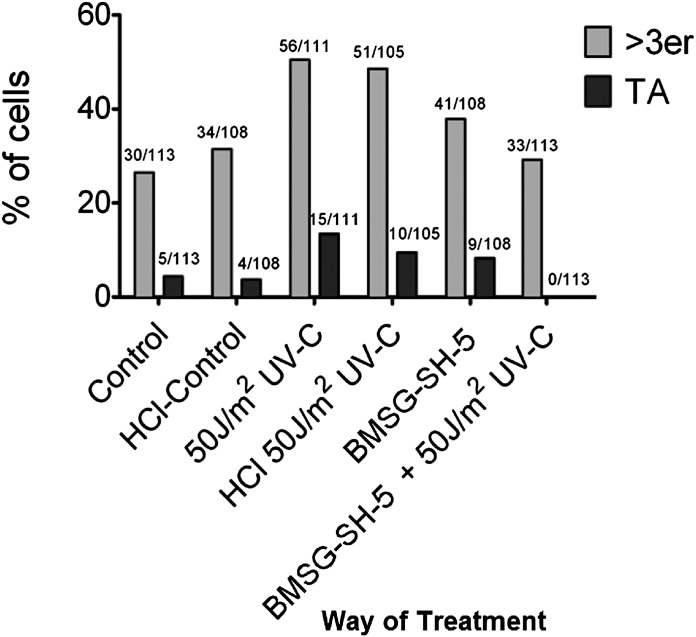
Telomeres, the ends of linear chromosomes, have long been known to induce genomic instability via breakage-fusion-breakage cycles as a result of short telomeres (Cowell and Miller, 1983; Murnane 2006). In these cases, telomeric fusions are causative for chromosomal breakage and the formation of translocations and other genetic changes. Nevertheless, it has more recently been shown that telomeres are also causative for genomic changes in a telomere-length– independent manner, namely, by changes in the 3D telomere organization in interphase nuclei (Mai, 2010). Here, telomeres do not fuse but form telomeric associations, which can be visualized as telomeric aggregates. It has been shown that the offspring of cells showing TAs also show chromosomal changes, which were not detected in control cells (Louis et al., 2005). Therefore, BMSG-SH-5 was examined for its ability to induce telomere aggregation in kerationzyte-derived HaCaT-myc cells (Boukamp et al., 1988; Schuermann 1990), which show a small but constant fraction of TAs (Ermler et al., 2004). Treatment of HaCaT-myc cells with BMSG-SH-5 (40 nM, 48 h) resulted in a significant increase of associates of three or more telomeres and TAs (Fig. 7, A and B). Both treated and untreated cells carried two associated telomeres. In the untreated sample, 28.8% of cells carried naturally occurring associates of more than three telomeres, and 1.7% carried TAs (Fig. 7B). In the sample treated with BMSG-SH-5, associates of more than three telomeres were found in 77.1% of cells, and TAs in 21.1% of cells. Additionally, the samples that had been investigated for telomere aggregate formation by FISH had been screened for signal intensity of the telomeric signals, which is a measure for telomere length (see Fig. 7C). This analysis showed that HaCaT-myc cells treated with the ligands results in an increase in telomere length 48 h after treatment. This effect is even enhanced in cells that had been exposed to the ligand and additionally been irradiated with 50 J/m2 UV-C. Interestingly, these double-treated samples did not show as many telomeric aggregates as the cell samples that were only UV irradiated (see Fig. 8).
Fig. 7.
(A) Pictures of single cells: 3D fluorescence in situ hybridization of HaCaT-myc cells applying a CY3 (red) labeled telomere-specific PNA probe (left: cell showing double and triple telomeres; right: cell showing a telomeric aggregate). (B) Telomere association (2er, 3er, > 3er) and telomere aggregates (TAs) in HaCaT-myc cells incubated with BMSG-SH-5 (40 nM, 48 h). (C) Representation of the telomeric signal intensity as a measure for telomere length. Each set of blue dots represents the median telomere length within a single cell; the black bar indicates the mean telomere length within each data set. Here, the ligand concentration was 40 nM, and the cells were irradiated with 5 J/m2 UV-C.
Fig. 8.
Telomere aggregate (TA) analysis. The figure shows the percentage of telomerically aberrant cells for the differently treated cell samples. Only cells with >3er and TAs are considered “aberrant” since double and triple telomeres can occur also in genomically stable cells. The first number above the bars represents the number of cells with the respective aberration per slide, and the second number represents the total number of observed cells per slide.
Compatibility with Published Data Sets.
Compounds BMSG-SH-3–5 were screened in the NCI 60 cell line panel, (http://www.dtp.nci.nih.gov/branches/btb/ivclsp.html) and showed activity against numerous cell lines, with LC50/GI50 ratios of up to 389 (see Supplementary Data). Their activity profiles were analyzed by the COMPARE algorithm against the NCI compound database (Paull et al., 1989). BMSG-SH-3 shows significant mechanistic correlation with other compounds of up to 82%, whereas BMSG-SH-4 and -5 only showed high mechanistic correlation with each other and BMSG-SH-3, and correlations with other compounds were only up to 59% Supplementary Table S2). A literature search revealed that correlated compounds have distinctive mechanisms of action:
DNA and G-quadruplex DNA binding, DNA damage
Inhibition of transcription and DNA replication
Inhibition of translation and protein synthesis
Targeting of mitochondria or antimitotic effect
A COMPARE analysis with the NCI microarray dataset was also performed (Supplementary Table S3). The genes expressed in the cell lines in which compounds BMSG-SH-3–5 show activity encode for proteins, of which several are nucleotide binding, such as zinc-finger proteins, or involved in alternative splicing. Genes encoding for proteins involved in protein transport and metabolic processes are correlated with ND compound activity, as well as proteins involved in cytokine or DNA damage signaling.
Discussion
The ability of compounds BMSG-SH-3–5 to stabilize telomeric G-quadruplexes has been demonstrated in previous studies using FRET, surface plasmon resonance, and circular dichroism techniques (Cuenca et al., 2008; Hampel et al., 2010). They are also confirmed to be inhibitors of the telomerase enzyme (Greulich-Bode K, unpublished observations). The present study shows that they are able to stabilize G-quadruplexes in promoter regions of the oncogenes c-kit and hif-1α, as well as a quadruplex formed in the mRNA of the bcl-2 gene. This gives them the potential to inhibit transcription and translation of these oncogenes or cell-survival genes, which may have antiproliferative or anticancer effects. Investigation of transcription or translation inhibition is beyond the scope of this study, although it is notable that we previously showed BMSG-SH-5 (Gunaratnam et al., 2011) to bind to a promoter quadruplex in the HSP90 gene and also to downregulate the expression of the HSP90 protein in BMSG-SH-5-treated pancreatic cancer tumors.
The accumulation of BMSG-SH-4 and -5 in nucleoli indicates interactions with rDNA, which may inhibit nucleolin binding, decrease the transcription of RNA polymerase I, and decrease ribosomal protein synthesis (Drygin et al., 2011). BMSG-SH-3 may be present in nucleoli, but its fluorescence is too weak for it to be visualized (see Fig. S1, Supplementary Data). The cytoplasmic accumulation of BMSG-SH-3–5 suggests interactions with extranuclear nucleic acids, such as mRNAs, ribosomal RNA, or mitochondrial DNA. Appropriate RNA sequences may form G-quadruplexes more readily than DNA sequences can, as much RNA is single-stranded and not associated with chromosomal proteins. Mitochondrial DNA contains 10 times more sequences that form putative G-quadruplexes than does nuclear DNA (Capra et al., 2010).
BMSG-SH-3 is able to initiate senescence in cancer cells, indicated by increasing growth inhibition in the long-term study, accompanied by an altered cell morphology and positive senescence-associated β-galactosidase staining of A549 cells treated with BMSG-SH-3 for 2 weeks. The ability of BMSG-SH-3 to uncap a telomeric DNA sequence from the hPOT1 protein and topoisomerase IIIα was previously demonstrated in vitro with a closely similar compound, (Hampel et al., 2010), so that the observed senescence is consistent with it being a result of telomere uncapping through G-quadruplex stabilization. This is in accordance with reports on human prostate cancer cells, where p16-associated senescence was detected after exposure to a G-quadruplex telomere-targeting agent (Incles et al., 2004).
BMSG-SH-3 initiates a DNA damage response in cancer cells, which has been demonstrated by the detection of γ-H2AX foci. This DNA damage signal is likely to be a response to telomere uncapping, which may result in telomere fusions, anaphase bridges, and chromosomal instability as a result of repeating fusion breakage cycles. Telomeric G-quadruplex targeting agents have previously been shown to produce chromosomal fusions (Incles et al., 2004), which indicates that ND compounds have the potential to promote the occurrence of telomeric end-to-end fusions and initiate telomere uncapping. They may additionally stabilize fused telomeres, as it has been proposed that telomeric ends are interconnected via G-quadruplexes (Maizels et al., 2006). BMSG-SH-3 causes G2/M arrest and apoptosis in cancer cells, which may be due to such telomeric chromosomal fusions, followed by anaphase bridges promoted by the ligand, leading to deregulation of mitosis and mitotic catastrophe. The hypothesis of compound-induced telomere fusions is strengthened by the increase in telomere length 48 h after G-quadruplex exposure. This increase can be explained by telomeric end-to-end fusions which result in higher signal intensities per telomeric signal.
Additionally and independent of the telomere length, BMSG-SH-5 induced changes in the 3D distribution of the telomeres in interphase nuclei, resulting in an increased number of telomeric associations (double, triple, and more than triple telomeres) and telomeric aggregates in HaCaT-myc cells. This altered telomere distribution has been linked to chromosomal instability by proving that cells that showed telomeric aggregates revealed chromosomal aberrations several population doublings later in metaphase (Louis et al., 2005; Mai, 2010). This is suggestive of induction of genomic instability via telomere aggregate formation by the action of BMSG-SH-5.
Formation of TAs in HaCaT-myc cells can also be induced by UV-C irradiation, where 48 and 72 h post irradiation approximately twice as many cells represent TAs (see Fig. 8). We used this knowledge to investigate whether aggregate formation is causative for the increase in telomere length in cells treated with BMSG-SH-5. Our results clearly show that the large increase in telomere length is not induced merely by the appearance of TAs: The number of aggregates is highest in the cells that had only been irradiated. Nevertheless, these cells do not show a large increase in telomere length. Vice versa, the ligand-treated cells show a very large increase in telomere length within 48 h of irradiation compared with nonirradiated cells, but have significantly lower numbers of TAs compared with the only UV-irradiated samples.
Taken together, the experimental results thus suggest four principal mechanisms of action for these ND compounds:
Telomere uncapping with subsequent DNA damage response and senescence
Inhibition of transcription/translation of oncogenes
Genomic instability though telomeric end fusions, (Boukamp, Popp and Krunic, 2005) resulting in mitotic catastrophe and apoptosis.
Induction of chromosomal instability by telomere aggregate formation
These four mechanisms are also found among the compounds that were correlated with these ND compounds in the mechanistic COMPARE study (Supplementary Table S2). BMSG-SH-4 and -5 have high mechanistic similarity with each other and correlate with other compounds only up to 59%, whereas there is a higher correlation for BMSG-SH-3 with compounds having the above distinctive mechanisms of action. This may be due to its higher selectivity for G-quadruplex over duplex DNA (possibly as a result of its shape and size not being compatible with duplex binding (Hampel et al., 2010)). A large number of DNA-binding and G-quadruplex–stabilizing compounds, such as anthracyclines, ellipticinium derivatives, and a porphyrin, were mechanistically correlated with the ND compounds. Both G-quadruplex stabilizing and classic DNA damage–inducing agents have been shown to cause senescence, for example, the anthracycline doxorubicin, which is able to induce senescence in cancer cells lacking p53 and p21 (Ewald et al., 2010). The COMPARE study against the microarray library shows that the NDs have effects on cell lines expressing proteins that bind to or interact with DNA and that may be involved in damage signaling. Compounds inhibiting protein expression were correlated with the ND mechanisms of action, as well as genes encoding for protein transporters. Antimitotic compounds and other compounds that target mitochondria were also correlated with the ND mechanisms of action; this antimitotic effect may be a result of mitotic catastrophe resulting from chromosomal instability after DNA damage, or it may be a direct effect of the compounds binding to mitochondrial DNA, which is indicated by the cell uptake studies finding that they were located in the cytoplasm.
These four mechanisms of action of the ND compounds may be relevant for the treatment of human cancer, as the likelihood of the occurrence of resistance is lower when multiple mechanisms of action (which may be synergistic) are present. It may also be possible to use NDs for combined therapy (polytherapy) together with other molecular targeted cancer therapeutics, (e.g., PARP inhibitors or antimitotic agents) (Salvati et al., 2010). NDs could also be used for targeting stromal and cancer stem cells for their ability to induce DNA damage and senescence. The ND compounds discussed here appear to be significantly more potent antiproliferative agents than others that have been previously reported (see, e.g., Nadai et al., 2011; Doria et al., 2012a,b; Wang et al., 2012), possibly as a result of their superior quadruplex-binding ability, although detailed mechanistic studies have not as yet been reported for these other ND compounds.
Supplementary Material
Abbreviations
- BMSG-SH-3
N,N'-bis(3-(4-methylpiperazin-1-yl)pentylamino)-2,6-bis(3-(4-methylpiperazin-1-yl)pentylamino)-1,2,5,8-naphthalenetetracarboxylic acid diimide
- BMSG-SH-4
N,N'-bis(3-(dimethylamine)propylamino)-2,6-bis(3-(dimethylamine)propylamino)-1,2,5,8-naphthalenetetracarboxylic acid diimide
- BMSG-SH-5
N,N'-bis(3-(pyrrolidine)propylamino)-2,6-bis(3-(pyrrolidine)propylamino)-1,2,5,8-naphthalenetetracarboxylic acid diimide
- BRACO-19
9-[4,N,N-dimethylamino) phenylamine]-3.6-box)3-[urrp;pdom-propionamio)acridine x 3HCl
- DAPI
4',6-diamidino-2-phenylindole
- 3D
three-dimensional
- DMEM
Dulbecco’s modified Eagle’s medium
- FISH
fluorescence in situ hybridization
- FRET
fluorescence resonance energy transfer
- ND
naphthalene diimide
- PBS
phosphate-buffered saline
- PARP
poly(ADP-ribose) polymerase
- PI
propidium iodide
- PNA
peptide nucleic acid
- TA
telomeric aggregates
- TMPyP4
meso-5,10,15,20-Tetrakis-(N-methyl-4-pyridyl)porphyrin
- 5′-UTR
5′-untranslated region
Authorship Contributions:
Participated in research design: Neidle, Hampel, Boukamp.
Conducted experiments: Hampel, Veith, Reszka, Greulich-Bode.
Performed data analysis: Hampel, Pepe, Reszka, Veith, Malhotra, Greulich-Bode.
Wrote or contributed to the writing of the manuscript: Hampel, Neidle, Greulich-Bode.
Footnotes
Work at the University College of London School of Pharmacy was supported by Cancer Research UK Program to S.N. [Grant C129/A4489]. S.M.H. was funded by the Philipp Brown Trust. Work at the German Cancer Research Center was funded by grants from the Deutsche Krebshilfe–Tumorstammzellverbund, and the BMBF (Bundesministerium für Forschung und Technik) UV-A Konsortium (both to P.B.). This research was supported by National Institutes of Health National Cancer Institute [Grant HHSN261200800001E.] The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Aspects of this work were previously presented in or at the following:
Hampel SM (2011) Design and Synthesis of G-Quadruplex Binding Small Molecules and their Evaluation as Anti-Cancer Agents. Ph.D. thesis. School of Pharmacy, University of London.
Hampel SM, de la Fuente M, Sidibe A, Schatzlein A, Riou JF, Neidle S (2011) Mechanistic studies of the anti-cancer activity of a G-quadruplex binding small molecule (poster). Royal Society of Chemistry Cancer Chemistry Group Symposium; 2011 June 01; the School of Pharmacy, London, United Kingdom.
Hampel SM, de la Fuente M, Sidibe A, Schatzlein A, Riou JF, Neidle S (2010) Targeting pancreatic cancers with a G-quadruplex binding small molecule (poster). 22nd EORTC-NCI-AACR Symposium on Molecular Targets and Cancer Therapeutics; 2010 Nov 16–19; Berlin, Germany.
Hampel SM, de la Fuente M, Schatzlein A, Neidle S (2010) Tetrasubstituted naphthalene diimides as selective telomeric G-quadruplex ligands for the treatment of pancreatic cancers (lecture). Ph.D. Research Day; 2010 Sept 24; the School of Pharmacy, Londonm United Kingdom.
Hampel SM, Sidibe A, Riou JF, Neidle S (2010) Tetrasubstituted naphthalene diimides with selectivity for telomeric G-quadruplex DNA and cancer cells (poster). European Cooperation in Science and Technology (COST) ACTION MP0802 Annual Meeting; 2010 Sept 15–17; The School of Pharmacy, London, United Kingdom
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Balasubramanian S, Hurley LH, Neidle S. (2011) Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat Rev Drug Discov 10:261–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejugam M, Gunaratnam M, Müller S, Sanders DA, Sewitz S, Fletcher JA, Neidle S, Balasubramanian S. (2010) Targeting the c-Kit promoter G-quadruplexes with 6-substituted indenoisoquinolines. ACS Med Chem Lett 1:306–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman ML, Paeschke K, Zakian VA. (2012) DNA secondary structures: stability and function of G-quadruplex structures. Nat Rev Genet 13:770–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. (1988) Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol 106:761–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukamp P, Popp S, Krunic D. (2005) Telomere-dependent chromosomal instability. J Investig Dermatol Symp Proc 10:89–94 [DOI] [PubMed] [Google Scholar]
- Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. (2006) Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res 34:5402–5415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger AM, Dai F, Schultes CM, Reszka AP, Moore MJ, Double JA, Neidle S. (2005) The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res 65:1489–1496 [DOI] [PubMed] [Google Scholar]
- Capra JA, Paeschke K, Singh M, Zakian VA. (2010) G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLOS Comput Biol 6:e1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang TC, Moshir S, Garini Y, et al. (2004) The three-dimensional organization of telomeres in the nucleus of mammalian cells. BMC Biol 2:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collie GW, Promontorio R, Hampel SM, Micco M, Neidle S, Parkinson GN. (2012) Structural basis for telomeric G-quadruplex targeting by naphthalene diimide ligands. J Am Chem Soc 134:2723–2731 [DOI] [PubMed] [Google Scholar]
- Cowell JK, Miller OJ. (1983) Occurrence and evolution of homogeneously staining regions may be due to breakage-fusion-bridge cycles following telomere loss. Chromosoma 88:216–221 [DOI] [PubMed] [Google Scholar]
- Cuenca F, Greciano O, Gunaratnam M, Haider S, Munnur D, Nanjunda R, Wilson WD, Neidle S (2008) Tri- and tetra-substituted naphthalene diimides as potent G-quadruplex ligands. Bioorg Med Chem Lett 18:1668–1673 DOI: PCT/GB 2008/051131. [DOI] [PubMed]
- Denchi EL, de Lange T. (2007) Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448:1068–1071 [DOI] [PubMed] [Google Scholar]
- Doria F, Nadai M, Folini M,, et al. (2012a) Hybrid ligand-alkylating agents targeting telomeric G-quadruplex structures. Org Biomol Chem 10:2798–2806 [DOI] [PubMed] [Google Scholar]
- Doria F, Nadai M, Sattin G, Pasotti L, Richter SN, Freccero M. (2012b) Water soluble extended naphthalene diimides as pH fluorescent sensors and G-quadruplex ligands. Org Biomol Chem 10:3830–3840 [DOI] [PubMed] [Google Scholar]
- Drygin D, Lin A, Bliesath J, Ho CB, O'Brien SE, Proffitt C, Omori M, Haddachi M, Schwaebe MK, Siddiqui-Jain A, et al. (2011) Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 71:1418–1430 [DOI] [PubMed] [Google Scholar]
- Ermler S, Krunic D, Knoch TA, Moshir S, Mai S, Greulich-Bode K-M, Boukamp P. (2004) Cell cycle-dependent 3D distribution of telomeres and telomere repeat-binding factor 2 (TRF2) in HaCaT and HaCaT-myc cells. Eur J Cell Biol 83:681–690 [DOI] [PubMed] [Google Scholar]
- Ewald JA, Desotelle JA, Wilding G, Jarrard DF. (2010) Therapy-induced senescence in cancer. J Natl Cancer Inst 102:1536–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldser DM, Greider CW. (2007) Short telomeres limit tumor progression in vivo by inducing senescence. Cancer Cell 11:461–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez D, Aouali N, Londoño-Vallejo A,, et al. (2003) Resistance to the short term antiproliferative activity of the G-quadruplex ligand 12459 is associated with telomerase overexpression and telomere capping alteration. J Biol Chem 278:50554–50562 [DOI] [PubMed] [Google Scholar]
- Gomez D, O’Donohue MF, Wenner T,, et al. (2006a) The G-quadruplex ligand telomestatin inhibits POT1 binding to telomeric sequences in vitro and induces GFP-POT1 dissociation from telomeres in human cells. Cancer Res 66:6908–6912 [DOI] [PubMed] [Google Scholar]
- Gomez D, Wenner T, Brassart B, Douarre C, O’Donohue MF, El Khoury V, Shin-Ya K, Morjani H, Trentesaux C, Riou JF. (2006b) Telomestatin-induced telomere uncapping is modulated by POT1 through G-overhang extension in HT1080 human tumor cells. J Biol Chem 281:38721–38729 [DOI] [PubMed] [Google Scholar]
- Gunaratnam M, Greciano O, Martins C, Reszka AP, Schultes CM, Morjani H, Riou JF, Neidle S. (2007) Mechanism of acridine-based telomerase inhibition and telomere shortening. Biochem Pharmacol 74:679–689 [DOI] [PubMed] [Google Scholar]
- Gunaratnam M, Swank S, Haider SM, Galesa K, Reszka AP, Beltran M, Cuenca F, Fletcher JA, Neidle S. (2009) Targeting human gastrointestinal stromal tumor cells with a quadruplex-binding small molecule. J Med Chem 52:3774–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaratnam M, de la Fuente M, Hampel SM, Todd AK, Reszka AP, Schätzlein A, Neidle S. (2011) Targeting pancreatic cancer with a G-quadruplex ligand. Bioorg Med Chem 19:7151–7157 [DOI] [PubMed] [Google Scholar]
- Hampel SM, Sidibe A, Gunaratnam M, Riou JF, Neidle S. (2010) Tetrasubstituted naphthalene diimide ligands with selectivity for telomeric G-quadruplexes and cancer cells. Bioorg Med Chem Lett 20:6459–6463 [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. (2011) Hallmarks of cancer: the next generation. Cell 144:646–674 [DOI] [PubMed] [Google Scholar]
- Huang FC, Chang CC, Lou PJ, Kuo IC, Chien CW, Chen CT, Shieh FY, Chang TC, Lin JJ. (2008) G-quadruplex stabilizer 3,6-bis(1-methyl-4-vinylpyridinium)carbazole diiodide induces accelerated senescence and inhibits tumorigenic properties in cancer cells. Mol Cancer Res 6:955–964 [DOI] [PubMed] [Google Scholar]
- Huang X, Darzynkiewicz Z. (2006) Cytometric assessment of histone H2AX phosphorylation: a reporter of DNA damage. Methods Mol Biol 314:73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incles CM, Schultes CM, Kempski H, Koehler H, Kelland LR, Neidle S. (2004) A G-quadruplex telomere targeting agent produces p16-associated senescence and chromosomal fusions in human prostate cancer cells. Mol Cancer Ther 3:1201–1206 [PubMed] [Google Scholar]
- Izbicka E, Wheelhouse RT, Raymond E, Davidson KK, Lawrence RA, Sun D, Windle BE, Hurley LH, Von Hoff DD. (1999) Effects of cationic porphyrins as G-quadruplex interactive agents in human tumor cells. Cancer Res 59:639–644 [PubMed] [Google Scholar]
- Karlseder J, Broccoli D, Dai YM, Hardy S, de Lange T. (1999) p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science 283:1321–1325 [DOI] [PubMed] [Google Scholar]
- Leonetti C, Amodei S, D’Angelo C, et al. (2004) Biological activity of the G-quadruplex ligand RHPS4 (3,11-difluoro-6,8,13-trimethyl-8H-quino[4,3,2-kl]acridinium methosulfate) is associated with telomere capping alteration. Mol Pharmacol 66:1138–1146 [DOI] [PubMed] [Google Scholar]
- Leonetti C, Scarsella M, Riggio G, et al. (2008) G-quadruplex ligand RHPS4 potentiates the antitumor activity of camptothecins in preclinical models of solid tumors. Clin Cancer Res 14:7284–7291 [DOI] [PubMed] [Google Scholar]
- Lipps HJ, Rhodes D. (2009) G-quadruplex structures: in vivo evidence and function. Trends Cell Biol 19:414–422 [DOI] [PubMed] [Google Scholar]
- Louis SF, Vermolen BJ, Garini Y, et al. (2005) c-Myc induces chromosomal rearrangements through telomere and chromosome remodeling in the interphase nucleus. Proc Natl Acad Sci USA 102:9613–9618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai S. (2010) Initiation of telomere-mediated chromosomal rearrangements in cancer. J Cell Biochem 109:1095–1102 [DOI] [PubMed] [Google Scholar]
- Maizels N. (2006) Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat Struct Mol Biol 13:1055–1059 [DOI] [PubMed] [Google Scholar]
- McLuckie KIE, Waller ZAE, Sanders DA, Alves D, Rodriguez R, Dash J, McKenzie GJ, Venkitaraman AR, Balasubramanian S. (2011) G-quadruplex-binding benzo[a]phenoxazines down-regulate c-KIT expression in human gastric carcinoma cells. J Am Chem Soc 133:2658–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murnane JP. (2006) Telomeres and chromosome instability. DNA Repair (Amst) 5:1082–1092 AMS [DOI] [PubMed] [Google Scholar]
- Nadai M, Doria F, Di Antonio M, Sattin G, Germani L, Percivalle C, Palumbo M, Richter SN, Freccero M. (2011) Naphthalene diimide scaffolds with dual reversible and covalent interaction properties towards G-quadruplex. Biochimie 93:1328–1340 [DOI] [PubMed] [Google Scholar]
- Paeschke K, Juranek S, Simonsson T, Hempel A, Rhodes D, Lipps HJ. (2008) Telomerase recruitment by the telomere end binding protein-beta facilitates G-quadruplex DNA unfolding in ciliates. Nat Struct Mol Biol 15:598–604 [DOI] [PubMed] [Google Scholar]
- Paull KD, Shoemaker RH, Hodes L, Monks A, Scudiero DA, Rubinstein L, Plowman J, Boyd MR. (1989) Display and analysis of patterns of differential activity of drugs against human tumor cell lines: development of mean graph and COMPARE algorithm. J Natl Cancer Inst 81:1088–1092 [DOI] [PubMed] [Google Scholar]
- Pennarun G, Granotier C, Gauthier LR, Gomez D, Hoffschir F, Mandine E, Riou JF, Mergny JL, Mailliet P, Boussin FD. (2005) Apoptosis related to telomere instability and cell cycle alterations in human glioma cells treated by new highly selective G-quadruplex ligands. Oncogene 24:2917–2928 [DOI] [PubMed] [Google Scholar]
- Pennarun G, Granotier C, Hoffschir F, Mandine E, Biard D, Gauthier LR, Boussin FD. (2008) Role of ATM in the telomere response to the G-quadruplex ligand 360A. Nucleic Acids Res 36:1741–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Lin CP, Fu X, Wood LM, Liu AA, Tsai YC, Chen Y, Barbieri CM, Pilch DS, Liu LF. (2006) G-quadruplexes induce apoptosis in tumor cells. Cancer Res 66:11808–11816 [DOI] [PubMed] [Google Scholar]
- Qin Y, Hurley LH. (2008) Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie 90:1149–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo A, Salvati E, Porru M, D’Angelo C, Stevens MF, D’Incalci M, Leonetti C, Gilson E, Zupi G, Biroccio A. (2009) Stabilization of quadruplex DNA perturbs telomere replication leading to the activation of an ATR-dependent ATM signaling pathway. Nucleic Acids Res 37:5353–5364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R, Müller S, Yeoman JA, Trentesaux C, Riou JF, Balasubramanian S. (2008) A novel small molecule that alters shelterin integrity and triggers a DNA-damage response at telomeres. J Am Chem Soc 130:15758–15759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R, Miller KM, Forment JV, Bradshaw CR, Nikan M, Britton S, Oelschlaegel T, Xhemalce B, Balasubramanian S, Jackson SP. (2012) Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat Chem Biol 8:301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvati E, Leonetti C, Rizzo A, et al. (2007) Telomere damage induced by the G-quadruplex ligand RHPS4 has an antitumor effect. J Clin Invest 117:3236–3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvati E, Scarsella M, Porru M, et al. (2010) PARP1 is activated at telomeres upon G4 stabilization: possible target for telomere-based therapy. Oncogene 29:6280–6293 [DOI] [PubMed] [Google Scholar]
- Schuermann M. (1990) An expression vector system for stable expression of oncogenes. Nucleic Acids Res 18:4945–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu N, Shingaki K, Kaneko-Sasaguri Y, Hashizume T, Kanda T. (2005) When, where and how the bridge breaks: anaphase bridge breakage plays a crucial role in gene amplification and HSR generation. Exp Cell Res 302:233–243 [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, de Lange T. (2002) Different telomere damage signaling pathways in human and mouse cells. EMBO J 21:4338–4348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara H, Shin-Ya K, Seimiya H, Yamada H, Tsuruo T, Ide T. (2006) G-Quadruplex stabilization by telomestatin induces TRF2 protein dissociation from telomeres and anaphase bridge formation accompanied by loss of the 3′ telomeric overhang in cancer cells. Oncogene 25:1955–1966 [DOI] [PubMed] [Google Scholar]
- Tanaka T, Halicka HD, Traganos F, Seiter K, Darzynkiewicz Z. (2007) Induction of ATM activation, histone H2AX phosphorylation and apoptosis by etoposide: relation to cell cycle phase. Cell Cycle 6:371–376 [DOI] [PubMed] [Google Scholar]
- Tauchi T, Shin-Ya K, Sashida G, Sumi M, Nakajima A, Shimamoto T, Ohyashiki JH, Ohyashiki K. (2003) Activity of a novel G-quadruplex-interactive telomerase inhibitor, telomestatin (SOT-095), against human leukemia cells: involvement of ATM-dependent DNA damage response pathways. Oncogene 22:5338–5347 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang X, Zhao J, Xie S, Wang C. (2012) Nonhematotoxic naphthalene diimide modified by polyamine: synthesis and biological evaluation. J Med Chem 55:3502–3512 [DOI] [PubMed] [Google Scholar]
- Zaug AJ, Podell ER, Cech TR. (2005) Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc Natl Acad Sci USA 102:10864–10869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou WJ, Deng R, Zhang XY, Feng GK, Gu LQ, Zhu XF. (2009) G-quadruplex ligand SYUIQ-5 induces autophagy by telomere damage and TRF2 delocalization in cancer cells. Mol Cancer Ther 8:3203–3213 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.