Abstract
Dihydrofolate reductase (DHFR), because of its essential role in DNA synthesis, has been targeted for the treatment of a wide variety of human diseases, including cancer, autoimmune diseases, and infectious diseases. Methotrexate (MTX), a tight binding inhibitor of DHFR, is one of the most widely used drugs in cancer treatment and is especially effective in the treatment of acute lymphocytic leukemia, non-Hodgkin’s lymphoma, and osteosarcoma. Limitations to its use in cancer include natural resistance and acquired resistance due to decreased cellular uptake and decreased retention due to impaired polyglutamylate formation and toxicity at higher doses. Here, we describe a novel mechanism to induce DHFR degradation through cofactor depletion in neoplastic cells by inhibition of NAD kinase, the only enzyme responsible for generating NADP, which is rapidly converted to NADPH by dehydrogenases/reductases. We identified an inhibitor of NAD kinase, thionicotinamide adenine dinucleotide phosphate (NADPS), which led to accelerated degradation of DHFR and to inhibition of cancer cell growth. Of importance, combination treatment of NADPS with MTX displayed significant synergy in a metastatic colon cancer cell line and was effective in a MTX-transport resistant leukemic cell line. We suggest that NAD kinase is a valid target for further inhibitor development for cancer treatment.
Introduction
Dihydrofolate reductase (DHFR) (5,6,7,8-tetrahydrofolate NADP oxidoreductase, EC 1.5.1.3), catalyzes the reduction of dihydrofolate (FH2) to tetrahydrofolate (FH4) with use of NADPH as a cofactor. The binding sites of FH2 and NADPH are at 2 different but connected regions of DHFR. They comprise a long channel in the active site of the enzyme, positioning the 2 ligands in close proximity to enable hydride transfer from NADPH to FH2, generating FH4. Tetrahydrofolate is converted to 10-formyl and 5-10 methylene FH4, essential cofactors in the synthesis of purines, thymidylate, and certain amino acids (Abali et al., 2008). Inhibition of DHFR results in a depletion of the reduced folate pool, inhibition of RNA and DNA synthesis, and cell death. For this reason, DHFR has been a critically important therapeutic drug target. DHFR inhibitors targeting the FH2 binding site have been used in the treatment of cancer, autoimmune diseases, and bacterial and fungal infections. However, DHFR inhibitors targeting the FH2 binding site have limitations primarily because of intrinsic and acquired resistance mechanisms (Gorlick et al., 1996; Cheok and Evans, 2006; Rego-Perez et al., 2008), including decreased uptake and, less commonly, gene amplification or changes in polyglutamylation. Thus, a search for new strategies that bypass these resistance mechanisms is warranted.
Strategies have been developed to reduce cellular levels of proteins that are involved in cell proliferation and cancer growth. Geldanamycin and its analogs, such as 17-allylamino-17-demethoxygeldanamycin, induce the degradation of several proteins. These proteins, such as mutant p53, bcr-abl, and Her2, are involved in cancer proliferation through complexing with the protein chaperone, heat shock protein 90. For example, geldanamycin destabilizes HER2 tyrosine kinase and suppresses Wnt/β-catenin signaling in HER2-overexpressing human breast cancer cells (Slack et al., 2001). However, degradation of these proteins by geldanamycin is not very selective and leads to hepatotoxicity (Latif et al., 2001). Another strategy reported is the induction of enzyme degradation through cofactor depletion in neoplastic cells. Methionine synthase, a vitamin B12 dependent enzyme, is an important enzyme in DNA synthesis. Inactivation of vitamin B12 by nitrous oxide, by rendering methionine synthase inactive, triggers apoptosis and results in inhibition of proliferation in a variety of cancer cell lines (Livak and Schmittgen, 2001). Recently, 5-aza-deoxycytidine has been shown to induce selective degradation of DNA methyltransferase 1 by the proteasomal pathway (Ghoshal et al., 2005) .
Ligand binding–induced conformational changes in proteins affect the thermodynamic stability of proteins (Ainavarapu et al., 2005). In fact, DHFR is stabilized by either its cofactor NADPH, or its potent inhibitor methotrexate (MTX), thus slowing down its degradation (Xiang, 2002). We hypothesized that the steady-state level of DHFR may be decreased by NAD(P) analogs by either acting as competitive inhibitors with NADPH for DHFR or reducing NADPH pools available for proper folding of DHFR, leading to increased degradation.
Recent studies demonstrated that the flux of anabolic pathways, such as nucleotide synthesis and fatty acid synthesis, which requires NADPH, is increased because of genetic alterations in HIF-1, Akt, and myc signaling pathways in cancer cells (Tong et al., 2009). Furthermore, NADPH is also important to maintain the reduced pools of glutathione (GSH). Although GSH is important for the removal of reactive oxygen species, which are elevated in cancers, GSH levels are also increased in many cancers, leading to resistance to chemo- or radiotherapy (Estrela et al., 2006). We hypothesized that reducing NADPH levels may be important both for the aforementioned pathways that are dysregulated in cancers and for regulation of DHFR.
There are several enzymes that reduce NADP to NADPH, but only one enzyme, NAD kinase, that generates NADP from NAD. Therefore, we initiated studies to identify inhibitors of NAD kinase that lead to decreased levels of NADPH, which might cause accelerated degradation of DHFR. Here, we identify 2 compounds, thionicotinamide adenine dinucleotide (NADS) and thionicotinamide adenine dinucleotide phosphate (NADPS), with potent cytotoxicity against cancer cells by decreasing the level of DHFR due to its inhibition of NAD kinase, which is necessary for NADPH generation. Although it has been previously shown in vitro that NADPH protects DHFR from degradation in cell-free extracts (Perkins et al., 1967; Wallace and Robert Matthews, 2002; Ainavarapu et al., 2005), the studies presented here for the first time show that NADPH also protects DHFR from degradation in a cellular context.
Materials and Methods
Cell Culture and Materials
RPMI-1640 medium (Invitrogen, Carlsbad, CA), Dulbecco’s modified Eagle medium, fetal bovine serum (FBS), dialyzed FBS, G418 Sulfate (Geneticin), penicillin, and streptomycin were purchased from Invitrogen. F-12 medium without glycine, hypoxanthine, and thymidine was obtained from the Media Core Facility at Sloan-Kettering Cancer Institute (New York, NY). Human mesenchymal stem cells were purchased from Lonza (Cologne, Germany). NAD and NADP analogs (Table 1) and MTX were purchased from Sigma-Aldrich.
TABLE 1.
Thirteen NAD/NADP analogs were screened for targeting NAPDH site in DHFR protein
| Variable | |
|---|---|
| A1046 | 3-Acetylpyridine adenine dinucleotide phosphate |
| A5137 | 3-Acetylpyridine adenine dinucleotide |
| A55403 | 3-Aminofluoranthene |
| N7256 | Nicotinamide hypoxanthine dinucleotide phosphate |
| N5881 | β-Nicotinamide adenine dinucleotide 3′-phosphate |
| N6756 | Nicotinamide hypoxanthine dinucleotide |
| A7266 | 3-Aminopyridine adenine dinucleotide |
| A5298 | 3-Acetylpyridine hypoxanthine dinucleotide |
| N4256 | Nicotinic acid adenine dinucleotide |
| N5257 | β-Nicotinamide adenine dinucleotide 2′:3′-cyclic monophosphate |
| P2171 | 3-Pyridinealdehyde adenine dinucleotide |
| T7375 | Thionicotinamide adenine dinucleotide |
| T551 | Thionicotinamide adenine dinucleotide phosphate |
C85 (a human colon cancer cell line) and CCRF-CEM (a human T-cell acute lymphoblastic leukemia cell line) were cultured in RPMI 1640 medium supplemented with 10% FBS, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. The parental CCRF-CEM cell line and 2 drug-resistant sublines derived from the parental line were used in this study. Although both of the sublines are resistant to methotrexate, CCRF-CEM/R resistance is attributable to 5-fold increase in DHFR because of gene amplification; CCRF-CEM/T is resistant to classic antifolates, such MTX and D1694 (raltitrexed, Tomudex) because of impaired transportation (Mini et al., 1985). DG44-(DHFR-enhanced green fluorescent protein[EGFP]) and DG44-(DHFR-S118A-EGFP) cell lines are Chinese hamster ovary cell lines with no DHFR expression, but contain stably transfected DHFR-EGFP or DHFR-S118A-EGFP fusion construct (Skacel et al., 2005) and were cultured in RPMI 1640 medium supplemented with 10% FBS and antibiotics.
siRNA Transfection
Double-stranded RNA complementary to NADK mRNA sequence 5′-GAG CGT CCT TGT CAT CAA GAA-3′ was generated using Integrated DNA Technologies Inc. (Coralville, IA). The sequence of the sense strand of the siRNA duplex targeting NADK is 5′-CUC GCA GGA ACA GUA GUU CUU-3′. C85 cells were seeded in a 6-well plate at a density of 6 × 105 cells/well in RPMI-1640 medium containing 10% FBS and no antibiotics the day before transfection. Cells (50–60% confluency) were transfected with 100 pmol siRNA with use of Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Cells were harvested 48 hours after transfection for protein analysis.
Western Blot Analysis
Total protein extracted from cell lines was resolved on SDS-PAGE, as described elsewhere (Hsieh et al., 2009). Protein concentration was determined using Bradford protein assay according to the manufacturer’s guidelines (Bio-Rad Laboratories Inc., Hercules, CA). Proteins were resolved by 15% SDS-PAGE and transferred onto a nitrocellulose membrane (Bio-Rad Laboratories Inc.). After blocking the membrane with 5% nonfat dry milk prepared in phosphate-buffered saline (PBS; pH 7.4) containing 0.1% Tween 20, the membrane was incubated with the desired primary antibody at the dilution suggested by the manufacturer’s protocol for 2 hours at room temperature or overnight at 4°C. The membrane was then incubated with peroxidase-conjugated secondary antibody, and the immunoreactive bands were visualized using the enhanced chemiluminescence kit (Pierce ECL Western Blotting Substrate, Rockford, IL) according to the manufacturer’s instructions. Anti-inosine-5′-monophosphate dehydrogenase (IMDPH) and anti- aldehyde dehydrogenase (ALDH1) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-α-tubulin and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies, purchased from Sigma-Aldrich, were used for loading controls. Anti-GFP antibody (Roche Diagnostics Corporation, Indianapolis, IN) was used to detect DHFR expression in DG44-(DHFR-EGFP) and DG44-(DHFR-S118A-EGFP) cell lines. The rabbit polyclonal antibody to human DHFR was custom-produced by Research Genetics/Invitrogen.
Cytotoxicity Assay
Two methods were used for the determination of cytotoxicity of antifolates and NAD(P) analogs. The 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) assay was performed according to the Cell Titer 96 Aqueous One Solution protocol (Promega, Madison, WI). Three thousand cells per well were plated in 96-well plates in RPMI 1640 media supplemented with 10% dialyzed FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin or in F-12 media lacking hypoxanthine, glycine, and thymidine supplemented with 10% dialyzed FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. After cells were incubated with the NAD analogs for 96 hours, the MTS assay was performed according to the manufacturer’s instructions. The absorbance at 490 nm was measured using a microplate reader. The percentage of viable cells was determined using SOFTmax Pro 2.6.1 software (Molecular Devices Corp., Sunnyvale, CA).
Cytotoxicity of NADS and NADPS was also determined using the Trypan blue exclusion assay. In brief, suspension cells with or without drug treatment were collected, and cell viability was determined using the Vi-CELL Series Cell Viability Analyzer (Beckman Coulter, Carlsbad, CA).
The cytotoxicity data were further analyzed using GraphPad Prism 4 software (GraphPad Software Inc., La Jolla, CA). The ED50 was determined using the nonlinear regression curve fit of the graphs drawn by GraphPad Prism 4 software. All experimental points were set up in replicate wells, and all experiments were repeated at least three times.
DHFR Half-Life Studies
To determine the half-life of DHFR in the absence or presence of NADPS, pulse-chase experiments were performed as described elsewhere (Noe et al., 2003) with minor modifications. In brief, two million CCRF-CEM/R cells were seeded on a 10 cm2 dish. Cells were incubated in methionine-free RPMI 1640 medium supplemented with 5% dialyzed FBS for 1 hour and then labeled with 500 μCi/plate of l-[35S]methionine-cysteine Express Cell Labeling Mix (PerkinElmer Life Sciences, Boston, MA) for 1 hour. Labeled cells were harvested and washed with phosphate-buffered saline (PBS) twice. Chase medium (RPMI 1640 medium supplemented 10% FBS and freshly prepared 2 mM methionine) in the presence or absence of 10 μM NADPS was added to cells at different time points, except for the control sample. Cells were lysed (150 mM NaCl, 5 mM EDTA, 50 mM HEPES, pH 7.5, and 1.0% Triton X-100), and DHFR was immunoprecipitated with anti-DHFR antibody and protein G-Sepharose 4 Fast Flow (GE Healthcare, Piscataway, NJ) at 4°C overnight. Beads were washed with lysis buffer three times and eluted with SDS-PAGE sample buffer, followed by electrophoresis. Gels were exposed to X-ray film and analyzed by densitometry with ImageJ software (National Institutes of Health, Bethesda, MD).
Clonogenic Assay
Two hundred cells per well of C85 cells were plated into 6-well culture plates. The following day, the medium was removed and replaced with fresh RPMI 1640 medium supplemented with 10% dialyzed FBS in the presence or absence of NADPS or MTX or the combination of the 2 drugs. After 2 weeks of incubation, the plates were washed with PBS (pH 7.4) and fixed with crystal violet solution (0.4% formaldehyde, 0.2% crystal violet), and colonies were counted.
Analysis of Drug Combination Studies
Exponentially growing C85 or CCRF-CEM cells were plated in 6-well dishes at a density of 200 cells per well. At 24 hours after the incubation, the cells were exposed to serial dilutions of MTX, NADPS, or both for 2 weeks. After incubation, the cells were stained, and viable colonies were counted. After calculating percentage inhibition, drug interactions were analyzed using the Calcusyn program (Biosoft, Cambridge, UK), which determines synergy or antagonism using the multiple drug effect equation (Chou and Talalay, 1984). Outcomes of the drug interactions were determined using the combination index (CI) values calculated by Calcusyn. If CI is greater than one, the combination is antagonistic; if CI is equal to one, the combination is additive; and if CI is less than one, the combination is synergistic.
Cell Cycle Analysis
C85 cells were incubated with or without NADPS or MTX for 24 hours. Cells were collected, washed with cold PBS twice, and then fixed with 70% ethanol overnight at −20°C. Cells were centrifuged at 1000g for 5 minutes, washed with PBS once, and then stained with 50 μg/ml propidium iodide staining solution (BD Biosciences, San Jose, CA) in the dark for 15 minutes. Distribution of the cells in the various phases of the cell cycle was determined using a FACScan flow cytometer (BD Biosciences). In each sample, 10,000 gated events were acquired. The cell cycle phases were analyzed using CELLQuest software (BD Biosciences).
Measurement of DHFR Activity
The specific activity of DHFR from lysates of hamster and the variants of human DHFR-EGFP was determined as described elsewhere (Ercikan-Abali et al., 1996).
NAD Kinase Activity and Inhibition Studies
Pure NAD kinase was purchased from Enzo Life Sciences (Farmingdale, NY). Effect of NADPS on NAD kinase activity was measured spectrophotometrically with a coupled assay that converts the NADP generated by NAD kinase into NADPH using glucose-6-phosphate dehydrogenase (G6PDH) (Lerner et al., 2001). In brief, NAD kinase activity was determined by measuring the increase in absorbance at 340 nm caused by the reduction of NADP to NADPH by G6PDH. The enzyme reaction for inhibition of NAD kinase by NADPS contains 2 mM NAD, 2 μg of human NAD kinase, 1 U G6PDH, and 1.5 mM or 3 mM NADPS, 10 mM ATP, 10 mM MgCl2, and 50 mM Tris-HCl (pH 7.8) in a final volume of 1 ml. Controls were without NAD or NADK or NADPS or reaction mix alone. The appearance of NADPH was measured at 340 nM using a Beckman spectrophotometer.
Quantitative Real-Time Reverse-Transcription Polymerase Chain Reaction
Quantitative RT-PCR was used to analyze DHFR mRNA levels in the presence and absence of 40 μM NADPS for 24 hours in C85 cells. DHFR and β-actin mRNAs were examined by quantitative real-time RT-PCR. After total RNA purification using TRIzol (Invitrogen), the integrity and the concentration of the RNA was determined using a NanoDrop spectrophotometer (NanoDrop Products, Wilmington, DE) and by 1% agarose gel. Quantitative PCR was performed with 50 ng of purified RNA TaqMan One Step RT-PCR according to the manufacturer’s protocol (Applied Biosystems, Foster City, CA) on an ABI Prism 7000 sequence detection system (Applied Biosystems) with TaqMan probe–based gene expression assays for DHFR and β-actin (Applied Biosystems). Gene expression levels of DHFR were normalized to β-actin. The reaction and cycling conditions for both genes were as follows: 48°C for 40 minutes and step 2, 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds, and 60°C for 1 minute. Reactions were terminated with incubation at 4° for 10 minutes. Each sample was run using three different wells, and each experiment was repeated for three times. The expression of DHFR in NADPS or MTX-treated cells relative to untreated control was calculated according to the 2−ΔΔCt method (Livak and Schmittgen, 2001) using β-actin as reference gene.
Computational Methods
The models of Human NADK were built from the X-ray crystal structure of human NAD kinase (3PFN) (Wang et al., 2010). The Jackal profix program was used to account for missing residues in the structure (Xiang, 2002). The models of human NAD kinase complexed to NAD+ and NADP+ were built from least squares fit of the protein backbone atoms of each model with 1Z0Z (Arch. fulgidus complex with NAD+) and 1Z0U (Arch. fulgidus complex with NADP+) (Liu et al., 2005). The STAMP structural alignment tool in the MultiSeq program in VMD was used to align the structures (Russell and Barton, 1992; Humphrey et al., 1996; Roberts et al., 2006). We used the UCSF Chimera Matchmaker (UCSF Chimera, Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco) and viewing program to map residue conservation to the cartoon plot of the human structure (Meng et al., 2006).
Dynamics.
Each model was refined using energy minimization and relaxed using molecular dynamics with the Amber 11 suite of biomolecular simulation programs (Case et al., 2005). The Amber99SB force-field was used in all molecular mechanics calculations (Hornak et al., 2006). Parameters for NAD+, NADP+, and NADPS+ were computed using the Amber Antechamber accessory program with AM1-BCC partial atomic charges (Wang et al., 2006). Each model was energy minimized in vacuo with use of 500 steps of steepest descents, followed by 2000 steps of conjugate gradient minimization with a nonbonded forces cutoff of 12.0 Å. The resulting model was solvated in a periodic octahedral box of TIP3P water, and the net formal charge was neutralized by addition of the appropriate number of Na+ ions. A short-range cutoff of 9.0 Å was used, and the PME method (Darden et al., 1993; Essmann et al., 1995) was used to account for long-range electrostatic interactions. Each model was energy minimized keeping the protein atoms restrained using 750 steps of steepest descent, followed by 750 steps of conjugate gradient minimization. The restrained minimization was followed by unrestrained minimization of the system using 1000 steps of steepest descents, followed by 1500 steps of conjugate gradient. A solvent equilibration dynamics run was performed keeping the protein atom positions restrained, using a Langevin thermostat with γ = 1.0 ps−1, random seed generation, and temperature equal to 300 K with no pressure coupling (an NVT ensemble) for a period of 100 ps. A 2-fs time step was used, and the shake constraint was used on all bonds to hydrogen. The solvent equilibration step was followed by an unrestrained production dynamics run using pressure coupling with a Berendsen barostat (Berendsen et al., 1984) set at 1 atm and temperature set at 300 K. The production dynamics was run for a period of 2 ns. The final snapshot of each trajectory was stripped of water and ions and refined using energy minimization with the generalized Born implicit solvation method (Hawkins et al., 1995, 1996) used to account for solvation. A 16.0 Å nonbonded forces cutoff was used for generalized Born minimization using OBC model I (Onufriev et al., 2000, 2004) with 500 steps of steepest descents, followed by 1000 steps of conjugate gradient minimization.
Molecular Mechanics/Generalized Born Surface Area Calculations.
The generalized Born OBC I model (Onufriev et al., 2000; Onufriev et al., 2004) with bondi2 radii was used with an external dielectric constant of 78.5 and an internal dielectric constant of 4.0 per the optimum Molecular Mechanics/Generalized Born Surface Area method (Hou et al., 2011)for proteins with polar binding sites (Wang et al., 2006).
Results
An NAD Analog, NADPS, Selectively Kills Cancer Cells and Spares Normal Cells
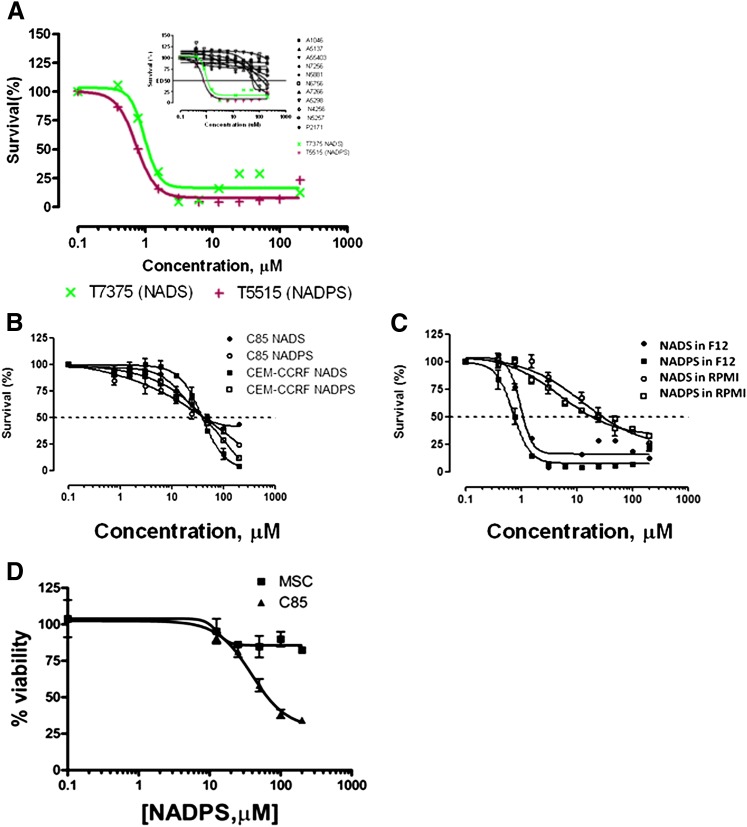
Thirteen NAD/NADP analogs were screened for cytotoxicity with use of the MTS assay in DG44-(DHFR-EGFP) cells. These cells are the DHFR-deficient Chinese hamster ovary cell line stably transfected with a DHFR-EGFP fusion construct driven by a cytomegalovirus promoter. Detection of the basal levels of DHFR in most of the cell lines is hard to detect; therefore, these cells were chosen for initial studies, because the transfected DHFR fusion protein with EGFP makes detection much easier. Moreover, DG44 cells with or without DHFR expression were used to verify that the cytotoxicity of the inhibitors is directly related to the expression of DHFR. To assess cytotoxicity of the analogs, Ham’s F-12 medium was used, because this medium has 28-fold less nicotinamide than the RPMI 1640 media (8.2 μM versus 0.295 μM), levels more comparable to those in human serum. Two compounds, NADS and NADPS, had potent cytotoxic effects when tested against the DG44-(DHFR-EGFP) cells. The ED50 for both compounds was 1 μM for DG44-(DHFR-EGFP) cells (Fig. 1A). The fact that both compounds were equally effective suggested that NADS was converted rapidly to NADPS by NAD kinase in cells. The other NAD(P) analogs tested (Table 1) had EC50 values greater than 200 μM (see the inset in Fig. 1A).
Fig. 1.
Screening of NAD/NADP analogs for cytotoxicity. (A) Two thousand DG44-(DGFR-EGFP) cells per well were plated and grown in Ham’s F12 medium supplemented with 10% dialyzed FBS containing no glycine, hypoxanthine, or thymidine. Concentrations of drugs tested ranged from 200 μM concentration. After incubation with NAD/NADP analogs for 96 hours, the viability of the transfectants was assessed using the MTS proliferation assay (see Materials and Methods for detail). (B) NADS and NADPS were tested in C85 and CCRF-CEM cells in RPMI 1640 medium for 96 hours. Although cell toxicity in C85 cells was determined using MTS assay, Trypan blue exclusion assay was used to determine cell viability in the presence of NADS and NADPS in CCRF-CEM cells. (C) Cytotoxicity of NADS and NADPS was tested for DG44-(DHFR-EGFP) cells in RPMI-1640 and Ham’s F12 using the MTS assay. (D) Seventy-five thousand C85 cells and mesenchymal stem cells (MSCs) were plated into 24-well tissue culture plates in triplicate in RPMI-1640 (C85) or Dulbecco's modified Eagle medium (DMEM) (mesenchymal stem cells) supplemented with 10% FBS. The following day, the medium was removed and replaced with fresh medium containing NADPS. After 96 hours of incubation, the cell viability was determined. The percentage cell survival was determined by Softmax Pro software, and GraphPad Prism 4 software was used to determine ED50 values using a sigmoidal dose-response curve fit.
To investigate whether the cytotoxicity observed with these analogs was cell specific, two different cell lines, a metastatic human colon cancer cell line (C85) and T cell lymphoblastic leukemia cells (CCRF-CEM), were treated with NADS and NADPS. Unlike DG44 cells, C85 and CCRF-CEM cells did not grow well in Ham’s F-12 media; therefore, RPMI 1640 medium supplemented with 10% dialyzed FBS was used to grow these cells. In contrast to DG44(DHFR-EGFP) cells grown in Ham’s F-12 medium, the ED50 for NADPS and NADS was 40 μM in C85 cells and CCRF-CEM cells grown in RPMI-1640 (Fig. 1B). To demonstrate that the higher ED50 for these two cell lines was attributable to the higher concentration of nicotinamide in RPMI-1640, DG44-(DHFR-EGFP) cells were also grown in RPMI-1640, and the cytotoxicity of NADS and NADPS were determined. As expected, the ED50 for both compounds was about 10-fold higher in RPMI-1640 than in Ham’s F-12 media (Fig. 1C), indicating that the lower toxicity of these analogs was attributable to higher nonphysiologic concentrations of nicotinamide, the precursor of NAD(P) in RPMI 1640 medium.
We also tested whether NADPS exhibits selective antitumor effect. Normal human cells, namely mesenchymal stem cells, were treated with NADPS. Seventy-five thousand C85 cells and mesenchymal stem cells were plated into 24-well tissue culture plates, and the following day, the medium was removed and replaced with fresh medium containing NADPS. After 96 hours of incubation, cell viability was determined. At ED50 and ED75 for NADPS in C85 cells, NADPS was only mildly cytotoxic to human mesenchymal stem cells (Fig. 1D). Thus, we identified an NAD analog, NADPS, which exhibited a potent and selective antitumor effect.
The Effect of NAD/NADP Analogs on DHFR Activity and Expression
ThioNADP(H) has been used as an inactive cofactor for NADPH in experiments using chicken, mouse, Escherichia coli, and Lactobacillus casei, and it has been shown to bind to the cofactor binding site (Dunn et al., 1978; Smith and Burchall, 1983; Thillet et al., 1990; McTigue et al., 1993). However, when we tested the effect of NADPS on DHFR enzyme activity, only slight inhibition of human DHFR was observed even with 180 μM NADS or NADPS (data not shown). We considered that the cytotoxicity by NADS and NADPS in the cell lines tested is attributable to a decrease in DHFR rather than inhibition of enzyme activity.
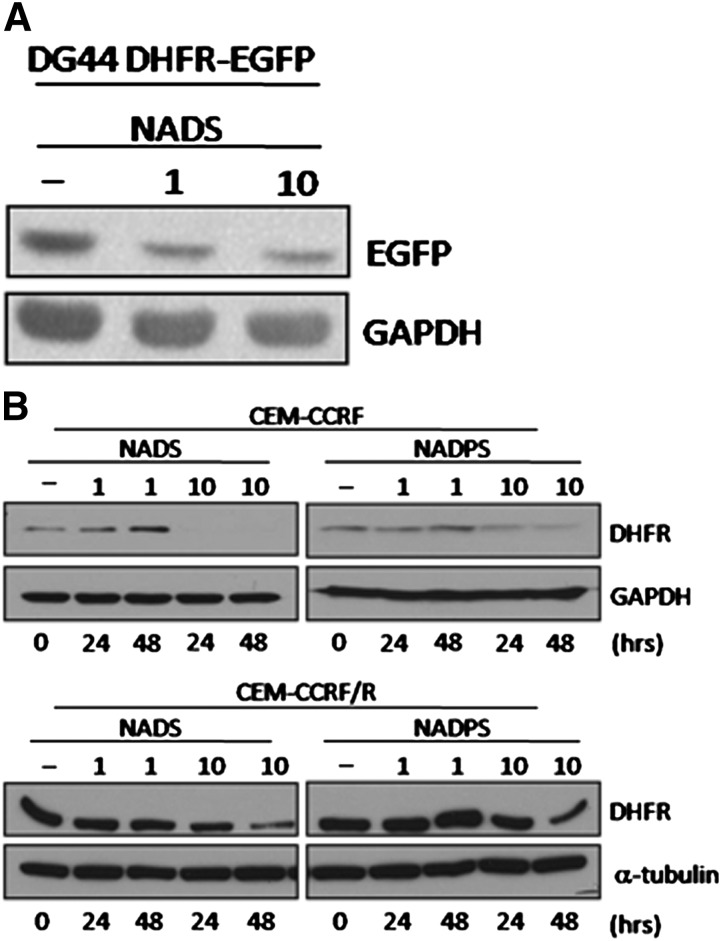
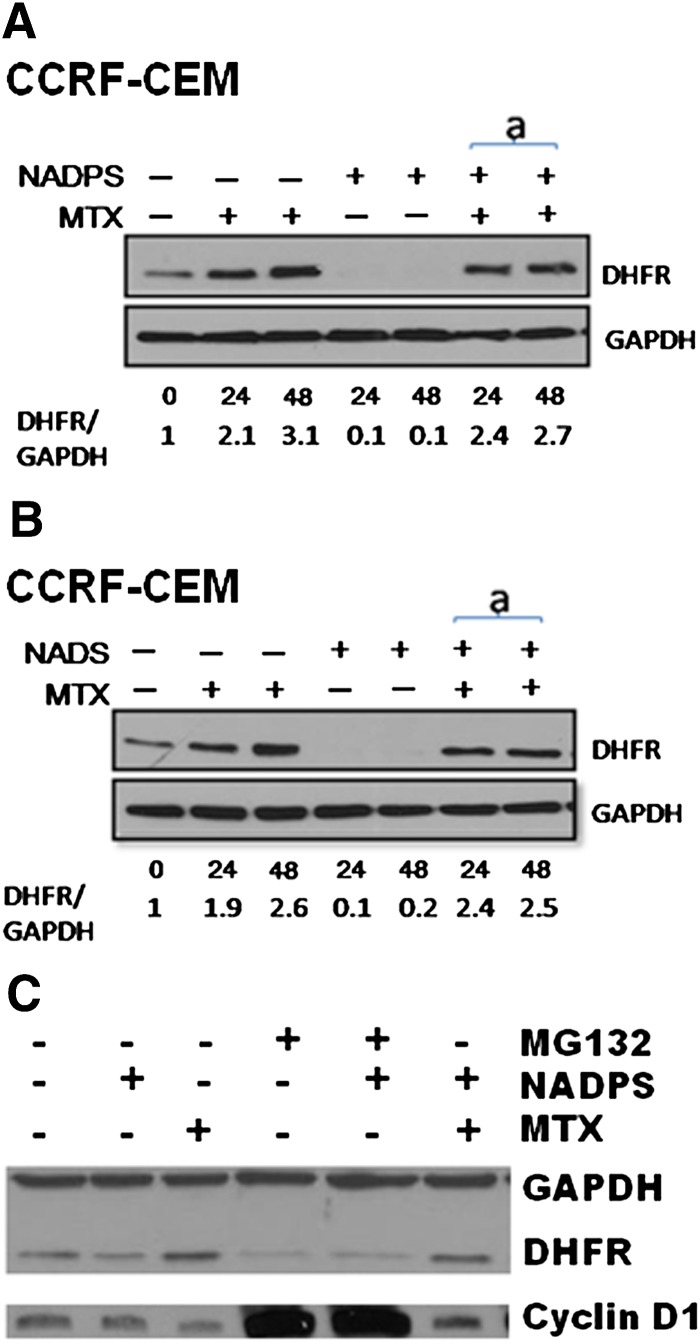
DHFR is stabilized in vitro by NADPH, FH2, and MTX in vitro ( Perkins et al., 1967; Johnston et al., 1995; Salvador et al., 2000; Ainavarapu et al., 2005). Therefore, a decrease in NADPH levels because of NADS and NADPS might lead to DHFR instability and degradation. We used DG44-(DHFR-EGFP) to determine whether DHFR levels are affected by NADS. After a 48-hour treatment with 1 and 10 μM NADS, DHFR fusion protein levels decreased, as measured by Western blot analysis (Fig. 2A). We further tested the effect of NADS and NADPS on DHFR levels in the CCRF-CEM and CCRF-CEM/R cell lines. The basal level of DHFR is significantly higher in CCRF-CEM/R cell line than in the parental CCRF-CEM cells; therefore, although DHFR levels decreased in CCRF-CEM/R cells, they were almost undetectable in the parental CCRF-CEM cell line when treated with NADS and NADPS (Fig. 2B).
Fig. 2.
NADS or NADPS treatment decreased steady-state levels of DHFR. (A) Western blot analysis of total lysates from DG44-(DHFR-EGFP) cells was performed after treatment with 0, 1, and 10 μM NADS for 48 hours. DHFR-EGFP fusion protein was detected using an antibody against EGFP. (B) CCRF-CEM and CCRF-CEM/R cells were exposed to 0, 1, and 10 μM NADS and NADPS for 0, 24, and 48 hours. The DHFR levels were detected using an anti-DHFR antibody. Equal loading was determined with GAPDH or α-tubulin, respectively.
Does NAD(P)S Primarily Target DHFR?
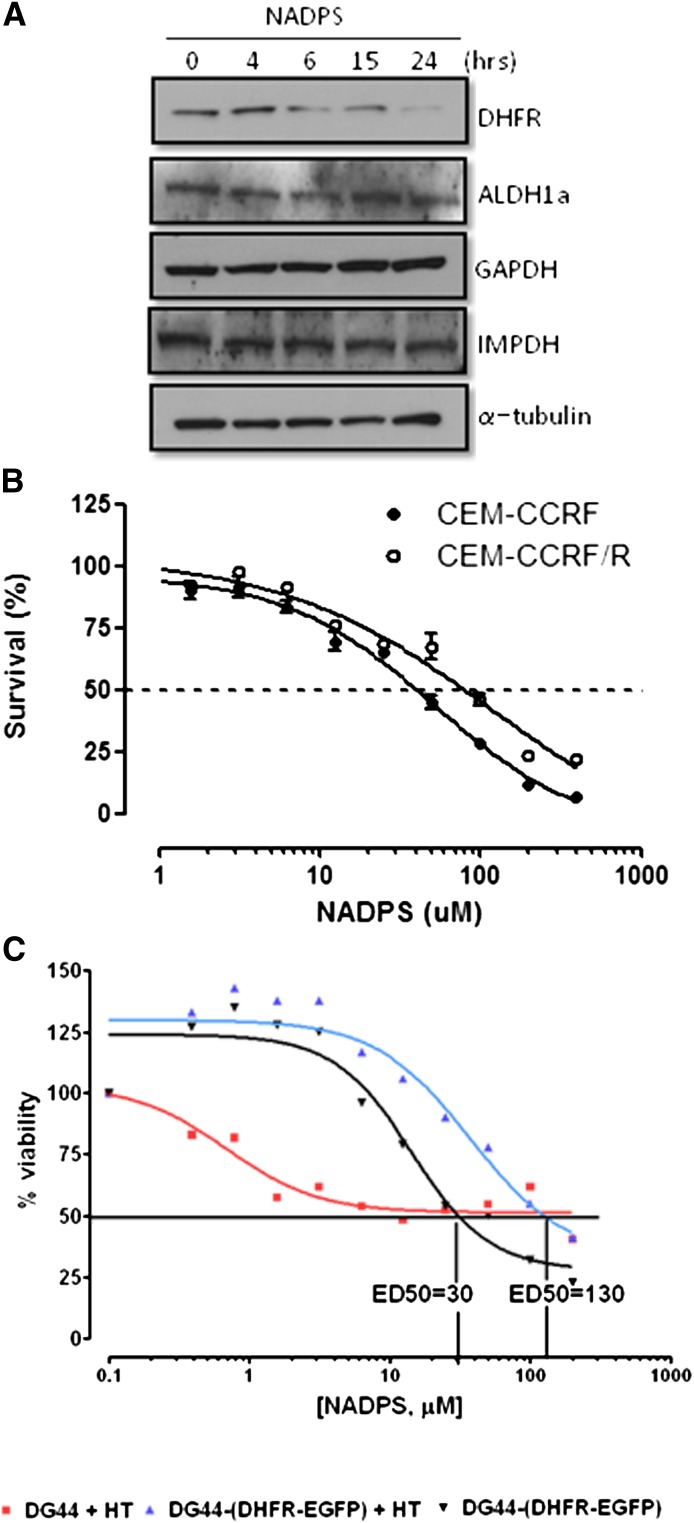
NADP and NADPH play multiple roles in cells and are involved in redox reactions, signal transduction, DNA repair, and posttranslational protein modifications (Bogan and Brenner, 2008). Although cell toxicity experiments demonstrated that cells overexpressing DHFR were less sensitive to NADPS (Fig. 3A), DHFR might not be the only target for these NAD/NADP analogs. Therefore, the effect of NADPS on other dehydrogenases was examined using C85 cells. Three different enzymes were selected for study, two of which are NAD-dependent dehydrogenases: GAPDH and IMPDH. The other is the NAD(P)-dependent dehydrogenase ALDH1a. IMPDH is the rate-limiting enzyme in the de novo purine biosynthesis, and NAD analogs have been used to inhibit the activity of IMPDH. For example, tiazofurin, an NAD analog is an orphan drug for the treatment of chronic myelogenous leukemia (Malek et al., 2004). NAD analog, N6-Naphthalenemethyl-2'-methoxybenzamido-beta-NAD+, is used to inhibit trypanosomal GAPDH. ALDH1a is a significant contributor to the reduction of NADP to NADPH. Protein levels of these three dehydrogenases were determined at different times after treatment with 10 μM NADPS (Fig. 3A). Although DHFR levels were significantly decreased even with 6 hours of treatment with NADPS, the protein levels of the other three dehydrogenases (ALDH1a, GAPDH, and IMPDH) remained the same.
Fig. 3.
Cytotoxic effect of NADPS is primarily attributable to targeting DHFR. (A) Western blot analysis of DHFR and other dehydrogenases (ALDH1a, GAPDH, and IMPDH). Protein levels were determined using antibodies specific to each dehydrogenase after treating C85 cells with 10 μM NADP-S for 0, 4, 6, 15, and 24 hours; α-tubulin was used as a loading control. (B) Trypan blue exclusion assay was used to determine cell viability of CCRF-CEM/R cells, containing a 5-fold increase of DHFR levels and CCRF-CEM parenteral cells after NADPS treatment for 96 hours. The cells were grown in RPMI-1640 medium supplemented with 10% dialyzed FBS. After harvesting cells, cell viability was determined using the Vi-CELL Series Cell Viability Analyzer. (C) MTS assay, as described in Materials and Methods, was performed to determine cytotoxic effect of NADPS in parental DG44 cells, a hamster ovary cell line with no DHFR expression and in DG44- (DHFR-EGFP), a subline of DG44 cells stably transfected with a DHFR-EGFP fusion construct. Parental DG44 cells were grown in RPMI 1640 medium that was supplemented with hypoxanthine and thymidine to allow these cells to grow in the absence of DHFR (▪, red). DG44-(DHFR-EGFP) cells were grown in RPMI 1640 media with (▴, blue) and without (▾, black) hypoxanthine and thymidine supplementation. Two thousand cells per well were plated, and cells were incubated with NADPS for 96 hours. The percentage cell survival was determined using Softmax Pro software, and GraphPad Prism 4 software was used to determine ED50 values using a sigmoidal dose-response curve fit.
Next, we determined whether the cytotoxicity observed with NADP analogs was attributable to specifically targeting DHFR. We compared the cytotoxicity of NADPS in cells overexpressing DHFR and in cells not expressing DHFR to cells with normal levels of DHFR. We hypothesized that the cells overexpressing DHFR should be resistant to NADPS. In the first experiment, CCRF-CEM, a human T cell lymphoblastic leukemia cell line, and a resistant subline (CCRF-CEM/R), due to a 5-fold increase in DHFR, were treated with NADPS. The EC50 of NADPS was 2-fold higher in the resistant cell line (CCRF-CEM/R) than in the parental CCRF-CEM cell line (Fig. 3B). Although the difference in EC50 between the resistant and sensitive cell line was not striking, this result was expected, because 5% of activity of the total basal levels of DHFR is sufficient for cell survival (Chattopadhyay et al., 2007). As shown in Fig. 2B, although NADPS treatment of CCRF-CEM/R cells leads to significant reduction in DHFR expression, these cells still had residual DHFR activity.
In the second experiment, both parental DG44 cells, which have no DHFR activity, and DG44-(DHFR-EGFP) cells stably transfected with wild-type DHFR were treated with NADPS. Parental DG44 cells were grown in RPMI 1640 medium that was supplemented with hypoxanthine and thymidine to allow these cells to grow in the absence of DHFR. If there is another target of NADPS, hypoxanthine and thymidine supplementation of these DHFR-null cells should not block the activity of NADPS, but these cells were resistant to NADPS treatment; there was more than 50% survival even at 200 μM NADPS (Fig. 3C). However, DHFR-transfected cells were sensitive to NADPS treatment with an EC50 of 30 μM (Fig. 3C), and the supplementation with hypoxanthine and thymidine led to an EC50 of 130 μM, which is 4-fold higher than in NADPS treatment alone (Fig. 3C).
Taken together, these experiments indicated that the cytotoxicity of NADPS was mainly attributable to decreased levels of DHFR. However, because DHFR-null cells were not completely resistant to NADPS treatment and the DG44-(DHFR-EGFP) were not rescued by NADPS cytotoxicity completely when the medium was supplemented with thymidine and hypoxanthine, there may be other targets of NADPS that contribute to the cytotoxicity observed.
The Effect of NADPS on DHFR Transcription
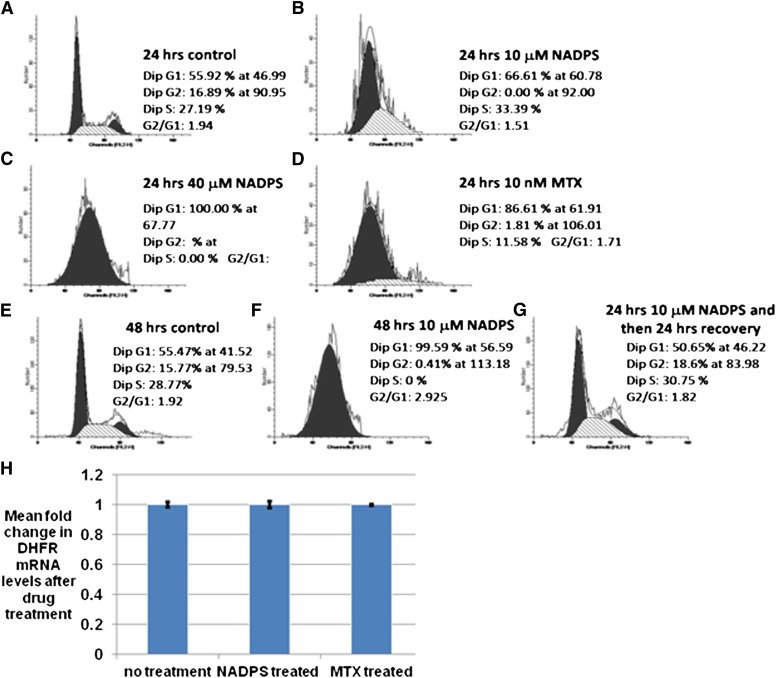
To investigate the possible mechanism of decreased DHFR protein levels, we first determined whether it is attributable to the G1/S cell cycle arrest with NADPS treatment. DHFR activity has been shown to increase during S-phase, because it is required for DNA synthesis (Abali et al., 2008). Because MTX blocks DNA synthesis and arrests cell growth in early S phase, we reasoned that the decrease in DHFR caused by NADPS might also block cells at the G1/S phase. C85 cells were incubated with NADPS for 24 hours and then stained with propidium iodide for DNA content analysis. Compared with the no-treatment control, the percentage of cells in the G1 phase increased to 11% at 10 μM and 44% at 40 μM NADPS concentrations (Fig. 4A–C). MTX was used as a positive control to show a G1/S phase block, which showed very similar results, albeit at lower concentration (Fig. 4D). Removal of NADPS from the cell culture after 24 hours reversed the G1/S block (Fig. 4E–G).
Fig. 4.
Transcriptional downregulation of DHFR mRNA through G1/S block cannot explain the decrease in DHFR levels of NADPS-treated cells. (A–C) Nonsynchronized C85 cells were treated with 0, 10, and 40 μM NADPS for 24 hours and processed for propidium iodide labeling and flow cytometry. (D) Nonsynchronized C85 cells were treated with 10 nM MTX for 24 hours and processed for PI-labeling and flow cytometry. (E–G) Recovery G1 block by removing NADPS in C85 cells. Nonsynchronized C85 cells were treated with 0 or 10 μM NADPS for 48hours (E and F). (G) C85 cells were incubated with 10 μM NADPS for 24 hours and then replaced with new medium in the absence of NADPS for 24 hours. Cells were harvested and processed for flow cytometry. (H) After treating CCRF-CEM cells with 10 μM NADPS and 10 nM MTX for 24 hours, total cytoplasmic RNA was extracted for quantitative RT-PCR analysis. DHFR mRNA levels were normalized to β-actin mRNA levels. The expression in the untreated sample was set to one. The y-axis is the mean of the fold changes in DHFR mRNA expression level; bars, standard deviation. The results were averaged from three independent experiments, and each experiment was performed in triplicate.
Although expression of DHFR is induced dramatically at the G1/S boundary after growth stimulation, dhfr mRNA is present throughout the cell cycle, suggesting that there is continuous transcription of DHFR at low levels throughout the cycle (Slansky and Farnham, 1996). Therefore, although MTX treatment leads to G1/S arrest, DHFR mRNA levels remain similar before and after MTX treatment (Sowers et al., 2003) . We investigated whether the same is true with NADPS treatment under the same conditions as the cell cycle experiments described above. CCRF-CEM cells were treated either with 10 μM NADPS or 10 nM MTX for 24 hours. After harvesting the cells, quantitative real-time RT-PCR was performed to compare DHFR mRNA levels between the drug-treated and untreated cells. There was no difference in the DHFR mRNA levels after either MTX treatment or NADPS treatment, compared with untreated cells (Fig. 4H), confirming that the decreased DHFR protein levels after NADPS treatment cannot be explained by transcriptional downregulation of DHFR mRNA through G1/S block .
The Effect of NAD(P)S on MTX-Mediated Translational Upregulation of DHFR
Administration of MTX and other antifolates increases the steady-state DHFR levels through a translation-dependent mechanism (Chu et al., 1993; Ercikan-Abali et al., 1997; Tai et al., 2004a,b; Skacel et al., 2005; Hsieh et al., 2009), and we proposed that DHFR mRNA is bound to the NADPH binding site of DHFR protein. After MTX administration, DHFR mRNA is released, thereby increasing DHFR translation (Hsieh et al., 2009). Because NADS and NADPS significantly decreased DHFR levels, we tested whether these compounds would interfere with MTX-mediated DHFR translational upregulation (Fig. 5). After treating CCRF-CEM cells with 10 μM NADS or NADPS alone for 24 hours and 48 hours or simultaneously combined with 10 nM MTX for 24 hours and 48 hours, DHFR levels were determined using Western blotting. The first lane inFig. 5, A and B are untreated cells. After treating the CCRF-CEM cells with MTX (lanes 2 and 3) for 24 and 48 hours, DHFR levels were increased; however, there was no detectable levels of DHFR after NADS or NADPS treatment (lanes 4 and 5 in Fig. 5, A and B). When the CCRF-CEM cells were treated simultaneously with NAD analogs and MTX, translational upregulation of DHFR by MTX was not hindered by the NAD analogs NADS or NADPS (Fig. 5, A and B).
Fig. 5.
A decreased DHFR level by NAD/NADP analogs is attributable to neither inhibition of MTX-mediated translational regulation of DHFR protein nor inhibition of ubiquitin proteasome pathway. DHFR protein levels were determined by Western blotting in CCRF-CEM cells. (A) CCRF-CEM cells were treated with 10 nM MTX alone, 10 μM NADPS alone, and simultaneously with 10 nM MTX and 10 μM NAPDS for 24 and 48 hours. (B) CCRF-CEM cells were treated with 10 nM MTX alone, 10 μM NADS alone, and simultaneously with 10 nM MTX and 10 μM NAPS for 24 and 48 hours. DHFR protein was detected by polyclonal anti-DHFR antibodies, and GAPDH was used as a loading control (A and B). Quantification of DHFR level was shown at the bottom of gels and measured by Image J program (provided by National Institutes of Health). (C) MCF-7 cells treated with 0.5 μM MG132, a proteasome inhibitor, in the presence and absence of 10 μM NADPS for 48 hours. GAPDH was used as the loading control, and Cyclin D1 was used as positive control for the activity of MG132. MCF-7 cells were also treated with MTX in the presence and absence of NADPS and MG132.
The Effect of NADPS on DHFR Degradation.
We further investigated whether proteasome inhibitors, such as MG132, could inhibit degradation of DHFR by NADPS. Because of the high toxicity of MG132 (carbobenzoxy-Leu-Leu-leucinal) in C85 cell, MCF-7 cells (human breast adenocarcinoma cell line) were treated with 0.5 μM MG132 in the presence of NADPS alone for 48 hours (Fig. 5C). To verify that MG132 was active, cyclin D1, which is known to be regulated by proteasomal degradation, was used as a positive control and GAPDH was used as a negative control (Wang et al., 2007). MG132 prevented cyclin D1 degradation but did not affect DHFR levels either in the presence or absence of NADPS. These results are in accordance with the published data. Noe et al. (2003) also demonstrated that treatment with the proteasome inhibitor MG132 did not affect the levels of DHFR. These findings provide evidence that the decreased DHFR protein levels after NADPS treatment is not attributable to inhibition of ubiquitin-mediated degradation.
Decreased Half-Life of DHFR due to NAD Kinase Inhibition by NADPS
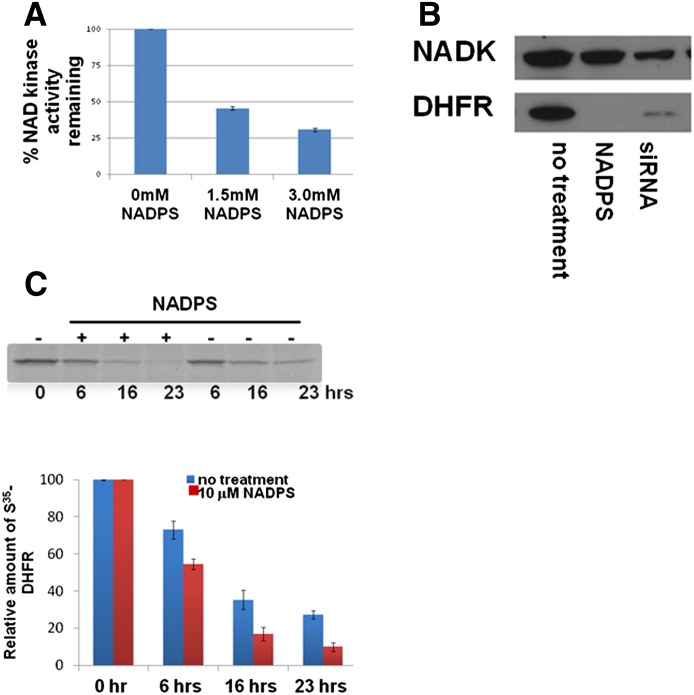
Because the decreased DHFR levels by NADPS could not be explained by any of the mechanisms described thus far, we hypothesized that the decreased steady-state levels of DHFR protein may be attributable to the decreased levels of the cofactor NADPH. DHFR is stabilized by its ligands, such as NADPH and MTX, thus inhibiting its degradation (Xiang, 2002). NAD kinase, which catalyzes phosphorylation of NAD to NADP, is the only enzyme in mammals that maintains the NADP pool (Pollak et al., 2007). Therefore, we tested whether NADPS inhibits the activity of NAD kinase. NAD kinase activity was determined spectrophotometrically using a coupled assay which generates NADP using NADK. NADP in turn acts as a substrate for glucose-6-phosphate dehydrogenase (Lerner et al., 2001). The activity of NAD kinase in the presence of 1.5 mM and 3.0 mM NADPS was reduced by 55 and 69%, respectively (Fig. 6A). We also determined the effect of NADS on NAD kinase. NADS, unlike NADPS, supported the activity of NAD kinase in the absence of NAD, suggesting that it is a substrate for NADK kinase. However, higher concentrations of NADS led to accumulation of NADPS inhibiting the reaction (data not shown). These results are in accordance with the rest of the results, because the outcome of treating the cells with either NADS or NADPS were identical.
Fig. 6.
NAD kinase is inhibited by NADPS, leading to decreased half-life of DHFR. (A) Inhibition of NAD kinase was determined spectrophotometrically by measuring the increase in absorbance at 340 nm caused by the reduction of NADP to NADPH by G6PDH using a coupled assay (see details in Materials and Methods). (B) Effect of NADK suppression on DHFR protein levels was determined by Western blotting in C85 cells after transfection for 48 hours with siRNA targeting NADK. DHFR protein levels were compared with untreated cells and with NADPS-treated cells. (C) The half-life of DHFR was measured by a 35S methionine radiolabeled pulse-chase experiment. CCRF-CEM/R cells were treated with 10 μM NADPS, and at the indicated times, samples were subjected to immunoprecipitation with an anti-DHFR antibody, followed by SDS-PAGE and autoradiography. The lower panel shows the quantification of results in the presence and absence of NADPS. Values shown are the means ± S.D. for three independent pulse-chase experiments. For statistical analysis, Student's t test was used; paired comparisons of cellular DHFR levels between NADPS treated and untreated were made. P values were <0.05 both at 16 and 23 hours, suggesting that there was a statistical difference in DHFR levels between treated and untreated cells.
To further demonstrate that the decrease in cellular DHFR levels in the presence of NADPS is indeed mediated by decreased NADPH levels through inhibition of NADK, we determined the effects of the knockdown of NADK expression on DHFR levels. C85 cells were transiently transfected with siRNA targeting NADK in C85 cells, and NADK and DHFR levels were determined using Western blotting. Compared with untreated cells, cells transfected with siRNAs specific for NADK exhibited significant decreases in DHFR protein levels (Fig. 6B) similar to the levels seen in cells treated with NADPS. These results provide further evidence of the role of NADPH levels in stabilizing DHFR levels.
The inhibition of NAD kinase by NADPS may provide an explanation for the lowered NADPH levels found after treatment of cells with NADPS; therefore, we considered whether the lower levels of DHFR in the presence of NADPS was attributable to the accelerated degradation of DHFR because of low levels of its cofactor, NADPH. A pulse-chase labeling experiment was performed to determine the half-life of DHFR protein in the presence and absence of NADPS. We chose CCRF-CEM/R cells for this study, because they have higher levels of DHFR due to gene amplification, making it easier to detect radiolabeled DHFR protein levels. The half-life of DHFR was reduced by 42% from 11.3 hours to 6.6 hours after treatment with 10 μM NADPS (Fig. 6C).
This experiment unequivocally demonstrated that the decrease in levels of DHFR is attributable neither to transcriptional nor translational inhibition but rather to accelerated degradation of DHFR protein after NADPS treatment, because transcriptional or translational inhibition of DHFR would not affect the DHFR half-life.
Modeling Studies of NADPS+ and NADP+ Binding to NAD Kinase
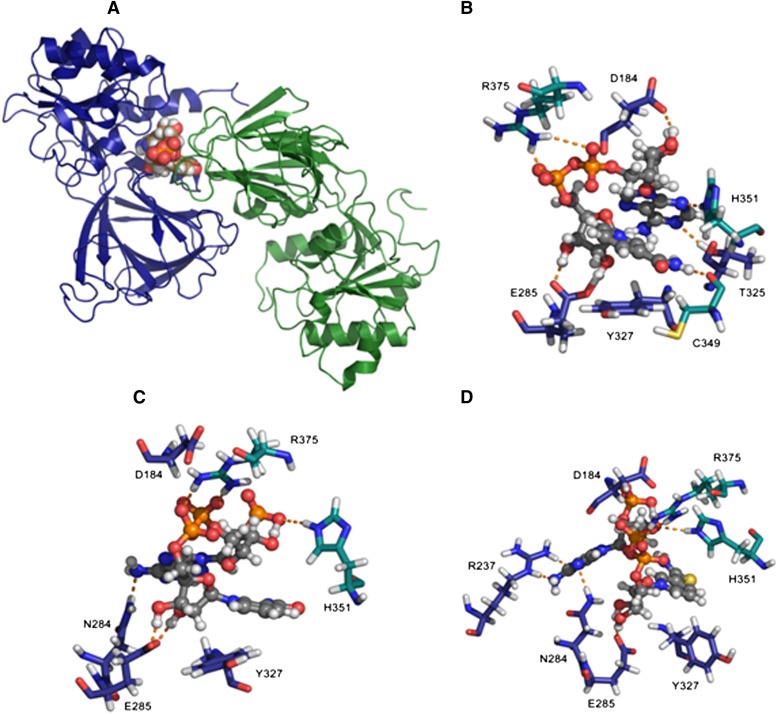
To better understand the binding of NADPS+ to NAD kinase, we developed models of NAD+, NADP+ bound to human structure as described in Materials and Methods to better understand the mechanism and explore potential for development of novel therapeutics based on inhibition of the kinase. The models produced for NAD+ and NADP+ complexes with NAD kinase were used to study NADPS+ via simple replacement of the nicotinamide amide oxygen with sulfur to study the impact of this replacement on NAD binding to NAD kinase. The model of the human NAD kinase complex with NAD+ is shown in Fig. 7, A and B. The nicotinamide ring in NAD+ leads to π-π stacking with the aromatic side chain of Y327. The 2′ and 3′ of the nicotinamide ribose sugar are anchored by hydrogen bonds to E285. The pyrophosphate linker between the sugars is stabilized by electrostatic interaction with R375 of chain B. The 2′- and 3′-hydroxyl groups of the adenine ribose sugar are in proximity to residue D184. The adenine ring is stabilized by hydrogen bonds to side chains of T325 (chain A) and H351 (chain B). Finally, the nicotinamide CONH2 N-H forms a strong hydrogen bond to the backbone C=O of C349 of chain B. All of these residues are well conserved.
Fig. 7.
NADPS+ has a more favorable binding to NAD kinase, compared with NADP+. (A) Human NAD kinase (cartoon) complexed with NAD+ (space-filling model). Chain A is colored deep blue, and chain B is colored forest green (last snapshot from 2 ns of dynamics in TIP3P water box). Built from Protein Data Bank codes 3PFN (Human) and 1Z0Z (Arch. fulgidus). (B) Close-up of binding region with NAD+ and key interacting residues depicted as stick models where NAD+ (ball and stick model) carbon atoms are colored gray and chain A protein residue carbon atoms are colored navy blue, with chain B residue carbon atoms colored teal. (C) Close-up of binding region with NADP+ and key interacting residues depicted as stick models where NADP+ (ball and stick model) carbon atoms are colored gray and chain A protein residue carbon atoms are colored navy blue, with chain B residue carbon atoms colored teal. (D) Close-up of binding region with NADPS+ and key interacting residues depicted as stick models where NADPS+ (ball and stick model) carbon atoms are colored gray and chain A protein residue carbon atoms are colored navy blue, with chain B residue carbon atoms colored teal.
In the model of NADP+ product in complex with NAD kinase (Fig. 7C), we see the impact that the phosphorylated 2′-hydroxyl of the adenine ribose ring has on bound conformation relative to the substrate NAD+. The 2′-phosphate benefits from hydrogen bond stabilization from H351 of chain B. The pyrophosphate linker remains stabilized by R375 of chain B, as in the case of NAD+; however, the adenine ring N6 nitrogen is now in proximity to hydrogen bond with residue N284 of chain A. The nicotinamide ring is canted further away from the plane of the tyrosine (Y327) aromatic ring. The glutamate residue (E285) serves a key role in stabilizing the 2′ and 3′ hydroxyls of the nicotinamide ribose sugar. The MM/GBSA results (Table 2) NAD+ binding compared with NADP+ indicates a loss in electrostatic energy stabilization at the expense of solvation. This might be a driving force for expulsion of the NADP+ product out of the binding site of the enzyme.
TABLE 2.
MM-GBSA results in kcal/mol, based on 100 evenly spaced snapshots from last 1 ns of 2 ns production run.
See computational methods section for calculation details. ε(ext) = 78.5; ε(int) = 4.0; ΔEgas = Evdw + Eelec; ΔEsolv = EGB + Esurf; ΔHbind = ΔEgas + ΔEsolv.
| Compound | Evdw | Eelec | EGB | Esurf | ΔEgas | ΔEsolv | ΔHbind |
|---|---|---|---|---|---|---|---|
| NAD+ | −66.2 ± 4.5 | −51.9 ± 7.3 | 51.1 ± 5.9 | −8.4 ± 0.2 | −118.0 ± 7.0 | 42.8 ± 5.9 | −75.3 ± 3.5 |
| NADP+ | −54.0 ± 4.4 | −14.3 ± 10.5 | 22.7 ± 9.4 | −8.2 ± 0.3 | −68.3 ± 10.6 | 14.5 ± 9.4 | −53.8 ± 3.0 |
| NADS+ | −56.1 ± 4.7 | −57.7 ± 5.6 | 55.5 ± 4.6 | −8.6 ± 0.2 | −113.8 ± 6.9 | 46.9 ± 4.5 | −66.9 ± .1 |
| NADPS+ | −70.0 ± 4.1 | 10.3 ± 7.6 | 1.1 ± 6.9 | −9.1 ± 0.2 | −59.7 ± 8.0 | −7.9 ± 6.9 | −67.6 ± 3.7 |
The NADPS+ bound NAD kinase (Fig. 7D) benefits from hydrogen bonds of the NADPS+ adenine ring with the side chain of R237 in addition to hydrogen bond with N284. The nicotinamide ring is now almost perpendicular in orientation to the side chain of Y327. The R375 and H351 residues of chain B stabilize the pyrophosphate linker. Residue E285 of chain A remains in proximity to stabilize the 2′ and 3′ hydroxyls of the nicotinamide ribose sugar. The MM/GBSA data in Table 2 indicate that NADPS+ enjoys a more favorable solvation energy in the lower dielectric environment of the NAD kinase binding site, compared with NADP+, most likely because of the more hydrophobic and less polar nature of the thioamide in NADPS+, compared with the amide in NADP+. This lower dielectric environment in the region of the nicotinamide in 10 Å is bordered by A329, A330, H351, and L353. The NADPS+ complex with NAD kinase has a much more favorable van der Waals energy in comparison with NADP+. The NADPS+ binding enthalpy is about 13.8 kcal/mol more favorable than NADP+.
Protection of DHFR by MTX from Degradation after NADPS Treatment
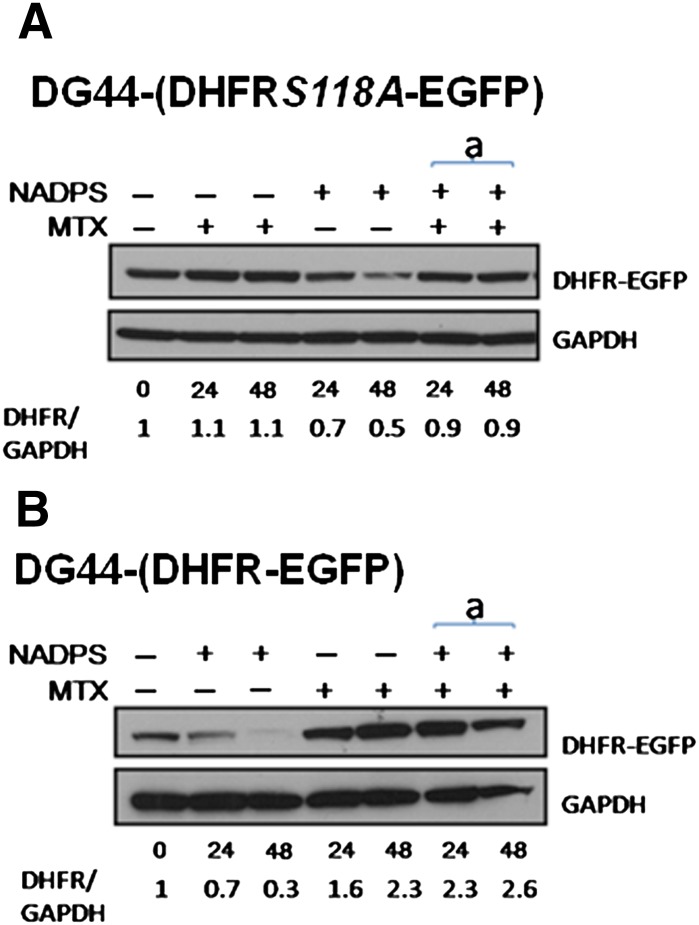
MTX has been shown to stabilize DHFR in vitro and, in particular, in ternary complex when DHFR is bound to NADPH/NADP (Nakano et al., 1994); however, it has not been previously shown in a cellular context. Demonstration of stabilization of DHFR by MTX in cells has been difficult because DHFR is always bound to NADPH. Because NADPS decreases the half-life of DHFR and, thus, the steady state levels of DHFR, we tested whether we can reverse this phenomenon with MTX. We used DG44-(DHFRS118A-EGFP) cells stably transfected with a variant of DHFR with a point mutation, S118A. This variant of DHFR is not upregulated by MTX, but kinetic properties are very similar to the wild type. Therefore, it allowed us to avoid interference with degradation (Hsieh et al., 2009; Skacel et al., 2005). DG44-(DHFRS118A-EGFP) cells were treated with NADPS alone or simultaneously with NADPS and MTX. Although NADPS alone decreased DHFR levels of the S118A mutant, the addition of MTX protected DHFR levels from degradation (Fig. 8A). DG44 Wild-Type DHFR-EGFP cells were used as a control to ensure that NADPS did not inhibit MTX-induced DHFR levels, as observed in CCRF-CEM cells (Fig. 8B). Thus, MTX indeed stabilizes DHFR in a cellular context and protects the enzyme from degradation because of cofactor depletion.
Fig. 8.
MTX protects cells from DHFR degradation by NADPS. DHFR protein levels were determined by Western blotting in DG44-(DHFR-EGFP) cells. (A and B) DG44 DHFR-EGFP cells or DG44 DHFRS118AEGFP cells were treated with 10 nM MTX alone, 10 μM NADPS alone, and the simultaneously addition of 10 nM MTX and 10 μM NAPDS for 24 and 48 hours. DHFR-EGFP and DHFRS118AEGFP protein were detected by a monoclonal anti-EGFP antibody. GAPDH was used as a loading control. Quantification of DHFR levels (shown at the bottom of gels) was measured by the Image J program (provided by NIH).
Combination Studies with NADPS and MTX
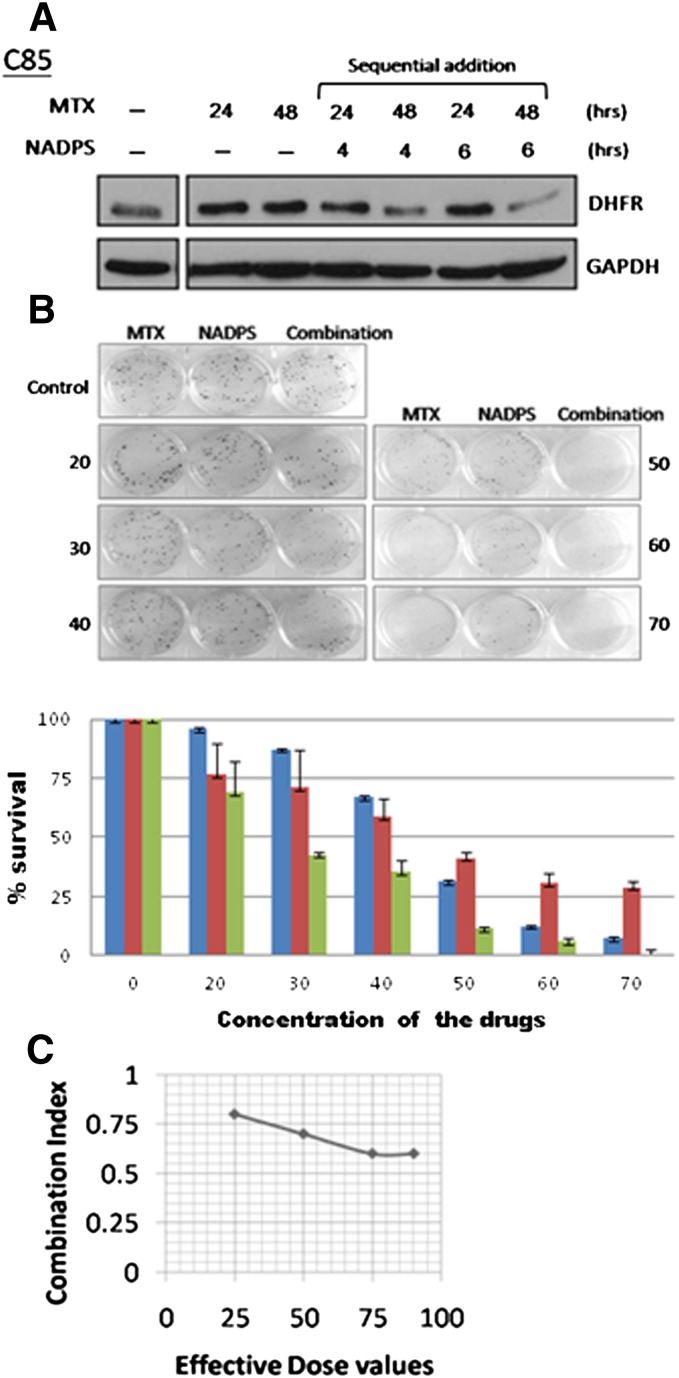
MTX and MTX polyglutamates tight-binding competitive inhibitors of DHFR and, at slightly acidic pH values, are pseudo-irreversible inhibitors. Lowering DHFR levels in the cell effectively decreases the concentration of MTX required to kill the cancer cells and reduces the toxicity of MTX to normal cells; therefore, we hypothesized that the addition of NADPS before MTX administration would enhance the cytotoxicity of MTX, because NADPS lowered DHFR levels. The effect of NADPS and MTX, compared with each drug alone, was determined using C85 cells. Ten micromoles of NADPS was added to these cells for 4 or 6 hours to decrease DHFR levels, followed by the addition of l and 10 nM MTX for 24 or 48 hours. DHFR levels were significantly decreased after sequential combination with NADPS and MTX in C85 cells (Fig. 9A). To determine whether the combination of MTX and NADPS was synergistic, several concentrations of a fixed ratio of 1000:1 (NADPS to MTX) were used with a clonogenic assay in C85 cells as the readout. Starting with 40 μM NADPS and 40 nM MTX, as the drug concentrations were increased, the percentage survival of the C85 cells dramatically decreased in combination treatment, in comparison with either drug alone (Fig. 9B). The Chou-Talalay method for drug combination was used to determine CI values for NADPS and MTX combination treatment. CI is a quantitative definition for additive effect (CI = 1), synergism (CI < 1), and antagonism (CI > 1) in drug combinations (Chou and Talalay, 1984). Figure 9C shows that, at ED that kills 25%, 50%, 70%, and 90% of the cells, the CI value was <1, indicating that NADPS treatment, followed by MTX, resulted in a significant synergy.
Fig. 9.
NADPS has a synergetic cytotoxic effect with MTX. DHFR levels were determined by Western blotting after sequential addition of NADPS and MTX. C85 (A) cells were treated with 10 nM MTX alone for 24 or 48 hours, pretreated with 10 mM NADPS for 4 or 6 hours, and followed by treatment with 10 nM MTX for 24 or 48 hours. DHFR protein was detected by an anti-DHFR antibody, and GAPDH was used as a loading control. (B) A clonogenic assay was performed to determine the cytotoxic effect of the combination of NADPS and MTX in C85 cells. Values were given as means ± SD for three independent experiments. (C) Combination index plots for C85 cells exposed to NADPS and MTX (see details in Materials and Methods).
The Effect of NADPS against MTX Transport–Resistant Cells
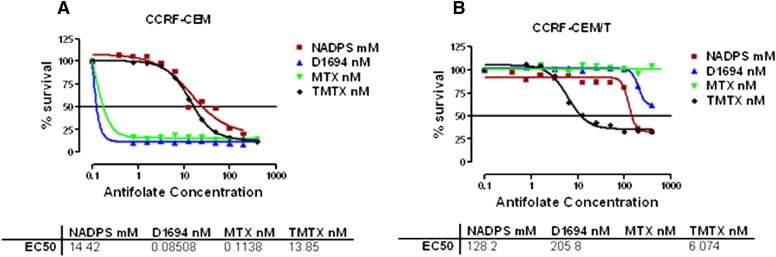
Impaired MTX accumulation, because of either decreased uptake or lack of retention because of poor polyglutamate formation, is the major factor limiting MTX effectiveness (Gorlick et al., 1996). However, in the clinic, an increase in MTX concentration is not an option to overcome resistance to MTX because of increased toxicity of MTX at high concentrations. NADPS by lowering DHFR levels would be predicted to be effective in tumors with intrinsic or acquired resistance to MTX. Therefore, we treated the CCRF-CEM /T cell line, resistant to classic antifolates, such MTX and D1694 (raltitrexed; Tomudex). because of impaired transport (Mini et al., 1985), and compared the results to the parent CCRF-CEM cells with NADPS. Trimetrexate (TMTX), a DHFR inhibitor, is lipid soluble and passively diffuses into cells and was used to demonstrate that both transport-resistant and sensitive cells would respond to TMTX treatment (Fig. 10, A and B). Both cell lines were also treated with either MTX or D1694, the latter an antifolate that targets thymidylate synthase primarily. Although the parent cells were sensitive to both of these classic antifolates (Fig. 10A), CCRF-CEM/T cells were extremely resistant to MTX (Fig. 10B) and D1694 (raltitrexed; Tomudex) (Fig. 10B), but not TMTX. CCRF-CEM/T cells were also sensitive to NADPS treatment, albeit 9-fold less sensitive, compared with the parental CCRF-CEM cells (Fig. 10. A and B). A moderate increase in resistance to TMTX and NADPS could be attributable to the slower growth rate of the CCRF-CEM/T cells than the parental CCRF-CEM cells.
Fig. 10.
NADPS is still effective in MTX transport–resistant cells. Parental CCRF-CEM (A) and CCRF- CEM /T cells, resistant to classic antifolates, such MTX and D1694 (raltitrexed; Tomudex) because of impaired transport (Mini et al., 1985) (B) were treated with MTX, D1694, and the lipophilic antifolate TMTX. The cells were grown in RPMI-1640 medium supplemented with 10% dialyzed FBS. After incubation with NADPS for 96 hours, cells were collected and viability was determined using Trypan blue exclusion assay with the Vi-CELL Series Cell Viability Analyzer. The percentage cell survival was determined by Softmax Pro software, and sigmoidal dose-response curve fit of the graphs drawn by GraphPad Prism 4 software was used to determine ED50 values. Although the concentrations of NADPS used in the experiment were micromolar, the antifolate concentrations were nanomolar.
Discussion
Modifying protein stability is a valid therapeutic target, as seen with the success of the proteasome inhibitors, such as bortezomib, which is currently used for the treatment of multiple myeloma and mantle cell lymphoma. However, there is not an approved drug that accelerates degradation of proteins important for cell proliferation. Geldanamycin and its analogs, such as 17-allylamino-17-demethoxygeldanamycin, inhibit the ATPase activity of heat shock protein Hsp90, leading to improper folding and activity of client proteins. Often, these misfolded proteins become the target of ubiquitin-dependent degradation (Miyata, 2005). However, degradation of these proteins by geldanamycin is not very selective and leads to hepatotoxicity (Latif et al., 2001). Recently, phase III trials with tanespimycin, a less toxic analog of geldanamycin for the treatment of multiple myeloma, were terminated because of lack of efficacy. In this study, we evaluated whether cofactor depletion, specifically NADPH, may also be an effective strategy to target cancer cells. NADPH pools are maintained in cells mainly through the hexose monophosphate pathway, reducing NADP to NADPH, the demand for NADPH in cancer cells is increased because of increased metabolic flux of nucleotide and fatty acid synthesis (Vander Heiden et al., 2009). In humans, the only enzyme that is capable of synthesizing NADP from NAD is NAD kinase; therefore, we initiated efforts to identify NAD(P)H analogs that may decrease the NADPH concentration in cells, rendering DHFR unstable and increasing its degradation.
To that effect, we identified two NAD analogs, NADS and NADPS, which inhibit NAD kinase, causing increased degradation of DHFR. These studies encourage development of additional novel analogs targeted to NAD kinase. The IC50 of both thionicotinamide adenine dinucleotides NADS and NADPS were very similar (10–40 μM) in RPMI-1640 media and were inversely dependent on the concentration of nicotinamide present in the media. Ham’s F-12 medium (0.295 μM) has 28-fold less nicotinamide than RPMI-1640 (8.2 μM), associated with 10-fold decrease in the IC50 of both NAD analogs. The concentration of nicotinamide in Ham’s F-12 media is very similar to the mammalian plasma concentration, suggesting that these two analogs would be more potent in vivo (O'Reilly and Niven, 2003). We excluded the following mechanisms for the cytotoxicity of NADS and NADPS. These analogs neither inhibit the activity of DHFR nor repressed the transcription of DHFR. They also did not interfere with the MTX-induced translational regulation of DHFR. Pulse-chase experiment clearly demonstrated that the decrease in the steady state levels of DHFR was attributable to accelerated degradation of DHFR in the presence of NADPS. We propose that a plausible mechanism for the degradation of DHFR by these nicotinamide analogs is attributable to lowered levels of NADPH, as a consequence of inhibition of NAD kinase. We recently demonstrated that benzamide riboside, a nicotinamide analog, reduces cellular NADP and NADPH levels by inhibiting NADK. In accordance with our findings here, NADK inhibition led to low levels of NADPH, and DHFR levels were significantly decreased (Roussel et al., 2012). Because of the similar absorbance and fluorescence of NADP and NADPS, we were unable to quantify the cellular NADP(H) pools in this study. To understand the binding of NADPS with NAD kinase, we performed a computer modeling study of the human NAD kinase with NADPS. Predictions pertaining to NADPS+ demonstrate a more stable complex with NAD kinase, compared with NADP+. The NADPS+ binding enthalpy is about 13.8 kcal/mol more favorable than NADP+ because of more stable van der Waals interactions with the enzyme and solvation energy. The binding enthalpies indicate that, when NAD+ is phosphorylated, its product NADP+ results in a less stable complex, leading to its expulsion from the enzyme, thus driving the process forward. However, in the case of NADS+, we see that formation of its phosphorylated product leads to a complex that is equally stable for the product NADPS+, which is more stable than the NADP+/kinase complex. Disruption of DHFR folding occurs when DHFR is not bound to NADPH. DHFR folding mechanisms involve formation of three ensembles. The first ensemble has considerable secondary structure with exposed hydrophobic core. The second ensemble is obligatory and begins to resemble the native state but is unable to bind either MTX or NADPH. The interconversion between the second ensemble and the third is slower in human DHFR than in E. coli or L. casei DHFR. It is possible that NADPS, by lowering NADPH levels, may be interfering with the interconversion of the second ensemble to third ensemble, which is the native state (Abali et al., 2008). Our results support this hypothesis, because synergy in combination studies was obtained only if the cells were treated with NADPS analogs first.
Earlier studies have shown that the level of DHFR determines whether cells would be sensitive to MTX treatment, and one mechanism of acquired resistance to MTX is an increase in DHFR level because of amplification of the DHFR gene (Alt et al., 1978; Banerjee et al., 2002). Because the NADS analogs significantly lowered DHFR levels, we anticipated that NADPS would potentiate MTX cytotoxicity. Combination studies with the C85 cells, a metastatic colon cancer cell line, demonstrated significant synergy when the cells were first treated with NADPS followed by MTX. CI values were lower than one at all EDs. Synergy is increased with a CI to 0.6 at 75% and 90% ED. Our data raise the hope that the resistance to MTX, one of the most widely used cancer chemotherapeutic agent, could be blocked with synergistic combinations of NADPS and MTX. We plan to continue our studies with preclinical studies.
Limited screening of other dehydrogenases indicated that NADPS lowered DHFR levels alone. However, NADPS may lower the steady-state levels of other NADP or NADPH requiring enzymes. Other targets are most likely the enzymes that participated in anabolic pathways, which are shown to be activated in cancer cells because of dysregulation of upstream signaling pathways, such as HIF-1, AMPK, and mTOR (Tong et al., 2009); therefore, it may be desirable to target several of these enzymes at once. In summary, our results demonstrate that, by inhibiting NAD kinase, steady-state levels of DHFR are lowered. Because NAD kinase is the only enzyme responsible for generating NADPH, this enzyme is a valid target for further inhibitor development and DHFR may be a marker for demonstrating the activity of inhibitors targeting NAD kinase in cancer cells.
Abbreviations
- ALDH1a
aldehyde dehydrogenase
- CI
combination index
- DHFR
dihydrofolate reductase
- EGFP
enhanced green fluorescent protein
- FBS
fetal bovine serum
- FH2
dihydrofolate
- FH4
tetrahydrofolate
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GSH
glutathione
- IMPDH
inosine-5′-monophosphate dehydrogenase
- MG132
carbobenzoxy-Leu-Leu-leucinal
- MTS
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt
- MTX
methotrexate; NADPS, thionicotinamide adenine dinucleotide phosphate
- NADS
thionicotinamide adenine dinucleotide
- PBS
phosphate-buffered saline
- RT-PCR
reverse-transcription polymerase chain reaction
- TMTX
trimetrexate
Authorship Contributions
Participated in research design: Hsieh, Tedeschi, Banerjee, Bertino, Abali.
Conducted experiments: Hsieh, Lawal, Kerrigan, Johnson Farley, Tedeschi, Lee.
Performed data analysis: Hsieh, Tedeschi, Kerrigan, Banerjee, Bertino, Abali.
Wrote or contributed to the writing of the manuscript: Kerrigan, Scotto, Banerjee, Tedeschi, Bertino, Abali.
Footnotes
This work was supported by the National Institutes of Health National Cancer Institute [Grant R01-CA08010].
References
- Abali EE, Skacel NE, Celikkaya H, Hsieh YC. (2008) Regulation of human dihydrofolate reductase activity and expression. Vitam Horm 79:267–292 [DOI] [PubMed] [Google Scholar]
- Ainavarapu SR, Li L, Badilla CL, Fernandez JM. (2005) Ligand binding modulates the mechanical stability of dihydrofolate reductase. Biophys J 89:3337–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt FW, Kellems RE, Bertino JR, Schimke RT. (1978) Selective multiplication of dihydrofolate reductase genes in methotrexate-resistant variants of cultured murine cells. J Biol Chem 253:1357–1370 [PubMed] [Google Scholar]
- Banerjee D, Mayer-Kuckuk P, Capiaux G, Budak-Alpdogan T, Gorlick R, Bertino JR. (2002) Novel aspects of resistance to drugs targeted to dihydrofolate reductase and thymidylate synthase. Biochim Biophys Acta 1587:164–173 [DOI] [PubMed] [Google Scholar]
- Berendsen HJC, Postma JPM, vanGunsteren WF, DiNola A, Haak JR. (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690 [Google Scholar]
- Bogan KL, Brenner C. (2008) Nicotinic acid, nicotinamide, and nicotinamide riboside: a molecular evaluation of NAD+ precursor vitamins in human nutrition. Annu Rev Nutr 28:115–130 [DOI] [PubMed] [Google Scholar]
- Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. (2005) The Amber biomolecular simulation programs. J Comput Chem 26:1668–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Moran RG, Goldman ID. (2007) Pemetrexed: biochemical and cellular pharmacology, mechanisms, and clinical applications. Mol Cancer Ther 6:404–417 [DOI] [PubMed] [Google Scholar]
- Cheok MH, Evans WE. (2006) Acute lymphoblastic leukaemia: a model for the pharmacogenomics of cancer therapy. Nat Rev Cancer 6:117–129 [DOI] [PubMed] [Google Scholar]
- Chou TC, Talalay P. (1984) Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul 22:27–55 [DOI] [PubMed] [Google Scholar]
- Chu E, Takimoto CH, Voeller D, Grem JL, Allegra CJ. (1993) Specific binding of human dihydrofolate reductase protein to dihydrofolate reductase messenger RNA in vitro. Biochemistry 32:4756–4760 [DOI] [PubMed] [Google Scholar]
- Darden T, York D, Pedersen L. (1993) Particle Mesh Ewald: An N-log(N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092 [Google Scholar]
- Dunn SM, Batchelor JG, King RW. (1978) Kinetics of ligand binding to dihydrofolate reductase: binary complex formation with NADPH and coenzyme analogues. Biochemistry 17:2356–2364 [DOI] [PubMed] [Google Scholar]
- Ercikan-Abali EA, Banerjee D, Waltham MC, Skacel N, Scotto KW, Bertino JR. (1997) Dihydrofolate reductase protein inhibits its own translation by binding to dihydrofolate reductase mRNA sequences within the coding region. Biochemistry 36:12317–12322 [DOI] [PubMed] [Google Scholar]
- Ercikan-Abali EA, Mineishi S, Tong Y, Nakahara S, Waltham MC, Banerjee D, Chen W, Sadelain M, Bertino JR. (1996) Active site-directed double mutants of dihydrofolate reductase. Cancer Res 56:4142–4145 [PubMed] [Google Scholar]
- Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen L. (1995) A smooth particle mesh ewald method. J Chem Phys 103:8577–8592 [Google Scholar]
- Estrela JM, Ortega A, Obrador E. (2006) Glutathione in cancer biology and therapy. Crit Rev Clin Lab Sci 43:143–181 [DOI] [PubMed] [Google Scholar]
- Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, Motiwala T, Jacob ST. (2005) 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol 25:4727–4741 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gorlick R, Goker E, Trippett T, Waltham M, Banerjee D, Bertino JR. (1996) Intrinsic and acquired resistance to methotrexate in acute leukemia. N Engl J Med 335:1041–1048 [DOI] [PubMed] [Google Scholar]
- Hawkins GD, Cramer CJ, Truhlar DG. (1995) Pairwise solute descreening of solute charges from a dielectric medium. Chem Phys Lett 246:122–129 [Google Scholar]
- Hawkins GD, Cramer CJ, Truhlar DG. (1996) Parametrized models of aqueous free energies of solvation based on pairwise descreening of solute atomic charges from a dielectric medium. J Phys Chem 100:19824–19839 [Google Scholar]
- Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65:712–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou T, Wang J, Li Y, Wang W. (2011) Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model 51:69–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh YC, Skacel NE, Bansal N, Scotto KW, Banerjee D, Bertino JR, Abali EE. (2009) Species-specific differences in translational regulation of dihydrofolate reductase. Mol Pharmacol 76:723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. (1996) VMD: visual molecular dynamics. J Mol Graph 14:33–38, 27–28 [DOI] [PubMed] [Google Scholar]
- Johnston JA, Johnson ES, Waller PR, Varshavsky A. (1995) Methotrexate inhibits proteolysis of dihydrofolate reductase by the N-end rule pathway. J Biol Chem 270:8172–8178 [DOI] [PubMed] [Google Scholar]
- Latif S, Bauer-Sardina I, Ranade K, Livak KJ, Kwok PY. (2001) Fluorescence polarization in homogeneous nucleic acid analysis II: 5′-nuclease assay. Genome Res 11:436–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner F, Niere M, Ludwig A, Ziegler M. (2001) Structural and functional characterization of human NAD kinase. Biochem Biophys Res Commun 288:69–74 [DOI] [PubMed] [Google Scholar]
- Liu J, Lou Y, Yokota H, Adams PD, Kim R, Kim SH. (2005) Crystal structures of an NAD kinase from Archaeoglobus fulgidus in complex with ATP, NAD, or NADP. J Mol Biol 354:289–303 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- Malek K, Boosalis MS, Waraska K, Mitchell BS, Wright DG. (2004) Effects of the IMP-dehydrogenase inhibitor, Tiazofurin, in bcr-abl positive acute myelogenous leukemia. Part I. In vivo studies. Leuk Res 28:1125–1136 [DOI] [PubMed] [Google Scholar]
- McTigue MA, Davies JF, 2nd, Kaufman BT, Kraut J. (1993) Crystal structures of chicken liver dihydrofolate reductase: binary thioNADP+ and ternary thioNADP+.biopterin complexes. Biochemistry 32:6855–6862 [DOI] [PubMed] [Google Scholar]
- Meng EC, Pettersen EF, Couch GS, Huang CC, Ferrin TE. (2006) Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinformatics 7:339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mini E, Moroson BA, Franco CT, Bertino JR. (1985) Cytotoxic effects of folate antagonists against methotrexate-resistant human leukemic lymphoblast CCRF-CEM cell lines. Cancer Res 45:325–330 [PubMed] [Google Scholar]
- Miyata Y. (2005) Hsp90 inhibitor geldanamycin and its derivatives as novel cancer chemotherapeutic agents. Curr Pharm Des 11:1131–1138 [DOI] [PubMed] [Google Scholar]
- Nakano T, Spencer HT, Appleman JR, Blakley RL. (1994) Critical role of phenylalanine 34 of human dihydrofolate reductase in substrate and inhibitor binding and in catalysis. Biochemistry 33:9945–9952 [DOI] [PubMed] [Google Scholar]
- Noé V, MacKenzie S, Ciudad CJ. (2003) An intron is required for dihydrofolate reductase protein stability. J Biol Chem 278:38292–38300 [DOI] [PubMed] [Google Scholar]
- O’Reilly T, Niven DF. (2003) Levels of nicotinamide adenine dinucleotide in extracellular body fluids of pigs may be growth-limiting for Actinobacillus pleuropneumoniae and Haemophilus parasuis. Can J Vet Res 67:229–231 [PMC free article] [PubMed] [Google Scholar]
- Onufriev A, Bashford D, Case DA. (2000) Modification of the generalized Born model suitable for macromolecules. J Phys Chem B 104:3712–3720 [Google Scholar]
- Onufriev A, Bashford D, Case DA. (2004) Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins 55:383–394 [DOI] [PubMed] [Google Scholar]
- Perkins JP, Hillcoat BL, Bertino JR. (1967) Dihydrofolate reductase from a resistant subline of the L1210 lymphoma. Purification and properties. J Biol Chem 242:4771–4776 [PubMed] [Google Scholar]
- Pollak N, Niere M, Ziegler M. (2007) NAD kinase levels control the NADPH concentration in human cells. J Biol Chem 282:33562–33571 [DOI] [PubMed] [Google Scholar]
- Rego-Pérez I, Fernández-Moreno M, Blanco FJ. (2008) Gene polymorphisms and pharmacogenetics in rheumatoid arthritis. Curr Genomics 9:381–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts E, Eargle J, Wright D, Luthey-Schulten Z. (2006) MultiSeq: unifying sequence and structure data for evolutionary analysis. BMC Bioinformatics 7:382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel B, Johnson-Farley N, Kerrigan JE, Scotto KW, Banerjee D, Felczak K, Pankiewicz KW, Gounder M, Lin HX, Abali EE, et al. (2012) A second target of benzamide riboside: Dihydrofolate reductase. Cancer Biol Ther 13:1290–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RB, Barton GJ. (1992) Multiple protein sequence alignment from tertiary structure comparison: assignment of global and residue confidence levels. Proteins 14:309–323 [DOI] [PubMed] [Google Scholar]
- Salvador N, Aguado C, Horst M, Knecht E. (2000) Import of a cytosolic protein into lysosomes by chaperone-mediated autophagy depends on its folding state. J Biol Chem 275:27447–27456 [DOI] [PubMed] [Google Scholar]
- Skacel N, Menon LG, Mishra PJ, Peters R, Banerjee D, Bertino JR, Abali EE. (2005) Identification of amino acids required for the functional up-regulation of human dihydrofolate reductase protein in response to antifolate Treatment. J Biol Chem 280:22721–22731 [DOI] [PubMed] [Google Scholar]
- Slack JL, Bi W, Livak KJ, Beaubier N, Yu M, Clark M, Kim SH, Gallagher RE, Willman CL. (2001) Pre-clinical validation of a novel, highly sensitive assay to detect PML-RARalpha mRNA using real-time reverse-transcription polymerase chain reaction. J Mol Diagn 3:141–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slansky JE, Farnham PJ. (1996) Transcriptional regulation of the dihydrofolate reductase gene. Bioessays 18:55–62 [DOI] [PubMed] [Google Scholar]
- Smith SL, Burchall JJ. (1983) Alpha-pyridine nucleotides as substrates for a plasmid-specified dihydrofolate reductase. Proc Natl Acad Sci USA 80:4619–4623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowers R, Toguchida J, Qin J, Meyers PA, Healey JH, Huvos A, Banerjee D, Bertino JR, Gorlick R. (2003) mRNA expression levels of E2F transcription factors correlate with dihydrofolate reductase, reduced folate carrier, and thymidylate synthase mRNA expression in osteosarcoma. Mol Cancer Ther 2:535–541 [PubMed] [Google Scholar]
- Tai N, Schmitz JC, Chen TM, Chu E. (2004a) Characterization of a cis-acting regulatory element in the protein-coding region of human dihydrofolate reductase mRNA. Biochem J 378:999–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai N, Schmitz JC, Liu J, Lin X, Bailly M, Chen TM, Chu E. (2004b) Translational autoregulation of thymidylate synthase and dihydrofolate reductase. Front Biosci 9:2521–2526 [DOI] [PubMed] [Google Scholar]
- Thillet J, Adams JA, Benkovic SJ. (1990) The kinetic mechanism of wild-type and mutant mouse dihydrofolate reductases. Biochemistry 29:5195–5202 [DOI] [PubMed] [Google Scholar]
- Tong X, Zhao F, Thompson CB. (2009) The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr Opin Genet Dev 19:32–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324:1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace LA, Robert Matthews C. (2002) Highly divergent dihydrofolate reductases conserve complex folding mechanisms. J Mol Biol 315:193–211 [DOI] [PubMed] [Google Scholar]
- Wang H, Tempei W, Wernimont AK, Tong Y, Guan X, Shen Y, Li Y, Arrowsmith CH, Edwards AM, Bountra C et al.(2010) Crystal structure of human NAD kinase, in RCSB PDB, 3PFN. DOI: 10.2210/pdb3pfn/pdb [Google Scholar]
- Wang J, Wang W, Kollman PA, Case DA. (2006) Antechamber, an accessory software package for molecular mechanics calculations. J Mol Graphics Model 25:247–260 [DOI] [PubMed] [Google Scholar]
- Wang K, Ma Q, Ren Y, He J, Zhang Y, Zhang Y, Chen W. (2007) Geldanamycin destabilizes HER2 tyrosine kinase and suppresses Wnt/beta-catenin signaling in HER2 overexpressing human breast cancer cells. Oncol Rep 17:89–96 [PubMed] [Google Scholar]
- Xiang J. (2002) JACKAL: A Protein Structure Modeling Package, Columbia University, New York, NY [Google Scholar]