Abstract
Interspecies differences in drug metabolism have made it difficult to use preclinical animal testing data to predict the drug metabolites or potential drug-drug interactions (DDIs) that will occur in humans. Although chimeric mice with humanized livers can produce known human metabolites for test substrates, we do not know whether chimeric mice can be used to prospectively predict human drug metabolism or a possible DDI. Therefore, we investigated whether they could provide a more predictive assessment for clemizole, a drug in clinical development for the treatment of hepatitis C virus (HCV) infection. Our results demonstrate, for the first time, that analyses performed in chimeric mice can correctly identify the predominant human drug metabolite before human testing. The differences in the rodent and human pathways for clemizole metabolism were of importance, because the predominant human metabolite was found to have synergistic anti-HCV activity. Moreover, studies in chimeric mice also correctly predicted that a DDI would occur in humans when clemizole was coadministered with a CYP3A4 inhibitor. These results demonstrate that using chimeric mice can improve the quality of preclinical drug assessment.
Introduction
Existing in vitro systems and in vivo testing in animal species have not always accurately predicted human pharmacokinetics or the human-specific drug metabolism pathways for candidate medications (Anderson et al., 2009; Leclercq et al., 2009; Walker et al., 2009). Interspecies differences in drug metabolism produce qualitative and quantitative differences between the drug metabolites produced in humans and animal species. The inability to preclinically identify the human-specific drug metabolites is particularly problematic, because it is most often a drug metabolite, and not the parent drug, that is responsible for an unexpected drug-induced toxicity (Guengerich and MacDonald, 2007; Smith and Obach, 2009). If a candidate drug has a human-specific (or more often, a human-predominant) drug metabolite, the use of preclinical toxicity testing in animal species is quite limited (Anderson et al., 2009; Leclercq et al., 2009).
To address this problem, we (Hasegawa et al., 2011) and others (reviewed in Yoshizato and Tateno, 2009; de Jong et al., 2010) have developed chimeric mice, in which mouse liver is replaced by transplanted human liver cells or tissue-engineered human liver (Chen et al., 2011). In one model system, uroplasminogen activator transgene expression facilitates the growth of transplanted human liver cells (Vyse et al., 1997; Tateno et al., 2004; Meuleman et al., 2005; Azuma et al., 2007; Katoh and Yokoi, 2007), whereas a fumarylacetoacetate hydrolase knockout mouse is used in the other system (Azuma et al., 2007) (Bissig et al., 2010). We recently produced a new model system for human liver replacement. In this new system, a herpes simplex virus type 1 thymidine kinase transgene was expressed in the liver of a highly immunodeficient mouse strain (NOG) (Ito et al., 2002) to produce the NOG mouse expressing a thymidine kinase transgene (TK-NOG) (Hasegawa et al., 2011). A brief exposure to a nontoxic dose of ganciclovir causes a rapid and temporally controlled ablation of mouse liver cells expressing the transgene, which enabled the transplanted human liver cells to develop into a mature human organ with a 3-dimensional architecture and gene expression pattern (including many human drug-metabolizing enzymes and transporters) characteristic of mature human liver. The absence of ongoing liver toxicity in the TK-NOG mice enabled the humanized liver to be stably maintained for >8 months without exogenous drug treatments. The humanized liver in chimeric TK-NOG mice was shown to express mRNAs encoding human cytochrome P450 (P450) enzymes, transporters, and transcription factors affecting drug metabolism at levels that were equivalent to those in the donor human hepatocytes. Moreover, there was extensive human CYP3A4 protein expression in the humanized livers, and chimeric TK-NOG mice could mediate human-specific drug biotransformation reactions (Hasegawa et al., 2011).
There are multiple examples in which chimeric mice have been shown to produce known human-specific metabolites for several test substrates (Tateno et al., 2004; Katoh and Yokoi, 2007; Chen et al., 2011; Hasegawa et al., 2011), including steroids (Katoh et al., 2007; Lootens et al., 2009; Pozo et al., 2009; Kamimura et al., 2010). However, we do not know whether chimeric mice can be used to predict the pattern of human drug metabolism for a candidate therapeutic before human clinical testing. In a recent study, chimeric mice produced mixed results when their ability to predict the pattern of human drug metabolism was assessed (De Serres et al., 2011). We also do not know whether chimeric mice can be used to prospectively evaluate the potential for a drug-drug interaction (DDI) involving a candidate therapeutic to occur in human subjects. Because more than 30% of the US population over 57 years of age take five or more prescription drugs at a given time, DDIs have created major problems for patients and for regulatory authorities (Zhang et al., 2010). However, with use of available in vitro or in vivo animal models, it has been difficult to predict many of the clinically important DDIs, which only became apparent after drug development was completed (Bode, 2010). Therefore, we investigated whether TK-NOG mice with humanized livers could provide more predictive information about the human metabolic pathways and metabolites for a candidate therapeutic, which is being developed as a novel treatment of hepatitis C virus (HCV) infection. Clemizole, an antihistamine drug that was once widely used for treatment of allergic disease, was recently discovered to be a potent inhibitor (IC50, 24 nM) of the interaction between an HCV protein (NS4B) and HCV RNA (Einav et al., 2008). Although clemizole was widely used during the 1950s and 1960s, this was before contemporary regulatory requirements were established for new drug development, and we have very minimal information about its pharmacokinetics and metabolism.
Materials and Methods
Chemicals and Reagents.
For in vitro and animal experiments, clemizole hydrochloride and omeprazole were purchased from Sigma (St. Louis, MO); ritonavir was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Pooled human liver microsomes and male rat and mouse liver microsomes were purchased from BD Gentest (Woburn, MA). Cryopreserved human hepatocytes and recombinant P450 enzymes were purchased from BD Biosciences (San Jose, CA). Rat hepatocytes were freshly isolated in accordance with standard procedures. All other chemicals were purchased from commercial sources and were of the highest purity available. Boceprevir was a gift from Leslie Holsinger (Virobay).
Mouse Pharmacokinetic Studies.
All animal experiments were performed using protocols approved by the Stanford Institutional Animal Care and Use Committee. Male C57BL6/J and Balb/c mice (8 weeks of age) were obtained from Jackson Laboratories and housed for 2 weeks before experimentation. NOG mice were obtained from In Vivo Sciences International (Sunnyvale, CA). Chimeric TK-NOG mice with humanized livers were prepared as described elsewhere (Hasegawa et al., 2011), except that the ganciclovir dose was increased to 25 mg/kg, which was administered 7 and 5 days before human liver cell transplantation. The human albumin levels were determined using previously described methods, and the human albumin concentration was shown to correlate with the extent of liver humanization (Hasegawa et al., 2011). The pharmacokinetic studies using these mice were performed 8–12 weeks after transplantation of human liver cells. Eight control NOG mice and eight humanized TK-NOG mice were administered 25 mg/kg by mouth clemizole, and blood samples were collected 30 minutes after administration. The C57BL/6J mice (3 per time point) were given 25 mg/kg by mouth clemizole, and blood samples were collected for analysis at 15 and 30 minutes and 1, 2, 4, and 6 hours after administration. For the DDI studies, eight humanized TK-NOG mice were given clemizole (25 mg/kg by mouth) with or without ritonavir (20 mg/kg by mouth), and blood samples were collected 30 minutes after administration. Six of these mice were also treated with debrisoquine (10 mg/kg by mouth) in the presence or absence of ritonavir (20 mg/kg by mouth), and plasma samples were obtained 2 hours later for analysis.
Quantitative Analysis of Clemizole and Metabolites in Plasma.
Mouse plasma (50 μl) was treated with acetonitrile with 0.1% formic acid (200 μl), vortexed, incubated at −20°C for 1 hour, and centrifuged at 10,000 rpm for 10 minutes. The supernatants were collected, dried, and resuspended in 50 μl 5% acetonitrile with 0.1% formic acid for the analysis by liquid chromatography and mass spectroscopy. High-performance liquid chromatography was performed using an Agilent 1200 column compartment (Agilent Technologies, Santa Clara, CA), capillary pump, and an autosampler on a Zorbax C18 column, 0.5 × 150 mm. The flow rate was 20 ml/min with a gradient from 5% solvent B (acetonitrile with 0.1% formic acid; solvent A is 0.1% formic acid in water) to 95% B in 30 minutes and held at 95% B for 5 minutes. Mass spectrometric analysis was performed on an Agilent Model 6520 qTOF mass spectrometer (Agilent Technologies) equipped with an electrospray ionization source. The heated capillary temperature in the source was held at 325°C. Full scan (m/z 110–1000) spectra or data-dependent tandem mass spectroscopy spectra were collected. The metabolites were identified on the basis of their collision-induced dissociation behavior in tandem mass spectrometry, accurate mass, and retention time. Quantitative analysis of clemizole was performed using a calibration curve at 9-1257 ng/ml clemizole spiked into blank mouse plasma and extracted as described above. An internal standard 1-(p-bromobenzyl)-2-(1-pyrrolidinylmethyl)-benzimidazole was also spiked at 1000 ng/ml. Relative amounts of clemizole and metabolites in each sample were calculated using the assumption that all compounds had the same mass spectroscopy response factor.
Statistical Analysis.
To assess the statistical significance of the differences in the relative amount of clemizole and its metabolites (M1, M2, M6, M12, M14, and M15) measured in plasma samples obtained from control and humanized TK-NOG mice (Table 1) or between control and ritonavir-treated mice, a two-sample two-sided t test was used.
TABLE 1.
Clemizole (25 mg/kg by mouth) was administered to eight control NOG mice (C1-8) and eight TK-NOG mice with humanized livers (Hu m1-8).
The relative amounts of six metabolites and clemizole in plasma 30 minutes after administration were measured. This table shows the human serum albumin (hAlb) concentration (mg/ml) in the humanized liver mice expressing a TK-NOG and the relative abundances of clemizole metabolites in plasma. The ratio indicates the average amount of each metabolite in humanized TK-NOG mice divided by that in control mice. Although both groups had equivalent relative amounts of three metabolites (P >0.3), the humanized mice had a significantly increased relative amount of the M1 and M6 metabolites and a significantly decreased relative amount of M14.
| Variable | hAlb | Clem | M1 | M2 | M6 | M12 | M14 | M15 |
|---|---|---|---|---|---|---|---|---|
| C1 | 4.1 | 4.1 | 10.4 | 14.1 | 28.3 | 37.5 | 1.4 | |
| C2 | 5.5 | 3.2 | 8.1 | 12.9 | 37.9 | 31.2 | 1.2 | |
| C3 | 6.8 | 2.3 | 3.3 | 8 | 24 | 50.6 | 4.8 | |
| C4 | 3.1 | 3.1 | 8 | 9.3 | 20.5 | 39.3 | 16.7 | |
| C5 | 2.8 | 8 | 5.7 | 12.1 | 27.5 | 33.3 | 10.7 | |
| C6 | 6.03 | 5.49 | 20.00 | 10.19 | 28.78 | 28.50 | 1.01 | |
| C7 | 3.81 | 1.79 | 6.52 | 12.17 | 27.04 | 47.22 | 1.46 | |
| C8 | 4.53 | 6.43 | 11.38 | 10.17 | 25.76 | 40.71 | 1.02 | |
| Hu m1 | 4.2 | 1.8 | 8.9 | 13.6 | 8.3 | 20.6 | 39.8 | 6.9 |
| Hu m2 | 1.5 | 4.4 | 7.1 | 8.3 | 13 | 29.6 | 35.5 | 2.4 |
| Hu m3 | 1.6 | 5.3 | 7.9 | 9.2 | 16.8 | 32.1 | 26.9 | 1.8 |
| Hu m4 | 2.2 | 4.3 | 7.1 | 14.2 | 5.9 | 14.8 | 48.5 | 5.2 |
| Hu m5 | 2.7 | 3.6 | 13.1 | 10 | 26 | 29 | 16.5 | 1.8 |
| Hu m6 | 1.3 | 6.25 | 12.25 | 18.24 | 23.10 | 26.37 | 12.33 | 1.46 |
| Hu m7 | 7 | 8.75 | 10.73 | 18.66 | 21.30 | 28.86 | 9.59 | 2.12 |
| Hu m8 | 1.6 | 7.02 | 9.04 | 13.59 | 19.92 | 28.77 | 20.69 | 0.97 |
| Ratio | 1.13 | 2.21 | 1.44 | 1.51 | 0.96 | 0.68 | 0.59 | |
| P value | 0.526 | 0.0004 | 0.095 | 0.049 | 0.659 | 0.046 | 0.389 |
Human Pharmacokinetic and Metabolite Studies.
Phase 1b studies (to be reported elsewhere) were conducted in patients with genotypes 1 and 2 HCV infection under Institutional Review Board–approved protocols to investigate the safety and tolerability, pharmacokinetics, and pharmacodynamics of clemizole HCl (see www.Clinicaltrials.gov and approval by the University of Ankara Medical School Ethics Committee; study sponsor, Eiger BioPharmaceuticals, Inc.). De-identified aliquots of excess material, which were not needed for clinical monitoring, were kindly provided by Wenjin Yang (Eiger BioPharmaceuticals, Inc.) for pharmacokinetic and metabolite analysis. The samples obtained were from human subjects who were administered 100 mg by mouth clemizole, with or without 100 mg by mouth ritonavir administered 1 hour before the clemizole, and blood samples were obtained 0–12 hours after administration. The relative abundance of clemizole and its metabolites in plasma were measured in samples obtained from 10 patients treated with clemizole alone and from three patients who participated in the subcomponent evaluating the effect of ritonavir coadministration, by liquid chromatography and mass spectroscopy analysis. as described below.
In Vitro HCV Replication Assay.
The plasmid FL-J6/JFH-5′C19Rluc2AUbi, which consists of the full-length HCV genome and expresses the Renilla luciferase (Tscherne et al., 2006), was a gift from Charles M. Rice. In vitro transcription of HCV RNA was performed as described elsewhere, with minor modifications (Einav et al., 2010). In brief, Huh 7.5 cells were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 1% l-glutamine (Invitrogen), 1% penicillin, 1% streptomycin (Invitrogen), 1× nonessential amino acids (Invitrogen), and 10% fetal-bovine serum (Omega Scientific, Tarzana, CA). Electroporation was performed by mixing 5 µg of RNA with 400 µl of ice-cold phosphate-buffered saline containing washed Huh 7.5 cells at a density of 1.5 × 107 cells/ml. The mixture was immediately pulsed (0.82 kV, five 99-µs pulses) with a BTX-830 electroporator. After a 10 minute recovery at 25°C, pulsed cells were diluted into 10 ml of prewarmed growth medium. Cells were passaged, their luciferase activity was verified, and they were seeded in 96-well plates (2 × 104 cells/well) 1 day before addition of the inhibitory compounds. Cells were grown in three replicates in the presence of serial dilutions of the inhibitory compounds. Untreated cells with or without corresponding concentrations of dimethyl sulfoxide were used as negative controls. Forty-eight hours after treatment with the inhibitory compounds, cells were subjected to Alamar Blue–based viability assays and luciferase assays.
Viability Assay and Luciferase Assay.
Cell viability was assessed by incubation with 10% Alamar Blue Reagent (Invitrogen) for 2 hours, the absorbance was measured at 570 nm and 630 nm with a plate reader (TECAN M1000), and viability was determined by comparing A570−A630. Luciferase activity was measured using the Renilla luciferase kit (Promega, Madison, WI) after assessing cell viability. The cells were washed with ice-cold phosphate-buffered saline and lysed with 20 µl of ice-cold Renilla lysis buffer (Promega). We injected 40 µl of the Renilla luciferase assay buffer containing assay substrate and measured luciferase activity with a 4-second integration using a plate reader. All experiments were done at least four times, and at least three replicates were performed each time.
Analysis of the Drug Synergy Data.
The in vitro drug efficacy data were analyzed using the The MacSynergy II program (kindly provided by M. N. Prichard) according to the Bliss independence model (Pritchard et al., 1992) (Prichard and Shipman, 1990). The effect of a drug combination was determined by subtracting the experimental values from theoretical additive values (Pritchard et al., 1992). A three-dimensional differential surface plot was used to evaluate the interaction among the tested drugs. Synergy was indicated if the plot peaked above a theoretical additive plane, and antagonism was indicated if depressions were formed below this plane (Prichard and Shipman, 1990). The data sets with four replicates were assessed at the 95% confidence level for each experiment (Pritchard et al., 1992; Prichard and Shipman, 1990; Prichard and Shipman, 1996). Synergy (volume under the curve) and log volume were calculated. As suggested (Pritchard et al., 1992), these data sets were interpreted in accordance with the following rules: volumes of synergy or antagonism at values of <25 µM2% are insignificant, 25–50 µM2% are minor but significant, 50–100 µM2% are moderate and probably important in vivo, and >100 µM2% are strong and likely to be important in vivo.
Results
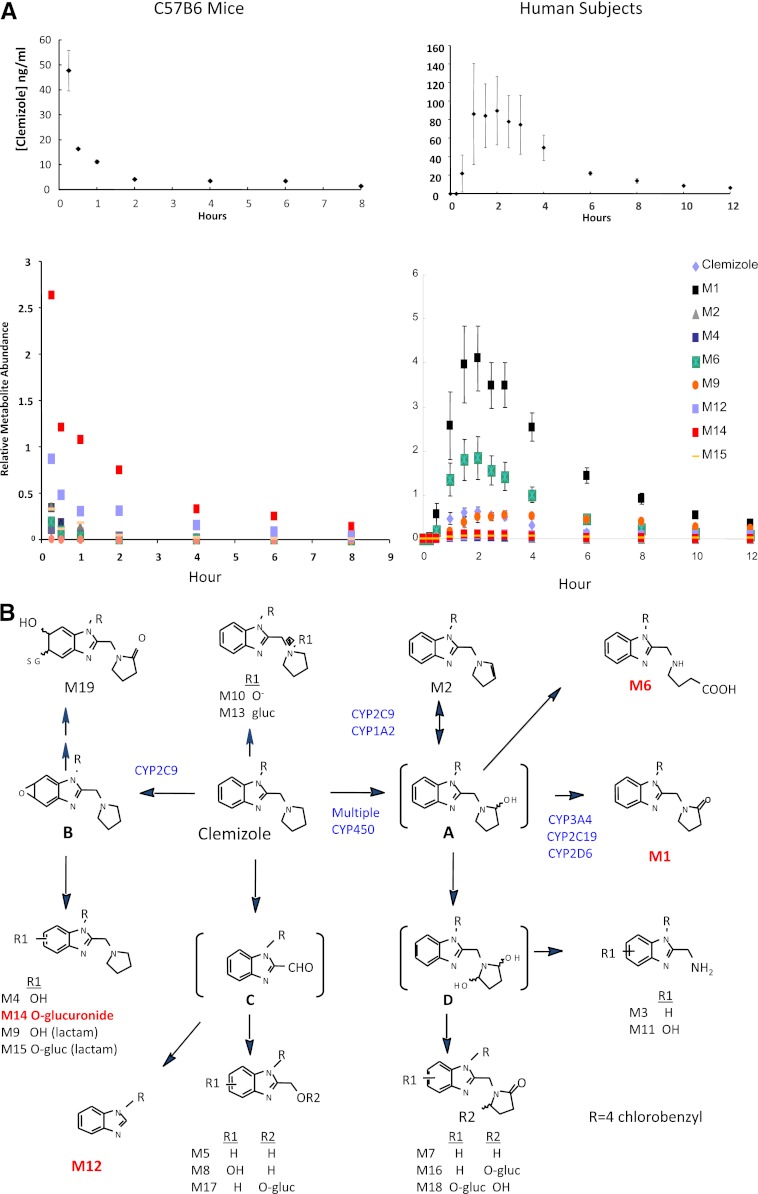
We first examined clemizole’s pharmacokinetic profile in a commonly used, conventional mouse strain. Clemizole had an unexpectedly short plasma half-life (measured at 0.15 hours); it was very rapidly biotransformed into a glucuronide (M14) and a dealkylated metabolite (M12) and into a variety of lesser metabolites in C57BL/6J mice (Fig. 1). Although we previously demonstrated that the pattern of metabolism for several drugs could differ among inbred strains (Guo et al., 2006, 2007), two other inbred strains exhibited the same ultra-rapid rate of clemizole metabolism and produced the same metabolites as C57BL/6J mice (Supplemental Fig. 1). Then, the pattern of clemizole metabolism in TK-NOG mice with humanized livers was compared with that in control mice with the same genetic background. On the basis of the human albumin concentration (range, 1.3–7.0 mg/ml) in their serum samples, the extent of liver humanization ranged from 13–70%. The relative amount of clemizole and six metabolites in plasma 30 minutes after administration of a single (25 mg/kg by mouth) dose of clemizole was measured (Table 1). The relative amounts of three metabolites did not significantly differ between control and humanized mice (P > 0.3). However, humanized TK-NOG mice had a substantially larger amount of metabolite M1 in their plasma (2.2-fold; P < 0.0004); along with a significantly increased level of M6 (1.5-fold; P < 0.049) and a decreased amount of M14 (0.68-fold; P < 0.046) in their plasma. Because these livers were only partially humanized, the remnant murine liver was also rapidly metabolizing this drug. However, by focusing on the differences in the drug metabolites that were produced by the humanized and conventional murine livers, drug metabolites that will predominate in humans could be identified. These data indicated that M1 and M6 could be important metabolites in humans and that clemizole metabolism in humans and mice could differ.
Fig. 1.
(A) Clemizole metabolism in mice and humans. The top graphs show the measured plasma clemizole concentration at the indicated times after a single oral dose of clemizole was administered to three C57BL/6 mice (25 mg/kg) or 10 human subjects (100 mg). Although clemizole was rapidly metabolized in mice (measured half-life, 0.15 hours), its measured half-life in humans was 3.4 hours. The bottom graphs show the relative normalized abundance of clemizole and metabolites in plasma at the indicated time after administration. In mice, clemizole was rapidly metabolized to two major (M12 and M14) metabolites, but only minimal amounts of M1 or M6 were produced, whereas in humans, clemizole was converted to two predominant metabolites (M1 and M6). (B) The structure of clemizole and the pathways for production of its metabolites. In humans, clemizole is converted to intermediate “A” by any of a number of P450s. Then, CYP3A4, CYP2C19, and CYP2D6 can further oxidize this intermediate to M1, although in the presence of CYP2C9 or CYP1A2, M2 is generated, but they cannot produce M1. Cyp2C9 appears to be the only source of M4, which is a minor metabolite in humans. The structures of the major human (M1, M6) and rodent (M12, M14) plasma metabolites are highlighted.
After the murine studies were completed, plasma samples were obtained from 10 human subjects after administration of a single (100 mg by mouth) dose of clemizole (Fig. 1). Clemizole had a much longer plasma half-life (measured at 3.4 hours) in humans and a very different pattern of drug metabolites, relative to mice. Clemizole was rapidly converted to a single major metabolite (M1), which accounted for 55% of all drug and metabolites present in human plasma. The area under the concentration time curve (AUC; 0–24 hours) was calculated to assess the relative level of drug or metabolite exposure over time. Comparison of the AUCs (0–24 hours) calculated for each metabolite indicated that mice and humans have very different levels of exposure to the major drug metabolites (Table 2). Although M1 and M6 account for 77% of human drug metabolite exposure, they account for only 6% of murine drug metabolite exposure. In contrast, M12 and M14 account for 77% of mouse drug metabolite exposure but only 2% of human metabolite exposure. The human metabolic profile was consistent with that observed in the humanized mice, which had indicated that the pathways mediating M1 and M6 production would be important.
TABLE 2.
The AUC (0–24 hours) for exposure to clemizole and the major in vivo metabolites were calculated for C57BL/6 mice and for human subjects using the data presented in Fig. 1.
For comparison with the in vitro results, the percentage of the total identified metabolites present after clemizole was incubated with human, rat, or mouse microsomes for 1 hour or after incubation with rat or human hepatocytes for 30 minutes are shown.
| Metabolite | % Total AUC |
Microsomes |
Hepatocytes |
||||
|---|---|---|---|---|---|---|---|
| Human | Mouse | Human | Mouse | Rat | Human | Rat | |
| Clemizole | 8.8 | 1.3 | 40 | 24 | 3 | 15 | 0 |
| M1 | 54.9 | 4.7 | 35 | 2 | 4 | 39 | 0 |
| M2 | 1.1 | 5.6 | 2 | 5 | 4 | 0.1 | 0 |
| M4 | 1.1 | 2.9 | 5 | 3 | 5 | 4 | 0 |
| M6 | 22.0 | 2.4 | 2 | 2 | 3 | 18 | 0 |
| M9 | 8.8 | 0.3 | 3 | 19 | 37 | 4 | 1 |
| M12 | 1.1 | 21.2 | 0.6 | 0 | 0 | 1 | 1 |
| M14 | 1.1 | 55.8 | 0 | 0 | 0 | 10 | 5 |
| M15 | 1.1 | 5.9 | 0 | 0 | 0 | 0 | 44 |
Clemizole Metabolism in Rodents and Humans.
A detailed in vitro characterization of the pathways for clemizole metabolism in rodents and humans was performed as described in the supplement (Supplemental Figs. 2 and 3; Supplemental Tables 1–3), and the interspecies differences are summarized in Fig. 1B. In human liver, clemizole is primarily converted to an intermediate A, which can be oxidized by several P450 enzymes (CYP3A4, CYP2C19, or CYP2D6) to M1. CYP3A4, which is the most abundantly expressed P450 enzyme in human liver, mediates the majority of this drug biotransformation reaction. The role of CYP3A4 is confirmed by the ability of ritonavir, which is an inhibitor of CYP3A4 activity, to inhibit clemizole metabolism in vitro (Supplemental Fig. 4). In contrast, a different type (CYP2C-like) of aromatic oxidation reaction produces the rodent-predominant metabolites (M12, M14, and M15), which is the dominant pathway for clemizole metabolism in rodent liver.
Predicting a Human DDI.
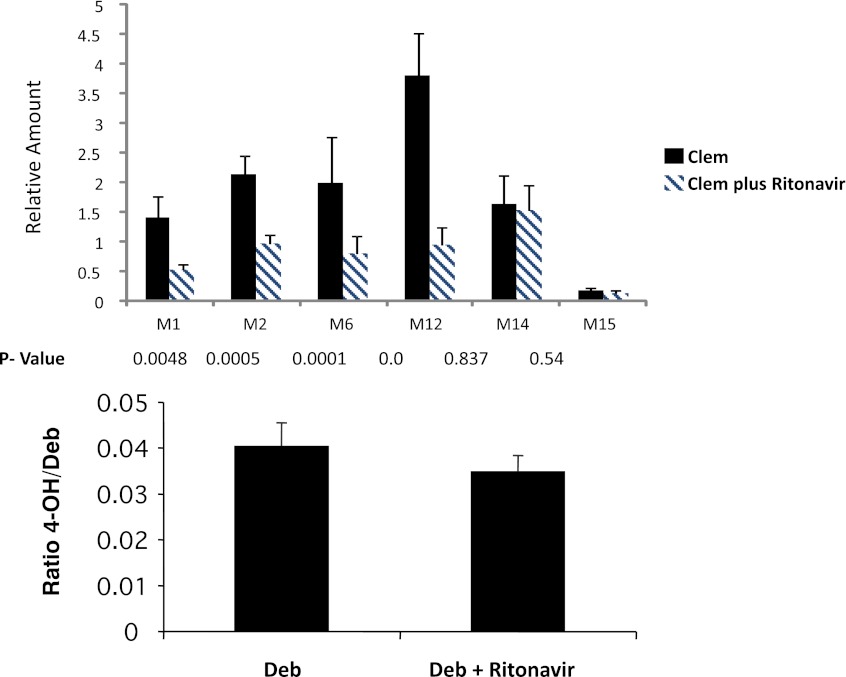
We wanted to determine whether studies using humanized TK-NOG mice could prospectively predict whether a potential DDI involving clemizole would occur. Ritonavir is a CYP3A4 inhibitor that has been coadministered with other drugs to increase their duration of action (Hsu et al., 1998). Given the identified pathways for clemizole metabolism in human liver, it was possible that ritonavir-mediated inhibition of CYP3A4 could alter clemizole pharmacokinetics. We hypothesized that, if a DDI of this type occurred in the humanized mice, a similar type of DDI would be observed in human patients. Therefore, clemizole was administered to eight humanized TK-NOG mice with or without ritonavir coadministration, and clemizole metabolites were analyzed in plasma samples. Ritonavir coadministration decreased the amount of the human-predominant clemizole metabolites (M1 and M6; P < 0.005) (Fig. 2). The concentration of M12, a mouse-predominant metabolite was also decreased by ritonavir coadministration, whereas the concentrations of two other metabolites (M14 and M15) that are produced by a mouse-specific biotransformation reaction were not altered. We have previously demonstrated that chimeric TK-NOG mice can metabolize debrisoquine to 4-OH-debrisoquine, which is a human-predominant route of drug metabolism that is catalyzed by CYP2D6 (Hasegawa et al., 2011). Ritonavir coadministration did not alter the CYP2D6-mediated conversion of debrisoquine to 4-OH debrisoquine in six humanized TK-NOG mice analyzed (P > 0.5) (Fig. 2). These results demonstrate that ritonavir coadministration specifically decreased the production of human CYP3A4-specific metabolites of clemizole and indicate that a DDI could occur if ritonavir was coadministered with clemizole in human subjects.
Fig. 2.
Top: Six humanized TK-NOG mice were first dosed with clemizole (Clem) alone (25 mg/kg by mouth) and then with clemizole (25 mg/kg by mouth) and ritonavir (20 mg/kg by mouth). The amount of six different clemizole metabolites in plasma was measured at 30 minutes after administration. The plasma concentrations of the human-predominant M1 and M6 metabolites were significantly decreased by ritonavir coadministration, as were the concentrations of M2 and M12, whereas those of M14 and M15 were unchanged. Bottom: Five humanized TK-NOG mice were first given debrisoquine (Deb) alone (10 mg/kg by mouth) and then debrisoquine (10 mg/kg by mouth) and ritonavir (20 mg/kg by mouth). The amount of its main metabolite, 4-OH-debrisoquine, was measured in plasma 2 hours after administration. The relative amount of 4-OH debrisoquine was not significantly changed by ritonavir coadministration.
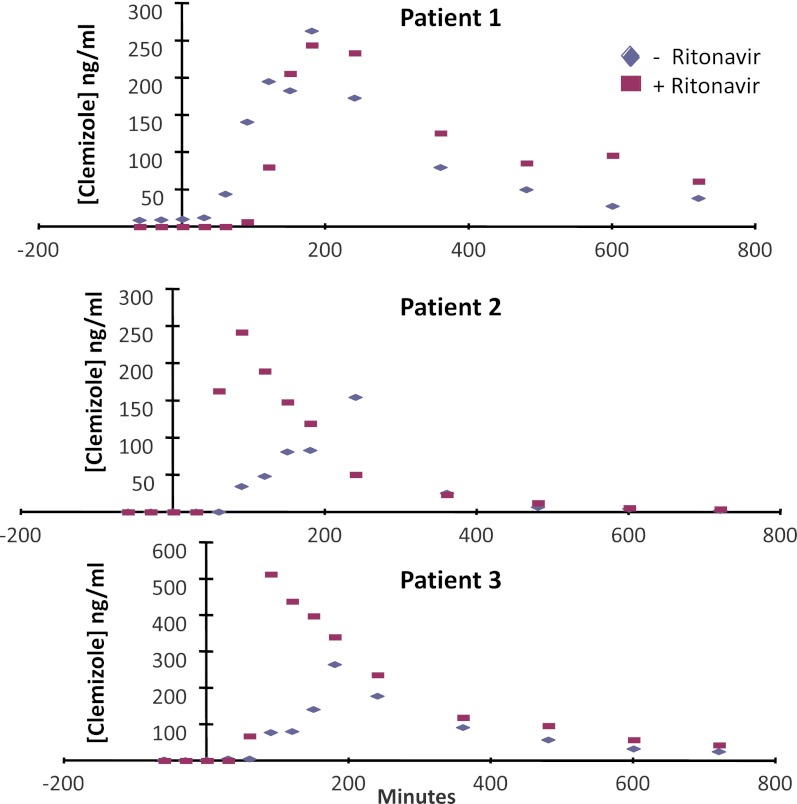
Because increasing the time that an HCV-infected individual maintains an effective plasma clemizole concentration could have a therapeutic benefit, a pilot clinical study was performed to determine whether ritonavir coadministration would increase plasma clemizole levels after administration. Three HCV-infected individuals were treated with clemizole (100 mg by mouth) alone and, on a separate day, with clemizole (100 mg by mouth) and ritonavir (100 mg by mouth, 1 hour before clemizole administration). The plasma clemizole concentration was measured over a 12-hour period after both treatments. Ritonavir coadministration increased the AUC (0–12 hours) for clemizole in all three treated individuals (by 22, 38, and 89%) (Fig. 3). Although ritonavir coadministration had a limited effect in the first patient, ritonavir coadministration substantially increased the Cmax and AUC for clemizole in patients 2 and 3. Thus, the humanized TK-NOG mouse results correctly predicted that a DDI associated with ritonavir and clemizole coadministration would occur, and this could be exploited to improve the efficacy of clemizole treatment of HCV infection.
Fig. 3.
Three HCV-infected individuals were first administered clemizole alone (100 mg by mouth) and then with clemizole (100 mg by mouth) and ritonavir (100 mg by mouth, twice daily). The amount of clemizole in the plasma was measured over a 12-hour period after administration.
Clemizole M1 Metabolite has Antiviral Activity.
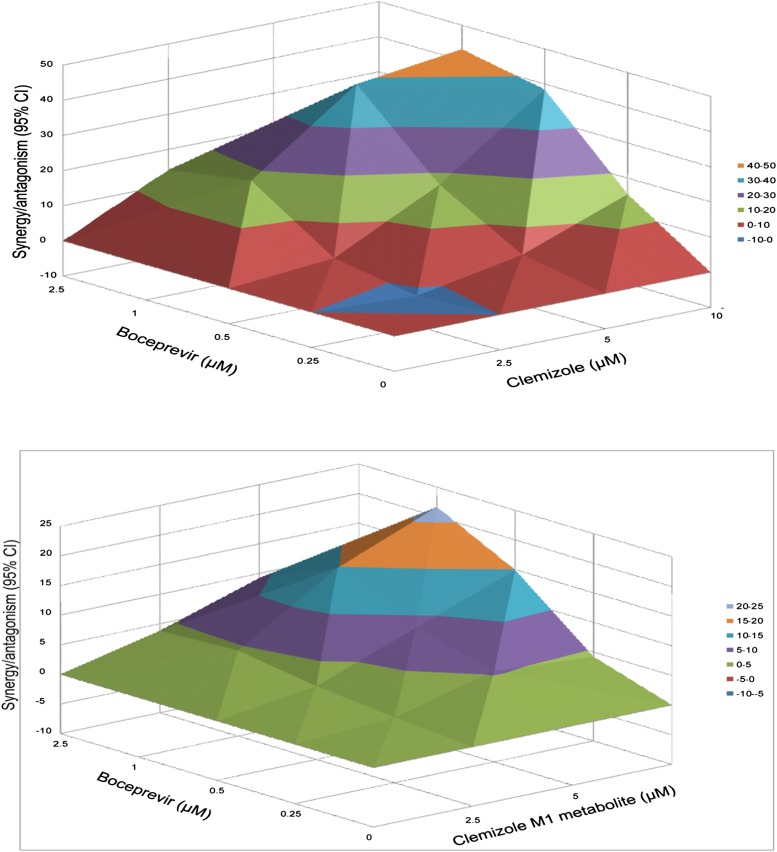
Because high levels of M1 were present in human plasma, it was of importance to determine whether this metabolite had antiviral activity. Clemizole has recently been shown to have a synergistic inhibitory effect on HCV replication in vitro when combined with protease inhibitors (Einav et al., 2010). Therefore, the effect of the M1 metabolite, either alone or in combination with a protease inhibitor (boceprevir) that was recently approved for treatment of HCV (Bacon et al., 2011; Poordad et al., 2011), was measured using an in vitro luciferase reporter-linked HCV replication assay. When M1 was combined with boceprevir, their combined antiviral effect in this assay was significantly more potent than the theoretical additive effects of either drug alone (Fig. 4). The calculated synergy volume was 101 µM2%; which (according to the established criteria [Pritchard et al., 1992]) is indicative of strong synergy that is likely to be important in vivo. There was no evidence of antiviral antagonism at any of the tested doses, and no cellular toxicity was noted after incubation with either drug alone, nor was cytotoxicity increased after exposure to the drug combinations. Thus, the observed synergy between boceprevir and M1 is specific, and it does not reflect a synergistic toxicity arising from the drug combination. Although the level of synergy observed with M1 was less than that obtained with clemizole (synergy volume 230 µM2%) (Fig. 4), these results indicate that clemizole’s major metabolite has significant antiviral activity.
Fig. 4.
Differential surface plot of the synergistic anti-HCV effect of boceprevir with either clemizole (top) or the M1 metabolite of clemizole (bottom). The 3-dimensional plot represents the differences between the actual antiviral effects and the theoretical additive effects at the indicated concentrations for the two compounds tested. Only statistically significant (95% confidence interval) differences between the two compounds were considered at any given concentration. Peaks above the theoretical additive plane indicate synergy, whereas depressions below it indicate antagonism. The colors indicate the level of synergy or antagonism. Both clemizole and the M1 metabolite exhibit a substantial synergistic antiviral effect with boceprevir.
Discussion
Differences between the rodent and human pathways used for metabolism of a candidate medication are not rare, which is why animal testing results have not always accurately predicted human pharmacokinetic parameters or the drug metabolism pathways for a candidate medication (Anderson et al., 2009; Leclercq et al., 2009; Walker et al., 2009). Similarly, the results from in vitro analyses using human hepatocytes or microsomal preparations have correctly predicted the in vivo human metabolite profile for about half (46–65%) of the 48 tested compounds (Dalvie et al., 2009). This study indicates how the use of humanized TK-NOG mice can enable human-predominant drug metabolites to be identified before human drug exposure. Although chimeric mice have both human and murine liver tissue, human drug metabolites could be predicted by identifying the differentially abundant metabolites produced in chimeric mice (relative to conventional mice). However, the variable extent of liver humanization (13–70%) (Supplemental Table 4) in the chimeric mice analyzed here could complicate the analysis of other drugs, especially when the pathways for murine and human drug metabolism are not as divergent as in the case examined here. The remaining mouse liver could produce metabolites that could confound toxicologic analyses performed with these chimeric mice. Therefore, methods are being developed to increase the extent of liver humanization that is obtained with this model system, which should improve the results obtained using humanized TK-NOG mice. Because three different inbred strains produced the same pattern of clemizole metabolites, there is a common murine (strain-independent) pattern of clemizole metabolism that differs from the pathways used in humans. The interspecies differences in clemizole metabolism highlight the difficulties in using data obtained from conventional rodent species to predict human drug metabolism and, consequently, potential drug-induced toxicity. For clemizole, if toxicities were caused by the human predominant metabolite M1, toxicology testing in rodents would provide a false assurance of drug safety. Similarly, rodent testing could raise a false drug safety concern if a rodent-predominant (M12, M14) metabolite or reactive intermediate B caused toxicity. Although some form of human testing is always required for assessment of a candidate drug, the information obtained from analyses performed in chimeric mice before human testing can be quite informative. The identified differences between the rodent and human pathways for clemizole metabolism assume an increased importance, because the major human clemizole metabolite had synergistic anti-HCV activity in vitro, which could extend the antiviral activity of clemizole when administered in combination with the recently approved HCV protease inhibitors. As demonstrated here, analyses in chimeric mice can reveal whether preclinical toxicity testing in animal species has adequately covered the metabolites that will be formed in humans. Although the interaction of ritonavir and clemizole provides a straightforward example, this study also demonstrates how chimeric mice can be used to assess whether a potential DDI is likely to occur.
Our analysis indicates that CYP3A4 plays a major role in clemizole metabolism. We previously demonstrated that there was abundant human CYP3A4 protein expression in the livers of chimeric TK-NOG mice (Hasegawa et al., 2011). Although the effect that genetic differences in CYP3A enzymes have on clemizole metabolism remains to be determined, any compound that is metabolized by CYP3A4 is usually also a CYP3A5 substrate. However, the high frequency of inactivating CYP3A5*3 alleles in white persons (up to 90%) and Asians (up to 80%) makes it likely that CYP3A5 may have a lesser role in clemizole metabolism (Lamba et al., 2002; Xie et al., 2004). It is more likely that inter-individual variability in CYP3A4 expression is responsible for the variable effect of ritonavir on the pharmacokinetic profiles in these three randomly selected white individuals.
Consistent with the results obtained from analysis of other drugs (Dalvie et al., 2009), the data in Table 2 indicate that the in vitro results (microsomes or hepatocytes) did not correctly predict the in vivo profile of clemizole metabolism. Because the rodent in vitro systems could not even predict how clemizole would be metabolized in vivo in rodents, it would have been very difficult to use human in vitro data to predict the clemizole metabolites that would be produced in human subjects. However, there are limiting factors affecting the use of these chimeric mice for human-specific pharmacokinetic and toxicologic studies. First, these mice cannot be used to analyze immune-mediated drug toxicities, because they are immunocompromised. Second, they cannot be used to identify extrahepatic human-specific factors affecting drug metabolism or clearance. Third, because we do not know the extent of biliary tract humanization in these chimeric mice, we do not know whether they can be used to predict the clearance of drugs in humans that depend on human-specific transporter-mediated hepatobiliary clearance. Until we know the extent of biliary tract humanization, we do not know whether the potential for drugs (e.g., bosentan [Fattinger et al., 2001]) to cause cholestatic liver toxicity in humans can be evaluated in these mice. Despite these limitations, this example demonstrates how the use of humanized mice can enhance our ability to predict human drug metabolism and the occurrence of a DDI for a candidate medication. As has occurred with other medications (Hsu et al., 1998), ritonavir coadministration could be used to increase the anti-HCV efficacy of clemizole. Although this is only one example, it indicates that it is likely that the use of chimeric mice could improve the quality of preclinical drug assessment.
Supplementary Material
Acknowledgments
The authors thank Wenjin Yang and Eiger BioPharmaceuticals, Inc., for providing the clinical samples used in this study.
Abbreviations:
- AUC
area under the concentration time curve
- P450
cytochrome P450
- DDI
drug-drug interaction
- HCV
hepatitis C virus
- NOG
highly immunodeficient mouse strain
- TK-NOG
NOG mouse expressing a thymidine kinase transgene
Authorship Contributions
Participated in research design: Nakamura, Nomura, Chen, Glen, Peltz.
Conducted experiments: Nishimura, Hu, Suemizu, Wu, Elazar, Liu, Idilman, Yurdaydin, Angus, Stedman, Murphy.
Performed data analysis: Zheng, Fitch.
Wrote or contributed to the writing of the manuscript: Hu, Peltz, Glenn.
Footnotes
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
This work was supported by the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases [Grant 1R01DK090992-01 to G.P.]; and the National Institutes of Health National Institute of Allergy and Infectious Diseases [Grant R42 AI088793 to J.G. and B.M.].
J.G. has an equity interest in Eiger BioPharmaceuticals and/or Eiger Group International. J.G., G.P., and B.M. are consultants for Eiger BioPharmaceuticals and/or Eiger Group International.
References
- Anderson S, Luffer-Atlas D, Knadler MP. (2009) Predicting circulating human metabolites: how good are we? Chem Res Toxicol 22:243–256 [DOI] [PubMed] [Google Scholar]
- Azuma H, Paulk N, Ranade A, Dorrell C, Al-Dhalimy M, Ellis E, Strom S, Kay MA, Finegold M, Grompe M. (2007) Robust expansion of human hepatocytes in Fah-/-/Rag2-/-/Il2rg-/- mice. Nat Biotechnol 25:903–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon BR, Gordon SC, Lawitz E, Marcellin P, Vierling JM, Zeuzem S, Poordad F, Goodman ZD, Sings HL, Boparai N, et al. (2011) Boceprevir for previously treated chronic HCV genotype 1 infection. N Engl J Med 364:1207–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissig K-D, Wieland SF, Tran P, Isogawa M, Le TT, Chisari FV, Verma IM. (2010) Human liver chimeric mice provide a model for hepatitis B and C virus infection and treatment. J Clin Invest 120:650–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode C. (2010) The nasty surprise of a complex drug-drug interaction. Drug Discov Today 15:391–395 [DOI] [PubMed] [Google Scholar]
- Chen AA, Thomas DK, Ong LL, Schwartz RE, Golub TR, Bhatia SN. (2011) Humanized mice with ectopic artificial liver tissues. Proc Natl Acad Sci USA 108:11842–11847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalvie D, Obach RS, Kang P, Prakash C, Loi CM, Hurst S, Nedderman A, Goulet L, Smith E, Bu HZ, et al. (2009) Assessment of three human in vitro systems in the generation of major human excretory and circulating metabolites. Chem Res Toxicol 22:357–368 [DOI] [PubMed] [Google Scholar]
- de Jong YP, Rice CM, Ploss A. (2010) New horizons for studying human hepatotropic infections. J Clin Invest 120:650–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Serres M, Bowers G, Boyle G, Beaumont C, Castellino S, Sigafoos J, Dave M, Roberts A, Shah V, Olson K, et al. (2011) Evaluation of a chimeric (uPA+/+)/SCID mouse model with a humanized liver for prediction of human metabolism. Xenobiotica 41:464–475 [DOI] [PubMed] [Google Scholar]
- Einav S, Gerber D, Bryson PD, Sklan EH, Elazar M, Maerkl SJ, Glenn JS, Quake SR. (2008) Discovery of a hepatitis C target and its pharmacological inhibitors by microfluidic affinity analysis. Nat Biotechnol 26:1019–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einav S, Sobol HD, Gehrig E, Glenn JS. (2010) The hepatitis C virus (HCV) NS4B RNA binding inhibitor clemizole is highly synergistic with HCV protease inhibitors. J Infect Dis 202:65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattinger K, Funk C, Pantze M, Weber C, Reichen J, Stieger B, Meier PJ. (2001) The endothelin antagonist bosentan inhibits the canalicular bile salt export pump: a potential mechanism for hepatic adverse reactions. Clin Pharmacol Ther 69:223–231 [DOI] [PubMed] [Google Scholar]
- Guengerich FP, MacDonald JS. (2007) Applying mechanisms of chemical toxicity to predict drug safety. Chem Res Toxicol 20:344–369 [DOI] [PubMed] [Google Scholar]
- Guo Y, Lu P, Farrell E, Zhang X, Weller P, Monshouwer M, Wang J, Liao G, Zhang Z, Hu S, et al. (2007) In silico and in vitro pharmacogenetic analysis in mice. Proc Natl Acad Sci USA 104:17735–17740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Lu P, Farrell E, Zhang X, Weller P, Monshouwer M, Wang J, Liao G, Zhang Z, Hu S, et al. (2006) In silico pharmacogenetics of warfarin metabolism. Nat Biotechnol 24:531–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Kawai K, Mitsui T, Taniguchi K, Monnai M, Wakui M, Ito M, Suematsu M, Peltz G, Nakamura M, Suemizu H, et al. (2011) The reconstituted ‘humanized liver’ in TK-NOG mice is mature and functional. Biochem Biophys Res Commun 405:405–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu A, Granneman GR, Bertz RJ. (1998) Ritonavir. Clinical pharmacokinetics and interactions with other anti-HIV agents. Clin Pharmacokinet 35:275–291 [DOI] [PubMed] [Google Scholar]
- Ito M, Hiramatsu H, Kobayashi K, Suzue K, Kawahata M, Hioki K, Ueyama Y, Koyanagi Y, Sugamura K, Tsuji K, et al. (2002) NOD/SCID/gamma(c)(null) mouse: an excellent recipient mouse model for engraftment of human cells. Blood 100:3175–3182 [DOI] [PubMed] [Google Scholar]
- Kamimura H, Nakada N, Suzuki K, Mera A, Souda K, Murakami Y, Tanaka K, Iwatsubo T, Kawamura A, Usui T. (2010) Assessment of chimeric mice with humanized liver as a tool for predicting circulating human metabolites. Drug Metab Pharmacokinet 25:223–235 [DOI] [PubMed] [Google Scholar]
- Katoh M, Sawada T, Soeno Y, Nakajima M, Tateno C, Yoshizato K, Yokoi T. (2007) In vivo drug metabolism model for human cytochrome P450 enzyme using chimeric mice with humanized liver. J Pharm Sci 96:428–437 [DOI] [PubMed] [Google Scholar]
- Katoh M, Yokoi T. (2007) Application of chimeric mice with humanized liver for predictive ADME. Drug Metab Rev 39:145–157 [DOI] [PubMed] [Google Scholar]
- Lamba JK, Lin YS, Schuetz EG, Thummel KE. (2002) Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev 54:1271–1294 [DOI] [PubMed] [Google Scholar]
- Leclercq L, Cuyckens F, Mannens GS, de Vries R, Timmerman P, Evans DC. (2009) Which human metabolites have we MIST? Retrospective analysis, practical aspects, and perspectives for metabolite identification and quantification in pharmaceutical development. Chem Res Toxicol 22:280–293 [DOI] [PubMed] [Google Scholar]
- Lootens L, Van Eenoo P, Meuleman P, Leroux-Roels G, Delbeke FT. (2009) The uPA(+/+)-SCID mouse with humanized liver as a model for in vivo metabolism of 4-androstene-3,17-dione. Drug Metab Dispos 37:2367–2374 [DOI] [PubMed] [Google Scholar]
- Meuleman P, Libbrecht L, De Vos R, de Hemptinne B, Gevaert K, Vandekerckhove J, Roskams T, Leroux-Roels G. (2005) Morphological and biochemical characterization of a human liver in a uPA-SCID mouse chimera. Hepatology 41:847–856 [DOI] [PubMed] [Google Scholar]
- Poordad F, McCone J, Jr, Bacon BR, Bruno S, Manns MP, Sulkowski MS, Jacobson IM, Reddy KR, Goodman ZD, Boparai N, et al. SPRINT-2 Investigators (2011) Boceprevir for untreated chronic HCV genotype 1 infection. N Engl J Med 364:1195–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo OJ, Van Eenoo P, Deventer K, Lootens L, Grimalt S, Sancho JV, Hernández F, Meuleman P, Leroux-Roels G, Delbeke FT. (2009) Detection and structural investigation of metabolites of stanozolol in human urine by liquid chromatography tandem mass spectrometry. Steroids 74:837–852 [DOI] [PubMed] [Google Scholar]
- Prichard MN, Shipman C., Jr (1990) A three-dimensional model to analyze drug-drug interactions. Antiviral Res 14:181–205 [DOI] [PubMed] [Google Scholar]
- Prichard MN, Shipman C., Jr (1996) Analysis of combinations of antiviral drugs and design of effective multidrug therapies. Antivir Ther 1:9–20 [PubMed] [Google Scholar]
- Pritchard MN, Prichard LE, Shipman C., Jr (1992) MacSynergy II version 1.0 user’s manual, University of Michigan, Ann Arbor [Google Scholar]
- Smith DA, Obach RS. (2009) Metabolites in safety testing (MIST): considerations of mechanisms of toxicity with dose, abundance, and duration of treatment. Chem Res Toxicol 22:267–279 [DOI] [PubMed] [Google Scholar]
- Tateno C, Yoshizane Y, Saito N, Kataoka M, Utoh R, Yamasaki C, Tachibana A, Soeno Y, Asahina K, Hino H, et al. (2004) Near completely humanized liver in mice shows human-type metabolic responses to drugs. Am J Pathol 165:901–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tscherne DM, Jones CT, Evans MJ, Lindenbach BD, McKeating JA, Rice CM. (2006) Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J Virol 80:1734–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyse TJ, Rozzo SJ, Drake CG, Izui S and Kotzin BL (1997) Control of multiple autoantibodies linked with a lupus nephritis susceptibility locus in New Zealand black mice. J Immunol 158:5566–5574. [PubMed]
- Walker D, Brady J, Dalvie D, Davis J, Dowty M, Duncan JN, Nedderman A, Obach RS, Wright P. (2009) A holistic strategy for characterizing the safety of metabolites through drug discovery and development. Chem Res Toxicol 22:1653–1662 [DOI] [PubMed] [Google Scholar]
- Xie HG, Wood AJ, Kim RB, Stein CM, Wilkinson GR. (2004) Genetic variability in CYP3A5 and its possible consequences. Pharmacogenomics 5:243–272 [DOI] [PubMed] [Google Scholar]
- Yoshizato K, Tateno C. (2009) In vivo modeling of human liver for pharmacological study using humanized mouse. Expert Opin Drug Metab Toxicol 5:1435–1446 [DOI] [PubMed] [Google Scholar]
- Zhang L, Reynolds KS, Zhao P, Huang SM. (2010) Drug interactions evaluation: an integrated part of risk assessment of therapeutics. Toxicol Appl Pharmacol 243:134–145 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.