Abstract
RNA interference (RNAi) is the major innate antiviral pathway in Aedes aegypti that responds to replicating arboviruses such as DENV and SINV. The mosquito’s RNAi machinery is capable of completely eliminating DENV2 from Ae. aegypti. On the other hand, transient silencing of key genes of the RNAi pathway increases replication of SINV and DENV2, allowing the viruses to temporally overcome dose-dependent midgut infection and –escape barriers at higher rates. Here we expressed FHV-B2 from the poly-ubiquitin (PUb) promoter in Ae. aegypti using the ΦC31 site-directed recombination system to investigate the impact of transgene-mediated RNAi pathway suppression on infections with SINV-TR339eGFP and DENV2-QR94, the latter of which has been shown to be confronted with a strong midgut escape barrier (MEB) in Ae. aegypti. FHV-B2 was constitutively expressed in midguts of sugar- and bloodfed mosquitoes of transgenic line PUbB2 P61. B2 over-expression suppressed RNA silencing of carboxypeptidase A-1 (AeCPA-1) in midgut tissue of PUbB2 P61 mosquitoes. Following oral challenge with SINV-TR339eGFP or DENV2-QR94, mean titers in midguts of PUbB2 P61 females were significantly higher at 7 days post-bloodmeal (pbm) than in those of non-transgenic control mosquitoes. At 14 days pbm, infection rates of carcasses were significantly increased in PubB2 P61 mosquitoes infected with SINV-TR339eGFP. Following infection with DENV2-QR94, midgut infection rates were significantly increased in the B2-expressing mosquitoes at 14 days pbm. However, B2 expression in PUbB2 P61 did not increase the DENV2-QR94 dissemination rate, indicating that the infection phenotype was not primarily controlled by RNAi.
Keywords: Aedes aegypti, mosquito, transgenic, ΦC31 recombination, dengue virus, Sindbis virus, FHV-B2, RNA interference, RNAi suppression
Introduction
The yellow fever mosquito, Aedes aegypti, is a vector of medically important arboviruses including the flaviviruses (Flaviviridae; Flavivirus) dengue virus type 1–4 (DENV1–4) and yellow fever virus (YFV) and alphaviruses (Togaviridae; Alphavirus) such as chikungunya virus (CHIKV) and under laboratory conditions Sindbis virus (SINV) (Sanchez-Vargas et al., 2004; Weaver and Reisen, 2010). Arboviruses often cause severe pathology in their vertebrate hosts but are usually not pathogenic to their invertebrate vectors (Blair, 2011). In the mosquito, arboviruses are confronted with several innate immune pathways, such as RNA interference (RNAi), Toll, immune deficiency (Imd), and Janus kinase-signal transducer and activator of transcription (JAK-STAT) (Sanchez-Vargas et al., 2004; Blair, 2011; Xi et al., 2008; Souza-Neto et al., 2009; Ramirez and Dimopoulos, 2010). Of these, RNAi is the major antiviral innate immune pathway in Ae. aegypti (Franz et al., 2006; Campbell et al., 2008; Sanchez-Vargas et al., 2009; Khoo et al., 2010; Mathur et al., 2010; Scott et al., 2010). RNA silencing is divided into three distinct, highly conserved small RNA pathways, micro RNA (miRNA), small interfering RNA (siRNA), and piwi-interacting RNA (piRNA) (Ghildiyal and Zamore, 2009). Based on the origin of the RNA substrate the siRNA pathway is further divided into endogenous small RNA (endo-RNAi) and exogenous small RNA (exo)-RNAi, the latter of which has a major impact on virus infections in mosquito vectors. RNAi can eliminate DENV2 from transgenic mosquitoes expressing an inverted-repeat (IR) RNA to trigger the RNAi pathway against the virus (Franz et al., 2006; Mathur et al., 2010). However, arboviruses are able to persistently infect vectors despite being targeted by the RNAi machinery as shown by the presence of 21 nt virus-derived small interfering RNAs (viRNAs) in arbovirus-infected, transmission-competent mosquito vectors (Scott et al., 2010; Hess et al., 2011). Moreover, temporally induced silencing of the RNAi machinery in Ae. aegypti led to significantly increased SINV and DENV2 titers combined with increased midgut infection and dissemination rates and a shortened extrinsic incubation period (EIP) without compromising insect survival (Campbell et al., 2008; Sanchez-Vargas et al., 2009; Khoo et al., 2010). Arboviruses must have evolved strategies to cope with the mosquito’s RNAi pathway. One such strategy could be a nucleic acid-based decoy mechanism (Siu et al., 2011). Plant viruses and several animal viruses encode proteins that suppress RNAi to protect their viral RNA from homology-dependent degradation (Burgyan and Havelda, 2011; Song et al., 2011). Effects of RNAi suppression in Ae. aegypti by a recombinant SINV have been recently investigated (Myles et al., 2008; Cirimotich et al., 2009). A recombinant SINV strain was engineered to express the Flock house virus (Nodaviridae; [FHV]) B2 protein, a potent suppressor of RNAi. The 12kDa B2 protein is essential for the replication of FHV (Chao et al., 2005; Wang et al., 2006; Li and Ding, 2005). B2 protein is a homodimer and indiscriminately binds to dsRNA molecules independent of their nucleotide sequences and sizes such as siRNAs duplexes and long dsRNAs, thereby protecting dsRNA from being accessed and processed by dicer2 of the RNAi machinery. When mosquitoes were orally infected with SINV-B2, virus titers, midgut infection and escape rates were significantly increased compared to the control virus. Strikingly, SINV-B2 caused high mortality among the mosquitoes at 4–6 days post-infection, suggesting that RNAi has the potential to protect the vector from pathogenic effects of replicating arboviruses (Myles et al., 2008; Cirimotich et al., 2009).
In previous studies key genes of the RNAi machinery in Ae. aegypti were temporarily and/or tissue specifically silenced. As a consequence, the effects on arbovirus replication were transient and did not affect mosquito fitness. The aim of this study was to generate transgenic mosquitoes expressing FHV-B2 as tools to study effects of constitutive RNAi suppression in trans on unrelated arboviruses such as SINV (recombinant strain: TR339eGFP) and DENV2 (strain: QR94). The QR94 strain belongs to the American genotype of DENV2 and has been shown to inefficiently disseminate from the mosquito midgut following oral acquisition (Salazar et al., 2010). SINV-TR339eGFP has been previously shown to encounter a dose-dependent MEB in Ae. aegypti, which was diminished when the virus titer in the midgut was increased (Kramer et al., 1981; Khoo et al., 2010). In this study, we wanted to assess whether both viruses would respond similarly to B2-mediated RNAi suppression in Ae. aegypti by increasing their replication efficiencies, eventually reaching concentrations high enough to become detrimental for the mosquito. We also wanted to observe whether stable RNAi suppression would allow SINV-TR339eGFP and DENV2-QR94 to disseminate more efficiently from the mosquito midgut, which would be an indication that the viruses are confronted with a dose-dependent MEB (Kramer et al., 1981).
Here we describe the generation of transgenic Ae. aegypti, which constitutively express FHV-B2 under control of the Ae. aegypti poly-ubiquitin (PUb) promoter (Anderson et al., 2010). To obtain a predictable transgene expression phenotype transgenic mosquitoes were generated using the ΦC31 site-directed integration system, which has recently been adapted for this purpose (Franz et al., 2011). SINV-TR339eGFP and DENV2-QR94 infection patterns in the RNAi impaired mosquitoes were evaluated and compared with those in non-transgenic control mosquitoes.
Results
Generation of ΦC31 recombinants of Aedes aegypti expressing FHV-B2 from the PUb promoter
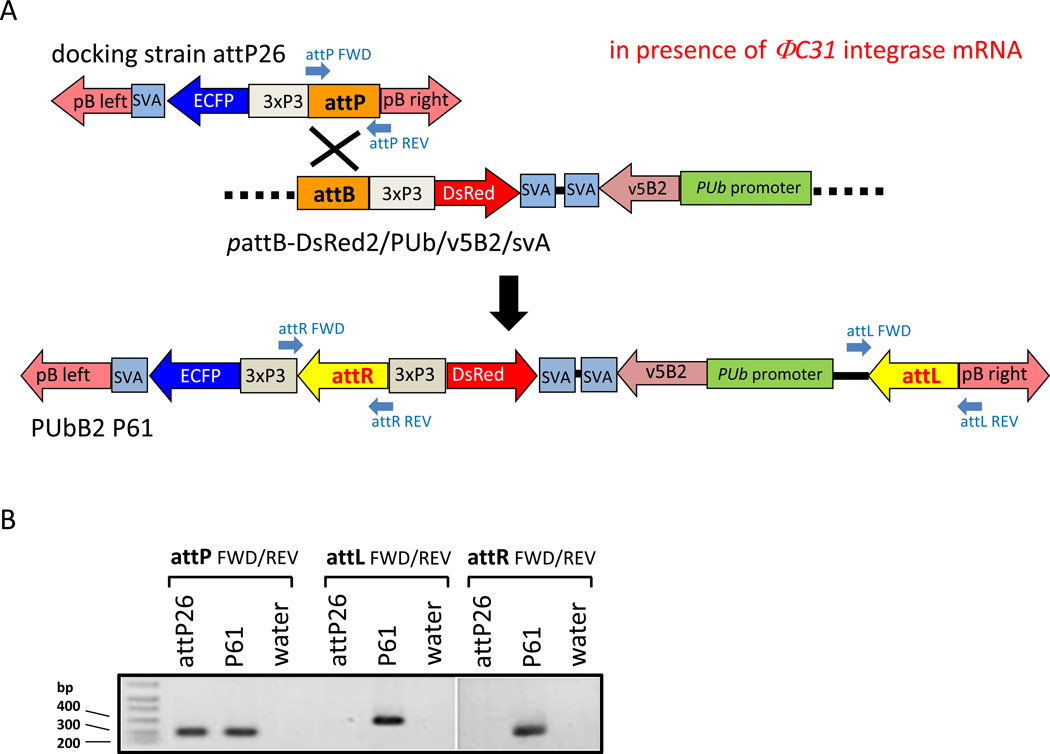
As shown in Fig. 1A, FHV-B2 expression was under control of the constitutive Ae. aegypti PUb promoter (Anderson et al., 2010). The gene-of-interest expression cassette was inserted into the attB site containing donor for site-specific integration into docking strain attP26 as described before (Franz et al., 2011). Following co-injection of 1828 preblastoderm embryos with in vitro transcribed ΦC31 integrase mRNA (600 ng/µl) and the modified attB donor plasmid (300 ng/µl) 196 G0 individuals (100 females, 96 males) survived. After outcrossing to the HWE recipient strain, 32 male and four female pools were established. One pool, P61, consisting of 29 G0 females produced offspring that showed both enhanced cyan fluorescent protein (ECFP) and DsRed eye marker expression indicating recombination between the attP site of the docking strain and the attB site of the donor plasmid (Fig. 1A). The estimated transformation frequency was 1%.
Figure 1.
Molecular characterization of ΦC31-generated recombinants of Ae. aegypti over-expressing FHV-B2 from the constitutive PUb promoter. (A) Diagram of the recombination event between the attP site of docking strain attP26 and the attB site of the donor pattB-DsRed2/PUb/v5B2/svA. (B) Confirmation of site-directed donor integration in PUbB2 P61 mosquitoes by conventional PCR. Annealing sites of PCR primers are shown in the diagram.
Molecular characterization of line PUbB2 P61
To confirm the ΦC31-based recombination event among PUbB2 P61 mosquitoes at the molecular level, we PCR amplified genomic DNA of regions encoding the recombination sites, which were expected to be converted from attP and attB into attR and attL. Previously, we reported that docking strain attP26 contains two attP site integrations (Franz et al., 2011). As shown in Fig. 1B, PCR data revealed that only one of the two attP sites in PUbB2 P61 mosquitoes was converted into attR and attL by recombination with the attB site of the donor whereas the other attP site remained intact. This indicates that there was a single donor integration event in PUbB2 P61 mosquitoes even though these mosquitoes could have potentially accommodated two ΦC31-mediated donor integrations.
FHV-B2 expression in PUbB2 P61 mosquitoes
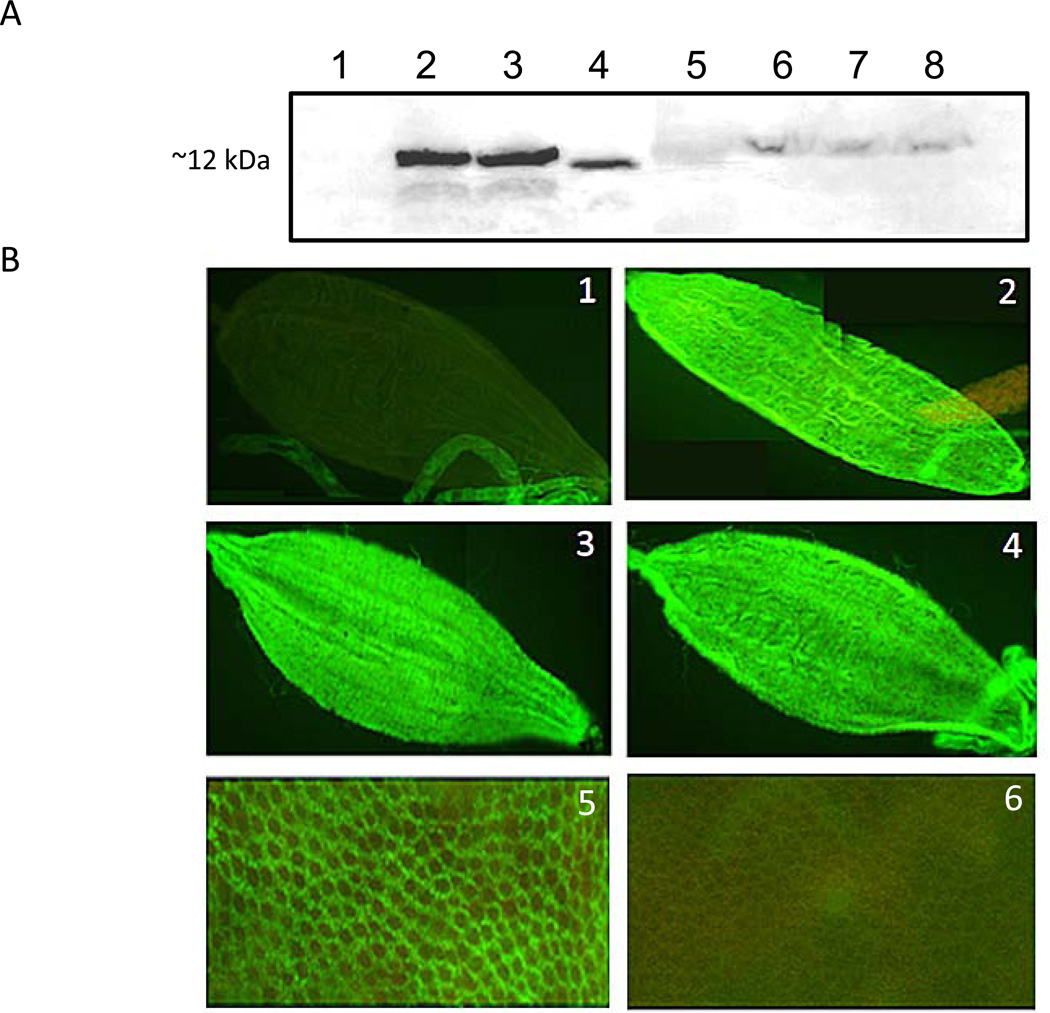
v5-tagged B2 protein was readily detected by Western blots in midguts of sugarfed PUbB2 P61 females at 5–7 days post-eclosion but not in the HWE (negative) control (Fig. 2A). In carcasses, however, B2 protein was only faintly detected at similar time points. Similarly, B2 was readily detected in midguts but not in carcasses of mosquitoes that had received a bloomeal before (data not shown). Presence of B2 antigen in midguts of sugar- or bloodfed females was confirmed in immunofluorescence assays (IFA). As shown in Fig. 2B, v5-tagged B2 antigen was detected in the entire midgut epithelium at different time points following sugarfeeding, bloodfeeding, or DENV2 infection. These results confirm that FHV-B2 was present in midguts of PUbB2 P61 mosquitoes regardless of sugar- or bloodfeeding, whereas in carcasses B2 was barely detectable.
Figure 2.
FHV-B2 over-expression in midguts of transgenic mosquitoes of line PUbB2 P61. (A) Western blot analysis of PUbB2 P61 females to detect FHV-B2 expression. Total protein extracted from five midguts or carcasses of sugarfed PubB2 P61 and HWE (negative control) mosquitoes was electrophoretically separated in 10% SDS-PAGE. B2 over-expression was detected with an IgG antibody specific for the v5 epitope tag. Total protein of SINV-TE3’2J-B2 infected cell culture medium was used as positive control. Lane 1: HWE midguts, 5 days post-eclosion; lane 2: PUbB2 P61 midguts, 5 days post-eclosion; lane 3: PUbB2 P61 midguts, 7 days post-eclosion; lane 4: SINV-TE3’2J-B2 culture supernatant, 36 h post-infection; lane 5: molecular size marker; lane 6: PUbB2 P61 carcasses, 1 day post-eclosion; lane 7: PUbB2 P61 carcasses, 5 days post-eclosion; lane 8: PUbB2 P61 carcasses, 7 days post-eclosion. (B) Detection of FHV-B2 protein in midguts by Immunofluorescence assay using anti-v5 IgG as detection antibody. (1) sugarfed HWE (negative control); (2) sugarfed PUbB2 P61 female; (3) PUbB2 P61 female at 5 days pbm; (4) DENV2-QR94 infected female at 5 days pbm; (5) close-up image (400×) of midgut epithelium over-expressing FHV-B2 in a sugarfed PUbB2 P61 female at 7 days post-eclosion; (6) close-up image (400×) of midgut epithelium of a sugarfed HWE female at 7 days post-eclosion.
FHV-B2 expression in PUbB2 P61 mosquitoes suppresses induced silencing of AeCPA-1 in midguts of bloodfed females
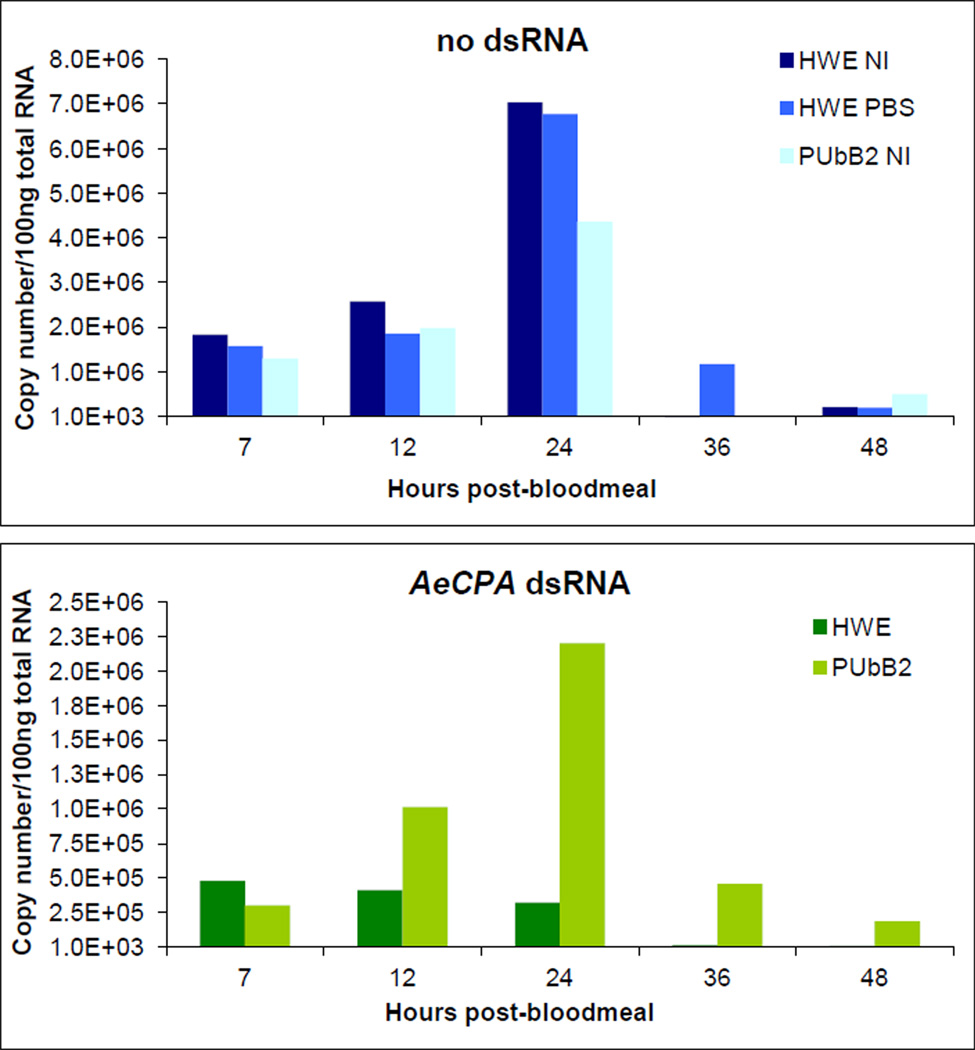
The ability of v5B2 to suppress RNAi in midguts of PUbB2 P61 females was analyzed in a functional assay in which induced silencing of the midgut-specific (Ae. aegypti) AeCPA-1 gene was monitored over time. Females were intrathoracically injected with 1µg dsRNA targeting AeCPA-1 (Isoe et al., 2009). Two days later, females received a non-infectious bloodmeal to induce AeCPA-1 transcription. Abundance of AeCPA-1 transcripts in midguts was monitored between 7–48 h post-bloodmeal (pbm). Within 24 h pbm, AeCPA-1 transcript copy numbers per 100 ng of total midgut RNA increased four-fold in PBS-injected or non-injected HWE and PUbB2 P61 females (Fig. 3). The highest AeCPA-1 transcript copy number (7.0×106) was detected at 24 h pbm among midgut RNA of non-injected HWE. After 24 h pbm, AeCPA-1 transcription levels steadily declined until reaching a base level of <10,000 copies by 48 h pbm. Following dsRNA injection, AeCPA-1 transcript copy numbers in midguts of HWE mosquitoes decreased at least 10-fold during the 48 h observation period, indicating robust silencing of the gene (Fig. 3). In midguts of similarly treated PUbB2 P61 females, AeCPA-1 transcription was reduced less than 2-fold at 24 h pbm, which indicates RNAi suppression through functional activity of B2.
Figure 3.
Functional assay to confirm suppression of induced silencing of the endogenous AeCPA-1 gene in midguts of PUbB2 P61 females. HWE and PUbB2 P61 females were intrathoracically injected with 1 µl of 1 µg/µl dsRNA targeting AeCPA-1 or with PBS (control). Twenty-four hours post-injection mosquitoes were given a bloodmeal. Following total RNA extraction from pools of 20 midguts at 7–48 h pbm, AeCPA-1 transcript copy number equivalents were quantified by qRT-PCR. NI = non-injected.
The results show that in midguts of PUbB2 P61 females, expression of v5B2 suppressed silencing of an endogenous gene with an efficiency of >50% during the observation period. This is a strong indication that in principle, B2 in midguts of PUbB2 P61 females was functional, which was confirmed in a second independent experiment (data not shown). We speculate that the inability of B2 to completely suppress silencing of an endogenous gene in PUbB2 P61 mosquitoes is an effect of B2 expression in these mosquitoes rather than being caused by the intrinsic nature of the B2 protein. It is possible that slight imbalances in the dsRNA : B2 protein ratio could affect the efficiency of gene silencing or silencing suppression.
Challenging of PUbB2 P61 mosquitoes with SINV and DENV2
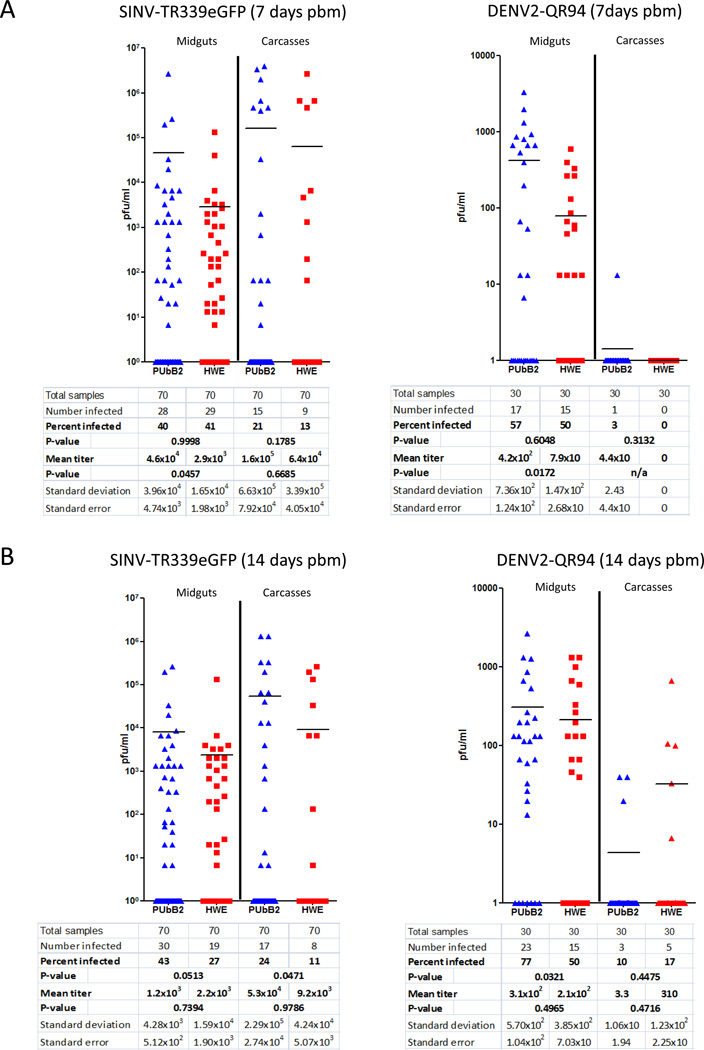
After oral challenge, SINV-TR339eGFP mean titers were significantly increased at 7 days pbm in midguts of PUbB2 P61 females (p-value: 0.0457; ANOVA, Tukey-Kramer test) as compared to the HWE control (Fig. 4A). Infection rates and mean virus titers of carcasses did not differ significantly at 7 days pbm. At 14 days pbm, viral infection rates were significantly increased in carcasses (p-value: 0.0471; chi-square test) but not in midguts (Fig. 4B), indicating that B2-mediated suppression allowed SINV-TR339eGFP to overcome the dose-dependent MEB more efficiently.
Figure 4.
Intensity of SINV-TR339eGFP and DENV2-QR94 infections in PUbB2 P61 and HWE mosquitoes. (A) virus titers and infection rates at 7 days pbm; (B) virus titers and infection rates at 14 days pbm. One-week post-eclosion, females were orally challenged with SINV-TR339eGFP (titer in the bloodmeal: 2.0×107 pfu/ml) or DENV2-QR94 (titer in the bloodmeal: 1.0×106 pfu/ml). Each data point represents the virus titer (pfu/ml) in an individual midgut or carcass as analyzed by plaque assays. P-values (α=0.05) for infection rates and mean virus titers are shown in the table.
We then challenged PUbB2 P61 females with the QR94 strain of DENV2, which belongs to the American genotype of DENV2 (Lorono-Pino et al., 2004). DENV2 strains of the American genotype have been shown to disseminate inefficiently from the midgut in Ae. aegypti (Armstrong and Rico-Hesse, 2003; Anderson and Rico-Hesse, 2006; Salazar et al., 2010). RNAi suppression in PUbB2 P61 females did not enable DENV2-QR94 to overcome the MEB since mean virus titers and infection rates in carcasses remained low for the transgenic and (HWE) control mosquitoes at both time points (Fig. 4A, 4B). However, similar to SINV-TR339eGFP, mean titers of DENV2-QR94 were significantly increased at 7 days pbm (p-value: 0.0172; ANOVA, Tukey-Kramer test) in midguts of PUbB2 P61 females when compared to the HWE control, even though mean DENV2-QR94 titers in midgut tissue of PUbB2 P61 mosquitoes remained 1–2 logs pfu/ml lower at both time points than mean titers of SINV-TR339eGFP. At 14 days pbm, DENV2-QR94 midgut infection rates were significantly increased in the transgenic mosquitoes (p-value: 0.0321; chi-square test). In summary, FHV-B2 expression in line PUbB2 P61 potentially increased the vector competence for SINV-TR339eGFP but not for DENV2-QR94. SINV-TR339eGFP mean titers were significantly increased in midgut tissue at 7 days pbm, which then led to increased disseminated infection rates at 14 days. In contrast, significantly increased DENV2-QR94 mean titers in mosquito midguts at 7 days pbm did not affect virus dissemination rates at 14 days pbm.
Discussion
The underlying hypothesis of this study was that FHV-B2-mediated suppression of the RNAi pathway in Ae. aegypti would strongly increase arboviruses titers and infection rates. As a result the viruses would become pathogenic to the vector if not kept in check by the antiviral immune pathway. Previously, we had shown that transgene-mediated silencing of Aa-dcr2 in the midgut of Ae. aegypti led to statistically significant higher SINV-TR339eGFP titers as well as midgut infection and dissemination rates (Khoo et al., 2010). However, due to the promoter used to drive effector gene expression in these transgenic mosquitoes, the RNAi impairment was temporal and tissue-specific. As a consequence, arboviruses did not produce titers in the mosquitoes high enough to affect overall vector fitness. We optimized our strategy regarding transgene design and expression in three ways to increase transgene-mediated suppression of RNAi in Ae. aegypti. 1) We chose to over-express a strong RNAi suppressor, FHV-B2, which had been shown before to inhibit siRNA biogenesis in Ae. aegypti (Myles et al., 2008; Cirimotich et al., 2009); 2) we used the Ae. aegypti PUb promoter to constitutively over-express B2 in mosquito midguts and secondary tissue; 3) we generated transgenic mosquitoes using the ΦC31 site-directed integration system to ensure predictable transgene expression in each mosquito generation. Constitutive over-expression of B2 in PUbB2 P61 mosquitoes suppressed the RNAi pathway in the mosquitoes as shown by the reduced efficiency of dsRNA targeting AeCPA-1 to silence its mRNA in female midguts after bloodfeeding. Though its original function is to render the replication intermediate (dsRNA) of FHV inaccessible to dicer2, the B2 protein is able to inhibit the activity of any dsRNA species present in the cytoplasm of an invertebrate cell (Lingel et al., 2005). Since the PUb promoter drives constitutive gene expression in midguts and to a lesser extent in secondary tissues as shown earlier and also in this study, B2-mediated suppression of RNA silencing of midgut-specific AeCPA-1 was not surprising (Anderson et al., 2010). Furthermore, we showed that two unrelated arboviruses, SINV-TR339eGFP and DENV2-QR94, responded similarly to RNAi suppression in midgut tissue of PUbB2 P61 mosquitoes by producing significantly increased mean virus titers at 7 days pbm. At 14 days pbm, PUbB2 P61 females infected with DENV2-QR94 had significantly increased midgut infection rates, whereas carcass infection rates of this virus were not affected. At the same time point, however, SINV-TR339eGFP had significantly increased carcass infection rates in the B2 expressing mosquitoes, indicating that the virus overcame the dose-dependent MEB more readily. It is possible that DENV2-QR94 failed to overcome the MEB in PUbB2 P61 mosquitoes because after all the overall virus dose in midgut tissue of the transgenic mosquitoes was still low by not exceeding 420 pfu/ml at 7 days pbm, which was ~5-fold higher than in the HWE control. Thus, the relatively low virus titer DENV2-QR94 generated in Ae. aegypti appeared to be a phenomenon, which was primarily not controlled by the mosquito’s RNAi response.
In conjunction with earlier observations described below the results of this study suggest that the efficiency of RNAi suppression depends on the concentration of B2 molecules being expressed in the cell. Following this scenario, the ratio between dicer2, B2 and viral RNA molecules in a cell would dictate how virus replication becomes affected. In an earlier germline transformation experiment we generated a transgenic mosquito line, Carb/B2-133, which over-expressed FHV-B2 from the midgut-specific, bloodmeal-inducible AeCPA promoter (Edwards et al., 2000; Moreira et al., 2000). Since this line was produced by using a TE instead of the ΦC31 site-directed recombination system, we had no control over the number of transgene integration events per cell. In early generations Carb/B2-133 showed an unusually strong eye marker expression level and when challenged with SINV-TR339eGFP, the virus produced significantly higher titers and infection rates in the transgenic line than in the HWE control (see Fig. S1). However, following three generations of outcrossing to the recipient strain, mosquitoes of this line displayed weak eye-marker expression levels and diminished effects on SINV-TR339eGFP titers and infection rates. We speculate that line Carb/B2-133 originally contained multiple transgene copies, which substantially enhanced B2 expression levels and as a consequence strongly increased virus infection rates and titers. However, the multi-copy transgene integration pattern was unstable in line Carb/B2-133 and therefore lost after three generations.
Previous reports showed that FHV-B2 expression from recombinant SINV-TE3’2J caused a dramatic increase in virus titers associated with significant mortality of infected HWE mosquitoes (Myles et al., 2008; Cirimotic et al., 2009). The authors showed that complete RNAi impairment due to in cis expression of the RNAi suppressor caused the recombinant arbovirus to become pathogenic for the mosquito. When we infected PUbB2 P61 mosquitoes with SINV or DENV2, we did not observe any increased levels of mortality among the mosquitoes (see also Fig. S2). The mode of B2 production in the two systems could account for the observed differences. B2 expression from a recombinant virus leads to RNAi pathway suppression in each virus-infected cell, since each viral RNA generating cell also produces equivalent quantities of B2 (Myles et al., 2008; Cirimotich et al., 2009). However, when expressing B2 in trans using PUbB2 P61 mosquitoes it is possible that not every cell, which is infected by a virus also produces sufficient concentrations of B2 to completely suppress RNAi, thereby enhancing virus replication.
The only other example of transgene-mediated FHV-B2 expression in insects is that of Drosophila, in which B2 was expressed from the Daughterless, actin5C, or hsp promoter (van Rij et al., 2006; Chou et al., 2007; Berry et al., 2009). Injection of the transgenic flies with Drosophila C virus (DCV; Dicistroviridae) lowered the 50% lethal dose of the virus by ~280-fold as compared to the similarly infected, non-transgenic control. The virus is lethal when injected into flies but not when orally acquired (Lautie-Harivel, 1992). In addition, DCV encodes a potent suppressor of RNAi, DCV-1A at the N-terminus of ORF 1 (van Rij et al., 2006). Thus, the presence of both virus-encoded and heterologous RNAi suppressors in the insect cell could have increased the pathogenic potential of DCV. Previous investigations suggest that arboviruses do not encode strong suppressors of RNAi (Myles et al., 2008; Cirimotich et al., 2009). Thus, effects of RNAi suppression in the Drosophila-DCV system might not be directly comparable to those in the mosquito-arbovirus system.
Experimental procedures
Transgene construction and generation of transgenic mosquitoes
The template of the v5-tagged FHV-B2 cDNA originated from recombinant SINV-TE3’2Jv5B2 (Cirimotich et al., 2009). In plasmid pSLfa1180fa-AeCPA/EGFP/svA (Franz et al., 2011), the AeCPA promoter and EGFP encoding fragments were exchanged for PUb and v5B2. The resulting PUb/v5B2/svA expression cassette was then transferred into modified pattB-DsRed2 (Nimmo et al., 2006) using the AscI sites to generate plasmid pattB-DsRed2/PUb/v5B2/svA. Transgenic Ae. aegypti were generated using the ΦC31 site-directed integration system and the Higgs White Eye (HWE)-based docking strain attP26 as described before (Franz et al., 2011). The transgenic Ae. aegypti line was named PUbB2 P61.
Molecular characterization
Site-directed donor integration by recombination between attB of the donor plasmid and attP of docking strain attP26 was confirmed in a conventional PCR assay. Total DNA was extracted from single females of lines attP26 and PUbB2 P61 using the DNeasy Blood & Tissue kit (Qiagen, Valencia, CA). For each PCR reaction 500 ng of total DNA were used. The (non-recombined) attP site was detected using primers attP FWD: 5’CAAATGTGTTCTGTGATGACCTG3’ and attP REV: 5’CTCCCTTGCTACTGACATTATGG3’. Recombined attL and attR sites were detected using primer pairs attL FWD 5’GAGGTCGACGATGTAGGTCAC3’, attL REV 5’ACCTTTTCTCCCTTGCTACTGAC3’ and attR FWD 5’TCAAACTAAGGCGGAGTGG3’, attR REV 5’GATGGGTGAGGTGGAGTACG3’, respectively (Nimmo et al., 2006).
Transient silencing of AeCPA-1
Total RNA was extracted from 10 Ae. aegypti females using TRIzol Reagent (Invitrogen, Carlsbad, CA). A 490 bp cDNA template of the coding region of the AeCPA-1 gene (VectorBase accession # AAEL010782) was amplified from total RNA by RT-PCR using primers CPA FWD (5’GACTTATGGAAACGAAGCCGCT3’) and CPA REV (5’ACAAATATGCCTGGATTTCCCGTTTTG3’) and the SuperScript One-Step RT-PCR kit with Platinum Taq (Invitrogen, Carlsbad, CA). The amplicon was cloned into pCR II-TOPO (Invitrogen). cDNA obtained from this cloning step was PCR amplified using primers that contained T7 promoter sequences at their 5’ends (T7-CPA FWD: 5’-TAA TAC GAC TCA CTA TAG GGG ACT TAT GGA AAC G-3’ and T7-CPA REV: 5’-TAA TAC GAC TCA CTA TAG GGA CAA ATA TGC CTG G-3’; T7 sequences are underlined). The resulting amplicon was in vitro transcribed using the Ambion MEGAscript Kit (Invitrogen) to produce AeCPA-derived dsRNA. Seven day-old HWE and PUbB2 P61 females were intrathoracically injected with 1 µl of 1 µg/µl AeCPA-1 dsRNA each. Controls were either not injected or injected with Phosphate Buffered Saline (PBS). Mosquito midguts were dissected at 7, 12, 24, 36, and 48 h post-injection. Total RNA was extracted from pools of 20 midguts using TRIzol Reagent and used for quantitative RT-PCR (qRT-PCR).
Quantitative RT-PCR
Using the AeCPA-1-specific primers CPA qRT FWD (5’CAT ACC GGT GAG CAC GCC TC3’) and CPA qRT REV (5’GTG TAG GCA ATC TTT ACG TCC AAA G3’), a 185 bp cDNA fragment of the gene was PCR-amplified from total RNA of Ae. aegypti, which differed from the cDNA fragment used as template for dsRNA production. Following pCR II-TOPO cloning of the amplicon, the resulting plasmid was serially diluted and used as the standard curve to calculate transcript copy number equivalents per ng of total RNA. Using the QuantiFast SYBR Green RT-PCR Kit (Qiagen, Valencia, CA), qRT-PCR reactions contained the above primers at 10 µM concentration and 100 ng of total RNA from mosquito midguts. qRT-PCR reactions were conducted in a IQ5 Real-Time PCR Detection system (Bio-Rad, Hercules, CA) according to the following protocol: 10 min at 50°C, 5 min at 95°C and 35 cycles each of 10 s at 95°C and 30 s at 60°C.
Immunofluorescence assay
Midguts of PUbB2 P61 and HWE mosquitoes were dissected and stored in 4% formaldehyde (Vector, Burlingame, CA) for 3 days at 4°C. Fixed midguts were washed in PBS and permeabilized with PBT (PBS with 1% 10X BSA and 2% Triton X-100) for 1 h at room temperature. The v5B2 protein was detected by incubating midguts with the mouse anti-v5 IgG antibody (Invitrogen Carlsbad, CA) at a dilution of 1:200 for 1.5 h at 37°C. Midguts were subsequently incubated with a biotin-labeled anti-mouse secondary antibody (Sigma-Aldrich, Carlsbad, CA) at a dilution of 1:200 for 1.5h at 37°C. Midguts were incubated with streptavidin-fluorescein at a dilution of 1:100 in PBS before being washed in PBS and mounted onto glass slides.
Immunoblot analysis
Immunoblot analysis was conducted according toCirimotich et al. (2009). Briefly, midguts (n=5) of PUbB2 P61 and HWE mosquitoes were dissected and homogenized in 10 µl of sample buffer [NuPAGE® LDS Sample Buffer with NuPAGE Reducing Agent (Invitrogen, Carlsbad, CA)] using a handheld tissue grinder with pestle. Ten microliters of the total protein (~180 ng/lane) was electrophoretically separated in a 10% SDS-polyacrylamide (SDS-PAGE) gel (Invitrogen) and transferred onto a nitrocellulose membrane at 30 volts for 1 h. Membranes were blocked for 1 h at room temperature in blocking buffer (PBS with 0.05% Tween 20 and 5% nonfat dry milk) and incubated at 4°C overnight with a mouse anti-v5 IgG antibody at a dilution of 1:5,000 in blocking buffer. Membranes were then incubated with a horseradish peroxidase-conjugated goat anti-mouse IgG secondary antibody (KPL Inc., Gaithersburg, MD) diluted 1:1,000 in blocking buffer for 1 h at room temperature. The Vector VIP substrate Kit for Peroxidase (Vector, Burlingame, CA) was used to develop the membrane.
Virus propagation
SINV-TR339eGFP stocks were generated according toKhoo et al. (2010). SINV was grown in Vero cells at 0.01 m.o.i and harvested 36–38 h post infection by removing the cell culture medium. DENV2-QR94 is an American genotype DENV2 strain, which was collected in 1994 from a patient in Quintana Roo State, Mexico (Lorono-Pino et al., 2004). Stocks of the virus were generated in Ae. albopictus (C6/36) cells by infecting cells at 0.01 m.o.i., changing the cell culture medium 6 days post infection and maintaining the cells until 12 days post infection. Virus was harvested by removing the cells culture medium and scraping off the cells.
Oral virus infection of mosquitoes
Until bloodfeeding, adult mosquitoes were reared on raisins and water. Each 2.5 L carton contained 100 females and five males. Raisins and water were removed from cartons 36 h and 5 h, respectively prior to bloodfeeding. To infect one week-old females with SINV-TR339eGFP or DENV2-QR94, defibrinated sheep blood was mixed at a 1:1 ratio with freshly harvested virus. The SINV titer in bloodmeal was 2.0×107 pfu/ml and that of DENV2 was 1.0×106 pfu/ml. Female mosquitoes were fed for 1 h with 2 ml of bloodmeal at 37°C using one glass feeder per carton. After bloodfeeding, only those females that were three quarters or fully engorged were selected. These females were reared in 470 ml cartons and fed with sucrose and water until further analysis.
Determination of virus titers by plaque assay
Virus titers from individual midguts and carcasses were determined by plaque assay at 7 and 14 days pbm. SINV-TR339eGFP- and DENV2-QR94-infected samples were homogenized in 0.5 ml 7% FBS-complemented MEM and DMEM medium, respectively. Homogenized samples were filtered with Acrodisc HT Tuffryn 0.2 µm syringe filters (Pall Life Sciences, East Hills, NY). Vero cells (for SINV) or LLC-MK2 cells (for DENV2) were seeded into 24-well plates and left for three days to achieve confluence. Cells were infected with 10-fold serial dilutions of individual midgut or carcass homogenates. Cells were incubated for 1 h at 37°C before overlaid with an agarose nutrient mixture [1x Medium 199 (Sigma-Aldrich, St. Louis, MO), 10% FBS, 4% NaHCO3, 0.5% MEM vitamins, 0.5% MEM amino acids (Mediatech Inc., Manassas, VA); 1% Hanks-DMEM medium (only used for DENV2)]. For SINV, plates were incubated at 37°C for 4 days and for DENV2, plates were incubated at 37°C for 12 days. Cells were then stained with MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) (Sigma-Aldrich, St. Louis, MO), incubated at 37°C for 24 h and the number of plaques was counted for each sample. Virus titers of individual mosquitoes were calculated as pfu/ml.
Statistical analysis
Statistical analyses were performed using SAS Statistical Analysis Software (SAS Institute Inc., Cary, NC). The MIXED procedure was used for restricted maximum likelihood parameter estimation with incomplete data followed by the Tukey-Kramer test. SINV-TR339eGFP and DENV2-QR94 infection levels were normalized by log10 transformation. Infection rates were analyzed with the Chi-square test.
Supplementary Material
Acknowledgements
We thank J. zumBrunnen for help with statistical analyses and C. Meridith for providing stocks of HWE eggs. This work was funded by R01 grant AI073298-01 from the National Institutes of Health.
Contributor Information
CCH Khoo, Email: ckhoo@colostate.edu.
JB Doty, Email: jdoty@cdc.gov.
MS Heersink, Email: monica.heersink@colostate.edu.
KE Olson, Email: kolson@colostate.edu.
AWE Franz, Email: afranz@colostate.edu.
References
- Anderson JR, Rico-Hesse R. Aedes aegypti vectorial capacity is determined by the infecting genotype of dengue virus. Am J Trop Med Hyg. 2006;75(5):886–892. [PMC free article] [PubMed] [Google Scholar]
- Anderson MAE, Gross TL, Myles KM, Adelman ZN. Validation of novel promoter sequences derived from two endogenous ubiquitin genes in transgenic Aedes aegypti. Insect Mol Biol. 2010;19(4):441–449. doi: 10.1111/j.1365-2583.2010.01005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong PM, Rico-Hesse R. Efficiency of dengue serotype 2 virus strains to infect and disseminate in Aedes aegypti. Am J Trop Med Hyg. 2003;68(5):539–544. doi: 10.4269/ajtmh.2003.68.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair CD. Mosquito RNAi is the major innate immune pathway controlling arbovirus infection and transmission. Future Microbiol. 2011;6(3):265–277. doi: 10.2217/fmb.11.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgyan J, Havelda Z. Viral Suppressors of RNA silencing. Trends Plant Sci. 2011;16(5):265–272. doi: 10.1016/j.tplants.2011.02.010. [DOI] [PubMed] [Google Scholar]
- Berry B, Deddouche S, Kirschner D, Imler JL, Antoniewski C. Viral suppressors of RNA silencing hinder exogenous and endogenous small RNA pathways in Drosophila. PLoS One. 2009;4(6):e5866. doi: 10.1371/journal.pone.0005866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CL, Keene KM, Brackney DE, Olson KE, Blair CD, Wilusz J, Foy BD. Aedes aegypti uses RNA interference in defense against Sindbis virus infection. BMC Microbiol. 2008;8:47. doi: 10.1186/1471-2180-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol. 2005;12(11):952–957. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- Chou YT, Tam B, Linay F, Lai EC. Transgenic inhibitors of RNA interference in Drosophila. Fly. 2007;1(6):311–316. doi: 10.4161/fly.5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirimotich CM, Scott JC, Phillips AT, Geiss BJ, Olson KE. Suppression of RNA interference increases alphavirus replication and virus-associated mortality in Aedes aegypti mosquitoes. BMC Microbiol. 2009;9:49. doi: 10.1186/1471-2180-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MJ, Moskalyk LA, Donelly-Doman M, Vlaskova M, Noriega FG, Walker VK, Jacobs-Lorena M. Characterization of a carboxypeptidase A gene from the mosquito, Aedes aegypti. Insect Mol Biol. 2000;9(1):33–38. doi: 10.1046/j.1365-2583.2000.00159.x. [DOI] [PubMed] [Google Scholar]
- Franz AWE, Sanchez-Vargas I, Adelman ZN, Blair CD, Beaty BJ, James AA, Olson KE. Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified Aedes aegypti. P Natl Acad Sci USA. 2006;103(11):4198–4203. doi: 10.1073/pnas.0600479103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz AWE, Jasinskiene N, Sanchez-Vargas I, Isaacs AT, Smith MR, Khoo CCH, Heersink MS, James AA, Olson KE. Comparison of transgene expression in Aedes aegypti generated by mariner Mos1 transposition and ΦC31 site-directed recombination. Insect Mol Biol. 2011;20(5):587–598. doi: 10.1111/j.1365-2583.2011.01089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10(2):94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess AM, Abhishek NP, Ptitsyn A, Ebel GD, Olson KE, Barbacioru C, Monighetti C, Campbell CL. Small RNA profiling of dengue virus-mosquito interactions implicates the PIWI RNA pathway in anti-viral defense. BMC Microbiol. 2011;11:45. doi: 10.1186/1471-2180-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isoe J, Zamora J, Miesfeld RL. Molecular analysis of the Aedes aegypti carboxypeptidase gene family. Insect Biochem Mol Biol. 2009;39(1):68–73. doi: 10.1016/j.ibmb.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo CCH, Piper J, Sanchez-Vargas I, Olson KE, Franz AWE. The RNA interference pathway affects midgut infection- and escape barriers for Sindbis virus in Aedes aegypti. BMC Microbiol. 2010;10:130. doi: 10.1186/1471-2180-10-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer LD, Hardy JL, Presser SB, Houk EJ. Dissemination barriers for western equine encephalomyelitis virus in Culex tarsalis infected after digestion of low viral doses. Am J Trop Med Hyg. 1981;30(1):190–197. doi: 10.4269/ajtmh.1981.30.190. [DOI] [PubMed] [Google Scholar]
- Lautie-Harivel N. Drosophila C virus cycle during the development of two Drosophila melanogaster strains (Charolles and Champetieres) after larval contamination by food. Biol Cell. 1992;76(2):151–157. doi: 10.1016/0248-4900(92)90207-h. [DOI] [PubMed] [Google Scholar]
- Li H-W, Ding S-W. Antiviral silencing in animals. FEBS Letters. 2005;579(26):5965–5973. doi: 10.1016/j.febslet.2005.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, Sattler M. The structure of the flock house virus B2 protein, a viral suppressor of RNA interference, shows a novel mode of double-stranded RNA recognition. EMBO Rep. 2005;6(12):1149–1155. doi: 10.1038/sj.embor.7400583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorono-Pino MA, Farfan-Ale JA, Zapapta-Peraza AL, Rosado-Peredes EP, Flores-Flores LF, Garcia-Rejon JE, Diaz F, Blitvich BJ, Andrade-Narvaez M, Jiminez-Rios E, Blair CD, Olson KE, Black WC, Barry BJ. Introduction of American/Asian genotype of Dengue 2 virus into the Yucatan State of Mexico. Am J Trop Med Hyg. 2004;71(4):485–492. [PubMed] [Google Scholar]
- Mathur G, Sanchez-Vargas I, Alvarez D, Olson KE, Marinotti O, James AA. Transgene-mediated suppression of dengue viruses in the salivary glands of the yellow fever mosquito, Aedes aegypti. Insect Mol Biol. 2010;19(6):753–763. doi: 10.1111/j.1365-2583.2010.01032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira L, Edwards MJ, Adhami F, Jasinskiene N, James AA, Jacobs-Lorena M. Robust gut-specific gene expression in transgenic Aedes aegypti mosquitoes. P Natl Acad Sci USA. 2000;97(2):10895–10898. doi: 10.1073/pnas.97.20.10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles KM, Wiley MR, Morazzani EM, Adelman ZN. Alphavirus-derived small RNAs modulate pathogenesis in disease vector mosquitoes. P Natl Acad Sci USA. 2008;105(50):19938–19943. doi: 10.1073/pnas.0803408105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo DD, Alphey L, Meredith JM, Eggleston P. High efficiency site-specific genetic engineering of the mosquito genome. Insect Mol Biol. 2006;15(2):129–136. doi: 10.1111/j.1365-2583.2006.00615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez JL, Dimopoulos G. The Toll immune signaling pathway control conserved anti-dengue defenses across diverse Ae. aegypti strains and against multiple dengue virus serotypes. Dev Comp Immunol. 2010;34(6):625–629. doi: 10.1016/j.dci.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar MI, Lorono-Pino MA, Farfan-Ale J, Olson KE, Beaty BJ. American and American/Asian genotypes of dengue virus differe in mosquito infection efficiency: candidate molecular determinants of productive vector infection. Revista Biomedica. 2010;21:121–135. [Google Scholar]
- Sanchez-Vargas I, Travanty EA, Keene KM, Franz AWE, Beaty BJ, Blair CD, Olson KE. RNA interference, arthropod-borne viruses, and mosquitoes. Virus Res. 2004;102(1):65–74. doi: 10.1016/j.virusres.2004.01.017. [DOI] [PubMed] [Google Scholar]
- Sanchez-Vargas I, Scott JC, Poole-Smith BK, Franz AWE, Barbosa-Solomieu V, Wilusz J, Olson KE, Blair CD. Dengue virus type 2 infections of Aedes aegypti are modulated by the mosquito’s RNA interference pathway. PLOS Pathog. 2009;5:e1000299. doi: 10.1371/journal.ppat.1000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JC, Brackney DE, Campbell CL, Bondu-Hawkins V, Hjelle B, Ebel GD, Olson KE, Blair CD. Comparison of dengue virus type 2-specific small RNAs from RNA interference-competent and-incompetent mosquito cells. PLoS Negl Trop Dis. 2010;4:e848. doi: 10.1371/journal.pntd.0000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu R, Fragkoudis R, Simmonds P, Donald CL, Chase-Topping ME, Barry G, Attarzadeh-Yazdi G, Rodriguez-Andres J, Nash AA, Merits A, Fazakerley JK, Kohl A. Antiviral RNA interference responses induced by Semliki Forest virus infection of mosquito cells: characterization, origin, and frequency-dependent functions of virus-derived small interfering RNAs. J Virol. 2011;85(6):2907–2917. doi: 10.1128/JVI.02052-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Gao S, Jiang W, Chen S, Liu Y, Zhou L, Huang W. Silencing suppressors: viral weapons for countering host cell defenses. Protein Cell. 2011;2(4):273–281. doi: 10.1007/s13238-011-1037-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza-Neto JA, Sim S, Dimopoulos G. An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. P Natl Acad Sci USA. 2009;106(42):17841–17846. doi: 10.1073/pnas.0905006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rij RP, Saleh MC, Berry B, Foo C, Houk A, Antoniewski C, Andino R. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster . Genes Dev. 2006;20(21):2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R, Atkinson P, Ding SW. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312(5772):452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Reisen WK. Present and future arboviral threats. Antiviral Res. 2010;85(2):328–345. doi: 10.1016/j.antiviral.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, Ramirez JL, Dimopoulos G. The Aedes aegypti Toll pathway controls dengue virus infection. PLOS Pathog. 2008;4:e1000098. doi: 10.1371/journal.ppat.1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.