Abstract
The acridinone derivates 5-dimethylaminopropylamino-8-hydroxytriazoloacridinone (C-1305) and 5-diethylaminoethylamino-8-hydroxyimidazoacridinone (C-1311) are promising antitumor agents with high activity against several experimental cellular and tumor models and are under evaluation in preclinical and early phase clinical trials. Recent evidence from our laboratories has indicated that both compounds were conjugated by several uridine diphosphate-glucuronyltransferase (UGT) isoforms, the most active being extrahepatic UGT1A10. The present studies were designed to test the ability and selectivity of UGT1A10 in the glucuronidation of acridinone antitumor agents in a cellular context. We show that in KB-3 cells, a HeLa subline lacking expression of any UGT isoforms, both C-1305 and C-1311 undergo metabolic transformation to the glucuronidated forms on overexpression of UGT1A10. Furthermore, UGT1A10 overexpression significantly increased the cytotoxicity of C-1305, but not C-1311, suggesting that the glucuronide was more potent than the C-1305 parent compound. These responses were selective for UGT1A10 because documented overexpression of UGT2B4 failed to produce glucuronide products and failed to alter the cytotoxicity for both compounds. These findings contribute to our understanding of the mechanisms of action of these agents and are of particular significance because data for C-1305 contradict the dogma that glucuronidation typically plays a role in detoxification or deactivation. In summary, these studies suggest that extrahepatic UGT1A10 plays an important role in the metabolism and the bioactivation of C-1305 and constitutes the basis for further mechanistic studies on the mode of action of this drug, as well as translational studies on the role of this enzyme in regulation of C-1305 toxicity in cancer.
Introduction
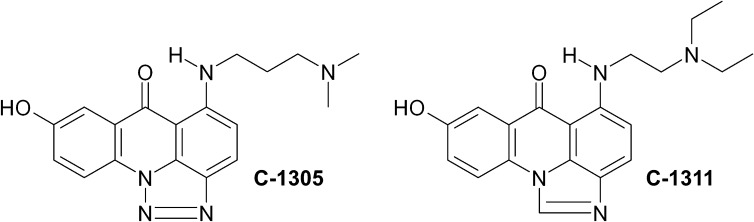
The triazoloacridinone derivative C-1305 (5-dimethylaminopropylamino-8-hydroxytriazoloacridinone) (Fig. 1) has shown high antitumor activity against several experimental tumors in mice, particularly leukemias and colon carcinomas (Cholody et al., 1990a; Kusnierczyk et al., 1994), and has been selected for extended preclinical trials. The imidazoacridinone analog C-1311 (5-diethylaminoethylamino-8-hydroxyimidazoacridinone) (Fig. 1) has shown potent activity against experimental models of murine and human colorectal cancer in vitro and in animals (Cholody et al., 1990b; Burger et al., 1996). It was evaluated in phase I clinical trials in patients with advanced solid tumors (Thomas et al., 2008; Isambert et al., 2010) and was effective in a phase II clinical trial in women with metastatic breast cancer (Capizzi et al., 2008). Despite their clinical potential, the biologic and biochemical mechanisms of C-1305 and C-1311 action are still under extensive study (Mazerska et al., 2001, 2003; Augustin et al., 2006; Skwarska et al., 2007). Both compounds intercalate to DNA and inhibit topoisomerase II activity (Skladanowski et al., 1996; Dziegielewski et al., 2002; Lemke et al., 2004; Koba and Konopa, 2007). Furthermore, the lethal actions of these drugs appear to be due to covalent DNA cross-linking occurring only after metabolic activation (Dziegielewski and Konopa, 1996; Koba and Konopa, 2007).
Fig. 1.
Chemical structures of the two compounds used in this study, C-1305 and C-1311, are shown.
The key role of metabolism in DNA covalent binding by imidazoacridinones and triazoloacridinones prompted us to study the enzymes involved in their metabolic transformation (Mazerska et al., 2001). We showed that cytochrome P450 enzymes were not involved in C-1305 or C-1311 activation, but both compounds were selective irreversible inhibitors of CYP1A2 and CYP3A4 but not of CYP2 family isoforms (Fedejko-Kap et al., 2011; Potega et al., 2011). The metabolites observed with rat and human microsomes, as well as with human hepatocellular liver carcinoma cell line (HepG2) cells, were shown to be flavin monooxygenase (FMO)-mediated Nω-oxide derivatives in the aminoalkyl side chain. Identical metabolites were found with the human recombinant flavin-containing monooxygenases FMO1 and FMO3 (Fedejko-Kap et al., 2011; Potega et al., 2011).
Our recent studies revealed that both C-1305 and C-1311 underwent UDP- glucuronyltransferase (UGT)-mediated metabolism in human microsomes to 8-hydroxyglucuronides (Fedejko-Kap et al., 2012). Furthermore, the formation of 8-hydroxyglucuronides with microsomes from human intestine was more efficient than that with liver microsomes (Fedejko-Kap et al., 2012). Because UGT1A10 shows selective expression in intestinal microsome preparation, this result implies that glucuronidation was catalyzed mainly by UGT1A10 of the UGT1A family, whereas UGT2B family isoforms did not participate. This finding was confirmed by testing the activities of 12 human recombinant UGT isoforms from the 1A and 2B families, where it was found that extrahepatic UGT1A10 was most active against both C-1305 and C-1311 (Fedejko-Kap et al., 2012).
In view of these in vitro results and lack of cellular data, the present studies were designed to test the ability and selectivity of UGT1A10 in the glucuronidation of acridinone antitumor agents in a cellular context. The consequences of metabolic transformation on cytotoxicity and cell-cycle distribution were assessed to clarify the downstream effects of treatment with these compounds. We show that in KB-3 cells, a HeLa subline lacking UGT isoforms, both C-1305 and C-1311 undergo metabolic transformation to the glucuronidated forms on overexpression of UGT1A10 but not on overexpression of UGT2B4. Furthermore, UGT1A10 overexpression significantly increased the cytotoxicity of C-1305, but not C-1311, suggesting that the glucuronide was more potent than the C-1305 parent compound. Our findings define a novel pathway and indicate that UGT enzymes, which are commonly viewed as solely a means of detoxification for a variety of anticancer drugs, in this case play a role in the bioactivation of C-1305.
Materials and Methods
Triazoloacridinone C-1305 and imidazoacridinone C-1311 derivatives were synthesized as the dihydrochloride salts in the laboratory of Prof. J. Konopa in the Department of Pharmaceutical Technology and Biochemistry at Gdańsk University of Technology (Gdańsk, Poland) (Cholody et al., 1990a,b, 1996). C-1305 and C-1311 were prepared as 10 mM stock solutions in dimethyl sulfoxide (DMSO) and kept at −20°C until use. Recombinant human UGT1A10 was produced in baculovirus-infected insect cells as described previously (Fedejko-Kap et al., 2012). High Pure RNA Isolation Kit, Transcriptor First Strand cDNA Synthesis Kit, and LightCycler FastStart DNA Master SYBR Green I were purchased from Roche Diagnostics (Mannheim, Germany). Forward and reverse primers for UGT1A10, UGT2B4, and glyceraldehyde-3-phosphate dehydrogenase (GADPH) genes were obtained from Genomed (Warsaw, Poland). Propidium iodide (PI)/RNase staining buffer was obtained from BD Pharmingen (San Jose, CA). Optima liquid chromatography-mass spectrometry–grade methanol and ammonium formate were from Thermo Fisher Scientific (Pittsburgh, PA). Cell culture media, supplements, and antibiotics were obtained from Cellgro Mediatech (Manassas, VA). All other chemicals, unless otherwise stated, were obtained from Sigma Chemical Co. (St. Louis, MO).
Cell Culture and Transfection.
Human KB-3 carcinoma cell line (a variant of HeLa) was maintained in monolayer culture at 37°C in a humidified 5% CO2 atmosphere in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 50 units/ml penicillin, and 50 mg/ml streptomycin. Cells were transiently transfected with 5 μg of UGT1A10-pcDNA3.39 (pcDNA3.1 vector with Kozak site 28), UGT2B4-pIRES, or pcDNA3.1 (Invitrogen, Carlsbad, CA) in some experiments, together with a plasmid encoding green fluorescent protein (GFP). Plasmids were amplified using XL1-Blue competent cells (Agilent Technologies, Santa Clara, CA) and purified using Endotoxin-free Maxi Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. For transfection, 106 cells at 80% confluence were harvested, washed with phosphate-buffered saline (PBS), and suspended in 100 μl of electroporation buffer (10 mM HEPES pH 7.2, 150 mM NaCl, 4 mM CaCI2 and 0.5 M mannitol), and 5 μg of plasmid DNA was added per sample. Cells were then electroporated in 1-mm gap cuvettes at 140 V, 250 μF, 13 Ω using an Electro Cell Manipulator BTX 600 (BTX; Harvard Apparatus, Inc., Holliston, MA). After electroporation, cells were immediately suspended in fresh media and used the next day. GFP-transfected cells were visualized by fluorescence microscopy using an Olympus BX60 microscope.
mRNA Isolation and Real-Time Quantitative Polymerase Chain Reaction.
KB-3 cells (106) 24 hours after transfection with UGT1A10 or UGT2B4 were collected, washed twice with ice-cold PBS, and suspended in 200 μl of PBS. mRNA was isolated using High Pure RNA Isolation Kit according to the manufacturer’s instructions (Roche, Basel, Switzerland). In brief, cells were lysed in lysis-binding buffer and transferred to a filter tube. Contaminating DNA was removed by 15-minute incubation of the filter tube with DNase I solution. The filter tube was washed once with buffer I and twice with buffer II, and after centrifugation, total RNA was eluted using elution buffer. The concentration of RNA was determined by NanoDrop 2000 (Thermo Fisher Scientific); 1 μg of RNA was reverse transcribed using Transcriptor First Strand cDNA Synthesis Kit according to the manufacturer’s instructions (Roche Diagnostics) in 20 μl of reaction containing 1 μg of RNA, 2.5 μM anchored-oligo(dT)18Primer, 8 mM MgCl2, 20 U of Protector RNase Inhibitor, deoxynucleotide mix (1 mM each) and 20 U of Transcriptor Reverse Transcriptase. The reverse transcription reaction was carried out for 30 minutes at 55°C and stopped by heating to 85°C for 5 minutes and placing on ice. The cDNA solution was stored at −20°C until use. Real-Time polymerase chain reaction (PCR) was performed on 2 μl of cDNA using 0.5 μM forward and reverse primers on LightCycler 1.5 using LightCycler FastStart DNA Master SYBR Green I according to manufacturer’s instructions (Roche Diagnostics). Primers were ordered from Genomed, and the sequences were as follows: UGT1A10 forward primer, CCTCTTTCCTATGTCCCCAATGA; UGT1A10 reverse primer, CCTTAGTCTCCATGCGCTTTGC; UGT2B4 forward primer, TCTTTCGATCCCAACAGCC; UGT2B4 reverse primer, CATCTCTTAACCAGCTGCTTGATA; for reference gene, GADPH forward primer, TGCACCACCAACTGCTTAGC; and GADPH reverse primer, GGCATGGACTGTGGTCATGAG. Cycling conditions were as follows: 1 cycle for 10 minutes at 95°C (preincubation), 45 cycles of amplification: 10 seconds at 95°C (denaturation), 15 seconds at 59°C (annealing), 20 seconds at 72°C (extension), and 1 cycle for 30 seconds at 40°C (cooling). Results were standardized to the reference gene GADPH. Relative expression level of UGT1A10 and UGT2B4 was quantified using comparative method 2−ΔΔCt (Livak and Schmittgen, 2001). Results were obtained from two independent experiments each analyzed twice.
HPLC and Mass Spectrometric Analyses of Metabolites.
To analyze the transformation products of C-1305 or C-1311 in cells expressing UGT1A10 or UGT2B4, transfected KB-3 cells were plated in 60-mm dishes; after reaching 80% confluence, cells were treated with 30 μM C-1305 or C-1311 for 24, 48, or 72 hours. Cells were collected with the media by gentle scraping, sedimented, washed with ice-cold PBS, suspended in ice-cold 60% methanol, and lysed by sonication for 15 minutes. After 1 hour on ice, insoluble material was removed by centrifugation at 16,000 × g for 15 minutes at 4°C. Aliquots (300 μl) of the supernatants were directly analyzed by reverse-phase high-performance liquid chromatography with UV-visible detection at 420 nm using a 5 μm of Suplex pKb-100 analytical column (0.46 cm × 25 cm, C18; Supelco, Bellefonte, PA) with an Agilent 1050 System (Santa Clara, CA). HPLC was carried out at a flow rate of 1 ml/min in 50 mM ammonium formate buffer, pH 3.2, with a linear gradient from 15% to 80% methanol for 25 minutes followed by a linear gradient from 80% to 100% methanol for 3 minutes. Mass spectrometric analysis of peaks from the reverse-phase column was performed by electrospray ionization with positive ion detection carried out with an Agilent 1100 LS-MSD mass spectrometer with a binary pump autosampler (Agilent Technologies).
Glucuronidation Assay with Human Recombinant UGT1A10.
C-1305 or C-1311 (0.1 mM) was preincubated with 5 μg of membrane fractions of baculovirus-infected Sf9 insect cells expressing recombinant His tag UGT1A10 in a reaction mixture of 30 μl containing 50 mM Tris-HCl, pH 7.5, 8 mM MgCl2, and 0.025 mg/ml alamethicin at 37°C for 5 minutes; the reaction was started by the addition of uridine 5′-diphospho-glucuronic acid to 30 mM. After 60 minutes, the reaction was terminated by the addition of 30 μl of ice-cold methanol. The mixture was centrifuged for 15 minutes at 12,000g and analyzed by HPLC as described above.
Cell Viability Assays.
Vector-, UGT1A10-, or UGT2B4-transfected KB-3 cells (2000/well) were seeded in 96-well plates. The following day, C-1305 or C-1311 was added at concentrations up to 10 μM in a fixed final concentration of 0.1% DMSO. Controls received vehicle (0.1% DMSO) alone. After 72 hours, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (50 μg/well) was added for 4 h, followed by DMSO solubilization of the cells and absorbance reading at 540 nm. Each point was conducted in triplicate, and data are expressed relative to vehicle-treated controls. The median inhibitory concentration (IC50) value is defined as the drug concentration required reducing absorbance, or relative cell survival, to 50% of the control value.
Cell-Cycle Distribution Analysis.
Vector-, UGT1A10-, or UGT2B4-transfected KB-3 cells were plated in 60-mm dishes. After reaching 80% confluence, cells were treated with 8 μM C-1305 or 3 μM C-1311 or vehicle (0.1% DMSO) for 24, 48, or 72 hours. Cells were collected by trypsinization and fixed in ice-cold 70% ethanol overnight at −20°C. On the day of analysis, cells were washed in PBS, resuspended in 1 ml of PI/RNase staining buffer, incubated for 30 minutes in the dark at room temperature, and analyzed by flow cytometry using a FACSCalibur (Becton Dickinson, San Jose, CA). The data were analyzed using the WinMDI (The Scripps Research Institute, La Jolla, CA).
Statistical Analysis.
Most of the assays were carried out minimally in duplicate. Data were expressed as mean ± S.D. or as a representative of at least two experiments. Analysis was carried out using Prism software (GraphPad Software, Inc., San Diego, CA) and performed using the Student’s t test with P values of ≤0.05 considered significantly different.
Results
Transfection and Expression of UGT1A10 in KB-3 Cells.
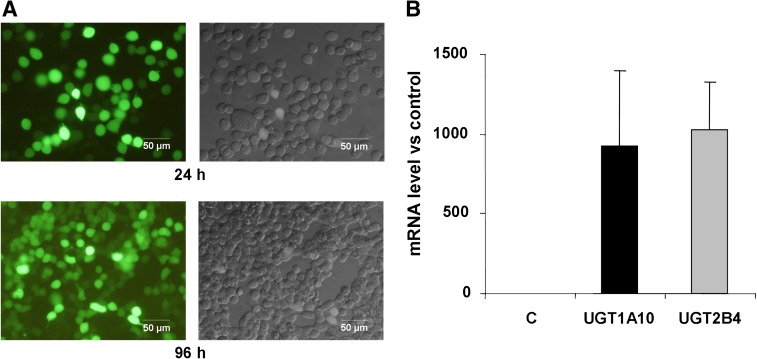
Our recent studies with human recombinant UGTs and microsomal preparations indicated that UGT1A10 is the most active in glucuronidation of C-1305 and C-1311 in vitro, with other UGT isoforms exhibiting little activity (Fedejko-Kap et al., 2012). To examine further the role of UGTs in transformation of these important agents and to investigate whether UGT1A10 showed activity in a cellular context, we expressed human UGT1A10 in KB-3 cells. The KB-3 cell line is a subline of HeLa and does not express any UGT isoforms (unpublished observations). Toward this end, KB-3 cells were transiently transfected via electroporation with an expression plasmid encoding human UGT1A10, as described under Materials and Methods. UGT2B4 was also overexpressed in KB-3 cells for comparison since this isoenzyme was inactive in the glucuronidation of these compounds in vitro (Fedejko-Kap et al., 2012) and thus served as a useful negative control. Because UGT1A10 and UGT2B4 antibodies currently available do not exhibit the desired level of specificity, with numerous bands observed on immunoblots, cells were cotransfected with a plasmid encoding GFP as a means to monitor transfection efficiency. In addition, mRNA levels of UGT1A10 and UGT2B4 were examined in transfected cells by reverse transcription PCR. Visualization of the cells by fluorescence and differential interference contrast microscopy revealed transfection efficiency (number of fluorescent cells versus total cells) of about 60% after 24 h and approximately 70% after 96 h of transfection. Representative images of cells transfected with UGT1A10 and GFP are shown in Fig. 2A. Note that whereas the intensity of fluorescent staining varied from cell to cell, cells with observable fluorescence were included in the calculation. Reverse transcription PCR analysis of transfected cells indicated greatly increased mRNA levels of UCT1A10 and UGT2B4 compared with control untransfected cells, confirming successful expression (Fig. 2B). The data demonstrate efficient transfection and expression of the UGT isoenzymes and provide an experimental platform and suitable model system to study the role of UGTs in biotransformation.
Fig. 2.
Efficiency of transfection. (A) KB-3 cells were transiently transfected with a plasmid encoding GFP and viewed after 24 h or 96 h by fluorescence (left panels) or differential interference contrast (right panels) microscopy, as described under Materials and Methods. The bar indicates 50 μm. (B) mRNA levels of UGTs after transfection. KB-3 cells were transiently transfected with plasmids encoding UGT1A10 or UGT2B4 and after 24 h mRNA was isolated and subjected to real-time quantitative PCR analysis, as described under Materials and Methods. Levels are expressed relative to the control gene GAPDH and represent mean ± S.D., n = 4. (C) mRNA isolated from control untransfected cells.
Metabolic Transformation of C-1305 and C-1311 by UGT1A10, but not UGT2B4, Expressed in Cells.
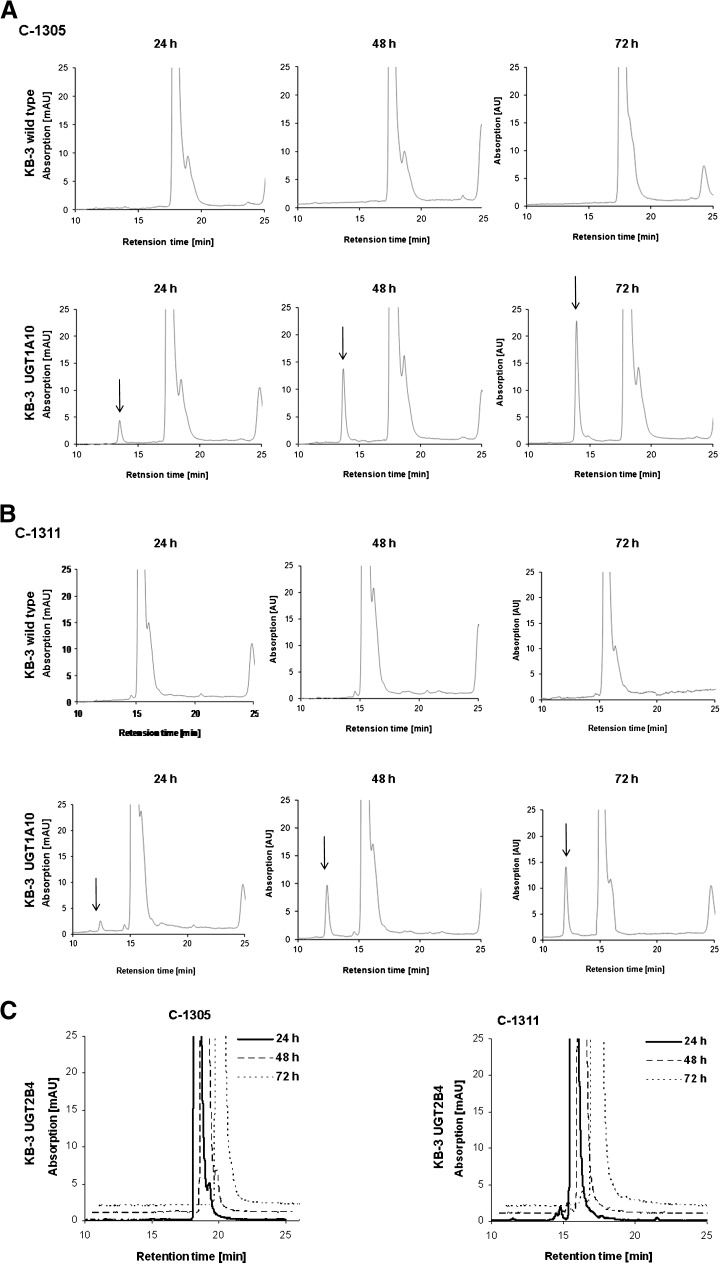
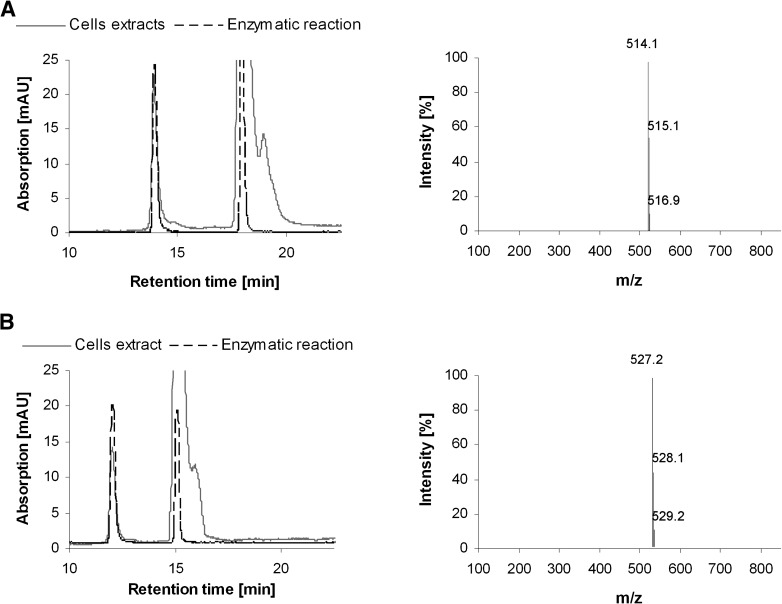
To determine whether C-1305 and C-1311 underwent UGT1A10-mediated transformation in a cellular context, control KB-3 cells or cells transiently transfected with UGT1A10 (KB-3/UGT1A10) or with UGT2B4 (KB-3/UGT2B4) for comparison were treated with 30 μM C-1305 or C-1311 for 24, 48, or 72 h, and cell pellets were washed, extracted in methanol, and analyzed by HPLC, as described under Materials and Methods. In extracts from KB-3 cells, only the parent compound was observed, eluting with retention times of 18–20 min for C-1305 (Fig. 3A) and 15−17 min for C-1311 (Fig. 3B). In extracts from KB-3/UGT1A10 cells, on the other hand, an additional species, eluting earlier than the parent compound, was observed (Fig. 3, A and B, arrows). The abundance of this metabolite increased with the time of incubation of the cells with the drug. In contrast, KB-3/UGT2B4 cells did not generate this metabolite from C-1305 or from C-1311 (Fig. 3C). Importantly, the retention times of the C-1305 metabolite and the C-1311 metabolite were identical to those generated after incubation of each compound with recombinant UGT1A10, as indicated by the superimposition of the HPLC profiles (Fig. 4, A and B). To confirm that the metabolic products generated in KB-3/UGT1A10 cells were the glucuronides, mass spectrometric analysis was performed on the HPLC peaks (Fig. 4, A and B). As shown in the adjacent panels of those figures, the m/z of the C-1305 and C-1311 metabolites were 514.1 and 527.2, respectively, which are the expected combined mass of C-1305 (337 g/mol) or C-1311 (350 g/mol) plus glucuronic acid (194 g/mol) under dehydration (−18 g/mol) with positive ion detection (+1). Thus, the data presented in Figs. 3 and 4 conclusively demonstrate the ability of UGT1A10 to glucuronidate C-1305 and C-1311 in the context of living cells, whereas UGT2B4 was inactive in this regard, consistent with expectations. The relative areas of the peaks in the HPLC profiles of the parent compounds versus the UGT1A10-generated glucuronides after 72-hour treatment of KB-3/UGT1A10 cells were calculated to assess the extent of conversion. On the basis of data from three independent experiments, it was determined that 12.4 ± 4.2% of C-1305 was present as the glucuronidated form and 12.6 ± 4.5% of C-1311 was present as the glucuronidated form.
Fig. 3.
HPLC analysis of C-1305 and C-1311 metabolic transformation. (A and B) Nontransfected KB-3 cells or cells transiently transfected for 24 hours with UGT1A10 (KB-3/UGT1A10) were treated with 30 μM C-1305 (A) or 30 μM C-1311 (B) for 24, 48, or 72 hours, and methanol extracts were subjected to HPLC analysis as described under Materials and Methods. Top panels in (A) and (B): KB-3 cells; Bottom panels in (A) and (B): KB-3/UGT1A10 cells. Glucuronide products indicated by arrows. (C) KB-3 cells transiently transfected for 24 h with UGT2B4 (KB-3/UGT2B4) were treated with 30 μM C-1305 (left panel) or 30 μM C-1311 (right panel) for 24, 48, or 72 hours, and methanol extracts were subjected to HPLC analysis. Note the lack of glucuronide product.
Fig. 4.
Cellularly derived metabolic transformation products correspond to those derived enzymatically. HPLC profiles of C-1305 (A) or C-1311 (B) after transformation by UGT1A10 in KB-3 cells (solid line) or in vitro with recombinant enzyme (dotted line) superimposed to confirm identity of glucuronide. Panels to the right show electrospray ionization-mass spectrometry analysis of the corresponding glucuronide products.
Effect of UGT1A10 Expression on Drug Cytotoxicity.
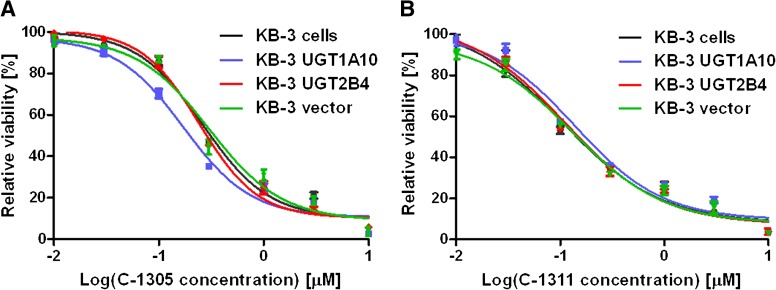
We next sought to determine the effect of UGT1A10-mediated biotransformation on the cellular cytotoxicity of the two acridinones under study. Toward this end, MTT cell viability assays were conducted, as described in Materials and Methods, with untransfected KB-3 cells or cells transiently transfected with either UGT1A10, UGT2B4, or pcDNA3.1 vector. Cells were treated with increasing concentrations of C-1305 or C-1311 for 72 hours, and the data presented are relative to vehicle-treated controls. Representative survival curves for C-1305 are presented in Fig. 5A and for C-1311 in Fig. 5B; note that the transfection procedure did not affect cell viability. These experiments were repeated eight times for C-1305 and five times for C-1311, and mean IC50 values, defined as the drug concentration required to reduce control values by 50%, were determined (Table 1). From these data, it is evident that expression of UGT1A10 resulted in a statistically significant decrease in IC50 value for C-1305 compared with the three control values, with P ≤ 0.0005 in each case (Table 1). Cells expressing UGT2B4 or empty vector were not altered in their response to C-1305, with IC50 values that were not significantly different from untransfected cells (P > 0.05). These are important controls that show that neither the transfection procedure itself nor the expression of a nonrelevant but closely related enzyme affect drug response. In contrast, the expression of UGT1A10 slightly but significantly lessened the response of cells to C-1311: the IC50 value was significantly different from each of the two controls (Table 1). Overall, these results strongly suggest that the glucuronidated product of C-1305 is more cytotoxic to KB-3 cells than the parent compound, whereas the glucuronidated product of C-1311 exhibits slightly lower cytotoxicity than the parent compound.
Fig. 5.
Cell viability assays. Vector-transfected KB-3 cells or cells transiently transfected for 24 h with UGT1A10, UGT2B4, or pcDNA3.1 vector were treated with the indicated concentrations of C-1305 (A) or C-1311 (B) for 72 hours and subjected to MTT cell viability assay as described under Methods and Materials. Cell viability is expressed relative to cells exposed to vehicle. The curves shown are representative of eight independent experiments for C-1305 and five independent experiments for C-1311 (see Table 1).
TABLE 1.
Relative sensitivities to C-1305 and C-1311 of untransfected or transfected KB-3 cells lines
The IC50 values shown represent mean ± S.D. (n = 8 for C-1305 and n = 5 for C-1311) and were derived from cell viability curves as in Fig. 5.
| Compound |
Cell Line |
IC50 |
|
|---|---|---|---|
| Mean |
SD |
||
| μM | |||
| C-1305 | KB-3 | 0.263 | 0.016*** |
| KB-3 UGT1A10 | 0.180 | 0.014 | |
| KB-3 pcDNA3.1 | 0.260 | 0.020*** | |
| KB-3 UGT2B4 | 0.261 | 0.028*** | |
| C-1311 | KB-3 | 0.106 | 0.014* |
| KB-3 UGT1A10 | 0.131 | 0.008 | |
| KB-3 pcDNA3.1 | 0.102 | 0.008* | |
| KB-3 UGT2B4 | 0.113 | 0.018a | |
Significantly different from KB-3 UGT1A10, P < 0.0005; *Significantly different from KB-3 UGT1A10, P < 0.01.
Not significantly different from KB-3 UGT1A10, P > 0.05.
Cell-Cycle Distribution Analysis.
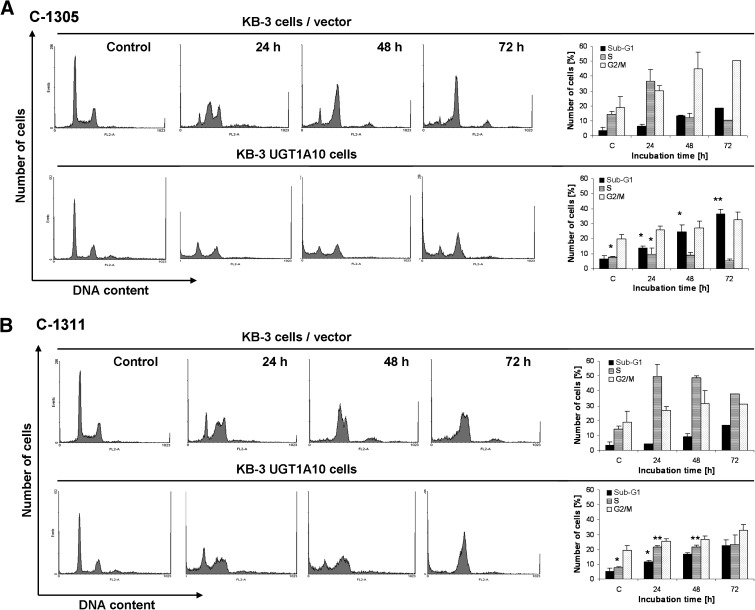
To determine the effects of C-1305 and C-1311 on cell-cycle distribution, it was important first to normalize the drug concentrations, such that equivalently toxic concentrations could be compared. MTT cell viability assays gave IC50 values of 0.260 μM for C-1305 and 0.112 μM for C-1311 (Fig. 5; Table 1). For the following experiments, equitoxic concentrations of 8 μM C-1305 and 3 μM C-1311, representing [IC50] × 30, were used. Vector transfected or UGT1A10 transfected KB-3 cells were treated with either 8 μM C-1305 or 3 μM C-1311 for 24, 48, or 72 hours, and DNA content was analyzed by propidium iodide staining and flow cytometry, as described under Methods and Materials. The data are presented in Fig. 6. C-1305 caused an increase in the proportion of untransfected cells in S and G2/M phases at 24 hours, largely at the expense of cells in G1 phase. After 48- and 72-hour treatment with C-1305, cells with 4 N DNA content were prominently represented, suggesting arrest in G2 and/or M phases, and there was an increase in cells with sub-G1 DNA content, representing an apoptotic population, from 3.4% (no treatment) to 18.7% after 72-hour treatment (Fig. 6A, top panels). Compared with C-1305, imidazoacridinone C-1311 caused S-phase arrest and, to a lesser extent, G2 and/or M arrest. Likewise, an increase in the proportion of cells with sub-G1 DNA content was observed from 3.4 to 16.9% (Fig. 6B, top panels). Cell-cycle distribution analysis of KB-3 cells overexpressing UGT1A10 showed certain differences versus results obtained with untransfected cells. The G2/M block caused by C-1305 and S-phase block induced by C-1311 observed in untransfected KB-3 cells was not as marked in UGT1A10 transfected cells, although both compounds induced accumulation of KB-3/UGT1A10 cells with 4 N DNA content, which was more prominent after treatment with C-1311 (Fig. 6, A and B, lower panels). The biggest difference on UGT1A10 expression was in the proportion of cells with sub-G1 DNA content after C-1305 treatment. This population reached 37.2% compared with 18.7% in untransfected cells after 72-hour treatment with C-1305 (Fig. 6A, bottom panels). This result is consistent with the cell viability data (Fig. 5; Table 1), and it confirms that UGT1A10 expression increases the cytotoxicity of C-1305. The sub-G1 population was 22.4% in KB-3/UGT1A10 cells versus 16.9% in untransfected cells treated for 72 hours with C-1311, consistent with the cell viability data indicating that UGT1A10 expression only slightly alters C-1311 cytotoxicity.
Fig. 6.
Cell-cycle distribution. Vector-transfected KB-3 cells or KB-3 cells transiently transfected for 24 hours with UGT1A10 were untreated (control) or treated with 8 μM C-1305 (A) or 3 μM C-1311 (B) for the times indicated and subjected to propidium iodide staining and flow cytometry as described under Methods and Materials. The histograms show the number of cells (y-axis) versus DNA content (x-axis) and are representative of at least two experiments for each condition. Bar graphs on the right show data quantitation with the percentage of cells with sub-G1, S, or G2/M DNA content as indicated. Significant differences in the cell percentages between vector-transfected and UGT1A10-transfected are indicated as follows: *P ≤ 0.05; **P ≤ 0.01.
Discussion
UGTs catalyze the transfer of the glucuronic acid moiety of UDP-glucuronic acid to a functional group of the substrate, either a phase I metabolite or the native compound (Ritter, 2000). Glucuronidation is considered to be the deactivation or detoxification mechanism of exogenous and endogenous compounds. The acridinone derivates C-1305 and C-1311 are promising antitumor agents and have been investigated in many aspects of their pharmacological properties, but information about their cellular metabolism and cytotoxic mechanism of action remains scarce. Recent evidence from our laboratories has indicated that both compounds were conjugated by several UGT isoforms, the most active being extrahepatic UGT1A10 (Fedejko-Kap et al., 2012). However, reactions that occur in vitro do not always occur in cells and as such are not necessarily of physiologic importance, and it is critical to verify in vitro findings with cell-based assays. The present studies were therefore undertaken to test the ability and selectivity of UGT1A10 in the glucuronidation of these agents in a cellular context and to examine the consequences of metabolic transformation on cytotoxicity. We chose the KB-3 cell line because it is well characterized in our laboratory, is relatively easy to transfect, and, most importantly for the current studies, it is devoid of UGT expression (unpublished observations).
The results presented here clearly demonstrate that both drugs under study, C-1305 and C-1311, undergo glucuronidation in cells expressing UGT1A10, with approximately 12% conversion after 72 h. Cell viability assays and analysis of sub-G1 DNA content revealed that expression of UGT1A10 significantly increased the lethality of C-1305 but only slightly altered the response to C-1311. This suggests the intriguing possibility that the glucuronide metabolite of C-1305 has higher cytotoxic activity than the parent compound. Increased activity of C-1305 was seen only with UGT1A10, not with UGT2B4 expression, further supporting the conclusion that it is the glucuronidated form of the drug that is responsible and not a nonspecific effect of UGT expression. On the other hand, UGT1A10 expression slightly but significantly decreased the cellular response to C-1311, suggesting that the glucuronide metabolite of C-1311 may have lower activity than the parent compound. Thus, despite the subtle differences in structure and chemical properties between imidazoacridinones and triazoloacridinones (Lemke et al., 2004; Fedejko-Kap et al., 2012), the finding that the glucuronide of C-1305 is more toxic than the parent compound may be an important factor to be considered in evaluation of the clinical value and application of C-1305 versus C-1311. The differential cytotoxicities of the respective glucuronides are certainly intriguing given that the 8-OH group is in the structurally conserved portion of the molecules (Fig. 1), and more detailed structure-function studies are needed to elucidate the mechanistic basis of this finding.
Only a limited number of other reports describe the glucuronide conjugates that have higher toxicity than the unconjugated parent compound. For example, NO-glucuronides of hydroxamic acids and acylglucuronides of carboxylic acids have been shown to have higher cytotoxicity compared with their respective parent aglycons, in both cases via electrophilic chemical reactivity (Ritter, 2000; Sallustio et al., 2006). In addition, glucuronidated forms of morphine, carboxylic acid drugs, and estrogens all appear to exhibit biologic effects distinct from the parent compounds that contribute to clinical toxicities (Gosland et al., 1993; Ritter, 2000; Sallustio et al., 2006; van Dorp et al., 2006; Wittwer and Kern, 2006). The data presented here for C-1305 represent another example of this phenomenon. Such data are of particular interest because they contradict the commonly held dogma that glucuronidation typically plays a role in detoxification or deactivation and may help in the prediction of patient response based on individual UGT expression profiles. The system we have developed, with highly efficient and persistent expression of UGT1A10 suitable for cell viability assays, can readily be extended to other UGT family members, facilitating studies on the metabolism of a wide variety of drugs and the biologic consequences of UGT action.
Acknowledgments
The authors thank Stacie Bratton for outstanding assistance in preparation of the manuscript.
Abbreviations
- C-1305
5-dimethylaminopropylamino-8-hydroxytriazoloacridinone
- C-1311
5-diethylaminoethylamino-8-hydroxyimidazoacridinone
- DMSO
dimethyl sulfoxide
- FMO
flavin monooxygenase
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GFP
green fluorescent protein
- HepG2
human hepatocellular liver carcinoma cell line
- IC50
median inhibitory concentration
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PI
propidium iodide
- UGT
uridine diphosphate-glucuronyltransferase
Authorship Contributions
Participated in research design: Pawlowska, Chu, Mazerska, Radominska-Pandya, Chambers.
Conducted experiments: Pawlowska, Chu, Fedejko-Kap.
Performed data analysis: Pawlowska, Chu, Mazerska, Augustin, Radominska-Pandya, Chambers.
Contributed to the writing of the manuscript: Pawlowska, Chu, Mazerska, Radominska-Pandya, Chambers.
Footnotes
This work was supported by grants from the National Institutes of Health National Cancer Institute [Grant CA109821] (to T.C.C.); the National Institutes of Health National Institute of General Medical Sciences [Grant GM075893] to A.R.-P.; and the Department of Defense [Grant X81XWH-11-1-0795] funded by U.S. Army Medical Research and Materiel Command to A.R.-P. M.P. was supported in part by the Development of Interdisciplinary Doctoral Studies at Gdansk University of Technology project in the Area of Novel Technologies, supported by the European Union within European Social Fund and by a Research and Development grant from Chemical Faculty of Gdansk University of Technology [Grant 019432/009]. R.C. was supported in part by funds from the Winthrop P. Rockefeller Cancer Institute, University of Arkansas for Medical Sciences.
References
- Augustin E, Moś-Rompa A, Skwarska A, Witkowski JM, Konopa J. (2006) Induction of G2/M phase arrest and apoptosis of human leukemia cells by potent antitumor triazoloacridinone C-1305. Biochem Pharmacol 72:1668–1679 [DOI] [PubMed] [Google Scholar]
- Burger AM, Double JA, Konopa J, Bibby MC. (1996) Preclinical evaluation of novel imidazoacridinone derivatives with potent activity against experimental colorectal cancer. Br J Cancer 74:1369–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capizzi RL, Roman LA, Tjulandin S, Smirnova I, Manikhas A, Paterson JS, Major A, Lundberg AS, Fumoleau P. (2008) Phase II trial of C1311, a novel inhibitor of topoisomerase II in advanced breast cancer. J Clin Oncol 26:1055 [Google Scholar]
- Cholody WM, Horowska B, Paradziej-Lukowicz J, Martelli S, Konopa J. (1996) Structure-activity relationship for antineoplastic imidazoacridinones: synthesis and antileukemic activity in vivo. J Med Chem 39:1028–1032 [DOI] [PubMed] [Google Scholar]
- Cholody WM, Martelli S, Konopa J. (1990a) 8-Substituted 5-[(aminoalkyl)amino]-6H-v-triazolo[4,5,1-de]acridin-6-ones as potential antineoplastic agents: synthesis and biological activity. J Med Chem 33:2852–2856 [DOI] [PubMed] [Google Scholar]
- Cholody WM, Martelli S, Paradziej-Lukowicz J, Konopa J. (1990b) 5-[(Aminoalkyl)amino]imidazo[4,5,1-de]acridin-6-ones as a novel class of antineoplastic agents: synthesis and biological activity. J Med Chem 33:49–52 [DOI] [PubMed] [Google Scholar]
- Davis FM, Tsao TY, Fowler SK, Rao PN. (1983) Monoclonal antibodies to mitotic cells. Proc Natl Acad Sci USA 80:2926–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziegielewski J, Konopa J. (1996) Interstrand crosslinking of DNA induced in tumor cells by a new group of antitumor imidazoacridinones. Proc Am Assoc Cancer Res 37:410 [Google Scholar]
- Dziegielewski J, Slusarski B, Konitz A, Skladanowski A, Konopa J. (2002) Intercalation of imidazoacridinones to DNA and its relevance to cytotoxic and antitumor activity. Biochem Pharmacol 63:1653–1662 [DOI] [PubMed] [Google Scholar]
- Fedejko-Kap B, Bratton SM, Finel M, Radominska-Pandya A, Mazerska Z. (2012) The role of human UDP-glucuronosyltransferases in the biotransformation of the triazoloacridinone and imidazoacridinone antitumor agents C-1305 and C-1311: highly selective substrates for UGT1A10. Drug Metab Dispos 40:1736–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedejko-Kap B, Niemira M, Radominska-Pandya A, Mazerska Z. (2011) Flavin monooxygenases, FMO1 and FMO3, not cytochrome P450 isoenzymes, contribute to metabolism of anti-tumour triazoloacridinone, C-1305, in liver microsomes and HepG2 cells. Xenobiotica 41:1044–1055 [DOI] [PubMed] [Google Scholar]
- Gosland M, Tsuboi C, Hoffman T, Goodin S, Vore M. (1993) 17 beta-estradiol glucuronide: an inducer of cholestasis and a physiological substrate for the multidrug resistance transporter. Cancer Res 53:5382–5385 [PubMed] [Google Scholar]
- Isambert N, Campone M, Bourbouloux E, Drouin M, Major A, Yin W, Loadman P, Capizzi R, Grieshaber C, Fumoleau P. (2010) Evaluation of the safety of C-1311 (SYMADEX) administered in a phase 1 dose escalation trial as a weekly infusion for 3 consecutive weeks in patients with advanced solid tumours. Eur J Cancer 46:729–734 [DOI] [PubMed] [Google Scholar]
- Koba M, Konopa J. (2007) Interactions of antitumor triazoloacridinones with DNA. Acta Biochim Pol 54:297–306 [PubMed] [Google Scholar]
- Kuśnierczyk H, Chołody WM, Paradziej-Lukowicz J, Radzikowski C, Konopa J. (1994) Experimental antitumor activity and toxicity of the selected triazolo- and imidazoacridinones. Arch Immunol Ther Exp (Warsz) 42:415–423 [PubMed] [Google Scholar]
- Lemke K, Poindessous V, Skladanowski A, Larsen AK. (2004) The antitumor triazoloacridone C-1305 is a topoisomerase II poison with unusual properties. Mol Pharmacol 66:1035–1042 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- Mazerska Z, Dziegielewski J, Konopa J. (2001) Enzymatic activation of a new antitumour drug, 5-diethylaminoethylamino-8-hydroxyimidazoacridinone, C-1311, observed after its intercalation into DNA. Biochem Pharmacol 61:685–694 [DOI] [PubMed] [Google Scholar]
- Mazerska Z, Sowiński P, Konopa J. (2003) Molecular mechanism of the enzymatic oxidation investigated for imidazoacridinone antitumor drug, C-1311. Biochem Pharmacol 66:1727–1736 [DOI] [PubMed] [Google Scholar]
- Potega A, Dabrowska E, Niemira M, Kot-Wasik A, Ronseaux S, Henderson CJ, Wolf CR, Mazerska Z. (2011) The imidazoacridinone antitumor drug, C-1311, is metabolized by flavin monooxygenases but not by cytochrome P450s. Drug Metab Dispos 39:1423–1432 [DOI] [PubMed] [Google Scholar]
- Ritter JK. (2000) Roles of glucuronidation and UDP-glucuronosyltransferases in xenobiotic bioactivation reactions. Chem Biol Interact 129:171–193 [DOI] [PubMed] [Google Scholar]
- Sallustio BC, Degraaf YC, Weekley JS, Burcham PC. (2006) Bioactivation of carboxylic acid compounds by UDP-glucuronosyltransferases to DNA-damaging intermediates: role of glycoxidation and oxidative stress in genotoxicity. Chem Res Toxicol 19:683–691 [DOI] [PubMed] [Google Scholar]
- Skladanowski A, Plisov SY, Konopa J, Larsen AK. (1996) Inhibition of DNA topoisomerase II by imidazoacridinones, new antineoplastic agents with strong activity against solid tumors. Mol Pharmacol 49:772–780 [PubMed] [Google Scholar]
- Skwarska A, Augustin E, Konopa J. (2007) Sequential induction of mitotic catastrophe followed by apoptosis in human leukemia MOLT4 cells by imidazoacridinone C-1311. Apoptosis 12:2245–2257 [DOI] [PubMed] [Google Scholar]
- Thomas AL, Anthoney A, Scott E, Ahmed S, Lundberg AS, Major A, Capizzi RL, Twelves CJ. (2008) C-1311, a novel inhibitor of FLT3 and topoisomerase II: a phase 1 trial of once every three week schedule in patients with advanced solid tumors. J Clin Oncol 26:S2576 [Google Scholar]
- van Dorp EL, Romberg R, Sarton E, Bovill JG, Dahan A. (2006) Morphine-6-glucuronide: morphine’s successor for postoperative pain relief? Anesth Analg 102:1789–1797 [DOI] [PubMed] [Google Scholar]
- Wittwer E, Kern SE. (2006) Role of morphine’s metabolites in analgesia: concepts and controversies. AAPS J 8:E348–E352 [DOI] [PMC free article] [PubMed] [Google Scholar]