A model combining clinical information with MR imaging, fluorine 18 fluorodeoxyglucose (FDG) PET, and cerebrospinal fluid markers yielded the highest accuracy for predicting future mild cognitive impairment conversion; however, the most efficient model included only FDG PET with the clinical covariates.
Abstract
Purpose:
To assess the extent to which multiple Alzheimer disease (AD) biomarkers improve the ability to predict future decline in subjects with mild cognitive impairment (MCI) compared with predictions based on clinical parameters alone.
Materials and Methods:
All protocols were approved by the institutional review board at each site, and written informed consent was obtained from all subjects. The study was HIPAA compliant. Alzheimer’s Disease Neuroimaging Initiative (ADNI) baseline magnetic resonance (MR) imaging and fluorine 18 fluorodeoxyglucose (FDG) positron emission tomography (PET) studies for 97 subjects with MCI were used. MR imaging–derived gray matter probability maps and FDG PET images were analyzed by using independent component analysis, an unbiased data-driven method to extract independent sources of information from whole-brain data. The loading parameters for all MR imaging and FDG components, along with cerebrospinal fluid (CSF) proteins, were entered into logistic regression models (dependent variable: conversion to AD within 4 years). Eight models were considered, including all combinations of MR imaging, PET, and CSF markers with the covariates (age, education, apolipoprotein E genotype, Alzheimer’s Disease Assessment Scale-Cognitive subscale score).
Results:
Combining MR imaging, FDG PET, and CSF data with routine clinical tests significantly increased the accuracy of predicting conversion to AD compared with clinical testing alone. The misclassification rate decreased from 41.3% to 28.4% (P < .00001). FDG PET contributed more information to routine tests (P < .00001) than CSF (P = .32) or MR imaging (P = .08).
Conclusion:
Imaging and CSF biomarkers can improve prediction of conversion from MCI to AD compared with baseline clinical testing. FDG PET appears to add the greatest prognostic information.
© RSNA, 2012
Supplemental material: http://radiology.rsna.org/lookup/suppl/doi:10.1148/radiol.12120010/-/DC1
Introduction
Alzheimer disease (AD) is a neurodegenerative disorder that results in progressive cognitive, functional, and behavioral changes (1,2). It affects more than 30 million people worldwide and that number is expected to triple by 2050 (3–6). Increasing evidence suggests that the pathologic timeline of AD may begin years to decades before clinical diagnosis, with an initial asymptomatic phase (preclinical AD) followed by a phase with mild cognitive impairment (MCI) (2,7,8). Approximately 10%–15% of subjects with MCI progress to meet criteria for AD yearly (9–11). Because new treatments are likely to be most effective at the earliest stages of AD, there is great urgency to develop sensitive markers that facilitate detection and monitoring of early brain changes in at-risk individuals. Such markers may also help speed development of novel therapies to prevent or slow brain loss.
A number of biomarkers are under investigation for prognostic utility in MCI, of which structural magnetic resonance (MR) imaging and fluorine 18 fluorodeoxyglucose (FDG) positron emission tomography (PET) are the two best-studied imaging markers (12). Structural MR imaging has shown medial temporal lobe atrophy in patients with MCI and AD, especially in the entorhinal cortex, amygdala, hippocampus, and parahippocampal gyrus (2). FDG PET has revealed hypometabolism in the temporoparietal regions, posterior cingulate cortex, and frontal lobe even prior to atrophy (2,13–15). Despite these findings, these modalities have not yet been proved accurate enough for prognostic use in the routine clinical setting or clinical drug trials. It is possible, however, that MR imaging and FDG PET are complementary (13,16) and yield additional information when interpreted jointly.
The Alzheimer’s Disease Neuroimaging Initiative (ADNI) was launched in 2003 and is a national multisite study with the goal of collecting a wide range of longitudinal data in 200 healthy elderly control subjects, 400 subjects with MCI, and 200 subjects with AD. ADNI is the result of efforts of many coinvestigators from academic institutions and private corporations, and subjects were recruited from more than 50 sites across the United States and Canada. The data include a wide array of neuropsychological test results, genetic data, cerebrospinal fluid (CSF) biomarkers, structural MR imaging results, and PET scan results, and subjects are followed up every 6 months (17). The primary goal of ADNI was to test whether biologic markers, clinical features, and neuropsychological assessment can be combined to measure progression of disease. Numerous studies of combined biomarker utility have been published from ADNI (11,18–24), but relatively few have examined the additive utility of such biomarkers above and beyond routine neuropsychological tests. This is critical because none of the biomarkers are likely to be used clinically before cognition is assessed.
In the current study, we analyzed whole-brain structural MR imaging and FDG PET results in subjects with MCI from the ADNI by using a multivariate analysis technique called independent component analysis (ICA). ICA is used to isolate unique features from complex biomarkers, potentially revealing hidden patterns underlying three-dimensional imaging data sets (25). This technique eliminates the need for a priori knowledge of the effects on underlying brain anatomy (26) and uses whole-brain data, in lieu of a region-of-interest approach (27). Furthermore, it allows one to incorporate, yet not overweight, the large amount of information in whole-brain MR and FDG PET imaging data by distilling the data set into a few essential patterns, or components, which account for the variability across subjects. With this in mind, the purpose of this study was to examine the additive value of biomarkers, such as CSF proteins, MR imaging, and FDG PET, above and beyond routine clinical tests to develop the best model that could differentiate subjects with MCI who converted to AD from those who did not.
Materials and Methods
Data used in the preparation of this article were de-identified and obtained from the ADNI database (www.loni.ucla.edu/ADNI). For up-to-date information, see www.adni-info.org. Eligibility criteria can be found in the ADNI Procedure Manual (28) at http://adni.loni.ucla.edu/research/protocols/. All protocols were approved by the institutional review boards at each site, and written informed consent was obtained from all subjects prior to enrollment. The study was Health Insurance Portability and Accountability Act compliant. The research reported in this study was not directly industry supported; the ADNI was supported, and these sponsors are listed in the Acknowledgments and Disclosures sections. The authors of this article who do not have any connection to the industry companies listed in the Acknowledgments and Disclosures had control over the inclusion of any data and information that may present a conflict of interest. This analysis was not supported by any specific external funds but was done by the authors on their own research time.
Sample
In this retrospective study, we identified 97 patients with MCI from the ADNI who had baseline clinical examination results, 1.5-T magnetization-prepared rapid acquisition gradient-echo MR imaging results, FDG PET scan results, CSF markers, apolipoprotein E (ApoE) genotype, and results from at least one follow-up clinical examination as of October 19, 2010 (Appendix E1 [online]).
MR Image Acquisition and Analysis
T1-weighted (1.5-T) magnetization-prepared rapid acquisition gradient-echo MR images were preprocessed, having undergone correction for gradient nonlinearity, intensity nonuniformity, and residual intensity nonuniformity. The images were scaled on the basis of phantom acquisitions (29). Whole-brain gray matter probabilities were then extracted from the MR imaging results by using a voxel-based morphometry analysis (FSL-VBM) (30,31) that utilizes FMRIB Software Library tools (Oxford Centre for Functional Magnetic Resonance Imaging of the Brain, University of Oxford, Oxford, England; http://www.fmrib.ox.ac.uk/fsl) (32) (Appendix E1 [online]).
FDG PET Image Acquisition and Analysis
FDG PET images were coregistered, averaged, reoriented, intensity corrected, and smoothed (29). These FDG PET images were registered to the subject’s corresponding brain-extracted MR imaging studies and then to standard MNI152 (Montreal Neurological Institute, Montreal, Québec, Canada) space by using an image registration tool (FLIRT, or FMRIB linear image registration tool; University of Oxford) and a nonlinear registration tool (FNIRT, or FMRIB nonlinear image registration tool; University of Oxford) (32,33) (Appendix E1 [online]).
CSF Acquisition and Analysis
CSF samples were obtained by using lumbar puncture and examined for tau, p-tau181P, and β–amyloid1–42 (Aβ1–42) as described previously (34). More detailed protocols can be found on the ADNI Web site (35). All three proteins were included in this study; ratios were not included. The CSF proteins were used as continuous variables in the logistic regression, so there were no cutoff values for CSF that labeled subjects as having normal or abnormal results.
Clinical and Cognitive Measures
Subjects with MCI from the ADNI were followed up for up to 4 years, with visits occurring every 6 months for 2 years and every 6–12 months after that. The cognitive measures compiled included the Wechsler Memory Scale-Logical Memory (delayed and immediate), Alzheimer’s Disease Assessment Scale-Cognitive subscale (ADAS-Cog) (70- and 85-point scales, which excluded and included delayed word recall and number cancellation, respectively), Clinical Dementia Rating-Sum of Boxes, and Mini-Mental State Examination. All cognitive scores used were baseline scores. Conversion to AD was based on clinical evaluation, with subjects having Mini-Mental State Examination scores between 20–26 (inclusive) and a Clinical Dementia Rating of 0.5 or 1.0; they also had to meet National Institute of Neurological Disorders and Stroke and Alzheimer’s Disease and Related Disorders Association criteria for probable AD (28).
Implementation of ICA on MR Imaging and FDG PET Data
MR imaging gray matter probability maps and FDG PET images were analyzed separately by using standard ICA in a toolbox (Fusion ICA Toolbox, or FIT, version 2.0b, 2009; MIALAB, The Mind Research Network, University of New Mexico, Albuquerque, NM) (27) to isolate unique sources that vary among subjects (25,26). Four MR imaging and nine FDG PET components were extracted, and the loading parameters were used for statistical analysis (Appendix E1 [online]). Notably, the components were not extracted on the basis of the known outcome of the study; rather, they were extracted to account for variance across the entire MCI study population without regard to future conversion status.
Statistical Analysis
The loading parameters for all components were analyzed by group (converters vs nonconverters) by using an independent t test to determine which MR imaging or FDG PET components were associated with conversion to AD. To determine which neuropsychological test was best associated with conversion, logistic regression models (dependent variable: conversion within 4 years) using one neuropsychological test in combination with age, education, ApoE genotype, and the three biomarkers (MR imaging, FDG PET, and CSF proteins) were made. This was compared with a model without any neuropsychological tests. By using these results, logistic regression models were constructed by using age, education, ApoE genotype, and ADAS-Cog as covariates. The biomarkers were then added sequentially to get every combination of variables to develop the best model that differentiated converters from nonconverters. To evaluate the different models, we used three different measures: the area under the receiver operating characteristic curve, the corrected Akaike information criterion, and the misclassification rate. The area under the receiver operating characteristic curve and the Akaike information criterion are both measures of goodness of fit, but the Akaike information criterion also takes efficiency into account by applying a penalty for each added variable (Appendix E1 [online]). We also performed an analysis incorporating time of follow-up in our model. Hypothesis testing by using a Χ2 test on the deviance statistic was performed to determine if the models were significantly different from one another (36). Tenfold cross-validation was performed on all the regression models; these cross-validated misclassification rates reflect a model trained on a subset of the data and tested on the remaining data. P values less than .05 were considered to indicate significant differences.
Results
Subject Characteristics
The mean age for all 97 subjects with MCI was 75.1 years ± 7.2 (standard deviation). Male-to-female ratio was 2.2, and 96.9% of subjects were white. A total of 35.1% of subjects had a family history of AD, and 54.6% had a positive finding for the ApoE4 genotype. The mean follow-up duration for all subjects with MCI was 31.5 months ± 10. Of these, 43 progressed to AD during follow-up (converters) and 54 did not (nonconverters), with converters tending to have longer follow-up times by about 4 months. The average time from the initial screening visit to conversion was 20.7 months ± 9.8. Converters did not differ from nonconverters in mean age, sex ratio, education, race, ethnicity, family history of AD, or ApoE4 gene prevalence. They did, however, have lower baseline delayed recall memory scores (P < .05) and higher ADAS-Cog scores (P < .05) than nonconverters (Table 1).
Table 1.
Baseline Characteristics of MCI Study Sample
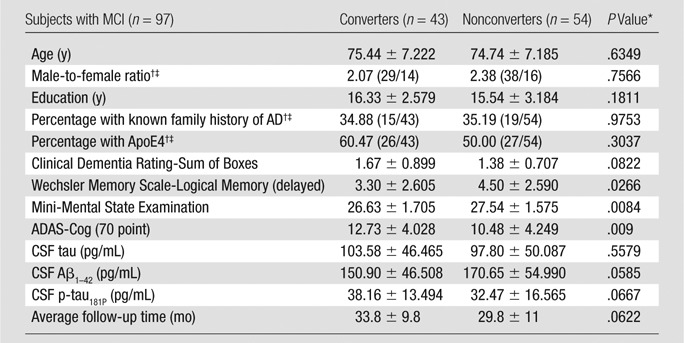
Note.—Unless otherwise indicated, data are means ± standard deviations.
Unless otherwise indicated, P values were obtained by performing t tests.
Data in parentheses are numbers of patients.
P values obtained by performing Χ2 tests.
MR Imaging and PET Components Extracted from ICA
Independent t tests of loading parameters revealed one significant MR imaging component that was significantly lower in converters than nonconverters (P = .009) and showed positive changes across all subjects in the bilateral medial temporal lobes, inferior and lateral temporal lobes, and anterior and inferior frontal lobes. Negative changes were seen in the periventricular white matter. Independent t tests also revealed three significant PET components that differentiated converters from nonconverters. The most significant of these showed positive changes across all subjects in the temporoparietal lobes and the posterior cingulate region (P = .0006). The MR imaging and PET components that most significantly differentiated converters from nonconverters are shown in Figure 1.
Figure 1:
Example components from separate ICAs. Z threshold was 1.5. Both components significantly differentiated converters from nonconverters. Left: The MR imaging component (Comp) highlights in red the bilateral medial temporal lobes, inferior and lateral temporal lobes, and anterior and inferior frontal lobes, consistent with atrophy in these regions in converters. Negative signal, noted in blue, is seen in the periventricular white matter, consistent with higher levels of white matter disease in converters. Right: The FDG PET component highlights in red the temporoparietal lobes, right greater than left, and the posterior cingulate region, consistent with hypometabolism in these regions in converters.
Finding the Best Neuropsychological Test Associated with Conversion
Without any neuropsychological tests, the model including age, education, ApoE genotype, and the three biomarkers resulted in a misclassification rate of 12.5%. Adding Mini-Mental State Examination did not change the misclassification rate. Clinical Dementia Rating-Sum of Boxes and ADAS-Cog (70-point scale) lowered the misclassification rates to 11.5% and 9.4%, respectively. Wechsler Memory Scale-Logical Memory (delayed recall) and ADAS-Cog (85-point scale) increased the misclassification rate to 15.6% and 13.7%, respectively. Overall, ADAS-Cog (70-point scale) had the lowest misclassification rate at 9.4% and was included as the representative neuropsychological test in all other models.
Best Model Separating Converters from Nonconverters
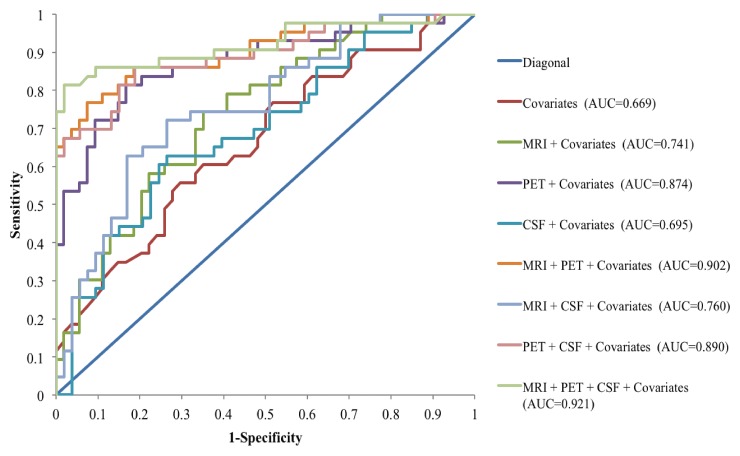
Misclassification rate of neuropsychological testing and other clinical data alone was relatively high (39.18%), which was confirmed with k-fold (k = 10) cross-validation (misclassification rate = 41.3%). The addition of each of the biomarkers reduced the misclassification rates (Table 2), but the model with the lowest misclassification rate included all three biomarkers and the clinical covariates (misclassification rate = 9.38%; area under the curve = 0.921). The model with the lowest Akaike information criterion included only PET with the covariates (Akaike information criterion = 116.6). k-Fold cross-validation (k = 10) showed that the misclassification rate with all three biomarkers was 11.7% in the training set and 28.4% in the validation set. Compared with clinical data alone, the addition of all three biomarkers reduced the misclassification rate by 25.1% in the training set (consistent with findings seen in the overall sample) but by only 12.9% in the validation set. The cross-validated misclassification rate for PET with the clinical covariates was 27.2%. MR imaging alone and CSF alone with the clinical covariates had cross-validation misclassification rates of 39.2% and 39.6%, respectively. Ultimately, cross-validation showed that MR imaging alone and CSF alone only reduced misclassification rates by about 2%, whereas PET alone reduced it by 14.1% (Table 2). Figure 2 shows the receiver operating characteristic curves for all of the models. Incorporating follow-up time (in months) in the model did not change the main findings (Table E1 [online]).
Table 2.
MCI Conversion: Effect of Various Test Combinations

Note.—MR imaging is all four MR imaging ICA components, PET is all nine PET ICA components, and CSF is all three CSF proteins (tau, p-tau181P, and Aβ1–42). Regression models were used to determine which combination of biomarkers was best at predicting conversion in subjects with MCI. The covariates for this analysis included age, education, ApoE genotype, and ADAS-Cog (70-point scale). For hypothesis testing, each model is compared with a submodel or a model with a subset of components. The submodel to which each model is compared is listed in the Comparison Model column. The P values indicate if there is a significant difference between the two models. k-Fold cross-validation of the largest model (k = 10) had misclassification rates of 11.7% and 28.4% for the training and validation sets, respectively. The average standard error of the validation sets is 3.6%. Of note, only 96 subjects were used in all of the models that included CSF because one subject was missing p-tau from the original ADNI database. AICc = corrected Akaike information criterion, Miscl-R = misclassification rate, ROC = receiver operating characteristic.
Figure 2:
Receiver operating characteristic curves for all of the logistic regression models for predicting conversion from MCI to AD. Of the three biomarkers alone, FDG PET added the most prognostic information with an area under the curve (AUC) of 0.874, compared with MR imaging (area under the curve = 0.741) and CSF proteins (area under the curve = 0.695).
Hypothesis testing revealed that the addition of PET data to the covariates significantly increased the predictive value of the model (P < .00001), whereas MR imaging and CSF alone did not (P = .08 and .32, respectively). All three biomarkers combined with the covariates, however, was a significantly better predictor of conversion compared with PET alone with the covariates (P = .02).
Discussion
In the current study, routine clinical tests (age, education, ApoE4 status, and cognitive tests) had the highest misclassification rate for predicting which subjects with MCI would convert to AD. The combination of all three biomarkers (MR imaging, FDG PET, and CSF proteins) with the covariates (age, education, ApoE status, and ADAS-Cog score) was the best predictive model; however, of the three biomarkers individually, FDG PET was the only biomarker that significantly improved the predictive value of the covariates.
Prior studies have also examined the usefulness of AD biomarkers alone and in combination. As expected, no single biomarker or combination of biomarkers has shown high discriminate values for subjects with MCI (37). This poor discrimination is because of the heterogeneous nature of MCI, many of which may not represent a prodromal AD condition. In addition, group discrimination is further complicated by a higher dropout rate of nonconverters. This makes their clinical outcomes more ambiguous, because it is possible that such subjects might have converted to AD after their last visit. One study by Heister et al (24), analyzing longitudinal data in subjects with MCI from the ADNI, showed that those who dropped out of the study early but retained the MCI diagnosis at their last visit had greater memory impairment than those who completed the study.
This emphasizes the risk that true MCI converters may be misclassified as nonconverters because of premature dropout and shorter follow-up duration. In our study, there was a slightly shorter follow-up time among nonconverters, although this was not significant.
A number of previous studies have shown that imaging modalities are better able to help predict clinical decline than baseline neuropsychological scores alone (7). This finding is consistent with the biomarker cascade model, which states that pathologic manifestations of AD begin years before symptoms arise when neuropsychological tests would detect them (7,8). Furthermore, while CSF markers have been shown to be predictive of conversion (8), some studies have suggested that imaging modalities may be more predictive of clinical change than CSF markers (13,38). In comparing the imaging modalities, FDG PET has been a better predictor of conversion than MR imaging (7,13,16).
When analyzing the modalities together, Walhovd et al (16) found that while there is some redundancy of data between MR imaging and PET in predicting memory change, inclusion of both explained more variance among subjects. Likewise, in other studies, MR imaging and CSF provided complementary information about time to conversion in subjects with MCI, and the combination of the two provided a better prediction than either source alone (11,38).
The potential reasons why FDG PET may offer greater accuracy or sensitivity than other biomarkers at the MCI stage are not fully known. There remains lack of consensus on the exact timeline of evolution of FDG PET deficits in relation to CSF biomarker changes, although there is growing consensus that metabolic deficits are greater in magnitude than volumetric changes earlier in the disease. FDG PET studies have shown subjects with MCI to have progressive metabolic deficits in posterior cingulate, parietal, temporal, and precuneus regions, and indeed some genetically susceptible subjects may have such metabolic reductions decades before the anticipated onset of symptoms (39). It has been suggested that early life metabolic deficits may lay the groundwork for disruption of resting brain networks that may increase the risk for cognitive progression to AD (40,41). On the other hand, one study performed on the ADNI data set demonstrated that hippocampal volumes showed larger effect sizes than FDG PET measures in subjects with MCI with isolated memory impairment (42). The reason for this discrepancy may lie in the methods used in these different studies and the higher precision associated with automated volumetric measurements made in subject-native space, which were used in the Karow et al (42) study; this is compared with voxel-based morphometry, used in this study, which warps all subjects’ images to a common template and assesses gray matter probability in this common template space.
There have been a number of recent reports published regarding the value of combining biomarkers to predict conversion to AD, and many suggest that combining modalities offers additional information. However, one study that also used the ADNI data set found that clinical (cognitive and functional) measures were a more robust predictor of conversion compared with MR imaging or CSF markers (19). Our study is different from these in a few important ways, however. First, we included FDG PET in our analysis, which is not true for most studies to date including those published by Gomar et al (19), Cui et al (18), Ewers et al (20), Davatzikos et al (11), and McEvoy et al (23). Second, we used whole-brain data rather than regions-of-interest data. While Landau et al (21) and Chen et al (22) included FDG PET in their analyses, they only included hippocampal volume from MR imaging. Using select regions of interest has the potential to lose valuable information, and using whole-brain data eliminates the need for a priori assumptions about the specific regions of the brain implicated in disease. It also does not presume that disease progression follows discrete anatomic boundaries (43).
In addition, to our knowledge, this was the first study using ICA to assess the extent to which multiple biomarkers improve the ability to identify future decline in subjects with MCI compared with cognitive testing and ApoE4. ICA of neuroimaging data is a data-driven approach that can be used to extract nonredundant sources of information from whole-brain data. The components highlight regions that have been implicated in the development of AD, including the medial temporal lobes at MR imaging and the temporoparietal lobes and posterior cingulate region at FDG PET (2,13,14). This technique has been used to identify disease-related regions in schizophrenia (44), but its use in early AD needs to be further investigated. Until these findings are replicated in other MCI samples, these results should be viewed as preliminary. In addition, the toolbox software (Fusion ICA Toolbox) used for this analysis is an open-source software program that is publically available.
In cross-validation, we trained the model by using subjects with MCI rather than control subjects and patients with AD because of possible intermediate or transient findings in the MCI population that might not be apparent in the extremes of the cognitive spectrum. We used “anytime conversion” because it avoids the problem of specifying an arbitrary window for subjects to convert that could result in inaccurate classification. We also did not examine the region-of-interest–based markers, such as hippocampal volumetry, the best-studied MR imaging predictor of future decline in subjects with MCI (45), because our goal was to use ICA to explore the relative contributions of all brain regions and identify new regions that might add value. Thus, it is possible that our research sample, known colinearity between ApoE4 and CSF Aβ1–42 and tau, use of ICA as a predictor variable, and the specific covariates we selected may have minimized the contributions of CSF or MR imaging in our cross-validation.
There were several limitations to the current study. First, the sample size of our study, although larger than most longitudinal MCI multibiomarker studies, was still relatively small, and there is a chance that the regression models overfit the data, yielding falsely low misclassification rates. This may explain why the misclassification rates obtained through validation were higher than on the entire sample. That being said, the purpose of ICA is to reduce the number of predictors to be much less than that of the voxel level, which can produce more than 100 000 predictors. Second, the MCI population of the ADNI was highly enriched with patients with prodromal AD because of the strict inclusion and exclusion criteria. This population is likely not representative of the patient population seen in clinical practice, so more studies are needed to investigate if these findings are generally applicable. A slight difference in follow-up time between the converters and nonconverters, while not statistically significant, was another potential limitation that must be considered. In addition, we did not perform a recalculation of a new set of ICA components on the basis of each fold of the cross-validation. The ICA components were not extracted to differentiate converters from nonconverters, but instead, to account for variance across the entire study population; nevertheless, these components need to be replicated in other populations. In the process of extracting gray matter probability maps with voxel-based morphometry (FSL-VBM), periventricular white matter disease was sometimes confused for gray matter. This would explain the appearance of the periventricular white matter on the MR imaging component (Fig 1) and makes sense in the context that white matter disease has been found in subjects with MCI and AD (46). As such, our findings should not be interpreted as confirming predictive utility but merely as initial findings that should be replicated and cross-validated in a prospective study.
In summary, a model combining clinical information with MR imaging, FDG PET, and CSF markers yielded the highest accuracy for predicting future MCI conversion. However, the most efficient model included only FDG PET with the clinical covariates. This is in part related to considerable shared variance between ApoE4 (part of the clinical covariate) and CSF Aβ1–42, and between ApoE4 and hippocampal atrophy. Thus, in patients with MCI in whom ApoE4 genotype and cognitive testing is already available, FDG PET would likely yield the greatest additional value. However, in patients in whom ApoE4 genotyping is not available, MR imaging and CSF proteins may add valuable information.
Advances in Knowledge.
• Adding data from three imaging and molecular biomarkers, MR imaging, fluorine 18 fluorodeoxyglucose (FDG) PET, and cerebrospinal fluid (CSF) proteins, to routine clinical tests in patients with mild cognitive impairment (MCI) reduces false classification rates in predicting conversion to Alzheimer disease (AD).
• Among these three imaging and molecular biomarkers, FDG PET appears to be the primary contributor, with misclassification rates for FDG PET, MR imaging, and CSF compared with clinical variables alone of 27.2% (P < .00001), 39.2% (P = .08), and 39.6% (P = .32), respectively.
Implications for Patient Care.
• Combining routine clinical tests with MR imaging, FDG PET, and CSF biomarkers yields the highest accuracy for predicting conversion to AD in subjects with MCI; however, the model with the highest efficiency includes only clinical tests and FDG PET, suggesting that the benefit of additional diagnostic tests is unclear.
• Further validation, standardization, and cost-effectiveness studies are needed to translate the most useful biomarkers into routine clinical practice.
Disclosures of Conflicts of Interest: J.L.S. Financial activities related to the present article: author received travel fellowship for poster presentation (July 2011, covered hotel for four nights and conference registration) for Alzheimer’s Association 2011 International Conference on Alzheimer’s Disease; author received travel fellowship for educational purposes (covered travel, hotel and conference registration) for 9th Annual Symposium on Mild Cognitive Impairment, 2011. Financial activities not related to the present article: none to disclose. Other relationships: none to disclose. J.R.P. Financial activities related to the present article: none to disclose. Financial activities not related to the present article: author serves on Janssen Alzheimer Immunotherapy Neuroradiology Advisory Board. Other relationships: none to disclose. F.C.S. No relevant conflicts of interest to disclose. K.R.C. No relevant conflicts of interest to disclose. V.D.C. Financial activities related to the present article: inventor of FIT software. Financial activities not related to the present article: none to disclose. Other relationships: none to disclose. R.E.C. Financial activities related to the present article: institution receives grant from ADNI. Financial activities not related to the present article: author serves on medical advisory board for Lilly and Avid, is a consultant for Bayer, and receives payment for lectures from Lilly; institution receives grants from Avid for clinical trial. Other relationships: none to disclose. P.M.D. Financial activities related to the present article: institution receives grants from ADNI. Financial activities not related to the present article: author is consultant for Avid/Lilly, Baxter, Neuroptix, TauRx, Sonexa, and BMS; institution receives grants from Lilly, Elan, BMS, and ADNI-GO; author receives payment for lecture from Lundbeck and Accera; author has stock from Sonexa and Clarimedix. Other relationships: none to disclose.
Supplementary Material
Acknowledgments
We thank Matthew MacCarthy, MD, for his help with data analysis. Data used in the preparation of this article were obtained from the ADNI database (adni.loni.ucla.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete list of ADNI investigators can be found at http://adni.loni.ucla.edu/wp-content/uploads/how_to_apply/ADNI_Authorship_List.pdf. Data collection and sharing for this project was funded by the ADNI (National Institutes of Health grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott, AstraZeneca, Bayer Schering Pharma, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson and Johnson, Eli Lilly, Medpace, Merck, Novartis, Pfizer, F. Hoffman-La Roche, Schering-Plough, Synarc, as well as nonprofit partners the Alzheimer’s Association and the Alzheimer’s Drug Discovery Foundation, with participation from the U.S. Food and Drug Administration. Private sector contributions to ADNI are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by National Institutes of Health grants P30 AG010129 and K01 AG030514 and the Dana Foundation.
Received January 17, 2012; revision requested February 22; revision received June 4; accepted June 8; final version accepted August 15.
V.D.C. supported by National Institutes of Health grant 1RO1EB005846 and the National Science Foundation-Science and Engineering Indicators grant NSF0612076 for the Fusion ICA Toolbox used in this analysis. P.M.D. and J.R.P. receive funding from the Alzheimer's Disease Neuroimaging Initiative (ADNI). The data were derived from the ADNI, whose funding sources are listed in the Acknowledgments.
Funding: This research was supported by the National Institutes of Health (grant NIH 1RO1EB005846).
Abbreviations:
- AD
- Alzheimer disease
- ADAS-Cog
- Alzheimer's Disease Assessment Scale-Cognitive subscale
- ADNI
- Alzheimer's Disease Neuroimaging Initiative
- ApoE
- apolipoprotein E
- CSF
- cerebrospinal fluid
- FDG
- fluorine 18 fluorodeoxyglucose
- ICA
- independent component analysis
- MCI
- mild cognitive impairment
References
- 1.Khachaturian ZS. Diagnosis of Alzheimer’s disease. Arch Neurol 1985;42(11):1097–1105 [DOI] [PubMed] [Google Scholar]
- 2.Petrella JR, Coleman RE, Doraiswamy PM. Neuroimaging and early diagnosis of Alzheimer disease: a look to the future. Radiology 2003;226(2):315–336 [DOI] [PubMed] [Google Scholar]
- 3.Weiner MW, Aisen PS, Jack CR, et al. The Alzheimer’s disease neuroimaging initiative: progress report and future plans. Alzheimers Dement 2010;6(3):202–211e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alzheimer’s Association 2010 Alzheimer’s disease facts and figures. Alzheimers Dement 2010;6(2):158–194 [DOI] [PubMed] [Google Scholar]
- 5.Hebert LE, Scherr PA, Bienias JL, Bennett DA, Evans DA. Alzheimer disease in the US population: prevalence estimates using the 2000 census. Arch Neurol 2003;60(8):1119–1122 [DOI] [PubMed] [Google Scholar]
- 6.Trojanowski JQ. Searching for the biomarkers of Alzheimer’s. Pract Neurol 2004;3:30–34 [Google Scholar]
- 7.Hinrichs C, Singh V, Xu G, Johnson SC; Alzheimers Disease Neuroimaging Initiative. Predictive markers for AD in a multi-modality framework: an analysis of MCI progression in the ADNI population. Neuroimage 2011;55(2):574–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jack CR, Jr, Wiste HJ, Vemuri P, et al. Brain beta-amyloid measures and magnetic resonance imaging atrophy both predict time-to-progression from mild cognitive impairment to Alzheimer’s disease. Brain 2010;133(11):3336–3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morris JC, Storandt M, Miller JP, et al. Mild cognitive impairment represents early-stage Alzheimer disease. Arch Neurol 2001;58(3):397–405 [DOI] [PubMed] [Google Scholar]
- 10.Petersen RC, Smith GE, Waring SC, Ivnik RJ, Kokmen E, Tangelos EG. Aging, memory, and mild cognitive impairment. Int Psychogeriatr 1997;9(suppl 1):65–69 [DOI] [PubMed] [Google Scholar]
- 11.Davatzikos C, Bhatt P, Shaw LM, Batmanghelich KN, Trojanowski JQ. Prediction of MCI to AD conversion, via MRI, CSF biomarkers, and pattern classification. Neurobiol Aging 2010;32(12):2322e19–e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plant C, Teipel SJ, Oswald A, et al. Automated detection of brain atrophy patterns based on MRI for the prediction of Alzheimer’s disease. Neuroimage 2010;50(1):162–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walhovd KB, Fjell AM, Brewer J, et al. Combining MR imaging, positron-emission tomography, and CSF biomarkers in the diagnosis and prognosis of Alzheimer disease. AJNR Am J Neuroradiol 2010;31(2):347–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jagust WJ, Eberling JL, Wu CC, et al. Brain function and cognition in a community sample of elderly Latinos. Neurology 2002;59(3):378–383 [DOI] [PubMed] [Google Scholar]
- 15.De Santi S, de Leon MJ, Rusinek H, et al. Hippocampal formation glucose metabolism and volume losses in MCI and AD. Neurobiol Aging 2001;22(4):529–539 [DOI] [PubMed] [Google Scholar]
- 16.Walhovd KB, Fjell AM, Dale AM, et al. Multi-modal imaging predicts memory performance in normal aging and cognitive decline. Neurobiol Aging 2010;31(7):1107–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alzheimer’s Disease Neuroimaging Initiative About the study. http://adni.loni.ucla.edu/about/about-the-study/. Published 2010. Accessed September 19, 2010
- 18.Cui Y, Liu B, Luo S, et al. Identification of conversion from mild cognitive impairment to Alzheimer’s disease using multivariate predictors. PLoS ONE 2011;6(7):e21896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomar JJ, Bobes-Bascaran MT, Conejero-Goldberg C, Davies P, Goldberg TE; Alzheimer’s Disease Neuroimaging Initiative. Utility of combinations of biomarkers, cognitive markers, and risk factors to predict conversion from mild cognitive impairment to Alzheimer disease in patients in the Alzheimer’s disease neuroimaging initiative. Arch Gen Psychiatry 2011;68(9):961–969 [DOI] [PubMed] [Google Scholar]
- 20.Ewers M, Walsh C, Trojanowski JQ, et al. Prediction of conversion from mild cognitive impairment to Alzheimer’s disease dementia based upon biomarkers and neuropsychological test performance. Neurobiol Aging 2012;33(7):1203–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landau SM, Harvey D, Madison CM, et al. Comparing predictors of conversion and decline in mild cognitive impairment. Neurology 2010;75(3):230–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen K, Ayutyanont N, Langbaum JBS, et al. Characterizing Alzheimer’s disease using a hypometabolic convergence index. Neuroimage 2011;56(1):52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McEvoy LK, Holland D, Hagler DJ, Jr, et al. Mild cognitive impairment: baseline and longitudinal structural MR imaging measures improve predictive prognosis. Radiology 2011;259(3):834–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heister D, Brewer JB, Magda S, Blennow K, McEvoy LK; Alzheimer’s Disease Neuroimaging Initiative. Predicting MCI outcome with clinically available MRI and CSF biomarkers. Neurology 2011;77(17):1619–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calhoun VD, Adali T, Giuliani NR, Pekar JJ, Kiehl KA, Pearlson GD. Method for multimodal analysis of independent source differences in schizophrenia: combining gray matter structural and auditory oddball functional data. Hum Brain Mapp 2006;27(1):47–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Pearlson G, Windemuth A, Ruano G, Perrone-Bizzozero NI, Calhoun V. Combining fMRI and SNP data to investigate connections between brain function and genetics using parallel ICA. Hum Brain Mapp 2009;30(1):241–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rachakonda S, Liu J, Calhoun V. Fusion ICA toolbox (FIT) manual. Albuquerque, NM: The MIND Research Network, University of New Mexico, 2008 [Google Scholar]
- 28.Alzheimer’s Disease Neuroimaging Initiative Procedures manual. http://adni.loni.ucla.edu/research/protocols/. Published 2011. Accessed February 8, 2011
- 29.Alzheimer’s Disease Neuroimaging Initiative Image data. http://adni.loni.ucla.edu/about-data-samples/image-data/. Published 2010. Accessed September 19, 2010
- 30.Ashburner J, Friston KJ. Voxel-based morphometry: the methods. Neuroimage 2000;11(6 Pt 1):805–821 [DOI] [PubMed] [Google Scholar]
- 31.Good CD, Johnsrude IS, Ashburner J, Henson RN, Friston KJ, Frackowiak RS. A voxel-based morphometric study of ageing in 465 normal adult human brains. Neuroimage 2001;14(1 Pt 1):21–36 [DOI] [PubMed] [Google Scholar]
- 32.Smith SM, Jenkinson M, Woolrich MW, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 2004;23(suppl 1):S208–S219 [DOI] [PubMed] [Google Scholar]
- 33.Woolrich MW, Jbabdi S, Patenaude B, et al. Bayesian analysis of neuroimaging data in FSL. Neuroimage 2009;45(1 suppl):S173–S186 [DOI] [PubMed] [Google Scholar]
- 34.Shaw LM, Vanderstichele H, Knapik-Czajka M, et al. Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Ann Neurol 2009;65(4):403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alzheimer’s Disease Neuroimaging Initiative Biospecimens protocols. http://adni.loni.ucla.edu/research/protocols/biospecimens-protocols/ Published 2010 AccessedSeptember 19, 2010
- 36.McCullagh P, Nelder J. Generalized linear models. 2nd ed. Boca Raton, Fla: CRC Press, 1989 [Google Scholar]
- 37.Nestor PJ, Scheltens P, Hodges JR. Advances in the early detection of Alzheimer’s disease. Nat Med 2004;10(suppl):S34–S41 [DOI] [PubMed] [Google Scholar]
- 38.Vemuri P, Wiste HJ, Weigand SD, et al. MRI and CSF biomarkers in normal, MCI, and AD subjects: predicting future clinical change. Neurology 2009;73(4):294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reiman EM, Chen K, Alexander GE, et al. Correlations between apolipoprotein E epsilon4 gene dose and brain-imaging measurements of regional hypometabolism. Proc Natl Acad Sci U S A 2005;102(23):8299–8302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buckner RL, Snyder AZ, Shannon BJ, et al. Molecular, structural, and functional characterization of Alzheimer’s disease: evidence for a relationship between default activity, amyloid, and memory. J Neurosci 2005;25(34):7709–7717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buckner RL, Vincent JL. Unrest at rest: default activity and spontaneous network correlations. Neuroimage 2007;37(4):1091–1096; discussion 1097–1099 [DOI] [PubMed] [Google Scholar]
- 42.Karow DS, McEvoy LK, Fennema-Notestine C, et al. Relative capability of MR imaging and FDG PET to depict changes associated with prodromal and early Alzheimer disease. Radiology 2010;256(3):932–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan Y, Resnick SM, Wu X, Davatzikos C. Structural and functional biomarkers of prodromal Alzheimer’s disease: a high-dimensional pattern classification study. Neuroimage 2008;41(2):277–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Calhoun VD, Adali T. Feature-based fusion of medical imaging data. IEEE Trans Inf Technol Biomed 2009;13(5):711–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jack CR, Barkhof F, Bernstein MA, et al. Steps to standardization and validation of hippocampal volumetry as a biomarker in clinical trials and diagnostic criterion for Alzheimer’s disease. Alzheimers Dement 2011;7(4):474–485e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Medina D, DeToledo-Morrell L, Urresta F, et al. White matter changes in mild cognitive impairment and AD: a diffusion tensor imaging study. Neurobiol Aging 2006;27(5):663–672 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.