Abstract
There is a wide gap between the generation of large-scale biological datasets and more detailed, structural and mechanistic studies. However, recent studies that explicitly combine data from systems and structural biological approaches are having a profound effect on our ability to predict how mutations and small molecules affect atomic-level mechanisms, disrupt systems-level networks and ultimately lead to changes in organismal fitness. In fact, we argue that a shared framework for analyzing non-additive genetic and thermodynamic responses to perturbations will accelerate the integration of reductionist and global approaches. A stronger bridge between these two areas will allow for a deeper and more complete understanding of complex biological phenomenon and ultimately provide needed breakthroughs in biomedical research.
As sequencing efforts reveal unprecedented levels of genetic diversity in populations, key challenges remain in linking heritable variation to organismal fitness (Lander, 2011). The functional effect of sequence changes are commonly predicted starting from the top-down (using a network from systems biology) or the bottom-up (using an atomic structure from structural biology). However, biologists now consciously (and unconsciously) straddle the line between global, systems and reductionist, structural approaches. Here, we review how the growing synergies between these two tactics are advancing our understanding of the mechanisms of phenotypic change. A theme emerging from both systems and structural biology is that multiple perturbations can function non-additively. We anticipate that further parallels between non-additive effects on both network and macromolecular dynamics will emerge as scientists working at the systems-to-structure interface probe complex biological phenomenon. Learning how mutations and small molecules affect both atomic interactions and network organizations will advance our abilities to predict the phenotypic responses to perturbations and to design new therapies that restore homeostasis.
The interface between systems and structure: conservation is king
A major goal of systems biology is to determine the abundance of each protein, nucleic acid, or metabolite component and all the interactions that exist between them. In protein-protein interaction networks, an edge represents more than just a construct of graph theory - a connection between nodes implies a direct atomic contact between proteins. Aside from using a co-crystal structure to provide the ultimate “true-positive” confirmation of an interaction, what can structural biology contribute to our knowledge of systems-level organization?
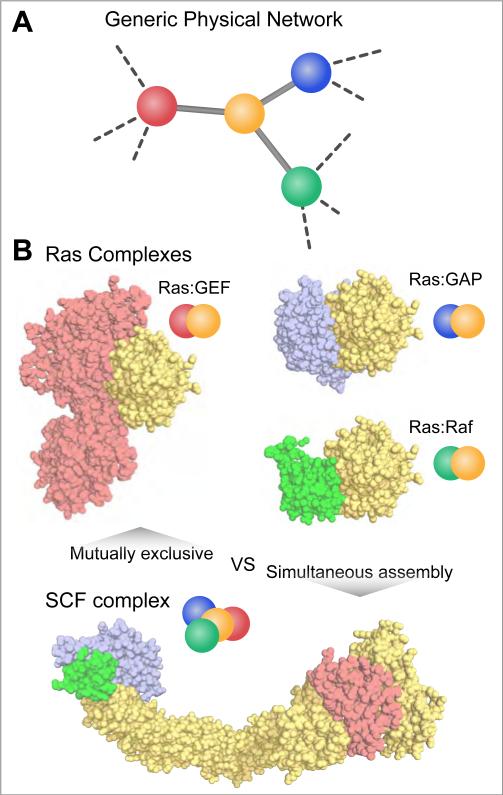
The number of interactions discovered by proteomics experiments (Havugimana et al., 2012) can dwarf the total number of protein structures available in the PDB (Rose et al., 2011). To bridge this gap, homology models can be used to approximate the interfaces of many interactions (Pieper et al., 2011; Zhang et al., 2012a). Using these principles, Kim and Gerstein developed a systems-level structural interaction network in yeast (Kim et al., 2006). The atomic-level details afforded by this structural network representation allowed them to distinguish interactions that can simultaneously assemble on a single receptor from those that are mutually exclusive (Figure 1A). For example, the GTPase Ras interacts with multiple proteins that compete for an overlapping interaction surface (Figure 1B). Only one of these proteins can therefore bind Ras at any given time (Wittinghofer and Vetter, 2011). In contrast, macromolecular machines often assemble many interaction partners simultaneously. Indeed, the SCF Ubiquitin Ligase complex must assemble Rbx1, Skp1 and a member of the F-box protein family along a Cullin scaffold to properly ubiquitinate target proteins (Zheng et al., 2002). Systematically characterizing overlapping and simultaneous binding within a protein-protein interaction network has resolved long-standing ambiguities about the relationship between a protein's evolutionary rate and its degree (i.e. number of interaction partners), by establishing a central role for the amount of accessible surface area buried upon protein-protein association (Kim et al., 2006). This approach can also be inverted: structural information can be leveraged to predict new protein-protein interactions. Remote structural alignments that consider geometric relationships between interacting proteins have recently been used to predict protein-protein interactions on a proteomic-scale (Zhang et al., 2012a).
Figure 1. Interpretation of physical networks.
A) A generic physical interaction network that could be derived from various systematic protein-protein interaction platforms. B) The interaction map in A) could represent a set mutually exclusive protein-protein interactions involving one common target (e.g. Ras complexes: Ras:GEF (1BKD), Ras:GAP (1WQ1), Ras:Raf (3KUD)) or a simultaneous assembly of proteins into a complex (e.g. SCF complex (1LDK)).
Similarly, structural insights can be used to assess the evolution and functional role of posttranslational modifications, such as phosphorylation, ubiquitination, and acetylation (Beltrao et al., 2012). The increased sensitivity of mass spectrometry to identify posttranslational modifications in proteomic samples has been a blessing and a curse: while new hypotheses can be generated based on the identification of modified sites, the functional significance of the vast majority of these modifications remains unknown. By examining posttranslational modifications in a structural context, Beltrao et al. found phosphosites located at protein-protein interaction interfaces are more highly conserved (Beltrao et al., 2012). Additionally, this study proposed and tested the hypothesis that posttranslational hotspots for controlling protein function are structurally conserved across family members. However, these data suggest that most post-translational modifications likely have no biological role. They proposed the near-neutrality of modifications probably increases the evolvability of interaction networks. Structural principles, therefore, can be applied to proteomic studies to pinpoint only those modifications that are highly conserved and likely to be functional.
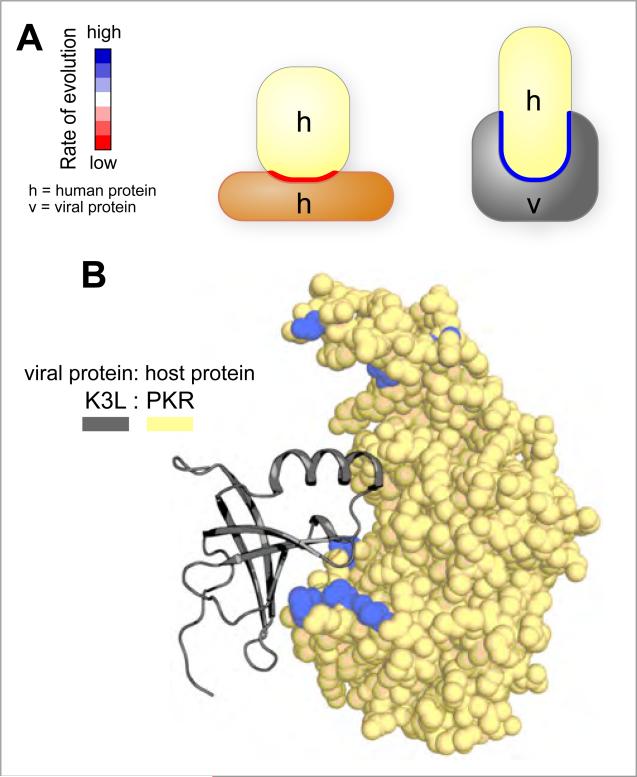
The recent construction of a human-viral structural interaction network revealed an exception to the principle that highly conserved surfaces are the most interesting regions (Franzosa and Xia, 2011). Consistent with the earlier work, these studies confirmed that interfaces involved in host protein-protein interactions evolve more slowly. However, interfaces exploited by pathogens tend to evolve much faster (Figure 2A). This result provides structural rationale for an evolutionary arms race between host and pathogen: there is selective pressure on host interfaces to evolve incompatibilities with their pathogen interaction partners. For example, the human kinase PKR has evolved under intense positive selection to avoid interactions with viral proteins such as poxvirus K3L, while maintaining interactions with endogenous interaction partners such as eIF2alpha (Elde et al., 2009) (Figure 2B). In the case of PKR, co-evolving mutations on multiple structural elements are likely required to decrease the affinity of binding the pathogenic protein while maintaining the proper connections to the host protein-protein interaction network.
Figure 2. Evolutionarily analysis of interfaces involved in mediating protein-protein interactions.
A) Regions of host proteins that are directly hijacked by viral factors are less conserved than interfaces that mediate interactions between host proteins. B) The human kinase PKR interacts with eIF2α and the viral protein K3L via a common surface. Residues coloured in blue line the interface region and display evidence for positive selection (elevated rates of amino acid substitutions). K3L (1LUZ) was docked to PKR based on a superposition of the PKR: eIF2α co crystal structure (2A1A).
The idea that co-evolution of interface residues across interacting proteins plays an important role in the evolution of network structure has also been tested in model systems. Using the yeast proliferating cell nuclear antigen (PCNA) DNA clamp as a model for studying protein-protein interaction network evolution, Aharoni and colleagues discovered that disrupting the delicate balance of affinities between different interaction partners can yield more severe phenotypes than gene deletions (Fridman et al., 2010). These studies suggest that large-scale genetic interaction studies, using deletion libraries or RNAi screens, should be supplemented with targeted mutations on specific components to tease apart the roles of multifunctional proteins. Because these mutations can, in principle, affect only one edge of a protein-protein interaction network, they have been termed “edgetic” mutations to contrast from gene deletions, which remove all edges associated with a given node (Amberg et al., 1995; Charloteaux et al., 2011). Similarly, structurally guided mutants could be used to dissect the roles of multifunctional macromolecular machines, such as RNA polymerase II (Braberg et al, submitted), which interact with different protein partners during complex functional cycles.
Mutations that tune the affinity of specific interactions or introduce new, unwanted interactions may have more severe phenotypic consequences than can be accessed with traditional gene knockouts. Mapping human disease-associated mutations onto structures has provided a structural rationale for pleiotropic genes, where distinct mutations are implicated in different diseases (Wang et al., 2012a). Mutations on a single interface are more commonly associated with a single disease, whereas mutations at different interfaces within the same protein tend to be associated with distinct diseases. Furthermore, mutations at interfaces with known disease implications provide a guilt-by-association indication for new genes, which may be more therapeutically tractable. For proteins with multiple domains or protein-protein interaction interfaces, these analyses can greatly narrow the scope of candidate genes to those most likely to be functionally relevant for the disease of interest. As both structural coverage of the human proteome and disease associated mutations discovered by exome sequencing studies increase, applying structural insights will likely increase the leverage of network-level observations to understand the genetic basis for disease (Aloy and Russell, 2003; Wang and Moult, 2001).
Making the complex whole: systems organize structure
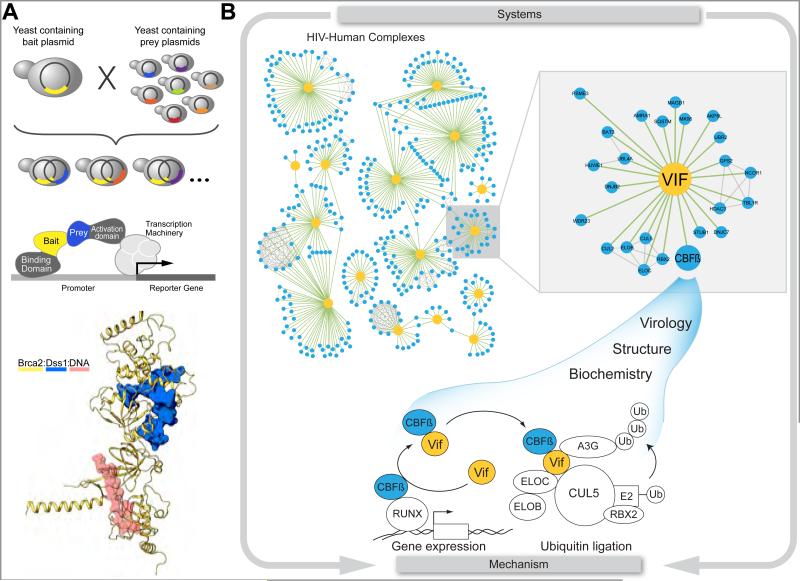
As structural biology shifts increasingly to characterizing complex, heterogeneous macromolecular assemblies, datasets generated from unbiased, global systems biology studies are playing prominent role. The influence of systems biology has already begun changing the normal course for identifying members of protein complexes from a “fishing trip” to a “mining expedition”. Because identifying all members of a protein complex is often a necessary pre-requisite for structural characterization, a common fishing approach is to use a yeast two-hybrid screen with single defined bait against a large, proteomic-scale prey library. An early example of how this approach can facilitate a deeper structural and mechanistic understanding of an important biological problem involves BRCA2, a DNA binding protein with important roles in breast cancer (Zheng et al., 2000). When structural studies of BRCA2 were initiated, baculovirus expression of BRCA2 in the absence of any interaction partners yielded insoluble protein (Yang et al., 2002). A two-hybrid screen based on cDNA derived from HeLa cells and mammary tissues identified DSS1 as a new interaction partner for BRCA2 (Marston et al., 1999). Co-expression of DSS1 and BRCA2 eventually yielded a co-crystal structure and the structure of a ternary complex with DNA demonstrating the importance of fishing out new interaction partners to enable structural studies (Yang et al., 2002) (Figure 3A). Interestingly, DSS1 has been shown to be part of multiple complexes, including an evolutionarily conserved component of the 26S proteasome (Wilmes et al., 2008).
Figure 3. Using unbiased systems approaches to solve, important biochemically intractable problems.
A) Use of a targeted two-hybrid approach for BRCA2 identified DSS1, which allowed for the structural elucidation (1MJE) of this important factor involved in breast cancer progression. B) An unbiased affinity tag-purification mass spectrometry approach on all HIV proteins revealed a connection between the accessory protein Vif and the human transcription factor CBF-β. This host protein, which is required for efficient HIV infection, allowed for the reconstitution and structural characterization of an active Vif-Cul-5 ubiquitination complex, which is required for the degradation of the host restriction factor APOBEC3G. The unbiased systems approach allowed for the discovery that by hijacking CBF-β, Vif is manipulating the ubquitination machinery and adversely effecting host transcriptional regulation.
In contrast to the BRCA2-focused yeast-two hybrid study that identified DSS1, most systems biology studies are performed with limited bias, systematically focusing on functionally related sets of proteins (Babu et al., 2012; Behrends et al., 2010; Krogan et al., 2004; Sowa et al., 2009) or even entire the entire proteome within an organism (Gavin et al., 2006; Havugimana et al., 2012; Hu et al., 2009; Krogan et al., 2006). These comprehensive approaches facilitate “mining expeditions” through a large database of interactions to identify multiprotein complexes that can be reconstituted biochemically and structurally (Figure 3B) (Aloy et al., 2004; Brooks et al., 2010). For example, in a recent global survey of the HIV proteome, Jager et al. identified interactions between human proteins and all 18 HIV-1 proteins and polyproteins (Jager et al., 2012a). The resulting 497 high confidence interactions provide many new opportunities to observe the atomic details of how a pathogen utilizes and subverts the normal function of host proteins. Examination of the interaction maps across two different cell lines revealed a surprising association of the transcription cofactor CBF-β, which heterodimerizes with Runx to regulate transcription in T-cells (Wong et al., 2011), with the HIV protein Vif. Although Vif was previously known to hijack an endogenous CUL-5 ubiquitin ligase complex to target for ubiquitination and degradation the host restriction factor APBOBEC3G (Yu et al., 2003), the architecture of this complex had remained elusive. Similar to DSS1 with BRCA2, co-expression of CBF-β provided the missing puzzle piece, enabled biochemical reconstitution and structural characterization of the Vif-Cul-5 complex (Jager et al., 2012b; Zhang et al., 2012b). Interestingly, Vif may be facilitating a dual hijack, one that affects a cullin-containing ubiquitination pathway as well as the rewiring of the transcriptional landscape regulated by the CBF-β -RUNX1 complex (Kim et al, Mol Cell, in press). Consistent with these results, a knockdown of CBF-β adversely affected HIV infectivity (Jager et al., 2012b; Zhang et al., 2012b). Future studies will investigate if and how both functional hijackings are required for efficient HIV infection. As further atomic-level details of this interaction emerge, it will likely serve as a paradigmatic example of how unexpected interaction partners can emerge from unbiased global studies to enable mechanistic studies.
Hybrid strategies that build on “-omics” techniques normally associated with systems biology are being used to uncover new binding partners and to accelerate biophysical studies for intrinsically disordered proteins (Dyson, 2011), macromolecular machines (Alber et al., 2007b) and transient complexes (Herzog et al., 2012). A particular challenge faced by these studies is to integrate and weight diverse data types into a self-consistent structural model (Alber et al., 2007a). For example, when performed on purified proteins, chemical cross-linking coupled with mass spectrometry provides distance restraints (Rappsilber, 2011). Crosslinking restraints can be incorporated into structure determination using well established methods similar to those used routinely in NMR (Havel et al., 1983); however, additional complications are encountered as crosslinking studies are extended to even more heterogeneous samples. In these studies, network context inferred from previous systems-level studies is essential for separating signals resulting from sterically incompatible complexes, allosteric distortion of individual subunits and entirely novel assemblies. The complexity of deconvoluting signals emerging from proteomic studies is exemplified by the Protein Phosphatase 2A (PP2A) holoenzymes, which consist of regulatory, catalytic, and scaffold subunits that provide combinatorial specificity to substrate proteins (Herzog et al., 2012). By purifying endogenous PP2A. Aebersold and colleagues revealed new players in the PP2A network, but also provided insights into the structural basis for how specific PP2A holoenzymes achieve interactions with a wide array of cellular processes. Proteomic-based crosslinking methods are a valuable source of structural data that can be obtained directly from heterogeneous mixtures and complement methods such as X-ray crystallography, NMR, and electron microscopy that rely on purified samples.
In addition to the natural links between structural biology and proteomics, other “-omics” technologies are beginning to generate interesting mechanistic problems. Synergies between genomics, metabolomics and structural studies are exemplified by an interesting mutation (R132H) in the metabolic enzyme isocitrate dehydrogenase-1 (IDH1). This mutation was first uncovered in a large genomics study, where it was present in 12% of Glioblastoma multiforme brain cancer patients (Parsons et al., 2008). Initial studies suggested that the mutation led to a dominant negative effect on the catalytic formation of the metabolite α-ketoglutarate (Zhao et al., 2009). Surprisingly, subsequent metabolomics profiling revealed no significant changes in levels of α-ketoglutarate between cells expressing mutant or wild type versions of the enzyme (Dang et al., 2009). However, levels of a different metabolite, R(-)-2-hydroxyglutarate, were significantly elevated. A crystal structure of the mutant enzyme revealed the key changes in active site geometry that enable this novel catalytic activity. The discovery of a new activity has motivated further systems-level studies into the role of R(-)-2-hydroxyglutarate in epigenetics (Sasaki et al., 2012) and the structural information provides additional hope that the mutant IDH1 enzyme represents a new target for therapeutic intervention in glioblastomas and acute myeloid leukaemias.
A new coupling of thermodynamics and genetic interactions
The examples discussed above represent only the initial cross-pollination between reductionist and global approaches. By recognizing the importance of non-additive interactions in genetics and biophysics, a common framework is being developed to more fully integrate systems and structural biology (Lehner, 2011). These interactions are commonly referred to as “epistatic” effects because the extent of the interaction depends on its genetic context (Phillips, 2008). In both genetics and biophysics, most perturbations act independently, yielding purely additive effects on fitness (Mani et al., 2008) or thermodynamic parameters (Wells, 1990). However, many functionally important perturbations may be benign in one context and deleterious in another (Cadwell et al., 2010). The non-additive effects of mutations may underlie “missing” genetic heritability - the limitation in our ability to explain functional consequences of sequence changes (Zuk et al., 2012). Exciting new efforts are underway to correlate genetic interaction measurements used in systems biology (Mani et al., 2008) with thermodynamic coupling analyses used in structural biology (Horovitz, 1996) to establish general principles of non-additivity and robustness that scale from systems to structure.
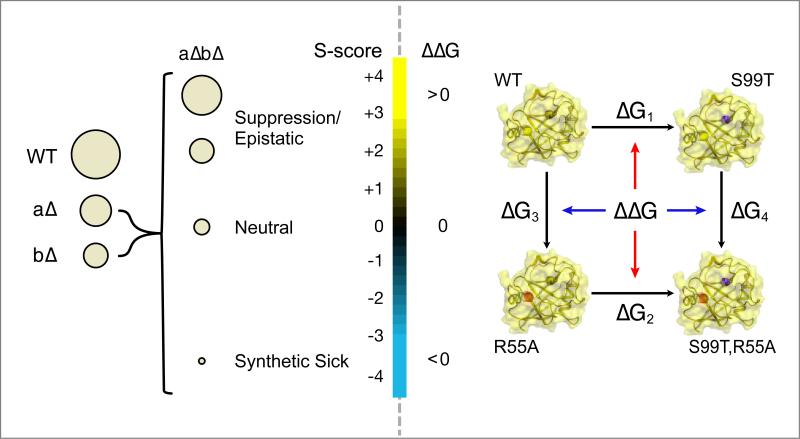
The most complete analyses of non-additive effects at a systems-level are from genetic interaction screens in the budding yeast, Saccharomyces cerevisiae. High throughput measurements, such as Synthetic Genetic Array (SGA) (Costanzo et al., 2010) or Epistatic Mini-Array Profile (E-MAP) experiments (Collins et al., 2010), compare quantitative measures of the fitness of individual gene knockouts to double knockouts (Figure 4). Positive genetic interactions occur when the fitness of the double mutant is higher than would be expected based on growth rates of the individual mutants. These alleviating interactions are enriched in gene products that physically interact or are part of a linear signaling pathway (Beltrao et al., 2010; Collins et al., 2007; Roguev et al., 2008). In contrast, cases where the double mutant is less fit than expected based on the individual mutations are termed negative genetic interactions. These aggravating interactions have been used to often identify proteins that function in parallel pathways of a given process (Tong et al., 2001). The extension of high-throughput genetic interaction methods to other organisms (Horn et al., 2011; Lin et al., 2012; Tischler et al., 2006; Typas et al., 2008) (Roguev et al., submitted) has created great interest in mining these datasets to generate combination therapies for human diseases. In general, genetic interactions are most conserved between species within protein complexes, less conserved within biological processes, and least conserved across distinct biological processes. This suggests a hierarchical modularity that governs how non-additive connections between proteins, complexes, and processes are functionally rewired and repurposed during evolution (Ryan et al., 2012). These genetic interaction approaches can extend across species to reveal dependencies between organisms in disease and symbiotic contexts (Fischbach and Krogan, 2010). A recent integration of insertional mutagenesis and depletion (iMAD) in the bacteria Legionella combined with RNAi in the host Drosophila melanogaster identified interesting cross-species synthetic lethal interactions that are relevant for pathogenesis (O'Connor et al., 2012). The fundamental principles of these genetic conflicts between species will likely be revealed by careful studies in classic host-pathogen systems such as bacteriophages and bacteria (Bondy-Denomy et al., 2012).
Figure 4. A common framework for understanding genetic and thermodynamic perturbations.
Non-additive effects are analyzed by similar methods and have similar distributions in a genetic (left) or biophysical context (right). For non-neutral genetic interactions and biophysical couplings, the effects of perturbations are context dependent. Quantitative genetic analysis distinguishes between negative ((a Δ b Δ) < (a Δ)(b Δ)), positive ((a Δ b Δ) > (a Δ)(b Δ)), and neutral ((a Δ b Δ) = (a Δ)(b Δ )) interactions based on the fitness of the organism, represented here by colony size. Biophysical studies measure the free energy of a process, for example protein folding, across different variants to distinguish between negative (Δ G3> Δ G4; Δ G1> Δ G2), positive (Δ G3< Δ G4; Δ G1< Δ G2), or the absence (Δ G3= Δ G4; Δ G1= Δ G2) of thermodynamic coupling (Δ Δ G).
The hierarchical design principles revealed by high-throughput genetic interaction studies have many parallels in the principles revealed by biophysical studies of protein function. Analysis of non-additive effects in biophysics studied in the context of protein folding (Horovitz and Fersht, 1990; Perry et al., 1989), enzyme mechanism (Carter et al., 1984), allostery (Sadovsky and Yifrach, 2007) and protein-ligand interactions (Baum et al., 2010; Schreiber and Fersht, 1995). As with genetic interaction studies, biophysical double mutant cycles reveal that most perturbations act independently (Wells, 1990) (Figure 4). However, both bioinformatics (Breen et al., 2012) and experimental (Bershtein et al., 2006; Tenaillon et al., 2012) studies suggest that non-additive (epistatic) effects play a dominant role in shaping the evolution of proteins. In the context of protein structure, non-additive effects of mutations often occur between residues that are in direct contact (Horovitz, 1987). For example, mutating two Alanine residues to form a new Lysine-Aspartate salt bridge results is only beneficial when both mutations are made as neither the Lysine-Alanine nor Aspartate-Alanine pair is likely to be stabilizing This “contact-based mechanism” of interactions parallels the positive genetic interactions observed between gene products that physically interact (Collins et al., 2007). A similar analogy of “parallel pathway mechanisms” exists between negative genetic interactions (Tong et al., 2001) and mutations that form new stabilizing intraprotein contacts or alternative intermolecular interaction binding modes. Promiscuous proteins with multiple binding modes may mask the negative effects of substitutions that only affect one interaction mechanism. Ancestral reconstruction of a nuclear hormone receptor revealed how two potential binding modes can be interconverted through conformational epistasis: where mutation at one site indirectly repositions the three-dimensional position of a remote second site and changes the functional effects of mutations at the second site (Ortlund et al., 2007). Similar to negative genetic interactions, conformational epistasis can lead to double mutations that affect both binding modes that are not apparent from the individual mutations.
However, despite general trends across atomic- and network-scales for positive and negative interactions, it remains difficult to predict the strength of non-additive interactions from network, structural, or evolutionary principles (Lehner, 2011). The simple contact-based or parallel pathway models can break down when long-range compensatory interactions occur (Lunzer et al., 2010), consistent with the fact that the correlation between direct protein-protein interactions and positive genetic interactions is not absolute, especially for complexes containing essential proteins (Costanzo et al., 2010). Mapping the genotype-phenotype-fitness relationship (Lunzer et al., 2005) can be additionally complicated by the ability of molecular chaperones to act as buffers that may enhance non-additive interactions as a protein nears its stability limit (Tokuriki and Tawfik, 2009) or a network is challenged by an external perturbation (Geller et al., 2007). To address the significance of these trends, comprehensive studies of protein mutations – on a scale similar to the genetic interaction studies – must be carried out. The advent of new sequencing methods now makes it possible to survey comprehensive sets of mutations under different selective pressures (Hietpas et al., 2011). One recent survey of all possible mutations in a PDZ domain revealed a strongly co-evolving set of amino acids within which two mutations act non-additively to change peptide binding specificity (McLaughlin Jr et al., 2012). Studies on this newly accessible scale have the potential to reveal new principles of non-additivity underlying protein stability and function (Araya et al., 2012). The extension of these principles to the co-evolution of protein-protein interactions will address the similarities in molecular mechanisms underlying epistatic interactions between and within individual gene products (Fridman et al., 2010; Skerker et al., 2008). Therefore, quantitative measures that intrinsically calibrate thermodynamic quantities to fitness measurements present new opportunities to discover unifying principles of buffering, non-additivity and adaptation across the atomic and network scales.
Looking dynamically forward
As these more comprehensive surveys of protein sequence and genetic interaction space continue to emerge, an additional challenge will be to adapt the common analytical models used in both systems and structural biology to include the fourth dimension: time. Much of the work we have highlighted here has focused on integrating multiple static representations, as exemplified by incorporating crystal structure information in a protein-protein interaction network diagram. Recently, structural interaction networks have been adapted to incorporate multiple protein conformations (Bhardwaj et al., 2011). Disease mutations that have only a minor effect on the dominant structure of a protein can exert a deleterious function by altering the ability to dynamically switch between near-native conformations (Shan et al., 2012). Indeed, mutations remote (Fraser et al., 2009; Wang et al., 2012b) and within (Bhabha et al., 2011) the active sites of enzymes can have large functional consequences without dramatically altering protein structure. Conservation of protein motions have been proposed to be a major constraint in protein evolution and may provide a mechanistic explanation for non-additive effects between residues that are not in direct contact (Reynolds et al., 2011). Microsecond-millisecond protein conformational dynamics can be exploited in the development of new protein therapeutics (Levin et al., 2012) and tuning the biophysical properties of small molecule inhibitors (Carroll et al., 2012).
At longer timescales, measuring the dynamics of non-additive responses in network contexts will also likely reveal new principles of modularity (Alexander et al., 2009). Indeed, profiling the changes in protein-protein interactions of the scaffold protein GRB2 after perturbations designed to activate different receptor tyrosine kinases revealed that the time-dependent rewiring and elaboration of new complexes centered around a conserved set of core interactions (Bisson et al., 2011). A comparison of genetic interactions in yeast in the presence and absence of DNA damaging agents revealed that although relationships within protein complexes are largely conserved, there is substantial rewiring in the functional relationships between complexes (Bandyopadhyay et al., 2010). These results suggest that the concept of a network “module” may become more diffuse as a time dimension and additional perturbations are incorporated. Similar to genetic interactions, current efforts are underway to discover non-additive effects in drug combinations (Yeh et al., 2009). Multiplex profiling of approved and clinically tested drugs uncovered new interactions that synergize to inhibit HIV replication (Tan et al., 2012). Varying the timing and dynamics of different perturbations may discover additional synergistic combinations. Methods, such as mass cytometry, that can dynamically read out the response of the cell across many parameters simultaneously can be used to guide the optimization of perturbation dynamics (Bodenmiller et al., 2012). Indeed, new therapeutic regimens are currently being developed around non-additive effects on the order and time course of drug combinations by exploiting network level principles (Lee et al., 2012). This study found that EGFR inhibition was ineffective when combined simultaneously with genotoxic drugs; however, staggered administration of the two perturbations dynamically rewires the cells and leading to a more efficient potential therapy. Collectively, these studies point to emerging parallels of the role of non-additivity on the sub-second timescale in protein dynamics, on the hours-weeks timescale in the cellular response to perturbations, and on the million-year timescale in the adaptations that shape interaction networks (Ideker and Krogan, 2012).
In this review, we highlighted how the use of systems and structural biological approaches are truly synergistic when used in combination to understand complex biological phenomenon. For example, unbiased, systems approaches can identify key components that facilitate structural analysis whereas structural information can provide insight into and even guide large-scale biological studies. Furthermore, we argue that there are striking similarities between the framework used to interpret quantitative genetic interactions and thermodynamic responses. Finally, detailed “small-scale” systems and synthetic biology efforts, which examine network motifs and quantitatively model signaling networks, are complimentary to the large-scale studies since they can be used to define core interactions and network principles (see review by Lim et al. in this issue). A challenge for future work will be to determine and integrate the quantitative relationships that exist at these different levels, including cell biology (Slack et al., 2008), pharmacology (Lounkine et al., 2012), and ecology (Hekstra and Leibler, 2012). In the near future, we expect methods that explicitly investigate these parallels from “systems to structure” will play an increasingly larger role in efforts to predict the phenotypic response to mutation, small molecules, pathogens, and other perturbations.
Acknowledgements
We thank Michael Shales for help preparing figures. JSF was supported by NIH (DP5OD009180) and QB3. JDG was supported by NIH (GM081879 and GM078360). NJK was supported by NIH (GM084448, GM082250, GM084279, GM081879, GM098101, AI090935, AI091575) and DARPA (HR0011-11-C-0094). NJK is a Searle Scholar and a Keck Young Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. Determining the architectures of macromolecular assemblies. Nature. 2007a;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007b;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Alexander RP, Kim PM, Emonet T, Gerstein MB. Understanding modularity in molecular networks requires dynamics. Sci Signal. 2009;2:e44. doi: 10.1126/scisignal.281pe44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloy P, Bottcher B, Ceulemans H, Leutwein C, Mellwig C, Fischer S, Gavin AC, Bork P, Superti-Furga G, Serrano L, et al. Structure-based assembly of protein complexes in yeast. Science. 2004;303:2026–2029. doi: 10.1126/science.1092645. [DOI] [PubMed] [Google Scholar]
- Aloy P, Russell RB. Understanding and predicting protein assemblies with 3D structures. Comp Funct Genomics. 2003;4:410–415. doi: 10.1002/cfg.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg DC, Basart E, Botstein D. Defining protein interactions with yeast actin in vivo. Nat Struct Biol. 1995;2:28–35. doi: 10.1038/nsb0195-28. [DOI] [PubMed] [Google Scholar]
- Araya CL, Fowler DM, Chen W, Muniez I, Kelly JW, Fields S. A fundamental protein property, thermodynamic stability, revealed solely from large-scale measurements of protein function. Proc Natl Acad Sci U S A. 2012;109:16858–16863. doi: 10.1073/pnas.1209751109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu M, Vlasblom J, Pu S, Guo X, Graham C, Bean BD, Burston HE, Vizeacoumar FJ, Snider J, Phanse S, et al. Interaction landscape of membrane-protein complexes in Saccharomyces cerevisiae. Nature. 2012;489:585–589. doi: 10.1038/nature11354. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S, Mehta M, Kuo D, Sung MK, Chuang R, Jaehnig EJ, Bodenmiller B, Licon K, Copeland W, Shales M, et al. Rewiring of genetic networks in response to DNA damage. Science. 2010;330:1385–1389. doi: 10.1126/science.1195618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum B, Muley L, Smolinski M, Heine A, Hangauer D, Klebe G. Non-additivity of functional group contributions in protein-ligand binding: a comprehensive study by crystallography and isothermal titration calorimetry. J Mol Biol. 2010;397:1042–1054. doi: 10.1016/j.jmb.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrao P, Albanese V, Kenner LR, Swaney DL, Burlingame A, Villen J, Lim WA, Fraser JS, Frydman J, Krogan NJ. Systematic functional prioritization of protein posttranslational modifications. Cell. 2012;150:413–425. doi: 10.1016/j.cell.2012.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrao P, Cagney G, Krogan NJ. Quantitative genetic interactions reveal biological modularity. Cell. 2010;141:739–745. doi: 10.1016/j.cell.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bershtein S, Segal M, Bekerman R, Tokuriki N, Tawfik DS. Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature. 2006;444:929–932. doi: 10.1038/nature05385. [DOI] [PubMed] [Google Scholar]
- Bhabha G, Lee J, Ekiert DC, Gam J, Wilson IA, Dyson HJ, Benkovic SJ, Wright PE. A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis. Science. 2011;332:234–238. doi: 10.1126/science.1198542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj N, Abyzov A, Clarke D, Shou C, Gerstein MB. Integration of protein motions with molecular networks reveals different mechanisms for permanent and transient interactions. Protein Sci. 2011;20:1745–1754. doi: 10.1002/pro.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson N, James DA, Ivosev G, Tate SA, Bonner R, Taylor L, Pawson T. Selected reaction monitoring mass spectrometry reveals the dynamics of signaling through the GRB2 adaptor. Nat Biotechnol. 2011;29:653–658. doi: 10.1038/nbt.1905. [DOI] [PubMed] [Google Scholar]
- Bodenmiller B, Zunder ER, Finck R, Chen TJ, Savig ES, Bruggner RV, Simonds EF, Bendall SC, Sachs K, Krutzik PO, et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat Biotechnol. 2012 doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2012 doi: 10.1038/nature11723. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen MS, Kemena C, Vlasov PK, Notredame C, Kondrashov FA. Epistasis as the primary factor in molecular evolution. Nature. 2012 doi: 10.1038/nature11510. [DOI] [PubMed] [Google Scholar]
- Brooks MA, Gewartowski K, Mitsiki E, Letoquart J, Pache RA, Billier Y, Bertero M, Correa M, Czarnocki-Cieciura M, Dadlez M, et al. Systematic bioinformatics and experimental validation of yeast complexes reduces the rate of attrition during structural investigations. Structure. 2010;18:1075–1082. doi: 10.1016/j.str.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll MJ, Mauldin RV, Gromova AV, Singleton SF, Collins EJ, Lee AL. Evidence for dynamics in proteins as a mechanism for ligand dissociation. Nat Chem Biol. 2012;8:246–252. doi: 10.1038/nchembio.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter PJ, Winter G, Wilkinson AJ, Fersht AR. The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus). Cell. 1984;38:835–840. doi: 10.1016/0092-8674(84)90278-2. [DOI] [PubMed] [Google Scholar]
- Charloteaux B, Zhong Q, Dreze M, Cusick ME, Hill DE, Vidal M. Protein-protein interactions and networks: forward and reverse edgetics. Methods Mol Biol. 2011;759:197–213. doi: 10.1007/978-1-61779-173-4_12. [DOI] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Collins SR, Roguev A, Krogan NJ. Quantitative genetic interaction mapping using the E-MAP approach. Methods Enzymol. 2010;470:205–231. doi: 10.1016/S0076-6879(10)70009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson HJ. Expanding the proteome: disordered and alternatively folded proteins. Q Rev Biophys. 2011;44:467–518. doi: 10.1017/S0033583511000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elde NC, Child SJ, Geballe AP, Malik HS. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature. 2009;457:485–489. doi: 10.1038/nature07529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, Krogan NJ. The next frontier of systems biology: higher-order and interspecies interactions. Genome Biol. 2010;11:208. doi: 10.1186/gb-2010-11-5-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa EA, Xia Y. Structural principles within the human-virus protein-protein interaction network. Proc Natl Acad Sci U S A. 2011;108:10538–10543. doi: 10.1073/pnas.1101440108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JS, Clarkson MW, Degnan SC, Erion R, Kern D, Alber T. Hidden alternative structures of proline isomerase essential for catalysis. Nature. 2009;462:669–673. doi: 10.1038/nature08615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman Y, Palgi N, Dovrat D, Ben-Aroya S, Hieter P, Aharoni A. Subtle alterations in PCNA-partner interactions severely impair DNA replication and repair. PLoS Biol. 2010;8:e1000507. doi: 10.1371/journal.pbio.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Geller R, Vignuzzi M, Andino R, Frydman J. Evolutionary constraints on chaperone-mediated folding provide an antiviral approach refractory to development of drug resistance. Genes Dev. 2007;21:195–205. doi: 10.1101/gad.1505307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havel TF, Kuntz ID, Crippen GM. The combinatorial distance geometry method for the calculation of molecular conformation. I. A new approach to an old problem. J Theor Biol. 1983;104:359–381. doi: 10.1016/0022-5193(83)90112-1. [DOI] [PubMed] [Google Scholar]
- Havugimana PC, Hart GT, Nepusz T, Yang H, Turinsky AL, Li Z, Wang PI, Boutz DR, Fong V, Phanse S, et al. A census of human soluble protein complexes. Cell. 2012;150:1068–1081. doi: 10.1016/j.cell.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekstra DR, Leibler S. Contingency and statistical laws in replicate microbial closed ecosystems. Cell. 2012;149:1164–1173. doi: 10.1016/j.cell.2012.03.040. [DOI] [PubMed] [Google Scholar]
- Herzog F, Kahraman A, Boehringer D, Mak R, Bracher A, Walzthoeni T, Leitner A, Beck M, Hartl FU, Ban N, et al. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science. 2012;337:1348–1352. doi: 10.1126/science.1221483. [DOI] [PubMed] [Google Scholar]
- Hietpas RT, Jensen JD, Bolon DN. Experimental illumination of a fitness landscape. Proc Natl Acad Sci U S A. 2011;108:7896–7901. doi: 10.1073/pnas.1016024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn T, Sandmann T, Fischer B, Axelsson E, Huber W, Boutros M. Mapping of signaling networks through synthetic genetic interaction analysis by RNAi. Nat Methods. 2011;8:341–346. doi: 10.1038/nmeth.1581. [DOI] [PubMed] [Google Scholar]
- Horovitz A. Non-additivity in protein-protein interactions. J Mol Biol. 1987;196:733–735. doi: 10.1016/0022-2836(87)90045-3. [DOI] [PubMed] [Google Scholar]
- Horovitz A. Double-mutant cycles: a powerful tool for analyzing protein structure and function. Fold Des. 1996;1:R121–126. doi: 10.1016/S1359-0278(96)00056-9. [DOI] [PubMed] [Google Scholar]
- Horovitz A, Fersht AR. Strategy for analysing the co-operativity of intramolecular interactions in peptides and proteins. J Mol Biol. 1990;214:613–617. doi: 10.1016/0022-2836(90)90275-Q. [DOI] [PubMed] [Google Scholar]
- Hu P, Janga SC, Babu M, Diaz-Mejia JJ, Butland G, Yang W, Pogoutse O, Guo X, Phanse S, Wong P, et al. Global functional atlas of Escherichia coli encompassing previously uncharacterized proteins. PLoS Biol. 2009;7:e96. doi: 10.1371/journal.pbio.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideker T, Krogan NJ. Differential network biology. Mol Syst Biol. 2012;8:565. doi: 10.1038/msb.2011.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, et al. Global landscape of HIV-human protein complexes. Nature. 2012a;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager S, Kim DY, Hultquist JF, Shindo K, LaRue RS, Kwon E, Li M, Anderson BD, Yen L, Stanley D, et al. Vif hijacks CBF-beta to degrade APOBEC3G and promote HIV-1 infection. Nature. 2012b;481:371–375. doi: 10.1038/nature10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PM, Lu LJ, Xia Y, Gerstein MB. Relating three-dimensional structures to protein networks provides evolutionary insights. Science. 2006;314:1938–1941. doi: 10.1126/science.1136174. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470:187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- Lee MJ, Ye AS, Gardino AK, Heijink AM, Sorger PK, Macbeath G, Yaffe MB. Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell. 2012;149:780–794. doi: 10.1016/j.cell.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B. Molecular mechanisms of epistasis within and between genes. Trends Genet. 2011;27:323–331. doi: 10.1016/j.tig.2011.05.007. [DOI] [PubMed] [Google Scholar]
- Levin AM, Bates DL, Ring AM, Krieg C, Lin JT, Su L, Moraga I, Raeber ME, Bowman GR, Novick P, et al. Exploiting a natural conformational switch to engineer an interleukin-2 ‘superkine’. Nature. 2012;484:529–533. doi: 10.1038/nature10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YY, Kiihl S, Suhail Y, Liu SY, Chou YH, Kuang Z, Lu JY, Khor CN, Lin CL, Bader JS, et al. Functional dissection of lysine deacetylases reveals that HDAC1 and p300 regulate AMPK. Nature. 2012;482:251–255. doi: 10.1038/nature10804. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lounkine E, Keiser MJ, Whitebread S, Mikhailov D, Hamon J, Jenkins JL, Lavan P, Weber E, Doak AK, Cote S, et al. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486:361–367. doi: 10.1038/nature11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunzer M, Golding GB, Dean AM. Pervasive cryptic epistasis in molecular evolution. PLoS Genet. 2010;6:e1001162. doi: 10.1371/journal.pgen.1001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunzer M, Miller SP, Felsheim R, Dean AM. The biochemical architecture of an ancient adaptive landscape. Science. 2005;310:499–501. doi: 10.1126/science.1115649. [DOI] [PubMed] [Google Scholar]
- Mani R, St Onge RP, Hartman J.L.t., Giaever G, Roth FP. Defining genetic interaction. Proc Natl Acad Sci U S A. 2008;105:3461–3466. doi: 10.1073/pnas.0712255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston NJ, Richards WJ, Hughes D, Bertwistle D, Marshall CJ, Ashworth A. Interaction between the product of the breast cancer susceptibility gene BRCA2 and DSS1, a protein functionally conserved from yeast to mammals. Mol Cell Biol. 1999;19:4633–4642. doi: 10.1128/mcb.19.7.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin RN, Jr, Poelwijk FJ, Raman A, Gosal WS, Ranganathan R. The spatial architecture of protein function and adaptation. Nature. 2012 doi: 10.1038/nature11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor TJ, Boyd D, Dorer MS, Isberg RR. Aggravating genetic interactions allow a solution to redundancy in a bacterial pathogen. Science. 2012;338:1440–1444. doi: 10.1126/science.1229556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW. Crystal structure of an ancient protein: evolution by conformational epistasis. Science. 2007;317:1544–1548. doi: 10.1126/science.1142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry KM, Onuffer JJ, Gittelman MS, Barmat L, Matthews CR. Long-range electrostatic interactions can influence the folding, stability, and cooperativity of dihydrofolate reductase. Biochemistry. 1989;28:7961–7968. doi: 10.1021/bi00445a061. [DOI] [PubMed] [Google Scholar]
- Phillips PC. Epistasis--the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet. 2008;9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper U, Webb BM, Barkan DT, Schneidman-Duhovny D, Schlessinger A, Braberg H, Yang Z, Meng EC, Pettersen EF, Huang CC, et al. ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 2011;39:D465–474. doi: 10.1093/nar/gkq1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J. The beginning of a beautiful friendship: cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J Struct Biol. 2011;173:530–540. doi: 10.1016/j.jsb.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds KA, McLaughlin RN, Ranganathan R. Hot spots for allosteric regulation on protein surfaces. Cell. 2011;147:1564–1575. doi: 10.1016/j.cell.2011.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Bandyopadhyay S, Zofall M, Zhang K, Fischer T, Collins SR, Qu H, Shales M, Park HO, Hayles J, et al. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science. 2008;322:405–410. doi: 10.1126/science.1162609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlic A, Quesada M, Quinn GB, Westbrook JD, et al. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CJ, Roguev A, Patrick K, Xu J, Jahari H, Tong Z, Beltrao P, Shales M, Qu H, Collins SR, et al. Hierarchical modularity and the evolution of genetic interactomes across species. Mol Cell. 2012;46:691–704. doi: 10.1016/j.molcel.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadovsky E, Yifrach O. Principles underlying energetic coupling along an allosteric communication trajectory of a voltage-activated K+ channel. Proc Natl Acad Sci U S A. 2007;104:19813–19818. doi: 10.1073/pnas.0708120104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brustle A, Harris IS, Holmes R, Wakeham A, Haight J, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488:656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber G, Fersht AR. Energetics of protein-protein interactions: analysis of the barnase-barstar interface by single mutations and double mutant cycles. J Mol Biol. 1995;248:478–486. doi: 10.1016/s0022-2836(95)80064-6. [DOI] [PubMed] [Google Scholar]
- Shan Y, Eastwood MP, Zhang X, Kim ET, Arkhipov A, Dror RO, Jumper J, Kuriyan J, Shaw DE. Oncogenic Mutations Counteract Intrinsic Disorder in the EGFR Kinase and Promote Receptor Dimerization. Cell. 2012;149:860–870. doi: 10.1016/j.cell.2012.02.063. [DOI] [PubMed] [Google Scholar]
- Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack MD, Martinez ED, Wu LF, Altschuler SJ. Characterizing heterogeneous cellular responses to perturbations. Proc Natl Acad Sci U S A. 2008;105:19306–19311. doi: 10.1073/pnas.0807038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Hu L, Luquette LJ, 3rd, Gao G, Liu Y, Qu H, Xi R, Lu ZJ, Park PJ, Elledge SJ. Systematic identification of synergistic drug pairs targeting HIV. Nat Biotechnol. 2012 doi: 10.1038/nbt.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O, Rodriguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD, Gaut BS. The molecular diversity of adaptive convergence. Science. 2012;335:457–461. doi: 10.1126/science.1212986. [DOI] [PubMed] [Google Scholar]
- Tischler J, Lehner B, Chen N, Fraser AG. Combinatorial RNA interference in Caenorhabditis elegans reveals that redundancy between gene duplicates can be maintained for more than 80 million years of evolution. Genome Biol. 2006;7:R69. doi: 10.1186/gb-2006-7-8-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature. 2009;459:668–673. doi: 10.1038/nature08009. [DOI] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Typas A, Nichols RJ, Siegele DA, Shales M, Collins SR, Lim B, Braberg H, Yamamoto N, Takeuchi R, Wanner BL, et al. High-throughput, quantitative analyses of genetic interactions in E. coli. Nat Methods. 2008;5:781–787. doi: 10.1038/nmeth.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wei X, Thijssen B, Das J, Lipkin SM, Yu H. Three-dimensional reconstruction of protein networks provides insight into human genetic disease. Nat Biotechnol. 2012a;30:159–164. doi: 10.1038/nbt.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Abeysinghe T, Finer-Moore JS, Stroud RM, Kohen A. A remote mutation affects the hydride transfer by disrupting concerted protein motions in thymidylate synthase. J Am Chem Soc. 2012b;134:17722–17730. doi: 10.1021/ja307859m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Moult J. SNPs, protein structure, and disease. Hum Mutat. 2001;17:263–270. doi: 10.1002/humu.22. [DOI] [PubMed] [Google Scholar]
- Wells JA. Additivity of mutational effects in proteins. Biochemistry. 1990;29:8509–8517. doi: 10.1021/bi00489a001. [DOI] [PubMed] [Google Scholar]
- Wilmes GM, Bergkessel M, Bandyopadhyay S, Shales M, Braberg H, Cagney G, Collins SR, Whitworth GB, Kress TL, Weissman JS, et al. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol Cell. 2008;32:735–746. doi: 10.1016/j.molcel.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittinghofer A, Vetter IR. Structure-function relationships of the G domain, a canonical switch motif. Annu Rev Biochem. 2011;80:943–971. doi: 10.1146/annurev-biochem-062708-134043. [DOI] [PubMed] [Google Scholar]
- Wong WF, Kohu K, Chiba T, Sato T, Satake M. Interplay of transcription factors in T-cell differentiation and function: the role of Runx. Immunology. 2011;132:157–164. doi: 10.1111/j.1365-2567.2010.03381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- Yeh PJ, Hegreness MJ, Aiden AP, Kishony R. Drug interactions and the evolution of antibiotic resistance. Nat Rev Microbiol. 2009;7:460–466. doi: 10.1038/nrmicro2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- Zhang QC, Petrey D, Deng L, Qiang L, Shi Y, Thu CA, Bisikirska B, Lefebvre C, Accili D, Hunter T, et al. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature. 2012a doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Du J, Evans SL, Yu Y, Yu XF. T-cell differentiation factor CBF-beta regulates HIV-1 Vif-mediated evasion of host restriction. Nature. 2012b;481:376–379. doi: 10.1038/nature10718. [DOI] [PubMed] [Google Scholar]
- Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, Yu W, Li Z, Gong L, Peng Y, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Li S, Boyer TG, Lee WH. Lessons learned from BRCA1 and BRCA2. Oncogene. 2000;19:6159–6175. doi: 10.1038/sj.onc.1203968. [DOI] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc Natl Acad Sci U S A. 2012;109:1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]