Abstract
Objectives
Neuropeptide Y (NPY) is a peptide involved in the regulation of appetite and energy homeostasis. Genetic data indicates that NPY decreases bone formation via central and peripheral activities. NPY is produced by various cell types including osteocytes and osteoblasts and there is evidence suggesting that peripheral NPY is important for regulation of bone formation. We sought to investigate the role of bone-derived NPY in bone metabolism.
Methods
We generated a mouse where NPY was over-expressed specifically in mature osteoblasts and osteocytes (Col2.3NPY) and characterized the bone phenotype of these mice in vivo and in vitro.
Results
Trabecular and cortical bone volume was reduced in 3-month-old animals, however bone formation rate and osteoclast activity were not significantly changed. Calvarial osteoblast cultures from Col2.3NPY mice also showed reduced mineralization and expression of osteogenic marker genes.
Conclusions
Our data suggest that osteoblast/osteocyte-derived NPY is capable of altering osteogenesis in vivo and in vitro and may represent an important source of NPY for regulation of bone formation. However, it is possible that other peripheral sources of NPY such as the sympathetic nervous system and vasculature also contribute to peripheral regulation of bone turnover.
Keywords: neuropeptide Y, osteoblast, osteocyte, Col2.3 promoter, osteogenesis
INTRODUCTION
Neuropeptide Y (NPY) is a neuronal regulator of energy homeostasis and has been identified as an important molecule involved in central-hypothalamic control of bone mass. NPY is an orexigenic peptide, and when its levels are increased in the hypothalamus of mice, it leads to increased food intake, weight gain and obesity [1, 2]. During starvation, when hypothalamic NPY levels are high, bone formation is reduced in order to save energy [1]. NPY is a 36-amino acid polypeptide and together with pancreatic polypeptide (PP) and peptide YY belongs to the pancreatic polypeptide hormone family [3, 4]. NPY signals through five different G-protein-coupled receptors: Y1, Y2, Y4, Y5 and Y6. Receptors Y1, Y2 and Y5 are found in the central nervous system, including the hypothalamus [5, 6]. Y1 mRNA has also been found in numerous peripheral tissues, including kidney, heart, lungs, bone marrow and bone [7]. Y1, but none of the other Y receptors are expressed in osteoblast lineage cells [8, 9]. However, Y2 receptor has been detected in the osteoblastic MC3T3 cell line [10].
A number of studies have investigated the central interaction of NPY with Y1 and Y2 receptors. Germline Y2 receptor deletion, or targeted deletion of Y2 in the hypothalamus resulted in almost identical phenotypes with increases in trabecular bone volume, due to elevated bone formation rate, demonstrating the importance of the Y2 receptor in the central actions of NPY with respect to bone mass [11, 12]. In addition, Y1 knockout mice showed increased cortical and trabecular bone mass and elevated bone formation, as well as a metabolic phenotype including increased adiposity and hyperinsulinemia [13–15]. However, targeted deletion of the Y1 receptor in the hypothalamus did not have any effect on bone mass [8]. Osteoblast-specific Y1 receptor deletion resulted in increased bone mass due to elevated osteoblast activity, without the metabolic perturbations observed in the global Y1 knockout [16]. This demonstrates a direct role for the Y1 receptor in osteoblast lineage cells and their activity and confirms that signaling through the Y1 receptor affects bone formation independently of central hypothalamic regulation.
NPY is expressed in numerous brain regions [17] but also in peripheral tissues such as liver, heart, spleen, endothelial cells of blood vessels and the sympathetic nervous system [18]. NPY is also expressed within bone tissue by preosteocytes/osteocytes and at lower levels in osteoblasts [9, 19]. In vitro studies have shown that NPY has a direct effect on the osteoblast lineage by inhibiting the differentiation of mesenchymal progenitors and mineral production by mature osteoblasts [9, 20].
Further studies are required to understand whether the effects of Y1 receptor signaling in the osteoblast lineage result from the effects of locally or centrally produced NPY. So far, targeted deletion of the NPY gene in bone or hypothalamus has not been achieved. Therefore, we have developed a mouse model of osteoblast lineage specific NPY overexpression using the 2.3kb fragment of the type I collagen (Col2.3) promoter to investigate the role of locally expressed NPY on osteogenesis. Here, we report the effects of bone-specific NPY overexpression on bone phenotype in vivo and in ex vivo primary osteoblast cultures.
MATERIALS AND METHODS
Generation of Col2.3NPY mice
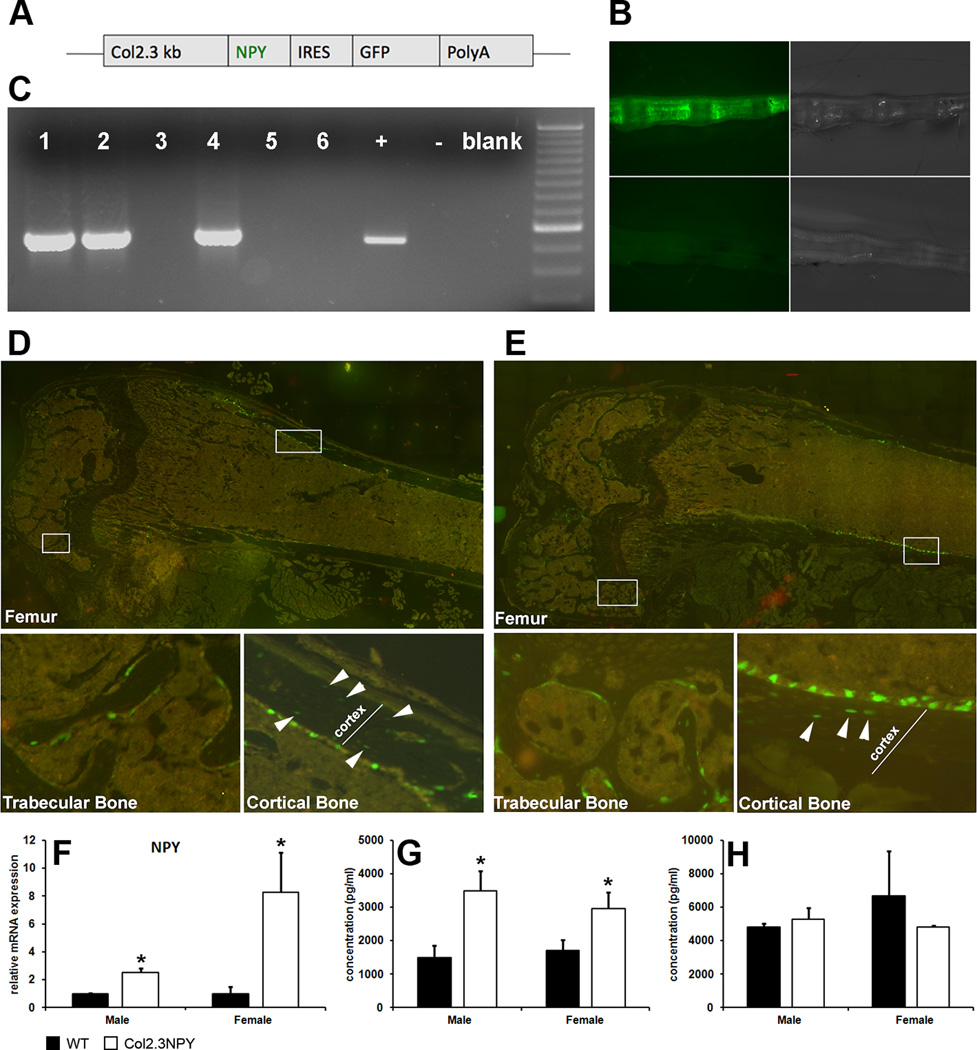
The 300-bp preproNPY cDNA was excised from the plasmid containing the DβH-NPY-IRES-LacZ transgene construct [21] with NsiI and inserted in an NsiI linearized plasmid (pLITIMUS 29) upstream of a viral 580-bp internal ribosomal entry site (IRES) and GFPtopaz sequence followed by a bovine growth hormone polyadenylation sequence (bGHpolyA). After the unique SalI restriction site had been removed, correct orientation of the NPY cDNA was confirmed using KpnI restriction digestion. The bicistronic NPY-IRES-GFPtopaz-bHGpolyA construct was inserted downstream of a 2.3-kb fragment of the rat collagen promoter (Col2.3) using NheI and XhoI. Thus, the Col2.3 promoter drives the expression of NPY and GFPtopaz genes (Figure 1A). The final 5.4 kb transgene construct was excised from the vector using SalI and microinjected into fertilized eggs at UCHC institutional gene targeting and transgenic facility and the oocytes were transferred to pseudopregnant C57BL6 females (Jackson Laboratories, Bar Harbor, ME, USA). Two transgenic lines were obtained and their expression characterized. Mice were genotyped by assessing GFPtopaz expression in tail vertebrae using fluorescent microscopy (Figure 1B). Genotypes were confirmed by PCR (forward primer: 5’-TCA TCT GCA CCA CCG GCA AGC-3’; reverse primer: 5’-AGC AGG ACC ATG TGA TCG CGC-3’) to amplify a 525 bp fragment of the GFPtopaz transgene (Figure 1C). PCR was performed with 35 cycles of 94°C for 30 s, 65°C for 30 s and 72°C for 40 s. Wild type littermates were used as experimental controls. Mice were sacrificed using CO2 asphyxiation, and procedures involving animals were approved by an institutional review board.
Fig. 1. Genotyping and confirmation of NPY overexpression Col2.3NPY mice.
Col2.3NPY mice have been developed where NPY and GFP expression are driven by the 2.3-kb fragment of collagen type I promoter (A). GFP fluorescence of tail vertebrae (B) is used to distinguish Col2.3NPY (upper panels) and wild type (lower panels) mice. PCR for GFP confirmed Col2.3NPY genotype (C). Histology shows GFPtopaz expression in a subset of osteoblasts and osteocytes (indicated by arrowheads) in both females (D) and males (E). Expression of NPY mRNA in bone tissue of 2–3 month old mice was measured using qPCR (F), and NPY protein concentration in bone tissue (G) and serum (H) was quantified using EIA.
Measurement of NPY protein concentration
Serum was collected from 3-month-old Col2.3NPY mice and their wild type littermates. Proteins were extracted from pooled tibias and femurs that had muscle and connective tissue removed, and bone marrow flushed. Bones were frozen in liquid nitrogen, powdered with a mortar and pestle and proteins were extracted using RIPA buffer containing Halt Protease Inhibitor Cocktail (Thermo Scientific). NPY levels were measured using a Human Neuropeptide Y EIA Kit (RayBiotech, Inc.) according to the manufacturer’s instructions. Serums, diluted 1:2 in the assay buffer, or 85 µg of protein extract were analyzed.
RNA extraction and analysis of gene expression
Total RNA was isolated from osteoblast cultures using TRIzol reagent (Life Technologies) according to the manufacturer’s instructions. Tibias and femurs from 2–3 month-old mice were excised, cleaned and bone marrow was flushed. Bones were snap frozen in liquid nitrogen and homogenised in TRIzol reagent using an ULTRA-TURRAX Basic Homogenizer (IKA). RNA was subjected to DNase treatment (Life Technologies) and 1µg of total RNA was used for cDNA synthesis using the Superscript III First-strand Synthesis System (Life Technologies). TaqMan® Gene expression Assays (Life Technologies) were used to analyze the expression of Npy (Mm00445771_m1), NPY-Y1 receptor (Mm00650798_g1), osteocalcin (OC, Mm03413826_mH), dentin matrix protein 1 (Dmp1, Mm00803833_g1) and bone sialoprotein (BSP, Mm00492355_m1) and quantitative real-time PCR (qPCR) was performed on the 7900HT Real-Time PCR System. GAPDH expression (Mm99999915_g1) was used as an internal standard. Relative expression of target genes was calculated using the ∆∆Ct method (RQ manager 1.2, Life Technologies). Expression in wild type samples was normalized to 1.
Micro computed tomography (µCT)
Femoral morphometry of 3-month-old mice (10–11/group) was assessed using conebeam micro-focus X-ray computed tomography (µCT40, Scanco Medical AG). Serial tomographic images were acquired at 55 kV and 145 µA, collecting 1000 projections per rotation at 300 ms integration time. Three-dimensional 16-bit grayscale images were reconstructed using standard convolution back-projection algorithms with Shepp and Logan filtering, and rendered within a 12.3 mm field of view at a discrete density of 578,704 voxels/mm3 (isometric 12-µm voxels). Segmentation of bone from marrow and soft tissue was performed in conjunction with a constrained Gaussian filter to reduce noise, applying calibrated hydroxyapatite-equivalent density thresholds of 495 and 740 mg/cm3 for trabecular and cortical bone, respectively. Trabecular morphometry was measured in the distal femur metaphysis, defining a volumetric region of interest within the endocortical surface of a 1 mm span referenced 1 mm from the growth plate. Cortical morphometry was measured within a 600 µm span referenced 5.1 mm from the growth plate, slightly distal of mid-diaphysis.
Histological analysis
Histological analysis of transgene expression
Soft tissues (brain, liver, spleen, kidney, lung, bladder, skin), calvaria and femurs were isolated from 1-month-old Col2.3NPY transgenic mice. All tissues were fixed in 4% paraformaldehyde solution for 2 days. Osseous tissue was decalcified in 14% ethylenediaminetetraacetic acid solution for 5 days. Tissues were placed in 30% sucrose overnight and immobilized in embedding media (Cryomatrix, Thermo Shandon). The embedded bones were longitudinally sectioned in 7-µm thick sections using adhesive tape (Finetec Ltd). Soft tissues were sectioned in 5-µm thick sections using CryoJane tape system (Instrumedics). Undecalcified femurs were processed in the same manner.
Histomorphometric analysis
Three-month-old Col2.3NPY and wild type mice were injected intraperitoneally with calcein (10 mg/kg) (Sigma-Aldrich Corp.) 7 days before sacrifice, and 90 mg/kg xylenol orange (Sigma-Aldrich) 2 days before sacrifice. Dynamic histomorphometry was performed on undecalcified femur sections using the Osteomeasure system (OsteoMetrics, Inc.). Trabecular bone measurements were initiated 250 µm away from the growth plate.
CTx Assay
Serum was collected from 3-month-old mice that were fasted overnight. CTx concentration was measured in undiluted serum using the RatLaps (CTX-I) EIA kit (Immunodiagnostic Systems Limited) according to the manufacturer’s instructions. Concentrations were interpolated from a standard curve using a 4-parametric logistic curve fit.
In vitro cultures
Mouse calvarial osteoblasts (mCOBs) were isolated from 6–8-day-old mice using a modified enzyme digestion protocol [22]. After excision, pups were genotyped using fluorescent microscopy. Calvariae were sequentially digested four times for 30 minutes in 0.05% trypsin and 0.5 mg/ml collagenase P digestion buffer at 37°C and cells from fractions 2–4 were pooled. Cells were plated at a density of 3 × 105/well in a 6-well plate in DMEM 10% FBS + non-essential amino acids + penicillin/streptomycin (P/S). Medium was switched to osteogenic medium (αMEM 10% FBS + P/S + 50 µg/ml ascorbic acid + 4 mM β-glycerophosphate) after seven days and cells were maintained 14 more days under differentiating conditions. Medium was replaced every 2 days. GFPtopaz expression of cells derived from Col2.3NPY mice was evaluated on a Zeiss Observer.Z1 fluorescent microscope using a filter cube optimized for GFPtopaz.
Alkaline phosphatase (ALP) activity and mineralization staining
Cell layers were rinsed in phosphate buffered saline, briefly fixed in 4% paraformaldehyde, and ALP activity was assessed using the ALP staining kit (Sigma-Aldrich Corp., St. Louis, MO, USA) according to the manufacturer's instructions. Von Kossa staining was performed after staining for ALP by incubating in 5% silver nitrate in an ultraviolet crosslinker. The results of staining were recorded using a flatbed scanner. The mineralized area was evaluated by setting a threshold for background and applying the same settings to all experimental samples (ImageJ, NIH).
Statistical analysis
All data are expressed as mean±SEM and unpaired t-test has been used to compare transgenic and wild type mice. EIA data were analyzed by SigmaPlot software (Systat Software, Inc.). For all analyses, p<0.05 was accepted as being statistically significant.
RESULTS
Bone directed overexpression of NPY
We have generated transgenic mice in which NPY is overexpressed under the control of a type I collagen promoter fragment (Col2.3) that directs expression in mature osteoblasts and osteocytes [23]. In addition to NPY cDNA, mice express the visual transgene GFPtopaz utilizing an IRES sequence (Figure 1A). Two independent transgenic lines were generated (line 09-5-5 and line 09-5-9) in C57BL6 background and transgene expression characterized. In both lines we have observed similar expression that is restricted to mineralized tissues. Analyses of the effects of NPY overexpression on the bone phenotype were completed using line 09-5-5. Heterozygous males were bred with C57BL6 mice and transgenic offspring were obtained in the expected ratio. We did not detect significant difference in weight between wild type mice and transgenic sex-matched littermates (males: WT 28.9±2.3g, Col2.3NPY 27.1±3.3g; females: WT 21.6±2.9g, Col2.3NPY 21.3±2.3g at 3 months of age). Histological analysis of various tissues in 1-month-old animals confirmed that GFP expression is restricted to osteoblasts and osteocytes within trabecular, cortical bone and calvaria (Figure 1D–E, and Suppl. Figure 1). Transgene expression was not detected in brain, spleen, kidney, liver, skin, bladder or lung (Suppl. Figure 2).
To evaluate bone-directed NPY overexpression, we measured the expression of NPY mRNA and protein in long bone tissue. Npy expression is increased 8 fold in bone samples of transgenic female and 2.5-fold in males compared with wild type littermates (Figure 1F). Bone protein isolates from Col2.3NPY males and females also showed significantly higher NPY concentrations than wild type littermates (Figure 1G). Serum NPY levels in were not elevated in Col2.3NPY mice indicating that bone-targeted overexpression does not alter the circulating levels of the NPY (Figure 1H).
In vivo effects of bone-specific overexpression of NPY
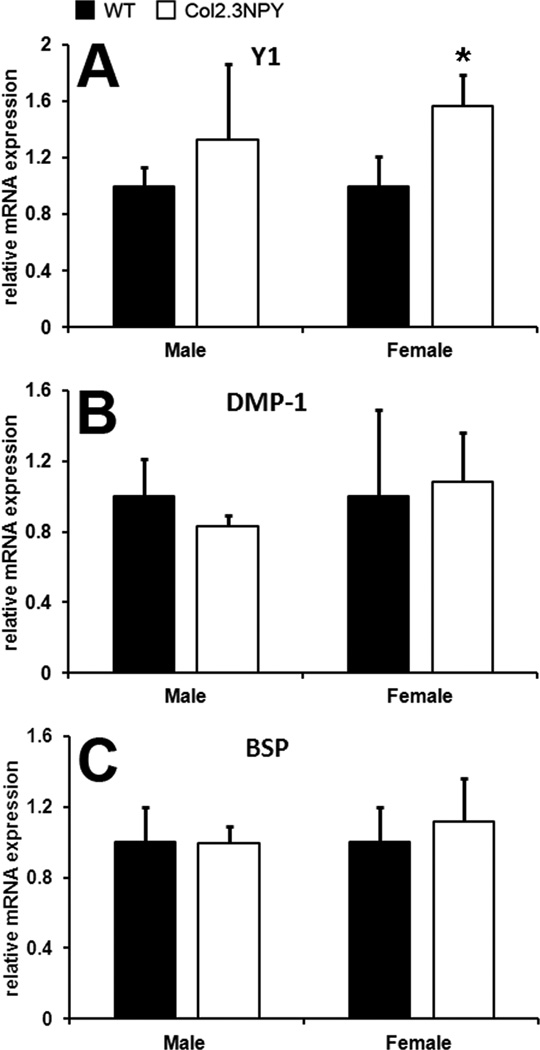
To investigate the effects of NPY overexpression on osteogenic marker expression in bones of Col2.3NPY transgenic mice we analyzed gene expression using real time PCR. While no statistically significant difference in Y1 receptor expression was observed in males, Y1 expression was elevated in Col2.3NPY female mice (Figure 2A). Y2 receptor expression was undetectable in all bone samples (data not shown). Gene expression analysis of osteogenic markers Dmp1 and OC did not show any statistically significant difference between transgenic males and females compared to sex-matched littermates (Figure 2B–C). The expression of osteogenic transcription factors Runx2 and osterix was also unchanged (data not shown).
Fig. 2. Real time PCR analysis of gene expression in bones.
RNA was extracted from long bone samples from 2-month-old male and 3 month-old female Col2.3NPY and wild type mice (n=3–11). Real time PCR was performed for the genes indicated and values shown are normalized to GAPDH levels and relative to wild type expression. * p<0.05 versus sex-matched wild type.
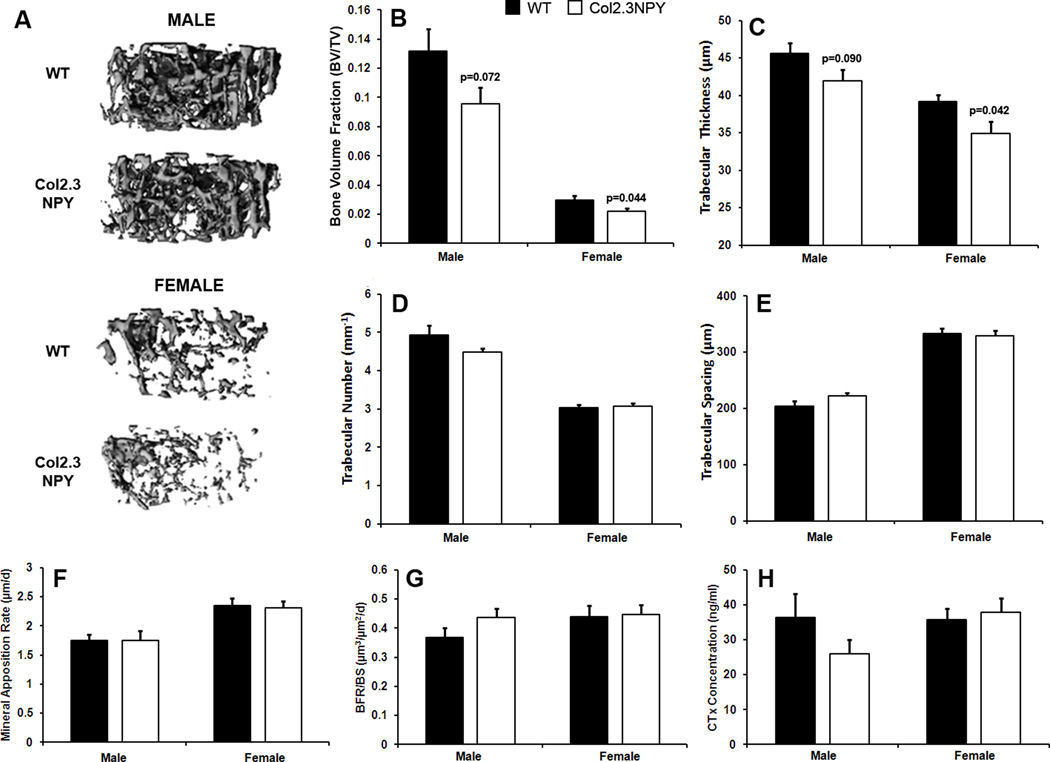
Analysis of quantitative indices of trabecular structure in the distal femur of 3-month-old mice demonstrated that bone-specific NPY overexpression led to a 25% reduction in trabecular bone volume compared to sex-matched littermates (Figure 3A–B). This corresponded with reduced trabecular thickness (Figure 3C), but no change in trabecular number (Figure 3D) or trabecular spacing (Figure 3E). NPY overexpression had no significant impact on mineral apposition rate or bone formation rate in trabecular bone in females or males (Figure 3F–G). The concentration of bone resorption marker CTx was also unchanged in the serum of transgenic mice (Figure 3H).
Fig. 3. Trabecular bone phenotype of Col2.3NPY mice.
Trabecular architecture in 3-month old Col2.3NPY mice and wild type littermates was determined using µCT (n=10–11) and is illustrated by representative 3D reconstructions (A). Parameters bone volume fraction (B), trabecular thickness (C), number (D) and spacing (E) are shown. Mineral apposition rate (F) and bone formation rate (G) were determined histomorphometrically (n=7–11). CTx levels in serum were also measured (H), (n=5–6).
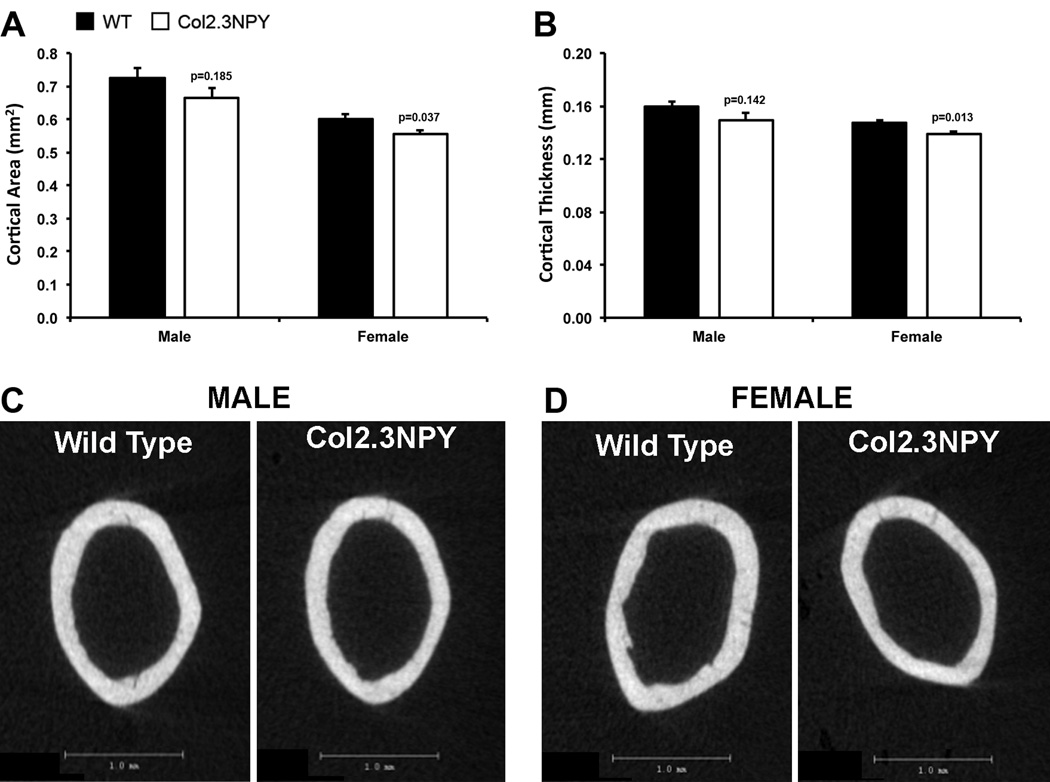
We also analyzed cortical bone parameters. Nominal femur length was unchanged in transgenic mice (data not shown), however, a there was a 7–8% reduction in cortical volume, reported as cross-sectional area (Figure 4A) and corresponding 6% reduction in cortical thickness (Figure 4B) in Col2.3NPY animals compared to wild type littermates. Two-dimensional µCT reconstructions of cortical bone from representative animals depicting these changes are shown (Figure 4C–D).
Fig. 4. Cortical bone phenotype of Col2.3NPY mice.
Cortical bone parameters were evaluated with µCT analysis. Cortical area (A), thickness (B) and 2D µCT images (C, D) are shown.
In vitro effects of bone-specific overexpression of NPY
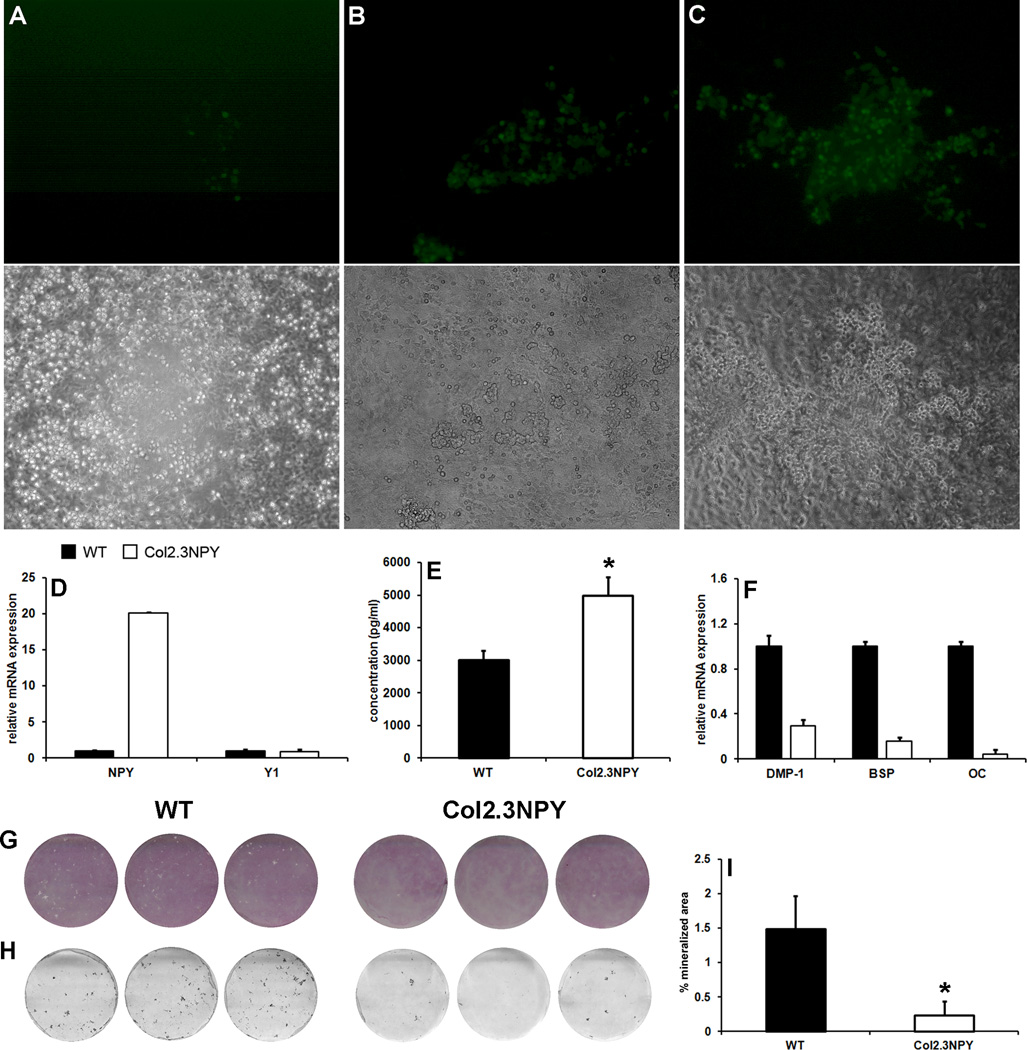
In vitro effects of osteoblast-specific NPY overexpression were evaluated in a primary osteoblast culture system. Using neonatal mouse calvarial osteoblasts (mCOB), we detected Col2.3 promoter driven GFP expression by day 15 (eighth day under osteogenic conditions). At this time point only a few cells within newly formed nodules expressed GFPtopaz (Figure 5A). The GFP fluorescence became stronger and more broadly expressed on day 17 (Figure 5B) and on day 21, the majority of the cells within mineralizing areas were GFP positive (Figure 5C). Analysis of NPY expression using qPCR and EIA confirmed increased NPY expression and release into the culture medium in mature Col2.3NPY cultures, while Y1 receptor expression was unchanged (Figure 5D–E). There were no clear changes in qualitative staining for the early osteogenic marker ALP in Col2.3NPY mCOB cultures (Figure 5G). However, markers of mature osteoblast differentiation (Bsp, Dmp1, and Oc) were decreased in Col2.3NPY cultures (Figure 5F). This observation correlated with a significantly reduced mineralized area (Figure 5H–I).
Fig. 5. In vitro differentiation in mCOB cultures.
Neonatal mouse calvarial osteoblasts were grown under osteogenic conditions. GFP expression in a representative culture is shown at day 15 (A), day 17 (B) and day 21 (C). Real time PCR analysis was performed on RNA extracted from day 21 cultures. Expression of the indicated genes in one representative experiment is shown (D, F). NPY protein expression was measured using EIA in the conditioned medium from the final 48h of culture (E). Cultures were stained for ALP expression on day 21 (G), and mineralization was assessed by von Kossa staining (H). Mineralized area was evaluated using image analysis (I). *, p<0.05 versus WT.
DISCUSSION
Our study evaluated the impact of osteoblast lineage derived NPY expression on bone. Overexpression of NPY was directed by the Col2.3 promoter which has previously been shown to have osteoblast and osteocyte-specific expression [22]. Overexpression was confirmed histologically, and at the RNA and protein levels. There was no increase in serum NPY levels in Col2.3NPY mice compared to wild type littermates indicating that changes in bone phenotype are due to local overexpression of NPY. Bone-directed NPY overexpression altered bone mass, including cortical and trabecular bone, as well as mineralization of osteoblasts in vitro.
The importance of NPY in central regulation of bone remodeling has been shown using NPY knockout mice, which have significantly increased bone mass due to an elevated bone formation rate [1]. Hypothalamus-directed NPY overexpression did not completely rescue the phenotype implying that other sources of NPY might play a role in bone homeostasis. Previous studies have reported NPY expression by osteolineage cells [9, 19] and our real-time PCR data confirmed that NPY is locally expressed in bone tissue. The pre-pro-NPY generated after translation is cleaved by several proteases and undergoes post-translational modification [24]. Protein levels of NPY were modestly increased in bone tissue. However, it should be noted that these bone preparations will not contain only osteolineage cells, and effects of the overexpression may be diluted by other sources of NPY in the bone such as vasculature and innervation [25, 26].
Bone-directed NPY overexpression results in a 25% reduction in trabecular bone mass at three months of age in both genders. This was associated with decreased trabecular thickness, but no change in trabecular number. Cortical volume was also lower in the transgenic mice. This data indicates that bone-derived NPY can alter bone mass in vivo. Despite these differences, we were unable to detect changes in osteogenic markers Dmp1, BSP and OC in bone tissue, or transcription factors Runx2 and Osterix. Nor did we find statistically significant changes in bone formation rate or mineral apposition rate that would explain the reduced bone volume. The osteoclast resorption marker CTx was also the same as wild type littermates. This is consistent with other models of perturbations in the NPY pathway primarily affecting the osteoblast lineage [1, 8, 11, 16]. It is possible that the absence of differences in bone formation parameters and marker genes is due to the age of the mice examined. Bone-directed NPY overexpression might have an impact on bone modeling during development or growth that is retained at three months of age. Since we were interested in the phenotype of skeletally mature mice, animals of other ages were not examined in this study. In addition, while some parameters appear to show larger or more significant changes in females, similar trends are present in the males.
Osteoblast-specific Y1 knockout mice have enhanced bone mass and bone formation implying that local signaling through this receptor regulates bone metabolism, however NPY is not the exclusive ligand of Y1. Peptide YY (PYY), a gut-derived member of the NPY family, also binds with high affinity to Y1 and Y2 receptors. PYY knockout mice have a similar bone phenotype to NPY deficient animals with increased bone volume and formation. Conversely, systemic overexpression of PYY causes lower osteoblast activity via a Y1 receptor-mediated mechanism [27]. Our transgenic model has a similar, albeit less robust phenotype than this PYY overexpression model. Presumably this is due to limited and localized NPY overexpression in osteoblasts and osteocytes, whereas PYY expression was elevated systemically including in the hypothalamus. This data suggests that both NPY and PYY levels may influence local Y1-mediated regulation of bone turnover.
Since Y1 receptor pathways have been shown to play an exclusive role in local NPY-mediated inhibition of bone formation [16, 20, 28], we examined Y1 receptor expression. Y1 receptor mRNA was present in bone tissue, and levels were subtly elevated in Col2.3NPY mice, but unchanged in cultures from Col2.3NPY animals. This contrasts with previously findings in vitro [10] and in vivo [28] that have shown that NPY is a negative regulator of Y1 receptor expression. We also confirmed that Y2 receptor expression was absent suggesting the effects seen in this study were mediated by Y1, in agreement with previous studies which show that osteoblast specific Y1 but not Y2 deletion in bone or peripheral tissue affects osteoblast activity and bone mass in vivo [16, 29]. In addition, a recent study indicated that pharmacological Y1 receptor antagonism using BIBO3304 also increases osteoblast activity in cortical and trabecular bone without any alternations in body weight, food intake or energy metabolism [30].
In order to confirm that osteoblast-derived NPY could exert autocrine/paracrine effects on the osteoblast lineage, we performed in vitro experiments. Treatment of mouse calvarial osteoblasts with exogenous NPY inhibits osteoblast differentiation [9]. Osteoblast cultures derived from Col2.3NPY mice were alkaline phosphatase positive, but mineralization and expression of osteogenic marker genes were markedly reduced compared to wild type controls. Confinement of differences to the late stages of culture coincides with overexpression of NPY beginning only in the final week of culture when mature osteoblasts begin to appear. These findings are consistent with increased osteoblast differentiation and mineralization in mCOB cultures from Y1 knockout mice where NPY signaling is ablated [8].
This novel mouse model has enabled us to demonstrate that osteoblast-derived NPY can have autocrine/paracrine effects on osteoblast differentiation and modify bone volume. Local changes in NPY levels were sufficient to alter bone mass in both genders. Further studies will be directed at evaluating bone parameters during development and in younger mice. NPY overexpression may have had effects primarily in younger animals that those examined in this study. The effects of this transgene on bone are subtler than the large reduction in bone formation parameters observed when NPY was overexpressed specifically in the hypothalamus [1], indicating that central NPY activity plays a critical role in NPY-related bone remodeling. In addition to local production by osteocytes and osteoblasts [8, 9, 20], NPY is also expressed in sympathetic nerves [25] and vascular smooth muscle cells [26]. These sources contribute to local control of bone remodeling [31] and given the subtle phenotype of the mice generated in this study, they may have a greater impact on bone turnover than osteolineage-derived NPY. Development of a floxed NPY allele that can be used to eliminate NPY from various peripheral sources will be critical to determining which cells are involved in NPY-mediated regulation of osteoblast activity.
Supplementary Material
Fluorescence microscopy images of the distal femur of a female Col2.3NPY mouse with a scanned overview of the bone (A), and higher magnification of trabecular (B) and cortical (C) bone showing GFP expression in most osteoblasts and osteocytes.
Fluorescence microscopy images of sections from calvaria and various soft tissues in Col2.3NPY mice are shown. GFP positive cells are present in calvaria, but are absent in all soft tissues examined.
Acknowledgments
Supported by: This work has been supported by NIH/NIAMS AR059315 grant to I.K.
Footnotes
DECLARATION OF INTEREST
The authors indicate no potential conflicts of interest.
References
- 1.Baldock PA, Lee NJ, Driessler F, Lin S, Allison S, Stehrer B, Lin EJ, Zhang L, Enriquez RF, Wong IP, McDonald MM, During M, Pierroz DD, Slack K, Shi YC, Yulyaningsih E, Aljanova A, Little DG, Ferrari SL, Sainsbury A, Eisman JA, Herzog H. Neuropeptide Y knockout mice reveal a central role of NPY in the coordination of bone mass to body weight. PLoS One. 2009;4:e8415. doi: 10.1371/journal.pone.0008415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stanley BG, Kyrkouli SE, Lampert S, Leibowitz SF. Neuropeptide Y chronically injected into the hypothalamus: a powerful neurochemical inducer of hyperphagia and obesity. Peptides. 1986;7:1189–1192. doi: 10.1016/0196-9781(86)90149-x. [DOI] [PubMed] [Google Scholar]
- 3.Tatemoto K, Carlquist M, Mutt V. Neuropeptide Y--a novel brain peptide with structural similarities to peptide YY and pancreatic polypeptide. Nature. 1982;296:659–660. doi: 10.1038/296659a0. [DOI] [PubMed] [Google Scholar]
- 4.Allen JM, Adrian TE, Tatemoto K, Polak JM, Hughes J, Bloom SR. Two novel related peptides, neuropeptide Y (NPY) and peptide YY (PYY) inhibit the contraction of the electrically stimulated mouse vas deferens. Neuropeptides. 1982;3:71–77. doi: 10.1016/0143-4179(82)90001-4. [DOI] [PubMed] [Google Scholar]
- 5.Parker RM, Herzog H. Regional distribution of Y-receptor subtype mRNAs in rat brain. Eur J Neurosci. 1999;11:1431–1448. doi: 10.1046/j.1460-9568.1999.00553.x. [DOI] [PubMed] [Google Scholar]
- 6.Sar M, Sahu A, Crowley WR, Kalra SP. Localization of neuropeptide-Y immunoreactivity in estradiol-concentrating cells in the hypothalamus. Endocrinology. 1990;127:2752–2756. doi: 10.1210/endo-127-6-2752. [DOI] [PubMed] [Google Scholar]
- 7.Nakamura M, Sakanaka C, Aoki Y, Ogasawara H, Tsuji T, Kodama H, Matsumoto T, Shimizu T, Noma M. Identification of two isoforms of mouse neuropeptide Y-Y1 receptor generated by alternative splicing. Isolation, genomic structure, and functional expression of the receptors. J Biol Chem. 1995;270:30102–30110. doi: 10.1074/jbc.270.50.30102. [DOI] [PubMed] [Google Scholar]
- 8.Baldock PA, Allison SJ, Lundberg P, Lee NJ, Slack K, Lin EJ, Enriquez RF, McDonald MM, Zhang L, During MJ, Little DG, Eisman JA, Gardiner EM, Yulyaningsih E, Lin S, Sainsbury A, Herzog H. Novel role of Y1 receptors in the coordinated regulation of bone and energy homeostasis. J Biol Chem. 2007;282:19092–19102. doi: 10.1074/jbc.M700644200. [DOI] [PubMed] [Google Scholar]
- 9.Igwe JC, Jiang X, Paic F, Ma L, Adams DJ, Baldock PA, Pilbeam CC, Kalajzic I. Neuropeptide Y is expressed by osteocytes and can inhibit osteoblastic activity. J Cell Biochem. 2009;108:621–630. doi: 10.1002/jcb.22294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teixeira L, Sousa DM, Nunes AF, Sousa MM, Herzog H, Lamghari M. NPY revealed as a critical modulator of osteoblast function in vitro: new insights into the role of Y1 and Y2 receptors. J Cell Biochem. 2009;107:908–916. doi: 10.1002/jcb.22194. [DOI] [PubMed] [Google Scholar]
- 11.Baldock PA, Sainsbury A, Couzens M, Enriquez RF, Thomas GP, Gardiner EM, Herzog H. Hypothalamic Y2 receptors regulate bone formation. J Clin Invest. 2002;109:915–921. doi: 10.1172/JCI14588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baldock PA, Sainsbury A, Allison S, Lin EJ, Couzens M, Boey D, Enriquez R, During M, Herzog H, Gardiner EM. Hypothalamic control of bone formation: distinct actions of leptin and y2 receptor pathways. J Bone Miner Res. 2005;20:1851–1857. doi: 10.1359/JBMR.050523. [DOI] [PubMed] [Google Scholar]
- 13.Kushi A, Sasai H, Koizumi H, Takeda N, Yokoyama M, Nakamura M. Obesity and mild hyperinsulinemia found in neuropeptide Y-Y1 receptor-deficient mice. Proc Natl Acad Sci USA. 1998;95:15659–15664. doi: 10.1073/pnas.95.26.15659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pedrazzini T, Seydoux J, Kunstner P, Aubert JF, Grouzmann E, Beermann F, Brunner HR. Cardiovascular response, feeding behavior and locomotor activity in mice lacking the NPY Y1 receptor. Nat Med. 1998;4:722–726. doi: 10.1038/nm0698-722. [DOI] [PubMed] [Google Scholar]
- 15.Burcelin R, Brunner H, Seydoux J, Thorensa B, Pedrazzini T. Increased insulin concentrations and glucose storage in neuropeptide Y Y1 receptor-deficient mice. Peptides. 2001;22:421–427. doi: 10.1016/s0196-9781(01)00357-6. [DOI] [PubMed] [Google Scholar]
- 16.Lee NJ, Nguyen AD, Enriquez RF, Doyle KL, Sainsbury A, Baldock PA, Herzog H. Osteoblast specific Y1 receptor deletion enhances bone mass. Bone. 2011;48:461–467. doi: 10.1016/j.bone.2010.10.174. [DOI] [PubMed] [Google Scholar]
- 17.Chronwall BM, DiMaggio DA, Massari VJ, Pickel VM, Ruggiero DA, O'Donohue TL. The anatomy of neuropeptide-Y-containing neurons in rat brain. Neuroscience. 1985;15:1159–1181. doi: 10.1016/0306-4522(85)90260-x. [DOI] [PubMed] [Google Scholar]
- 18.Ekblad E, Edvinsson L, Wahlestedt C, Uddman R, Hakanson R, Sundler F. Neuropeptide Y co-exists and co-operates with noradrenaline in perivascular nerve fibers. Regul Pept. 1984;8:225–235. doi: 10.1016/0167-0115(84)90064-8. [DOI] [PubMed] [Google Scholar]
- 19.Nunes AF, Liz MA, Franquinho F, Teixeira L, Sousa V, Chenu C, Lamghari M, Sousa MM. Neuropeptide Y expression and function during osteoblast differentiation--insights from transthyretin knockout mice. FEBS J. 2010;277:263–275. doi: 10.1111/j.1742-4658.2009.07482.x. [DOI] [PubMed] [Google Scholar]
- 20.Lee NJ, Doyle KL, Sainsbury A, Enriquez RF, Hort YJ, Riepler SJ, Baldock PA, Herzog H. Critical role for Y1 receptors in mesenchymal progenitor cell differentiation and osteoblast activity. J Bone Miner Res. 2010;25:1736–1747. doi: 10.1002/jbmr.61. [DOI] [PubMed] [Google Scholar]
- 21.Ruohonen ST, Pesonen U, Moritz N, Kaipio K, Roytta M, Koulu M, Savontaus E. Transgenic mice overexpressing neuropeptide Y in noradrenergic neurons: a novel model of increased adiposity and impaired glucose tolerance. Diabetes. 2008;57:1517–1525. doi: 10.2337/db07-0722. [DOI] [PubMed] [Google Scholar]
- 22.Kalajzic I, Kalajzic Z, Kaliterna M, Gronowicz G, Clark SH, Lichtler AC, Rowe D. Use of type I collagen green fluorescent protein transgenes to identify subpopulations of cells at different stages of the osteoblast lineage. J Bone Miner Res. 2002;17:15–25. doi: 10.1359/jbmr.2002.17.1.15. [DOI] [PubMed] [Google Scholar]
- 23.Marijanovic I, Jiang X, Kronenberg MS, Stover ML, Erceg I, Lichtler AC, Rowe DW. Dual reporter transgene driven by 2.3Col1a1 promoter is active in differentiated osteoblasts. Croat Med J. 2003;44:412–417. [PubMed] [Google Scholar]
- 24.Silva AP, Cavadas C, Grouzmann E. Neuropeptide Y and its receptors as potential therapeutic drug targets. Clin Chim Acta. 2002;326:3–25. doi: 10.1016/s0009-8981(02)00301-7. [DOI] [PubMed] [Google Scholar]
- 25.Lundberg JM, Martinsson A, Hemsen A, Theodorsson-Norheim E, Svedenhag J, Ekblom B, Hjemdahl P. Co-release of neuropeptide Y and catecholamines during physical exercise in man. Biochem Biophys Res Commun. 1985;133:30–36. doi: 10.1016/0006-291x(85)91837-6. [DOI] [PubMed] [Google Scholar]
- 26.Morgan DG, Kulkarni RN, Hurley JD, Wang ZL, Wang RM, Ghatei MA, Karlsen AE, Bloom SR, Smith DM. Inhibition of glucose stimulated insulin secretion by neuropeptide Y is mediated via the Y1 receptor and inhibition of adenylyl cyclase in RIN 5AH rat insulinoma cells. Diabetologia. 1998;41:1482–1491. doi: 10.1007/s001250051095. [DOI] [PubMed] [Google Scholar]
- 27.Wong IPL, Driessler F, Khor EC, Shi YC, Hormer B, Nguyen AD, Enriquez RF, Eisman JA, Sainsbury A, Herzog H, Baldock PA. Peptide YY regulates bone remodeling in mice: a link between gut and skeletal biology. PLoS One. 2012;7:e40038. doi: 10.1371/journal.pone.0040038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lundberg P, Allison SJ, Lee NJ, Baldock PA, Brouard N, Rost S, Enriquez RF, Sainsbury A, Lamghari M, Simmons P, Eisman JA, Gardiner EM, Herzog H. Greater bone formation of Y2 knockout mice is associated with increased osteoprogenitor numbers and altered Y1 receptor expression. J Biol Chem. 2007;282:19082–19091. doi: 10.1074/jbc.M609629200. [DOI] [PubMed] [Google Scholar]
- 29.Shi YC, Lin S, Castillo L, Aljanova A, Enriquez RF, Nguyen AD, Baldock PA, Zhang L, Bijker MS, Macia L, Yulyaningsih E, Zhang H, Lau J, Sainsbury A, Herzog H. Peripheral-specific Y2 receptor knockdown protects mice from high-fat diet-induced obesity. Obesity. 2011;19:2137–2148. doi: 10.1038/oby.2011.99. [DOI] [PubMed] [Google Scholar]
- 30.Sousa DM, Baldock PA, Enriquez RF, Zhang L, Sainsbury A, Lamghari M, Herzog H. Neuropeptide Y Y1 receptor antagonism increases bone mass in mice. Bone. 2012;51:8–16. doi: 10.1016/j.bone.2012.03.020. [DOI] [PubMed] [Google Scholar]
- 31.Shi YC, Baldock PA. Central and peripheral mechanisms of the NPY system in the regulation of bone and adipose tissue. Bone. 2012;50:430–436. doi: 10.1016/j.bone.2011.10.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fluorescence microscopy images of the distal femur of a female Col2.3NPY mouse with a scanned overview of the bone (A), and higher magnification of trabecular (B) and cortical (C) bone showing GFP expression in most osteoblasts and osteocytes.
Fluorescence microscopy images of sections from calvaria and various soft tissues in Col2.3NPY mice are shown. GFP positive cells are present in calvaria, but are absent in all soft tissues examined.