Summary
Computational prediction of protein function is frequently error-prone and incomplete. In Mycobacterium tuberculosis (Mtb), ~25% of all genes have no predicted function and are annotated as hypothetical proteins, severely limiting our understanding of Mtb pathogenicity. Here, we utilize a high throughput, quantitative, activity-based protein profiling (ABPP) platform to probe, annotate, and validate ATP-binding proteins in Mtb. We experimentally validate prior in silico predictions of >250 proteins and identify 72 hypothetical proteins as novel ATP binders. ATP interacts with proteins with diverse and unrelated sequences, providing a new and expanded view of adenosine nucleotide binding in Mtb. Several hypothetical ATP binders are essential or taxonomically limited, suggesting specialized functions in mycobacterial physiology and pathogenicity.
Introduction
Determining the function of protein-coding genes in a genome remains one of the most challenging problems in the post-genomic era. For most newly sequenced bacterial genomes, 50–70% of the protein coding genes are assigned a function derived by inference (i.e., by sequence similarity with previously characterized proteins), rather than by experiment, but these inferences are frequently inaccurate (Bork, 2000). Additionally, 30–50% of genes cannot be assigned function and are referred to as hypotheticals, severely limiting our ability to fundamentally understand microbial systems, and to manipulate them for human benefit.
While many excellent computational methods have been developed to predict and assign function to protein-coding genes, including homology-based (Bork and Koonin, 1998) and genomic context-based (Huynen, et al., 2000) approaches, the rate at which these genes are experimentally characterized is exceedingly slow. To address this challenge, we established a chemical biology platform combining activity-based protein profiling (ABPP) with quantitative mass spectrometry-based proteomics to facilitate high throughput experimental functional annotation. ABPP is a developing technology in functional proteomics that uses active site-directed chemical probes, termed activity-based probes (ABPs), to report directly on the functional state of enzymes within a complex biological sample (Cravatt, et al., 2008; Simon and Cravatt, 2010). By specifically probing protein function in a select portion of the proteome based upon shared principles of binding and reactivity, potentially all active members of a protein family can be identified simultaneously (Cravatt, et al., 2008; Simon and Cravatt, 2010). Herein, we apply our ABPP approach to experimentally annotate protein function across an entire protein family in the medically important pathogen Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis.
One of the largest functional classes of proteins is the ATP-binding proteins, which share ATP binding and hydrolysis as their unifying functional feature. ATP hydrolysis is a common reaction that profoundly shapes the cell’s physiology. Many ATP-binding proteins can be readily identified by sequence signatures, such as the Walker A and Walker B motifs, or structurally by common folds such as the Rossman fold. However, more divergent ATP-binding proteins are difficult to identify through sequence-based annotation, and many members of the class are likely still unknown. ATP-dependent enzymes, including chaperones, kinases, and transporters, play essential roles in Mtb viability, infection, pathogenesis, and drug resistance (Magnet, et al., 2010; Schreiber, et al., 2009). The importance of ATP-dependent enzyme functions and pathways make annotation of this class of proteins particularly relevant for guiding the discovery of new therapeutic targets to treat tuberculosis.
To improve the quality of the current Mtb genome annotation by experimental validation of in silico protein function assignments, and to discover new protein function not detectable by sequence-based methods, we combined activity-based protein profiling (ABPP) and quantitative LC-MS-based proteomics to establish a novel experimental annotation platform, accurate mass and time (AMT) tag-ABPP. We apply this technology to the broad assignment of function to the ATP-binding protein guild in Mtb. We identify a total of 317 ATP-binding proteins. For >70% of these proteins, our data provide experimental validation of prior in silico prediction. Importantly, we also identify a large number of proteins previously annotated as hypothetical proteins. These represent several new ATP-binding proteins, and highlight the diversity of ATP-binding sequences in Mtb and other bacterial species. Our survey of the ATP binding space in Mtb experimentally refines the functional annotation of the Mtb genome, and provides leads to new ATP-binding protein function in Mtb and other bacteria. As many of the identified hypothetical proteins are both unique to Mycobacteria and essential for in vitro growth or infection, they reveal new ATP-dependent functional proteins that could serve as therapeutic targets for the treatment of tuberculosis.
Results
Probe Design and Synthesis
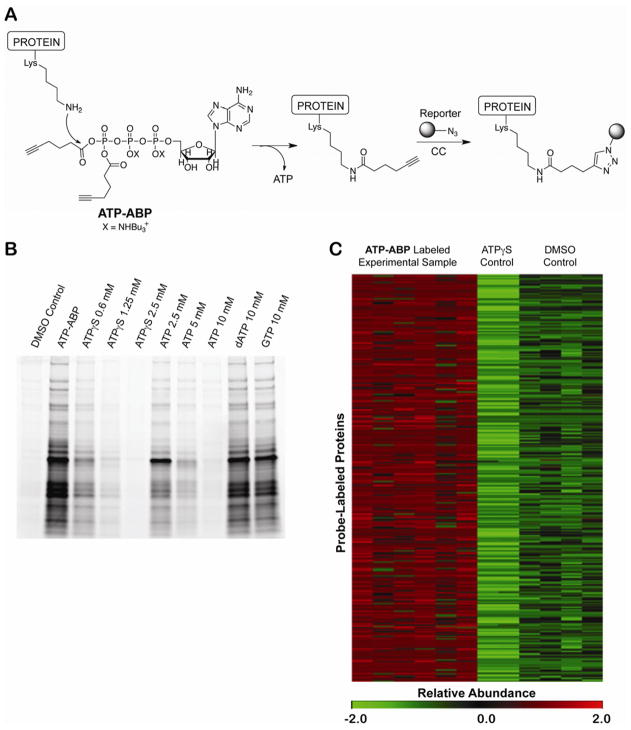
ATP-binding proteins constitute a large and centrally important protein guild in all organisms. Previously, a nucleotide acyl phosphate probe was developed for the labeling and characterization of ATP-binding proteins in eukaryotic proteomes by coupling ATP to biotin through a mixed anhydride on the terminal phosphate group of ATP (Patricelli, et al., 2007; Qiu and Wang, 2007). This probe binds to functional ATP-binding sites and facilitates covalent labeling through a reaction between the ε-amino groups of lysine residues and the mixed carboxylic phosphoric anhydride moiety of the probe to form a stable acetamide (Figure 1A) (DiSabato and Jencks, 1961; Kluger, 2000). A unique advantage of this probe is that labeling is inherently linked to the hydrolysis of the ATP analog. Thus, labeling by the probe is direct evidence of phosphate hydrolysis. Although specifically designed for the labeling of kinases and ATPases, the probe was found to broadly label other ATP-binding proteins (Patricelli, et al., 2007; Qiu and Wang, 2007). Other probe targets include nucleotide-binding proteins, CoA-binding proteins, and phosphate hydrolase/transfer enzymes. Reaction with the probe requires the presence of a nucleophilic amino acid residue. To minimize steric interference and improve binding, we removed the bulky biotin group from the terminal phosphate of ATP and replaced it with a click chemistry-compatible alkyne moiety giving ATP-ABP (Figure 1A) (Sadler, et. al., 2012). The alkyne group allows for the Cu(I)-catalyzed click chemistry addition of multifunctional tags for fluorescent detection, biotin tagging, and tagging for direct characterization of the probe-labeled amino acid residue(s) (Speers, et al., 2003; Speers and Cravatt, 2005) (Figure 1A).
Figure 1. Probe structure, selective labeling, and identification of Mtb proteins by ATP-ABP.
(A) ATP-ABP structure and labeling of proteins. The ε-amino group of Lys residues reacts with one of two acyl phosphate moieties on the probe transferring the click-chemistry (CC) compatible alkyne unit to the protein and releasing ATP. Biotin (MS analysis) or Cy5.5 (fluorescent gel analysis) is appended to the probe-labeled protein via CC. (B) In-gel analysis of ATP-ABP labeled Mtb lysate. Proteomes were labeled with ATP-ABP (20 μM) alone and in the presence of ATPγs, ATP, dATP, and GTP. Labeled proteins were visualized after SDS-PAGE. (C) Heat map illustration of quantitative functional activity profile for 317 Mtb proteins, demonstrating reproducibility within probe-labeled sample replicates (ATP-ABP), no-probe control sample replicates (DMSO Control) and ATPγS-pretreated control sample replicates (ATPγS Control). The MS-measured protein abundances are listed in Supplemental Table S1. The abundance values were converted in MultiExperiment Viewer (MeV) (Saeed, et al., 2006) to normalized score (z-score) for visualization. The scale is MeV normalized score (z-score) from low (green) to high (red).
Global quantitative activity profiling
To test the activity and selectivity of ATP-ABP, we labeled native Mtb proteome with ATP-ABP, appended a fluorescent Cy5.5 dye by click-chemistry, separated samples by SDS-PAGE, and visualized fluorescence of labeled proteins (Figure 1B). In the context of the Mtb proteome, the ATP-ABP showed labeling of distinct bands in the ABP-treated but not the untreated control sample. The non-hydrolyzable ATP analogue ATPγS competed with probe labeling in a concentration-dependent manner, completely blocking probe labeling at concentrations above 1mM. Similarly, ATP competed with probe labeling, requiring ~10mM ATP for complete blocking of probe binding. The 10-fold higher ATP concentration required for inhibition is likely due to hydrolysis of ATP, but not ATPγS, during the labeling reaction, effectively reducing the ATP concentration in the competitive inhibition study. To test the selectivity of ATP-ABP, we also tested the effect of dATP and another nucleotide, GTP, on probe binding. Even at concentrations that lead to complete probe inhibition with ATP, dATP and GTP did not affect ATP-ABP labeling, showing that ATP-ABP is selective for ATP-binding proteins (Figure 1B) in the Mtb proteome.
To identify probe-labeled proteins by mass spectrometry, lysates from exponentially growing Mtb were labeled with ATP-ABP, followed by covalent attachment of biotin by click-chemistry and enrichment of labeled proteins on streptavidin agarose resin. Resin-bound proteins were washed to remove non probe-labeled proteins and digested with trypsin. Peptides were analyzed by high-resolution LC-MS(/MS) and quantitative analyses performed using the AMT tag approach as described in Methods (Zimmer, et al., 2006). Our AMT-ABPP (accurate mass and time tag-activity based protein profiling) platform provides several advantages over conventional MudPIT approaches. These include quantitation directly from peptide signal intensities, accurate and statistically rigorous discrimination between true and false hits as described below, and no need for isotopic labeling procedures. Additionally, the AMT-tag approach is more sensitive due to utilization of LC-MS features, which alleviate the under-sampling problem encountered by traditional “shotgun” proteomics, allowing for deeper proteome coverage (Zimmer, et al., 2006).
To control for nonspecific probe binding, we analyzed DMSO-treated Mtb samples and Mtb samples treated with ATPγS prior to labeling. ATPγS controlled for adenosine-independent binding of the probe, while the DMSO-treated sample controlled for general nonspecific binding during streptavidin-based enrichment. LC–MS analyses of six probe-labeled sample replicates (ATP-ABP treated), four no-probe control sample replicates (DMSO-treated), and two ATPγS-pretreated control sample replicates (ATPγS-treated) identified a total of 794 proteins for which at least two unique peptides were measured per protein. We set the following criteria for inclusion in our further analysis: (i) a significant difference across the probe-labeled sample and the two negative control conditions as judged by ANOVA (p<0.05), (ii) a ≥5-fold higher abundance in the probe-labeled sample relative to the control samples, and (iii) reproducibility of peptide measurements across probe-labeled sample replicates. In typical quantitative proteomics analyses, a ≥2-fold change in abundance (p<0.05) is a generally accepted threshold for significant difference. Here, we apply a stringent threshold of ≥5-fold change in abundance between controls and probe-labeled samples, thereby reducing false discoveries and increasing our data confidence. Using these criteria, 317 proteins were identified for further analysis, as shown in Supplemental Table S1. This group of proteins represents a high-confidence set of ATP binding proteins. Because the 5-fold cutoff is stringent compared to comparable studies, we also assembled a second group of 277 hits with 2–5-fold enrichment of probe-labeled versus control (Supplemental Table S2). This group also contains many known ATP binding proteins, suggesting that this group, although with lower statistical confidence, contains true ATP binding proteins. Figure 1C shows a heat-map representation of the quantitative functional probe-labeling profile of the 317 Mtb proteins with >5-fold enrichment expressed as Z-scores. The heat map clearly shows high reproducibility within the six probe-labeled sample replicates (ATP-ABP treated; R2 = 0.89 ± 0.05), within the four no-probe control sample replicates (DMSO-treated; R2 = 0.87 ± 0.01), and within the two ATPγS-pretreated control sample replicates (ATPγS-treated; R2 = 0.98). Competition with ATPγS shows that all binding events are dependent on adenosine
Assessing performance of the AMT-ABPP approach using the ATP-ABP
Based on the proposed chemistry of probe-target interaction and prior studies on nucleotide acyl phosphate probes (Sadler, et. al., 2012; Patricelli, et al., 2007; Qiu and Wang, 2007), the ATP-ABP is expected to be reactive toward a select class of proteins including ATP-phosphohydrolases (ATPases/kinases), nucleotide (adenine, adenosine, NAD and/or FAD) and DNA/RNA binding proteins, acyl-phosphate reactive proteins, and acyl-CoA binding proteins. This a priori knowledge provides an opportunity to assess specificity of our AMT-ABPP approach using the ATP-ABP. To determine ATP-ABP target specificity, we surveyed the hits for functional characteristics according to existing annotation. Classification of labeled proteins, as shown in Figure 2 and Supplemental Table S1, was cross-validated by comparison of hits to proteins annotated as ATP-binders in PATRIC (Gillespie, et al., 2011) and TBDB (Reddy, et al., 2009), by literature text mining, and by bioinformatics analysis using Hidden Markov models (HMM) of protein families (TIGRFAM and PFAM) (Haft, et al., 2003). ATP-ABP labeling was observed among previously annotated ATPases, kinases, nucleotide binders, and acyl phosphate-reactive proteins. Of the proteins labeled, 68 proteins (~20%) are annotated as ATP-phosphohydrolases, including several well-known ATP-interacting proteins such as kinases, ATP-dependent proteases, and ATP-binding cassette (ABC) transporters. These proteins bind the ATP moiety of the probe and react directly with the mixed anhydride. The assignments of 48 of 68 proteins were confirmed by alignment with the PATRIC database’s ATPase/ATP-dependent category (Gillespie, et al., 2011). Assignment of the remaining 20 were supported by HMM analysis and/or the literature (Doerks, et al., 2012). A large portion of the labeled proteome consisted of nucleotide (adenine, adenosine, NAD and/or FAD) binding proteins with a conserved adenine binding motif capable of recognizing a structural element of the probe as well as containing a reactive amino acid capable of reacting with the mixed anhydride of ATP-ABP. This is in agreement with the previously reported reactivity profile of nucleotide acyl phosphate probes (Patricelli, et al., 2007; Qiu and Wang, 2007).
Figure 2. Specificity of ATP-ABP labeling in the Mtb proteome.
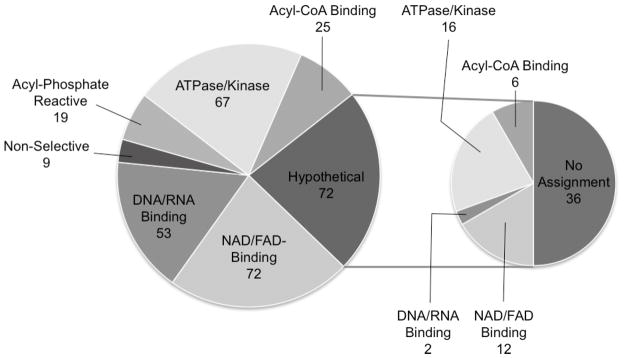
Pie chart shows functional classification of 317 proteins confidently identified as interacting with the ATP-ABP based on chemistry of the ATP-ABP, literature mining, and in silico prediction. The insert pie chart shows the further functional classification of 72 hypothetical proteins identified. Approximately 45% of the hypothetical proteins were confirmed to be ATP-binding proteins by additional bioinformatics analyses.
We also detected ATP-ABP binding of a large group of DNA and RNA binding proteins. To further define the ATP-ABP binding activity within this group, we compared the number of DNA to RNA binding proteins. Only 9 of the 51 proteins in this group are annotated as DNA binding proteins. While some DNA and RNA binding proteins, such as topoisomerase, bind ATP as a cofactor, all proteins in this family recognize adenosine in the context of DNA or RNA. However, consistent with the lack of competition of dATP with ATP-ABP (Figure 1B), the bias towards RNA binding proteins suggests that ATP-ABP does not recognize DNA deoxynucleotide binding sites. Thus, while their general adenosine binding propensity likely explains the identification of RNA binding proteins, probe binding to DNA binding proteins is more likely due to binding of ATP as a cofactor independent of DNA binding.
Additionally, 25 proteins known to bind or react with acyl-CoA molecules were labeled. This reactivity is likely governed by both adenine recognition and acyl phosphate reactivity, as these proteins are often responsible for hydrolysis of acyl-CoA. The probe also labeled 19 proteins that recognize non-nucleotide phosphate, such as pyridoxal phosphate, and that hydrolyze phosphate bonds (Figure 2, “Acyl-Phosphate Reactive”). These enzymes recognize and react with the probe directly through the phosphate and mixed anhydride moieties. Of the proteins for which annotation is available, the remaining nine labeled proteins have no known nucleotide binding capability or phosphohydrolase activity. Seventy-three proteins annotated as hypothetical were also labeled by ATP-ABP and are discussed in greater detail below. Assigning nine of 317 proteins as non-selective, we estimate a false labeling rate of ~3%, which is likely due to non-selective acylation of surface lysine residues by ATP-ABP (Patricelli, et al., 2007). However, we note the possibility that these proteins may have evolved allosteric interactions or are in complex with ATP-binding proteins. Together, our AMT-ABPP approach provides high-throughput experimental validation of functional annotation for ~250 in silico annotated members of the ATP-binding protein family, and provides experimental evidence to functionally annotate ~70 hypothetical proteins.
Pathway annotation of ATP-ABP labeled proteins
To test which pathways are particularly tractable for study by our ATP-ABP in terms of pathway coverage or representation of key components, the gene locus tags for ATP-ABP labeled proteins were uploaded into the gene cluster algorithm within the TB Database integrated platform (www.TBDB.org). This software clusters genes into broad functional (pathway) categories. Not surprisingly, proteins classified as conserved hypothetical or unknown represented one of the largest functional classes observed, ranking second in number (Figure 3). Proteins involved in lipid metabolism, intermediary metabolism, and respiration represented the largest functional cluster. Additionally, a significant number of ATP-ABP labeled proteins were assigned to the functional category, “Virulence, Detoxification, Adaptation,” in line with ATP-dependent enzymes playing essential roles in Mtb viability, infection, pathogenesis, and drug resistance (Magnet, et al., 2010; Schreiber, et al., 2009). To assess any biases towards a particular functional category (i.e., over-representation of a particular functional category), we compared our experimentally observed classification to that from genome annotation (Table 1). We define over-representation as a ~2-fold greater representation of a functional category by probe-labeled compared to annotated proteins.
Figure 3. Pathway distribution of 317 ATP-ABP labeled proteins from Mtb.
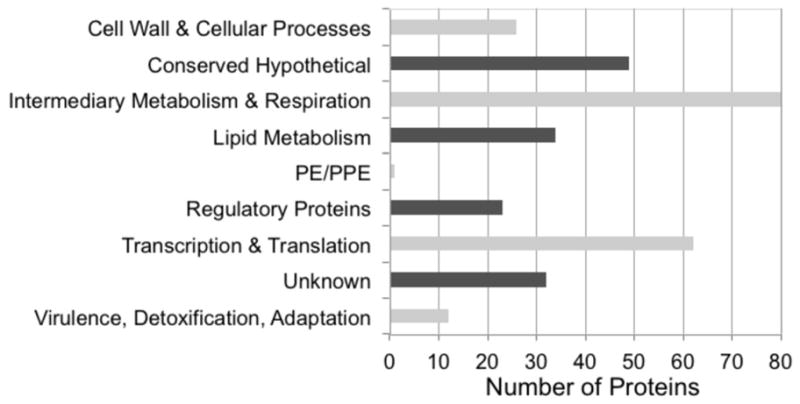
Proteins were mapped into functional processes and pathways using TBDB.org gene mapping programs.
Table 1.
Functional categories of observed ATP-ABP labeled proteins relative to functional categories predicted from genome. Fisher’s exact test was used to calculate p-values utilizing a 2×2 contingency table and the observation counts. Two tailed p-values were calculated. Over-represented categories are defined as ~2-fold greater (p<0.01) representation of a functional category by probe-labeled compared to prediction from the genome.
| Functional categories | Total from genome | Percent of genome | Total in observed proteome | Percent of observed proteome | p-value |
|---|---|---|---|---|---|
| Virulence, Detoxification, Adaptation | 90 | 2.3 | 12 | 3.8 | 0.1221 |
| Cell wall and Cell processes | 513 | 13 | 26 | 8.2 | 0.0109 |
| Conserved hypotheticals | 907 | 22.9 | 49 | 15.4 | 0.0016 |
| Transcription and Translation | 206 | 5.2 | 62 | 19.4 | 0.0001 |
| Insertion seqs and phages | 136 | 3.4 | 0 | 0 | 0.0001 |
| Intermediary metabolism and Respiration | 877 | 22.1 | 80 | 25.1 | 0.2351 |
| Lipid metabolism | 225 | 5.7 | 34 | 10.7 | 0.0009 |
| PE/PPE | 165 | 4.2 | 1 | 0.3 | 0.0001 |
| Regulatory proteins | 188 | 4.7 | 23 | 7.2 | 0.0589 |
| Unknown/Unclassified | 653 | 16.5 | 32 | 10 | 0.0019 |
| Total | 3960 | 100 | 319 | 100 |
Using this criterion, we observed the “Transcription and Translation” functional category to be over-represented. This is not unexpected since many of the proteins in this category play roles in cellular processes requiring interactions with adenosine nucleotide-like molecules. More interesting, our results also suggest the “Lipid metabolism” functional category to be over-represented under the experimental conditions tested in this study. The particularly good coverage of this pathway is a direct result of the acyl-CoA binding activity of our probe: 34 proteins in this category can be tracked by ATP-ABP, 24 of those through their acyl-CoA binding properties. Thus, the reactivity of ATP-ABP towards acyl-CoA binding proteins provides a unique advantage for the probing of lipid metabolism. Lipid metabolism is thought to play important roles in Mtb virulence, but the mechanism(s) remain largely unclear. By offering broad coverage of the “Lipid metabolism” functional category, our AMT-ABPP approach offers the opportunity to experimentally probe aspects of lipid metabolism in Mtb in a variety of conditions, including infection.
Assigning function to hypothetical proteins
Contributing to the experimental functional annotation of hypothetical proteins may be the most significant impact of this study. Approximately 25% of the Mtb coding sequences are still classified as hypothetical proteins (Lew, et al., 2011). Of the ATP-ABP labeled proteins, 33% (72 proteins), are annotated as hypothetical. These can be further organized into groups for which our functional annotation, combined with bioinformatic approaches, provides different levels of detail and novel functional insight (Figure 2). Hidden Markov model searches identified some level of homology for 33 of the 72 hypothetical proteins labeled by the ATP-ABP, showing homology consistent with nucleotide binding and/or ATPase activity (Supplemental Table S1). To further examine and verify hypothetical ATP-binders, we compared our hits to a recent in silico analysis of Mtb hypothetical genes by the Bork group (Doerks, et al., 2012). For 16 of 33 ATP-ABP labeled hypothetical proteins, our HMM analysis matches the Bork group in silico annotation (Doerks, et al., 2012) (Supplemental Table S1). Three additional proteins missed by our HMM analysis, but in agreement with our experimental assignment, were found by the Bork annotation to have ATPase or ATP-binding function. This analysis highlights the problem of conflicting protein function predictions that result from the use of different in silico annotation methods. Experimental data from the AMT-ABPP approach provides an excellent opportunity to resolve these discrepancies by validating protein function predictions. The experimental probe-labeling data provides critical evidence to validate otherwise purely predicted computational functional assignments.
A specific example of comparing our ATP-ABP target proteins identified in our screen to hypothetical proteins annotated as ATP binders by Bork is the hypothetical protein Rv0941c. In the Bork study, Rv0941c was annotated as a protein Ser/Thr kinase by orthology using genome context approaches. In our study, Rv0941c showed consistent ATP-ABP binding 15-fold over the control. Further sequence analysis and a literature search revealed that the Rv0941c C-terminal domain is similar to bacterial anti-sigma factors, a family with protein Ser/Thr kinase activity critically involved in regulating transcription (Hughes and Mathee, 1998). In agreement with these findings, a structure prediction using the Phyre2 (Kelley and Sternberg, 2009) server predicts a similar fold for the Rv0941c C-terminus to the anti-sigma factor SpoIIAB from B. subtilis, further supporting previous bioinformatic predictions (Doerks, et al., 2012; Greenstein, et al., 2007) that Rv0941c is an anti-sigma factor protein Ser/Thr kinase. In contrast to the C-terminal domain, the N-terminal domain of Rv0941c shows sequence similarity to anti-anti-sigma factors, suggesting that Rv0941c is a functional gene regulatory module that regulates alternative sigma factors.
A second group of 36 hypotheticals do not have discernible homology to known nucleotide binding domains. Interestingly, among the 36, four are predicted to be essential for optimal growth in vitro (Sassetti, et al., 2003), and four are predicted to be essential in infection (Sassetti and Rubin, 2003) (Supplemental Table S1). Moreover, most of these proteins are taxonomically limited: 22 are found only in Actinobacteria, nine are limited to Mycobacteria, and three proteins are found only in M. tuberculosis (Rv0394c, Rv0831c, Rv1507c). With the absence of any sequence motifs that link the 36 unknowns to nucleotide binding, this group provides a large and completely unexplored set of highly likely novel nucleotide binding proteins that may reveal new nucleotide binding sequences, domains, or folds.
Further analysis of hypotheticals not previously identified by in silico prediction revealed that three genes, Rv3614c-3616c, likely form an operon. All three proteins were labeled by ATP-ABP; we validated ATP-binding of one of these three proteins, Rv3614c, by recombinant expression and labeling (Figure 4A). All three are annotated as hypothetical and lack homology to any known protein domains, all three are essential for growth during infection (Sassetti and Rubin, 2003), and all three are taxonomically restricted to Mycobacteria. Recently, they have been shown to be essential for ESX-1-dependent protein secretion (type VII secretion system, or T7SS) and Mtb virulence (Fortune, et al., 2005; MacGurn, et al., 2005). Mycobacterial paralogs to these three genes occur next to the AAA family ATPase of T7SS, suggesting that these may be ATP-binding proteins or part of a complex that includes ATPases. Their essential function in T7SS, and lack of sequence homology, make them a particularly exciting set of priority targets for further functional characterization.
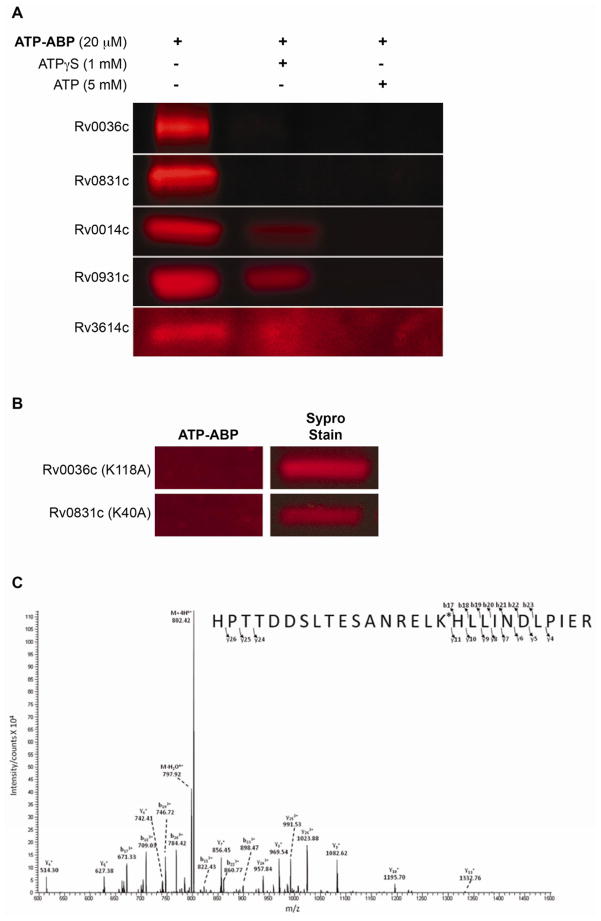
Figure 4. Validation of hypothetical protein labeling.
(A) ATP-ABP (20 μM) labeling of recombinantly expressed hypothetical proteins Rv0036c and Rv0831c, T7SS protein Rv3614c, and Ser/Thr protein kinases Rv0014c and Rv0931c (MS spectra showing site of probe labeling are shown in Supplemental Figure S1). Labeling of the hypothetical proteins was competitively inhibited by ATPγS (1 mM) and ATP (5 mM), showing adenosine-dependent probe binding. (B) ATP-ABP (20 μM) labeling of the K118A mutant of Rv0036c, and the K40A mutant of Rv0831c, revealing no probe labeling following click chemistry addition of a Cy5.5 dye and SDS-PAGE separation of proteins. Sypro Ruby Red stain indicates total protein used for labeling. (C) Annotated experimental MS/MS spectra showing labeling at lysine 40 of the peptide, “R.HPTTDSLTESANRELK*HLLINDLPIER.Q,” from the probe-labeled hypothetical protein RV0831c.
ATP-ABP labeling of T7SS-associated hypothetical proteins also occurs in proteins encoded in the eleven-gene operon Rv0282- Rv0292, one of four extended T7SS cassettes in the genome. Rv0282 is the AAA family ATPase of this T7SS cassette and was identified in our screen with 4-fold enrichment over the negative control (Supplemental Table S2). The ATP-ABP label also decorates proteins Rv0283 and Rv0284, encoded by genes that follow Rv0282 in tandem with overlapping stop/start codons that strongly suggest formation of a physical complex. Additional evidence for ATP binding of the protein complex is that Rv0284 contains three ATP/GTP binding site P-loop motifs.
In summary, our experimental results and computational analyses now allow for a more confident functional classification of a large number of hypothetical proteins as adenosine nucleotide-binding proteins, providing the first clues to the function of these Mtb proteins.
Targeted analysis of hypothetical proteins
To confirm and further explore the nucleotide binding properties of hypothetical proteins identified in our screen, we expressed Rv0036c and Rv0831c in E. coli, labeled the expressed proteins with ATP-ABP, and identified the probe-labeled amino acid residues by LC-MS/MS analysis. Both recombinant proteins were readily labeled with ATP-ABP, and ATP and ATPγS competed with probe binding (Figure 4A). Rv0036c is part of the TIGR03084 protein family, which is part of a larger set of probable enzymes, TIGR03083. Members of these protein families are found primarily in Actinobacteria. The function of these enzymes is uncharacterized, despite sharing sequence homology with other members of the protein family. Three out of nine members of family TIGR03083 encoded in Mtb H37Rv were labeled. Labeling of recombinant Rv0036c by ATP-ABP and subsequent tandem mass spectrometry analysis revealed the modification to occur at lysine 118. To confirm this assignment, we expressed a K118A mutant of Rv0036c. Mutation of K118 abrogated probe labeling, confirming K118 as the labeled residue (Figure 4B). Although this lysine is chemically suitable for labeling, it is not a conserved residue as shown by a multiple sequence alignment with its paralogs. Regions toward the N-terminus of this family show local sequence similarity, indicating remote homology to members of a protein family, DinB, which includes mycothiol, bacillithiol, and glutathione S-transferases (Newton, et al., 2011). Adenylyltransferase or CoA-transferase activity of Rv0036c, and other members of TIGR03083, is therefore likely.
Rv0831c is annotated as a hypothetical protein of unknown function. Of particular interest, Rv0831c has no discernible domain homology and is distributed almost exclusively within Mycobacteria, with only a few other distant sequence similarities outside the genus. Labeling of Rv0831c with ATP-ABP and subsequent tandem MS analysis revealed labeling at lysine 40 (Figure 4C). A K40A mutant of Rv0831c lost probe binding ability, confirming K40 as the reactive nucleophile (Figure 4B). Thus, Rv0036c and Rv0831c both contain reactive lysine residues, confirming their labeling by hydrolysis of the acyl phosphate moiety of ATP-ABP, and suggesting more pervasive presence of reactive lysine-based ATPases with new and previously unrecognized sequences (Figure 4). Finally, as an additional control for the reliability of MS- based assignment of reactive residues, we tested labeling, competition with ATPγS and ATP (Figure 4A), and identification of the site of probe labeling by MS for two serine/threonine protein kinases identified in our ATP-ABP screen, Rv0014c (PknB) and Rv0931c (PknD). Labeling of PknB was found at lysine 40, and PknD was labeled at lysine 44 (Figure 4A, Supplemental Figure S1). These sites of labeling match the expected ATP-binding site in PknB, and the equivalent PknD site (Lombana, et. al., 2010).
Experimental validation of structural annotation
To complement our experimental functional annotation, we performed an experimental structural annotation (Ansong, et al., 2008) to further improve the Mtb genome annotation. Using a previously described bacterial proteogenomics pipeline (Venter, et al., 2011), we analyzed global proteomic measurements of Mtb H37Rv to identify novel coding regions in the genome (Methods). These data validated ~50% of the predicted Mtb proteome at the protein level, corrected 40 translational start site errors (Supplemental Table S3), and identified 15 new protein-coding genes (Supplemental Table S4). An example of a novel protein-coding gene identified by our analysis is shown in Figure 5. The novel ORF now annotated as Rv4010 is defined by three peptides mapping to the genomic region 1113888 to 1114109, where no gene had been predicted. Note the presence of a canonical start codon ATG upstream of peptides defining the putative translational start site. Homology analysis of the novel genes revealed that most of the proteins are unannotated hypotheticals. Moreover, most are short (median length of 64 aa), and not annotated outside of Mycobacteria. Potentially due to either their length or their exclusive taxonomic distribution, the annotation of these 15 genes is sporadic within Mtb genomes. As of the writing of this report, NCBI lists 132 Mtb genome projects. Some of the novel genes identified here are annotated in the genomes of numerous Mtb strains (e.g. Rv4007 annotated in >60 strains), while some are only annotated in a few (Rv4014 annotated in < 10). This lack of annotation in other Mtb genomes typically represents false negatives missed during the annotation process, rather than strain diversity. The 15 newly identified protein coding-genes, and the 40 corrected gene models have been added to the RefSeq annotation with locus IDs Rv4000-Rv4014. The data can be downloaded directly through the RefSeq FTP site hosted by NCBI (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/Mycobacterium_tuberculosis_H37Rv_uid57777/), and results already appear in all NCBI tools such as BLAST.
Figure 5. Identification of novel Mtb H37Rv genes.
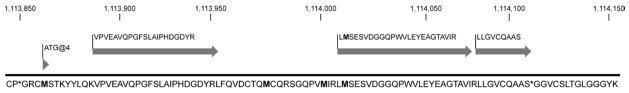
Three peptides identified by MS-based proteomics map to the genomic region 1113888 to 1114109 on the forward strand, where no gene had been previously predicted by computational approaches. Note canonical start codon ATG upstream of peptides.
Data Access
The 15 newly identified protein coding-genes, and the 40 corrected gene models have been added to the RefSeq annotation with locus IDs Rv4000-Rv4014. The data can be downloaded directly through the RefSeq FTP site hosted by NCBI (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/Mycobacterium_tuberculosis_H37Rv_uid57777/), and results already appear in all NCBI tools such as BLAST. All proteomics data has been deposited into the publically available, omics.pnl.gov, website. The newly identified protein coding-genes, corrected gene models, and proteomics data can also be found in Supplemental Tables S1–S4.
Discussion
Functional annotation of bacterial genomes has been exceedingly challenging, and simultaneous global annotation across an entire protein functional class remains largely intractable (Galperin and Koonin, 2010). High throughput experimental methods are needed for functional annotation to systematically characterize bacterial genomes. Computational prediction of function in bacteria is often incomplete or wrong (Deutschbauer, et al., 2011), and even in highly studied model systems such as E. coli, hundreds of genes remain poorly annotated or entirely hypothetical (Keseler, et al., 2011). Most experimentally characterized bacterial genes are derived from a small number of representative bacteria. This limits computational analyses to characterization of functions within gene families from a small set of bacteria for which a priori knowledge already exists. This leaves large areas of bacterial functional pathways in the dark (Rost, 2002). Other gene classes, such as transcription factors and transport proteins, have little conserved sequence homology, and computational approaches are unreliable for their identification (Price, et al., 2007; Ren and Paulsen, 2005). Moreover, the high GC content and dissimilarity of Mtb to other prokaryotes has made computational functional annotation of its genome particularly challenging (Kelkar, et al., 2011); as a consequence, no functional information is available for ~25% of Mtb proteins, highlighting the experimental challenges of protein function assignment in Mtb. Even using inference methods, which assign function via gene neighborhood analyses and other exhaustive informatics approaches (Doerks, et al., 2012), much of the Mtb genome remains functionally undefined. Therefore, systematic experimental methods for elucidating gene function in Mtb are needed, and these may lead to novel therapeutic targets, and targeted mapping of the biological pathways associated with Mtb viability, pathogenesis, and drug resistance.
Chemical biology approaches, in particular activity-based protein profiling (ABPP), have been developed to address these shortcomings (Barglow and Cravatt, 2007; Gomez, et al., 2011). While ABPP is emerging as a powerful approach to comprehensively identify protein function across a defined enzyme class in a proteome, this approach has not been applied to bacterial annotation. Here, we establish an ABPP-MS platform towards the functional annotation of protein functional classes in mycobacteria, and apply the strategy towards defining the Mtb adenosine nucleotide binding family. We chose the ATP-binding protein functional class because of its large size, central role in Mtb physiology and pathogenicity, the close similarity of the probe to the natural ligand, and because probe labeling is direct experimental evidence for hydrolysis by the target protein.
Our study identified 317 proteins, a majority of which was previously annotated as ATP-binding proteins, and using a less stringent cutoff, another 277 ATP binding proteins (Supplemental Tables S1 and S2). Although in some cases the ATPase activity of proteins is well documented, such as the serine/threonine protein kinases, most annotation is still inferred from sequence homology. Our data validate such inferences through direct experimental measurement. In many cases, our study provides the first experimental data available on the function of these enzymes. The “true positives” for which our data confirms previous annotation provide a benchmark for the selectivity and reliability of our approach. Among the targets identified here, we estimate a false positive rate of ~3%, as nine of 317 proteins were labeled, but were annotated as something other than ATP binding proteins (Figure 2).
The number of potential ATP-binding protein families in Mycobacteria is anticipated to be large. When analyzing the total complement of probe-labeled proteins, many of the ATP-binding families were represented by only one or two probe-labeled proteins. Of the 317 probe-labeled proteins, 279 did not have at least 20% sequence homology to the other probe-labeled proteins, implying the presence of numerous different ATP-binding protein families. The most common homologous domains identified in the probe-labeled proteins were the protein kinase domain, PF00069, and the ABC transporter ATP-binding protein domain, PF00005. Our data suggest that there is a great deal of unexplored ATP-binding protein space yet to be discovered.
Coverage of the entire complement of ATP-binding proteins in Mtb by our ABPP approach is not expected. ATP-ABP only labels proteins active under the given experimental condition. In evaluating a single growth phase in a defined medium, many proteins are likely not expressed, or the conditions may not render them functionally active. Indeed, this selectivity of the ABPP approach for functional enzymes facilitates new experimental design and functional discovery. Future efforts will profile protein functional changes under different experimental conditions specific to infection and drug resistance.
Although any chemical activity probe differing from the natural ligand has the potential for off-target labeling, our probe comes very close to the natural ligand and should minimize off-target binding. The only chemical differences between the ATP-ABP and ATP are in the extended triphosphate moiety, carrying a mixed anhydride (acylphosphate) reactive group containing the click-chemistry compatible alkyne. Beside this one modification, the adenosine of ATP is unchanged, allowing the probe to work as a faithful ATP mimetic. The reactive acyl phosphate moiety of ATP-ABP raises the possibility that surface residues react with the probe independent of adenosine binding. To identify and exclude these labeling events, we used ATPγS to test if the adenosine moiety can compete for all probe binding. Our quantitative MS approach allowed for precise determination of ATPγS binding relative to probe binding and led to confident detection of off-target binding across all targets. Binding in the absence of concomitant adenosine binding was rare, and was excluded from our analysis if competition with ATPγS was below a five-fold cut-off (p <0.05).
One major finding of this study is the identification and re-classification of 72 hypothetical proteins as ATP-binding proteins. These included 36 hypothetical proteins for which subsequent HMM profile analysis identified sequence similarity to nucleotide binding domains, and 36 hypothetical proteins that do not have discernible homology to known nucleotide binding domains, including eight that are essential for growth and infection. This latter set of 36 hypothetical proteins likely represents novel families of ATP-binding proteins. Recombinant expression and sequence analysis of two hypotheticals shows that they indeed label at residues consistent with ATP binding, suggesting shared reaction mechanisms with known ATP-binding proteins. Further experimental analysis will be necessary to fully confirm their role in ATP binding and hydrolysis, but our initial sample suggests that many of these are indeed functional ATP-binding proteins. Thus, ATP binding appears to be more widespread than previously thought, and can be facilitated by a much larger number of proteins with highly varied and novel sequences. Identification of these new members of the ATP binding family will aid in the annotation of other bacterial genomes and provide starting points for more generally defining the possible evolutionary solutions for ATP binding of proteins.
Significance
Mycobacterium tuberculosis, the causative agent of tuberculosis, is the main cause of death from bacterial infections. Our understanding of Mtb pathogenesis is limited by a lack of information on even the most basic functions of >25% of Mtb proteins. Although computational tools can predict protein function, these predictions are often incomplete and error-prone. Approaches for high-throughput experimental annotation are urgently needed.
We introduce a high-throughput approach for functional annotation of bacterial proteins that combines activity-based protein profiling and quantitative mass spectrometry. Probing the binding of the most prevalent protein cofactor, ATP, in the Mtb proteome, we confirm predictions on >250 ATP-binding proteins, and identify 72 hypothetical proteins as novel ATP-binding proteins, including proteins essential to ESX-1 secretion, a major virulence determinant of Mtb. We confirm lysine-based ATPase activity of hypothetical proteins with highly divergent sequences and, together with bioinformatic sequence analysis, determine that the probe-labeled hypothetical proteins contain a diversity of unrelated sequences, providing a new and expanded view of adenosine nucleotide binding in Mtb. Many of these hypothetical proteins are both unique to Mycobacteria and essential for infection, suggesting specialized functions in mycobacterial physiology and pathogenicity. Our ABPP platform provides a generally applicable approach for high-throughput protein function discovery and validation, and provides a large set of previously unrecognized ATP binding proteins.
Experimental Procedures
Probe Synthesis
See the Supplemental Data.
Preparation of M. tuberculosis H37Rv Cell Lysates
Mtb strain H37Rv was grown in 7H9 medium to an optical density of 1 measured at 600nm. Cells were harvested by centrifugation, washed in phosphate buffered saline, and lysed by bead-beating. Insoluble material was pelleted by centrifugation and the lysates were passed twice through a 0.2 μm filter for sterilization.
Probe Labeling and Sample Preparation for SDS-PAGE analysis
Log-phase Mtb H37Rv cell lysates (1 mg protein) in PBS were treated with ATP-ABP (20 μM), vortexed, and incubated for 1 hr at 37 °C.
Following probe incubation, proteomes were treated with an azide-derivatized Cy5.5 fluorescent reporter group (75 μM), tris(2-carboxyethyl) phosphine (TCEP, 1 mM), tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA, prepared in 4:1 tert-butanol:DMSO, 100 μM), and CuSO4 (1 mM). The samples were vortexed and incubated at room temperature in the dark for 1 hr. SDS-PAGE loading buffer (reducing) was added to the samples, heated at 85 °C for 2 minutes, and loaded onto a 10% Tris-Glycine gel. Gels were imaged using a Protein Simple FluorchemQ system.
Probe Labeling and Sample Preparation for LC-MS analysis
Mtb cell lysates (1 mg protein) were treated with ATP-ABP (20 μM), DMSO (no probe control), or ATPγS (inhibition control, 1 mM). Following addition of ATPγS, ATP-ABP (20 μM) was added. All samples were incubated for 1 hour at 37°C. Following probe incubation, pro teomes were treated with biotin-azide (36 μM), TCEP (2.5 mM), TBTA (250 μM), and CuSO4 (0.50 mM). The samples were vortexed and incubated at room temperature in the dark for 1.5 hours. Probe-labeled proteins were then enriched on streptavidin resin, reduced with TCEP, and alkylated with iodoacetamide. Proteins were digested on-resin with trypsin, and the resulting peptides collected for LC-MS analysis. For full details see the Supplemental Data.
LC-MS Analysis of Probe-Labeled Samples Utilizing AMT tag Approach
Proteomics data for unlabeled, probe-labeled, and inhibitor-pretreated probe-labeled samples were generated and analyzed using the accurate mass and time (AMT) tag proteomics approach (Zimmer, et al., 2006). See Supplemental Data for details.
To identify a protein as specifically labeled by the ATP-ABP, we required the following criteria: (i) the protein exhibits a significant difference across the probe labeled sample, and the two negative control conditions as judged by ANOVA (p<0.05), and (ii) the protein exhibits ≥5-fold more abundance in the probe labeled sample relative to both the no label negative control, and the inhibitor negative control sample.
Expression and Purification of Mtb Hypothetical Proteins
The full-length Rv0036 and Rv0831c genes were amplified from genomic Mtb H37Rv DNA and cloned into the pET28b expression vector in-frame with the N-terminal six-histidine tag. The vector was transformed into BL21 (DE3)-CodonPlus cells, and protein expression was induced at A600 of 0.6 by adding 100 μM isopropyl-1-thio-β-d-galactopyranoside. Protein was expressed for 20 hours at 20°C, cells were harvested, re-susp ended in 20 mM Tris (pH 7.5, 150 mM NaCl), and lysed by sonication. The lysate was cleared by centrifugation, and loaded on a metal-chelating affinity column. Fractions were pooled, loaded on a gel filtration column, and eluted in 20 mM Tris (pH 7.5, 150 mM NaCl).
Analysis of ATP-ABP Labeled Hypothetical Proteins
Purified proteins (30 μg) were labeled with ATP-ABP (20 μM), vortexed, and incubated for 1 h at 37 °C on a thermal mixer with mild agitation. The protein was denatured in 8M urea, digested with trypsin, and the peptides analyzed by LC-MS; see Supplemental Data for details.
Global Proteome Profiling for Structural Annotation
Mtb H37Rv whole cell lysate was prepared in PBS by French press (Mawuenyega, et al., 2005). Cell lysate was separated into cytosolic, light membrane, and cell wall subcellular fractions by centrifugation. Three replicates of whole cell lysate and subcellular fractions were processed, and the tryptic peptides analyzed by LC-MS. MS Spectra were analyzed by the bacterial proteogenomics pipeline using default values (Venter, et al., 2011). Briefly, tandem mass spectra were searched by Inspect against a translation of the genome (NC_000962), and subsequently rescored with PepNovo and MSGF. Searches did not include any post-translational modifications, but in accord with Inspect’s searching paradigm did not require tryptic specificity. Each stop-to-stop open reading frame (ORF) was included regardless of coding potential. We concatenated decoy records by shuffling each ORF. Significant peptide/spectrum matches (PSM) were those with an E-value of e−10 or smaller, which led to a peptide level FDR of ~0.1% (spectrum level FDR ~ 0.024%).
Supplementary Material
Highlights.
Over 250 Mtb ATP-binding proteins validated experimentally.
72 hypothetical Mtb proteins are novel adenosine binders.
The essential ESX-1 proteins Rv3614c-3616c have ATP binding activity.
Acknowledgments
We thank the Biological Separations & Mass Spectrometry group for helpful discussions and critical reading of the document. This work was supported in part by the Laboratory Directed Research and Development Program at PNNL, a national laboratory operated by Battelle for the U.S. DOE under contract DE-AC05-76RL01830. CA and JNA are supported by the NIAID NIH/DHHS through Interagency agreement Y1-AI-8401. This work used instrumentation and capabilities developed under support from the NIGMS (8P41GM103493-10), and the U.S. DOE. Proteomic measurements were made in the Environmental Molecular Sciences Laboratory, a DOE-BER national scientific user facility at PNNL. SHP was supported by the National Science Foundation (EF- 0949047). CG was supported by the Paul G. Allen Family Foundation Grant #8999, the American Lung Association, and a New Investigator Award by the University of Washington Center for AIDS Research, an NIH funded program (P30AI027757) which is supported by the following NIH Institutes and Centers (NIAID, NCI, NIMH, NIDA, NICHD, NHLBI, NIA). CO is the recipient of an American Society of Microbiology Robert D. Watkins Graduate Research Fellowship and a Bank of America Endowed Minority Fellowship. DHH was supported by the NHGRI (R01 HG004881).
Footnotes
Supplemental Data, Figures and Experimental Procedures, including probe synthesis, are available at http://www.chembiol.com.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ansong C, Purvine SO, Adkins JN, Lipton MS, Smith RD. Proteogenomics: needs and roles to be filled by proteomics in genome annotation. Brief Funct Genomic Proteomic. 2008;7:50–62. doi: 10.1093/bfgp/eln010. [DOI] [PubMed] [Google Scholar]
- Barglow KT, Cravatt BF. Activity-based protein profiling for the functional annotation of enzymes. Nat Methods. 2007;4:822–827. doi: 10.1038/nmeth1092. [DOI] [PubMed] [Google Scholar]
- Bork P. Powers and pitfalls in sequence analysis: the 70% hurdle. Genome Res. 2000;10:398–400. doi: 10.1101/gr.10.4.398. [DOI] [PubMed] [Google Scholar]
- Bork P, Koonin EV. Predicting functions from protein sequences--where are the bottlenecks? Nat Genet. 1998;18:313–318. doi: 10.1038/ng0498-313. [DOI] [PubMed] [Google Scholar]
- Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu Rev Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- Deutschbauer A, Price MN, Wetmore KM, Shao W, Baumohl JK, Xu Z, Nguyen M, Tamse R, Davis RW, Arkin AP. Evidence-based annotation of gene function in Shewanella oneidensis MR-1 using genome-wide fitness profiling across 121 conditions. PLoS Genet. 2011;7:e1002385. doi: 10.1371/journal.pgen.1002385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiSabato G, Jencks WP. J Am Chem Soc. 1961;83:4393–4400. [Google Scholar]
- Doerks T, van Noort V, Minguez P, Bork P. Annotation of the M. tuberculosis hypothetical orfeome: adding functional information to more than half of the uncharacterized proteins. PLoS One. 2012;7:e34302. doi: 10.1371/journal.pone.0034302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune SM, Jaeger A, Sarracino DA, Chase MR, Sassetti CM, Sherman DR, Bloom BR, Rubin EJ. Mutually dependent secretion of proteins required for mycobacterial virulence. Proc Natl Acad Sci U S A. 2005;102:10676–10681. doi: 10.1073/pnas.0504922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Koonin EV. From complete genome sequence to ‘complete’ understanding? Trends Biotechnol. 2010;28:398–406. doi: 10.1016/j.tibtech.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JJ, Wattam AR, Cammer SA, Gabbard JL, Shukla MP, Dalay O, Driscoll T, Hix D, Mane SP, Mao C, et al. PATRIC: the comprehensive bacterial bioinformatics resource with a focus on human pathogenic species. Infect Immun. 2011;79:4286–4298. doi: 10.1128/IAI.00207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez JE, Clatworthy A, Hung DT. Probing bacterial pathogenesis with genetics, genomics, and chemical biology: past, present, and future approaches. Crit Rev Biochem Mol Biol. 2011;46:41–66. doi: 10.3109/10409238.2010.538663. [DOI] [PubMed] [Google Scholar]
- Greenstein AE, MacGurn JA, Baer CE, Falick AM, Cox JS, Alber T. M. tuberculosis Ser/Thr protein kinase D phosphorylates an anti-anti-sigma factor homolog. PLoS Pathog. 2007;3:e49. doi: 10.1371/journal.ppat.0030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Selengut JD, White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes KT, Mathee K. The anti-sigma factors. Annu Rev Microbiol. 1998;52:231–286. doi: 10.1146/annurev.micro.52.1.231. [DOI] [PubMed] [Google Scholar]
- Huynen M, Snel B, Lathe W, 3rd, Bork P. Predicting protein function by genomic context: quantitative evaluation and qualitative inferences. Genome Res. 2000;10:1204–1210. doi: 10.1101/gr.10.8.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelkar DS, Kumar D, Kumar P, Balakrishnan L, Muthusamy B, Yadav AK, Shrivastava P, Marimuthu A, Anand S, Sundaram H, et al. Proteogenomic analysis of Mycobacterium tuberculosis by high resolution mass spectrometry. Mol Cell Proteomics. 2011;10:M111 011627. doi: 10.1074/mcp.M111.011627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Keseler IM, Collado-Vides J, Santos-Zavaleta A, Peralta-Gil M, Gama-Castro S, Muniz-Rascado L, Bonavides-Martinez C, Paley S, Krummenacker M, Altman T, et al. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res. 2011;39:D583–590. doi: 10.1093/nar/gkq1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluger R. Acyl Phosphate Esters: Charge-Directed Acylation and Artificial Blood. Synlett. 2000:1708–1720. [Google Scholar]
- Lombana TN, Echols N, Good MC, Thomsen ND, Ng HL, Greenstein AE, Falick AM, King DS, Alber T. Allosteric activation mechanism of the Mycobacterium tuberculosis receptor Ser/Thr protein kinase, PknB. Structure. 2010;18:1667–1677. doi: 10.1016/j.str.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew JM, Kapopoulou A, Jones LM, Cole ST. TubercuList--10 years after. Tuberculosis (Edinb) 2011;91:1–7. doi: 10.1016/j.tube.2010.09.008. [DOI] [PubMed] [Google Scholar]
- MacGurn JA, Raghavan S, Stanley SA, Cox JS. A non-RD1 gene cluster is required for Snm secretion in Mycobacterium tuberculosis. Mol Microbiol. 2005;57:1653–1663. doi: 10.1111/j.1365-2958.2005.04800.x. [DOI] [PubMed] [Google Scholar]
- Magnet S, Hartkoorn RC, Szekely R, Pato J, Triccas JA, Schneider P, Szantai-Kis C, Orfi L, Chambon M, Banfi D, et al. Leads for antitubercular compounds from kinase inhibitor library screens. Tuberculosis (Edinb) 2010;90:354–360. doi: 10.1016/j.tube.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Mawuenyega KG, Forst CV, Dobos KM, Belisle JT, Chen J, Bradbury EM, Bradbury AR, Chen X. Mycobacterium tuberculosis functional network analysis by global subcellular protein profiling. Mol Biol Cell. 2005;16:396–404. doi: 10.1091/mbc.E04-04-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton GL, Leung SS, Wakabayashi JI, Rawat M, Fahey RC. The DinB superfamily includes novel mycothiol, bacillithiol, and glutathione S-transferases. Biochemistry. 2011;50:10751–10760. doi: 10.1021/bi201460j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patricelli MP, Szardenings AK, Liyanage M, Nomanbhoy TK, Wu M, Weissig H, Aban A, Chun D, Tanner S, Kozarich JW. Functional interrogation of the kinome using nucleotide acyl phosphates. Biochemistry. 2007;46:350–358. doi: 10.1021/bi062142x. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. Orthologous transcription factors in bacteria have different functions and regulate different genes. PLoS Comput Biol. 2007;3:1739–1750. doi: 10.1371/journal.pcbi.0030175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Wang Y. Probing adenosine nucleotide-binding proteins with an affinity-labeled nucleotide probe and mass spectrometry. Anal Chem. 2007;79:5547–5556. doi: 10.1021/ac0622375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy TB, Riley R, Wymore F, Montgomery P, DeCaprio D, Engels R, Gellesch M, Hubble J, Jen D, Jin H, et al. TB database: an integrated platform for tuberculosis research. Nucleic Acids Res. 2009;37:D499–508. doi: 10.1093/nar/gkn652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Q, Paulsen IT. Comparative analyses of fundamental differences in membrane transport capabilities in prokaryotes and eukaryotes. PLoS Comput Biol. 2005;1:e27. doi: 10.1371/journal.pcbi.0010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B. Enzyme function less conserved than anticipated. J Mol Biol. 2002;318:595–608. doi: 10.1016/S0022-2836(02)00016-5. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. TM4 microarray software suite. Methods in enzymology. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- Sadler NC, Angel TE, Lewis MP, Pederson LM, Chauvigne-Hines LM, Wiedner SD, Zink EM, Smith RD, Wright AT. Activity-based protein profiling reveals mitochondrial oxidative enzyme impairment and restoration in diet-induced obese mice. PLoS One. 2012;7:e47996. doi: 10.1371/journal.pone.0047996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber M, Res I, Matter A. Protein kinases as antibacterial targets. Curr Opin Cell Biol. 2009;21:325–330. doi: 10.1016/j.ceb.2009.01.026. [DOI] [PubMed] [Google Scholar]
- Simon GM, Cravatt BF. Activity-based proteomics of enzyme superfamilies: serine hydrolases as a case study. J Biol Chem. 2010;285:11051–11055. doi: 10.1074/jbc.R109.097600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speers AE, Adam GC, Cravatt BF. Activity-based protein profiling in vivo using a copper(i)-catalyzed azide-alkyne [3 + 2] cycloaddition. J Am Chem Soc. 2003;125:4686–4687. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- Speers AE, Cravatt BF. A tandem orthogonal proteolysis strategy for high-content chemical proteomics. J Am Chem Soc. 2005;127:10018–10019. doi: 10.1021/ja0532842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter E, Smith RD, Payne SH. Proteogenomic analysis of bacteria and archaea: a 46 organism case study. PLoS One. 2011;6:e27587. doi: 10.1371/journal.pone.0027587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer JS, Monroe ME, Qian WJ, Smith RD. Advances in proteomics data analysis and display using an accurate mass and time tag approach. Mass Spectrom Rev. 2006;25:450–482. doi: 10.1002/mas.20071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.