Abstract
Engineered transcription activator-like effector nucleases (TALENs) have shown promise as facile and broadly applicable genome editing tools. However, no publicly available high-throughput method for constructing TALENs has been published and large-scale assessments of the success rate and targeting range of the technology remain lacking. Here we describe the Fast Ligation-based Automatable Solid-phase High-throughput (FLASH) platform, a rapid and cost-effective method we developed to enable large-scale assembly of TALENs. We tested 48 FLASH-assembled TALEN pairs in a human cell-based EGFP reporter system and found that all 48 possessed efficient gene modification activities. We also used FLASH to assemble TALENs for 96 endogenous human genes implicated in cancer and/or epigenetic regulation and found that 84 pairs were able to efficiently introduce targeted alterations. Our results establish the robustness of TALEN technology and demonstrate that FLASH facilitates high-throughput genome editing at a scale not currently possible with engineered zinc-finger nucleases or meganucleases.
Engineered Transcription Activator-Like Effector (TALE) repeat domains have generated much interest as a new platform for creating customized DNA-binding proteins.1–3 TALE repeats are highly conserved 33-35 amino acid sequences found in naturally occurring TALEs encoded by Xanthamonas bacteria. An individual TALE repeat binds to a single base pair of DNA and the identities of amino acids at two positions (known as repeat variable di-residues or RVDs) have been associated with specificities for different nucleotides.4, 5 TALE repeats can be joined together into more extended arrays capable of recognizing novel target DNA sequences. Such engineered arrays have been fused to gene regulatory domains to create customized transcription factors4, 6–11 and to non-specific nuclease domains to create targeted TALE nucleases (TALENs).6, 12–22 Repair of TALEN-induced double-strand breaks (DSBs) by either non-homologous end-joining (NHEJ) or homology-directed repair can induce efficient alteration of endogenous genes in yeast,16 plants,17 nematodes,18 zebrafish,21, 22 rats,20 and human somatic6, 15, 17 and pluripotent stem cells.19
Although the ability to design TALENs to nearly any DNA sequence of interest has been highlighted as an important potential advantage of the technology,1–3 only a very limited number of endogenous genes (17 in total) have been altered using TALENs in the published literature to date.6, 15–22 The TALENs used in these studies were constructed on different architectures and composed of variable numbers of TALE repeats making it difficult to ascertain whether these parameters affect the efficiencies of nuclease activity. In addition, a recent report has suggested limits to the targeting range of TALENs based on a computational analysis of naturally occurring TALE binding sites.17.
The simplicity of TALEN design raises the exciting prospect that large-scale pathway-and genome-wide gene modification projects might be possible but currently no publicly available, cost-effective, and high-throughput method for constructing these nucleases exists. Many different methods for constructing TALENs have been described,8–11, 16, 17, 21, 22 most of these utilizing variations on the Golden Gate cloning method. However, none of these methods are readily adaptable for automated high-throughput production due to requirements for PCR, gel isolation of fragments, and/or passage and characterization of intermediate constructs.8–11, 16, 17, 21, 22 In addition, many of these methods only enable production of TALE repeat arrays composed of certain fixed numbers of repeats.8, 11, 16, 23 A commercial high-throughput platform exists – Cellectis Bioresearch has advertised the capability to produce 7200 TALENs per year (or ~96 TALENs every 5 days) – but details of this proprietary method are not publicly available.
Here we describe development and optimization of the Fast Ligation-based Automatable Solid-phase High-throughput (FLASH) assembly method for rapid construction of large numbers of TALE repeat arrays. We used FLASH to construct 48 TALEN pairs targeted to a diverse range of EGFP reporter gene sequences and found that 100% of these nucleases were active in a human cell-based assay. We also made FLASH TALEN pairs targeted to 96 human genes involved in cancer or epigenetics and were able to use these nucleases to rapidly introduce targeted alterations into 84 of these genes. Our results provide large-scale experimental support for the broad and robust targeting range of TALEN technology and proof-of-principle that the FLASH platform can enable rapid, high-throughput gene editing not currently possible with engineered zinc-finger nucleases (ZFNs) or meganucleases.
Results
FLASH: An Automated, High-Throughput Method for Assembling TALE Repeat Arrays
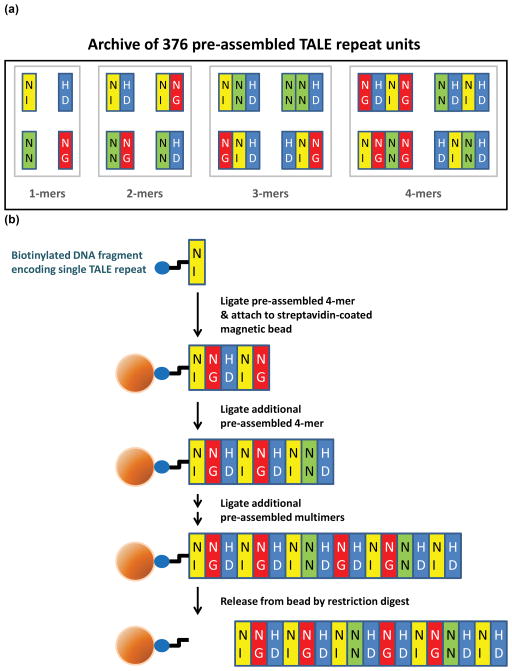
To enable large-scale construction of TALE repeat arrays, we devised the Fast Ligation-based Automatable Solid-phase High-throughput (FLASH) assembly method. Practice of FLASH relies on an archive of 376 plasmids that encode one, two, three, or four TALE repeats consisting of all possible combinations of the NI, NN, HD, or NG RVDs (Methods, Figure 1a, and Supplementary Tables 1 and 2). DNA fragments encoding TALE repeats are assembled in an iterative fashion on solid-phase magnetic beads, an innovation that permits automation of serial restriction digest, purification, and ligation steps on a liquid-handling platform (Methods, Supplementary Methods, and Figure 1b). Because the final fragment to be ligated can encode one, two, or three TALE repeats, arrays consisting of any desired number of TALE repeats can be assembled (Supplementary Figure 1). DNA fragments encoding the final full-length TALE repeat array are released from the beads by restriction enzyme digestion (Methods, Supplementary Methods and Figure 1b).
Figure 1. Graphical overview of the FLASH assembly method.
(a) Archive of 376 TALE repeat encoding plasmids required to practice FLASH. Plasmids encoding one, two, three, and four TALE repeats (colored rectangles) harboring various RVDs (represented by two upper-case letters within the rectangles) were constructed as described in Methods. (b) Schematic overview of the FLASH assembly process. A DNA fragment encoding a single TALE repeat and labeled on its 5′ end with biotin (blue oval) is initially ligated to a second DNA fragment encoding four specific TALE repeats and then attached to a streptavidin-coated magnetic bead (orange sphere). Additional DNA fragments encoding pre-assembled TALE repeats are ligated until an array of the desired length is assembled. The DNA fragment encoding the full-length TALE repeat array is then cleaved from the bead by restriction digestion.
We optimized FLASH so that it can be efficiently practiced in 96-well format using a robotic liquid-handling workstation (Methods and Supplementary Methods). With automation, we can assemble DNA fragments encoding up to 96 different TALE repeat arrays in less than one day. We also adapted FLASH so that medium-throughput assembly can be performed manually in one to two days using multi-channel pipets (data not shown). Fragments assembled using either approach can be cloned into expression vectors (e.g.—for expression as a TALEN) to generate sequence-verified plasmids in less than one week (Methods and Supplementary Methods). Using automated FLASH, we can make sequence-verified TALE expression plasmids for less than $100 each including the cost of labor.
Large-scale Testing of FLASH-Assembled TALENs Using a Human Cell-based Reporter Assay
To perform a large-scale test of the robustness of TALENs in human cells, we used FLASH to construct plasmids encoding 48 TALEN pairs targeted to different sites in the EGFP reporter gene. Monomers in each TALEN pair contained the same number of repeats (ranging from 8.5 to 19.5) and pairs were targeted to sites possessing a 16 bp “spacer” sequence between the “half-sites” (Supplementary Table 3).
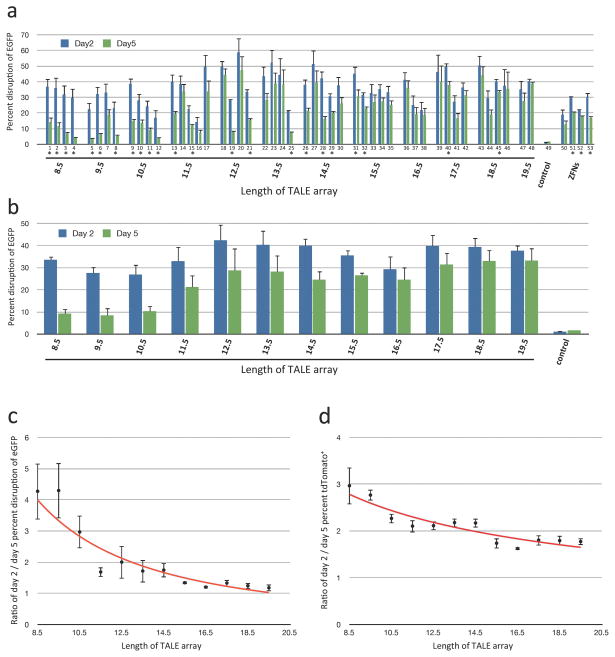
We tested each of these 48 TALEN pairs in human cells for its ability to disrupt the coding sequence of a chromosomally integrated EGFP reporter gene. In this assay, NHEJ-mediated repair of TALEN-induced breaks leads to loss of EGFP expression, which we quantitatively assessed two and five days following transfection (Methods). (All TALEN pairs we made targeted sites located at or upstream of nucleotide position 503 in EGFP, a position we have previously shown will disrupt EGFP function when mutated with a ZFN.24) Strikingly, we found that all 48 TALEN pairs showed significant EGFP gene-disruption activities comparable to those of four EGFP-targeted ZFN pairs previously made by the Oligomerized Pool Engineering (OPEN) method (Figure 2a). These results demonstrate that TALENs containing as few as 8.5 TALE repeats possess significant nuclease activities and provide large-scale evidence of the robustness of TALENs in human cells.
Figure 2. Gene-disruption activities of 48 FLASH-assembled TALENs targeted to EGFP.
(a) Activities of 48 TALEN pairs and four ZFN pairs in the EGFP gene-disruption assay. Percentages of EGFP-disrupted cells as measured 2 and 5 days following transfection of U2OS cells bearing a chromosomally integrated EGFP reporter gene with nuclease-encoding plasmids are shown. Target sites for the 48 TALEN pairs are shown in Supplementary Table 3. The four ZFN pairs were made by OPEN and have been previously described.24 Mean percent disruption of EGFP and standard error of the mean from three independent transfections are shown. All 48 TALEN pairs and all four ZFN pairs tested induced statistically significant (p<0.05) increases in EGFP-disrupted cells relative to control transfected cells on both day 2 and day 5. Asterisks indicate TALEN and ZFN pairs for which statistically significant differences exist between values of EGFP-disrupted cells on day 2 and day 5 (p<0.05). (b) Mean EGFP-disruption activities from (a) grouped by length of the TALENs. (c) Ratio of mean percent EGFP disruption values from day 2 to day 5. Ratios were calculated for groups of each length TALEN using the data from (b). Values greater than 1 indicate a decrease in the average percentage of EGFP-disrupted cells at day 5 relative to day 2. (d) Ratio of mean tdTomato-positive cells on day 2 and day 5 grouped by various lengths of TALENs. tdTomato-encoding control plasmids were transfected together with nuclease-encoding plasmids on day 0.
Interestingly, re-quantification of the percentage of EGFP-disrupted cells at day 5 post-transfection revealed that a greater number of shorter-length TALENs (e.g. those composed of 8.5 to 10.5 repeats) show significant reductions in the percentage of EGFP-disrupted cells than longer-length TALENs (Figure 2a). This trend can also be observed by comparing mean percentages of EGFP-disrupted cells for various length TALENs at day 2 and day 5 post-transfection (Figure 2b) and by plotting mean ratios of day 2/day 5 EGFP-disrupted cells for each TALEN length (Figure 2c). An analysis of variance (ANOVA) of data from Figure 2c (Methods) demonstrated a significant effect of TALEN length on the day 2/day 5 ratio of EGFP-disrupted cells (p = 8.9 × 10−8). One potential explanation for this effect is cellular toxicity associated with expression of shorter-length TALENs. Consistent with this hypothesis, in cells transfected with plasmids encoding shorter-length TALENs, we also observed greater reductions in the percentage of tdTomato-positive cells from day 2 to day 5 post-transfection (Figure 2d) (a tdTomato-encoding plasmid was co-transfected together with the TALEN expression plasmids on day 0). ANOVA of data from Figure 2d (Methods) also showed a significant effect of TALEN length on the day 2/day 5 ratio of tdTomato-positive cells (p = 1.5 × 10−8). Taken together, our results suggest that although shorter-length TALENs are as active as longer-length TALENs, the former can cause greater cytotoxicity in human cells.
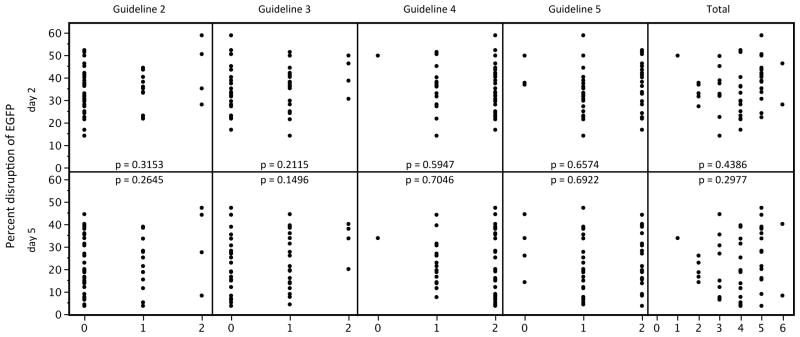
Our EGFP experiments also provided an opportunity to assess previously described computationally-derived design guidelines.17 All 48 of the sequences we targeted in EGFP failed to meet one or more of these guidelines (Supplementary Discussion; Supplementary Table 3; although all of these targets did meet the requirement for a 5′ T in both half-sites). Our 100% success rate for these 48 sites demonstrates that TALENs can be readily obtained for target sequences that do not follow four of these guidelines. In addition, for each of the four design guidelines, we did not find any statistically significant correlation between guideline violation and the level of TALEN activities on either day 2 or 5 post-transfection (Figure 3). We also failed to find a significant correlation between the total number of guideline violations and the level of TALEN activity (Figure 3). Thus, failure to meet four of the five previously described design guidelines when choosing target sequences does not adversely affect activities of TALENs made for those sites.
Figure 3. Computationally-derived design guidelines do not show statistically significant correlation with TALEN activity levels.
Each graph plots the mean EGFP-disruption activity of each of our 48 EGFP-targeted TALEN pairs against the number of half-sites within each target site that fail to meet one of four previously described design guidelines (Guidelines 2, 3, 4, or 5; see Supplementary Discussion17) or against the total number of guideline violations in each target site (Total Guideline Violations). Plots are shown for EGFP-disruption activities measured on day 2 and day 5 post-transfection. Guideline violation data are from Supplementary Table 3. Correlation p-values are shown for each plot.
High-throughput alteration of endogenous human genes using FLASH-assembled TALENs
We next sought to test the efficiency of TALENs for modifying endogenous genes in human cells. To do this, we used FLASH to engineer TALEN pairs for targets in 96 different human genes: 78 genes implicated in human cancer25 and 18 genes involved in epigenetics (Supplementary Table 4). For each gene, we designed TALENs that cleave near the amino-terminal end of the protein coding sequence although in a small number of cases the presence of repetitive sequences led us to target alternate sites in downstream exons or introns (Supplementary Table 4). Guided by our results with the EGFP TALENs and by spacer lengths defined in earlier studies,6 we constructed TALENs composed of 14.5, 15.5, or 16.5 repeats designed to cleave sites with 16, 17, 18, 19 or 21 bp spacers.
We tested the activities of our 96 TALEN pairs at their intended endogenous gene targets using a modified T7 Endonuclease I (T7EI) assay (Methods and Supplementary Figure 2).15, 26 In this assay, 84 of the 96 TALEN pairs showed efficient NHEJ-mediated mutagenesis at their intended target sites, an overall success rate of ~88% (Table 1 and Supplementary Figure 3). Efficiencies of TALEN-induced mutagenesis ranged from 2.5% to 55.8% with a mean of 22.2%. To provide molecular confirmation of the mutations we identified by T7EI assay, we sequenced target loci for 11 different TALEN pairs that induced varying efficiencies of mutagenesis (Figure 4). As expected, this experiment revealed characteristic insertions or deletion mutations (indels) at the expected target gene sites with frequencies similar to those observed by T7EI assay (compare Figure 4 and Table 1).
Table 1.
TALEN-induced mutation frequencies for 96 endogenous human genes involved in cancer and epigenetic regulation
| Gene | Mean indel mutation frequency (%) ± SEM |
|---|---|
| ABL1 | 22.5 ± 7.1 |
| AKT2 | 14.1 ± 7.3 |
| ALK | 12.7 ± 2.9 |
| APC | 48.8 ± 9.8 |
| ATM | 35.5 ± 15.6 |
| AXIN2 | 2.5 ± 0.6 |
| BAX | 14.7 ± 11.6 |
| BCL6 | 14.9 ± 5.9 |
| BMPR1A | 50.4 ± 16.4 |
| BRCA1 | 44.5 ± 15.5 |
| BRCA2 | 41.6 ± 10.5 |
| CBX3 | 35.2 ± 22.6 |
| CBX8 | 13.5 ± 3.4 |
| CCND1 | 40.5 ± 2.2 |
| CDC73 | 36.3 ± 7.7 |
| CDH1 | none |
| CDK4 | 21.5 ± 17.4 |
| CHD4 | 9.6 ± 0.1 |
| CHD7 | 11.4 ± 2.7 |
| CTNNB1 | 26.0 ± 8.1 |
| CYLD | 24.7 ± 2.3 |
| DDB2 | 15.8 ± 7.2 |
| ERCC2 | 55.8± 12.7 |
| ERCC5 | none |
| EWSR1 | 14.3 ± 8.2 |
| EXT1 | 9.5 ± 3.0 |
| EXT2 | 4.0 ± 1.2 |
| EZH2 | 41.3 ± 2.6 |
| FANCA | 9.7 ± 5.0 |
| FANCC | 23.7 ± 17.8 |
| FANCE | none |
| FANCF | 46.0 ± 7.7 |
| FANCG | 26.9 ± 16.2 |
| FES | 12.6 ± 10.6 |
| FGFR1 | 17.4 ± 6.2 |
| FH | 20.9 ± 11.8 |
| FLCN | 11.1 ± 4.4 |
| FLT3 | none |
| FLT4 | 9.9 ± 5.0 |
| FOXO1 | 8.5 ± 1.1 |
| FOXO3 | 7.3 ± 2.3 |
| GLI1 | 21.5 ± 12.4 |
| HDAC1 | 10.8 ± 3.0 |
| HDAC2 | 4.2 ± 0.9 |
| HDAC6 | 21.4 ± 2.1 |
| HMGA2 | 3.0 ± 1.5 |
| HOXA13 | 7.6 ± 3.1 |
| HOXA9 | 6.4 ± 2.7 |
| HOXC13 | 10.5 ± 0.3 |
| HOXD11 | none |
| HOXD13 | none |
| JAK2 | 44.9 ± 16.9 |
| KIT | none |
| KRAS | 9.4 ± 0.9 |
| MAP2K4 | 11.9 ± 7.1 |
| MDM2 | 33.0 ± 20.2 |
| MET | 40.4 ± 10.7 |
| MLH1 | 44.9 ± 6.3 |
| MSH2 | 27.5 ± 10.4 |
| MUTYH | 24.9 ± 8.4 |
| MYCL1 | 17.3 ± 0.6 |
| MYC | 13.4 ± 4.0 |
| MYCN | 16.3 ± 11.6 |
| NBN | 46.3 ± 15.5 |
| NCOR1 | 29.6 ± 13.1 |
| NCOR2 | 3.3 ± 0.6 |
| NTRK1 | none |
| PDGFRA | 16.0 ± 4.3 |
| PDGFRB | 16.0 ± 3.2 |
| PHF8 | 22.2 ± 6.1 |
| PMS2 | 26.9 ± 9.5 |
| PTCH1 | 27.5 ± 15.9 |
| PTEN | 31.5 ± 11.7 |
| RARA | 13.4 ± 6.1 |
| RBBP5 | 15.7 ± 9.5 |
| RECQL4 | 22.1 ± 16.2 |
| REST | none |
| RET | 5.4 ± 1.8 |
| RNF2 | none |
| RUNX1 | 25.1 ± 6.9 |
| SDHB | 36.4 ± 19.2 |
| SDHC | 13.7 ± 3.4 |
| SDHD | 42.0 ± 7.8 |
| SETDB1 | 33.5 ± 6.1 |
| SIRT6 | 43.3 ± 3.1 |
| SMAD2 | 3.9 ± 1.6 |
| SS18 | 31.4 ± 7.9 |
| SUZ12 | 13.1 ± 0.4 |
| TFE3 | 17.3 ± 2.4 |
| TGFBR2 | none |
| TLX3 | none |
| TP53 | 19.9 ± 3.6 |
| TSC2 | 30.7 ± 22.7 |
| VHL | 19.4 ± 1.1 |
| XPA | 12.9 ± 2.2 |
| XPC | 31.4 ± 4.2 |
Figure 4. DNA sequences and frequencies of FLASH TALEN-induced mutations at endogenous human genes.
For each endogenous gene target, the wild-type (WT) sequence is shown at the top with the TALEN target half-sites highlighted in yellow and the translation start codon of the gene (ATG) underlined and highlighted in bold type. Deletions are indicated by red dashes against a grey background and insertions by lowercase letters against a light blue background. The sizes of the insertions (+) or deletions (▵) are indicated to the right of each mutated site. The number of times that each mutant was isolated is shown in parentheses. Mutation frequencies are calculated as the number of mutants identified divided by the total number of sequences analyzed. Note that for several of the genes, we also identified larger deletions that extend beyond the sequences of the TALEN target sites. For each gene, sequencing was performed with the same genomic DNA used to perform the corresponding T7EI assays shown in Supplementary Figure 3.
Discussion
The novel FLASH platform we describe here will enable any interested researcher, core facility, or institution to produce TALE repeat arrays in an inexpensive and high-throughput manner. With FLASH, DNA fragments are assembled on solid phase magnetic beads rather than in solution, thereby enabling serial enzymatic reactions to be performed without the need for column-based wash or purification steps. FLASH also avoids the need for gel isolation or analysis of intermediate constructs, both of which are labor-intensive and difficult to automate. When performed with a liquid-handling robotic platform, FLASH can be used to assemble DNA fragments encoding 96 arrays in less than a day.
Our large-scale testing of 144 FLASH-assembled TALEN pairs provides the most comprehensive test of TALEN technology performed to date. 100% of the 48 EGFP-targeted TALEN pairs and ~88% of the 96 endogenous gene-targeted TALEN pairs we produced with FLASH can cleave their targets in human cells with mutation efficiencies similar to those induced by ZFNs engineered by the selection-based OPEN method. The nucleotide composition of the 96 EGFP TALEN half-sites and the 168 endogenous gene TALEN half-sites for which we successfully made TALENs is quite diverse, reflecting DNA sequences composed of variable numbers and percentages of G, A, T, and C bases (Supplementary Figures 4 and 5). We do not know the precise reason(s) why 12 TALEN pairs targeted to endogenous human genes failed to show activities in our T7EI assay. Possible explanations include inhibitory effects of chromatin structure or DNA modification or inefficient expression and/or folding of particular TALENs. Nonetheless, the high success rate we observe suggests that pre-screening TALENs using other surrogate assays (e.g. yeast-based reporter assays12, 17) may be unnecessary and that TALENs might be tested directly at the endogenous gene target in the cell type or organism of interest.
We note that all of the TALENs we assembled using FLASH were made using a particular framework of TALE repeats and amino- and carboxy-terminal sequences first described by Miller and colleagues.6 This framework has been used to construct nucleases that function efficiently in nematodes,18 zebrafish,21, 22 rats,20 and human somatic6, 15, 17 and pluripotent stem cells,19 and we therefore predict that FLASH-assembled TALENs will also likely show high activities and high success rates in other cell types and organisms. A related question to test in future experiments will be whether TALENs made on any of the other various architectures described in the literature8–11, 16, 17 will also exhibit the robustness that we observe with the particular framework used in our FLASH platform.
Our results demonstrate that the targeting range of TALENs is actually substantially higher than previously suggested by the Bogdanove and Voytas groups who have described five design guidelines for choosing potential cleavage sites (Supplementary Discussion). These guidelines limit the targeting range of TALENs to approximately one site in every 35 bps of DNA sequence17 and have been implemented in their web-based TALE-NT software17 (http://boglabx.plp.iastate.edu/TALENT/TALENT/). We found that we were able to successfully make active TALENs for 131 full target sites that fail to meet one or more of four of these design guidelines. Furthermore, we did not find any statistically significant correlation between failure to meet these four guidelines and the activity levels of our 48 EGFP-targeted TALENs. The discrepancy between these computationally-derived guidelines and our experimental results may be because the rules were derived from sites bound by monomeric TALEs whereas TALENs function as dimers.
By systematically making and testing TALENs for target sites of various lengths, we uncovered an inverse correlation between the number of TALE repeats in a TALEN and the degree of associated cytotoxicity. In our EGFP reporter experiments, we found that shorter TALENs are just as active as longer ones but these shorter TALENs also tend to be more cytotoxic, presumably due to their greater potential for binding to off-target sites elsewhere in the genome. Our findings suggest that cytotoxicity might be minimized by constructing longer TALENs (e.g.—those that harbor 14.5 to 19.5 TALE repeats), a hypothesis that can be tested in future experiments. Even with this restriction and other limitations on the length of the spacer sequence, we estimate that on average more than three TALEN pairs can be targeted per base pair of random DNA sequence (see Supplementary Discussion).
Production-scale use of FLASH should enable the construction of thousands of TALEN pairs per year. We have already made a total of more than 600 TALEN-encoding plasmids using FLASH (this manuscript and data not shown). In production mode, it should be straightforward for two scientists to construct a set of 96 TALE repeat arrays at least three times per week, enabling the generation of more than 7200 TALEN pairs per year. Our cost for making a pair of sequence-verified TALEN plasmids using FLASH (including labor) is less than $200. This low per-unit cost will be particularly important for large-scale gene editing projects or for academic core facilities interested in making large numbers of nucleases. Importantly, because we have not yet fully optimized the FLASH method, we believe the cost of producing a pair of TALENs could easily be further reduced.
We have also adapted the FLASH method so that it can be performed in medium-throughput. In this modified protocol, the overall approach remains unchanged but manipulations are carried out manually using a multi-channel pipet rather than with a liquid-handling robot. We have successfully used this protocol to assemble dozens of TALE repeat arrays in one to two days (data not shown). This alternative smaller-scale protocol provides access to FLASH for laboratories who do not have automated liquid-handling equipment.
An important issue for future investigation is the extent of undesired off-target alterations introduced by TALENs. Off-target sites for one TALEN pair in the human genome have already been identified using a computational approach.19 Application of improved methods for identification of nuclease off-target cleavage events27, 28 to TALENs may reveal additional off-target sites. Whole exome or genome sequencing, as recently done with human induced pluripotent stem cells modified by ZFNs29 and with yeast modified by TALENs,16 might also be informative. All TALENs we constructed by FLASH harbor the wild-type FokI domain and therefore may form unwanted homodimers capable of inducing off-target mutations. As previously demonstrated by others, using obligate heterodimeric FokI domains may reduce formation of undesirable homodimers.28, 30–32 Until off-target sites can be comprehensively identified, users of TALENs will need to account for these undesired potential effects, just as they currently do for ZFNs.
Our large-scale demonstration of the high success rate and near limitless targeting range of TALEN technology combined with our development of the high-throughput FLASH method represent important advances for the genome engineering field. FLASH will encourage and enable any researcher to rapidly, efficiently, and precisely alter any gene or DNA sequence of interest without the need for specialized protein engineering expertise or for extensive screening to identify active nucleases. The capability of FLASH to produce TALENs in high- or medium-throughput will change the scope of gene modification experiments that can be performed by both individual laboratories and core facilities (e.g. enabling pathway- or genome-wide projects). In this regard, we note that in this study we modified more endogenous genes than any other individual report using ZFNs, meganucleases, or TALENs of the past nine years. In addition, although we have focused on TALENs in this report, FLASH should also inspire innovative applications involving the fusion of engineered TALE repeat arrays to other functional domains to create novel targetable chimeric proteins. All reagents needed to practice FLASH and all TALEN expression plasmids we assembled will be available by request to members of the academic research community (http://www.talengineering.org). We expect that the FLASH TALEN platform should enhance the adoption and application of genome engineering technologies by a broad range of researchers.
Methods
Construction of a plasmid archive encoding pre-assembled TALE repeats
We sought to construct TALE repeat arrays using the same architecture first described by Miller et al.6 in which four distinct TALE repeat backbones that differ slightly in their amino acid and DNA sequences occur in a repeated pattern. We designated the first, amino-terminal TALE repeat in an array as the α unit. This is followed by β, γ, and δ units and then an ε unit that is essentially identical to the α unit except for the different positioning of a Type IIS restriction site on the 5′ end (required to enable creation of a unique overhang on the α unit needed for cloning). The ε unit is then followed again by repeats of β, γ, δ, and ε units. Due to constraints related to creation of a 3′ end required for cloning, slightly modified DNA sequences were required for TALE repeat arrays that end with a carboxy-terminal γ or ε unit. We designated these variant units as γ* and ε*.
For each type of TALE repeat unit (i.e.—α, β, γ, δ, ε, γ*, and ε*), we commercially synthesized (Genscript) a series of four plasmids, each harboring one of the four repeat variable di-residues (RVDs) that specifies one of the four DNA bases (NI = A; HD=C; NN=G; NG=T). Full DNA sequences of these plasmids are provided in Supplementary Table 1 and Supplementary Figure 6). For all 28 of these plasmids, the sequence encoding the TALE repeat domain is flanked on the 5′ end by unique XbaI and BbsI restriction sites and on the 3′ end by unique BsaI and BamHI restriction sites. Additionally, the overhangs generated by digestion of any plasmids encoding units designed to be adjacent to one another (e.g.—β and γ, or δ and ε) with BsaI and BbsI are complementary. Using these 28 different plasmids and serial ligation via the BsaI and BbsI restriction sites as previously described,21 we assembled an archive of all possible combinations of βγδε, βγδ, βγ, βγ*, and δε* repeats. In total, this archive consisted of 368 different plasmids encoding 256 βγδε combinations, 64 βγδ combinations, 16 βγ combinations, 16 βγ* combinations, and 16 δε* combinations (Supplementary Table 2). These 368 plasmids plus eight of the original 28 plasmids encoding single TALE repeats (four α and four β plasmids) are required to practice FLASH. With this archive of 376 plasmids listed in Supplementary Table 2, FLASH can be used to construct TALE repeat arrays of any desired length and composition (Supplementary Figure 1).
Preparation of TALE repeat-encoding DNA fragments for FLASH assembly
We provide here an overview of the FLASH assembly method and of the process for subcloning of FLASH-assembled DNA fragments into TALEN expression vectors but more detailed step-by-step protocols can be found in the Supplementary Methods. To prepare DNA fragments encoding α units for use in FLASH assembly, we performed 20 rounds of PCR with each α unit plasmid as a template using primers oJS2581(5′-Biotin–TCTAGAGAAGACAAGAACCTGACC–3′) and oJS2582(5′–GGATCCGGTCTCTTAAGGCCGTGG–3′). The resulting PCR products are biotinylated on the 5′ end. Each α PCR product was then digested with 40 units of BsaI-HF restriction enzyme to generate 4bp overhangs, purified using the QIAquick PCR purification kit (QIAGEN) according to manufacturer’s instructions except that the final product was eluted in 50 μl of 0.1X EB.
To prepare DNA fragments encoding β, βγδε, βγδ, βγ, βγ*, and δε* repeats, we digested 10 μg of each of these plasmids with 50 units of BbsI restriction enzyme in NEBuffer 2 for 2 hours at 37°C followed by serial restriction digests performed in NEBuffer 4 at 37°C using 100 units each of XbaI, BamHI-HF, and SalI-HF enzymes that were added at 5 minute intervals. The latter set of restriction digestions are designed to cleave the plasmid backbone to ensure that this larger DNA fragment does not interfere with subsequent ligations performed during the FLASH assembly process. These restriction digest reactions were then purified using the QIAquick PCR purification kit (QIAGEN) according to manufacturer’s instructions except that the final product was eluted in 180 μl of 0.1X EB.
Automated FLASH assembly
All steps of FLASH assembly were performed using a Sciclone G3 liquid-handling workstation (Caliper) in 96-well plates and using a SPRIplate 96-ring magnet (Beckman Coulter Genomics) and a DynaMag-96 Side magnet (Life Technologies). In the first step of FLASH, a biotinylated α unit fragment is ligated to the first βγδε fragment and then the resulting αβγδε fragments are bound to Dynabeads MyOne C1 streptavidin-coated magnetic beads (Life Technologies) in 2X B&W Buffer. Beads are then drawn to the side of the well by placing the plate on the magnet and then washed with 100 μl B&W buffer with 0.005% Tween 20 (Sigma) and again with 100 μl 0.1 mg/ml bovine serum albumin (BSA) (New England Biolabs). Additional βγδε fragments are ligated by removing the plate from the magnet, resuspending the beads in solution in each well, digesting the bead-bound fragment with BsaI-HF restriction enzyme, placing the plate on the magnet, washing with 100 μl B&W/Tween20 followed by 100μl of 0.1 mg/ml BSA, and then ligating the next fragment. This process is repeated multiple times with additional βγδε units to extend the bead-bound fragment. The last fragment to be ligated is always a β, βγ*, βγδ, or δε* unit to enable cloning of the full-length fragment into expression vectors (note that fragments that end with a δε* unit are always preceded by ligation of a βγ unit).
The final full-length bead-bound fragment is digested with 40 units of BsaI-HF restriction enzyme followed by 25 units of BbsI restriction enzyme (New England Biolabs). Digestion with BbsI releases the fragment from the beads and generates a unique 5′ overhang for cloning of the fragment. Digestion with BsaI-HF results in creation of a unique 3′ overhang for cloning.
Subcloning of TALE repeat array-encoding DNA fragments into TALEN expression vectors
We subcloned DNA fragments encoding our FLASH assembled TALE repeat arrays into one of four TALEN expression vectors. Each of these vectors includes a CMV promoter, a translational start codon optimized for mammalian cell expression, a triple FLAG epitope tag, a nuclear localization signal, amino acids 153 to 288 from the TALE 13 protein (as numbered by Miller et al.6), two unique and closely positioned Type IIS BsmBI restriction sites, a 0.5 TALE repeat domain encoding one of four possible RVDs (NI, HD, NN, or NG for recognition of an A, C, G, or T nucleotide, respectively), amino acids 715 to 777 from the TALE 13 protein, and the wild-type FokI cleavage domain.
All DNA fragments assembled by FLASH possess overhangs that enable directional cloning into any of the four TALEN expression vectors that has been digested with BsmBI. All four of the TALEN expression vectors (each possessing a different 0.5 TALE repeat) are already available from Addgene and full sequences of these plasmids are freely available on a web page dedicated to these constructs: http://www.addgene.org/talengineering/expressionvectors/.
To prepare a TALEN expression vector for subcloning, we digested 5 μg of plasmid DNA with 50 units of BsmBI restriction enzyme (New England Biolabs) in NEBuffer 3 for 8 hours at 55 degrees C. Digested DNA was purified using 90 μl of Ampure XP beads (Agencourt) according to manufacturer’s instructions and diluted to a final concentration of 5ng/μl in 1 mM TrisHCl. FLASH-assembled TALE repeat arrays were ligated into TALEN expression vectors using 400 U of T4 DNA Ligase (New England Biolabs). Ligation products were transformed into chemically competent XL-1 Blue cells. Typically, six colonies were picked for each ligation and plasmid DNA isolated by an alkaline lysis miniprep procedure. Simultaneously, the same colonies were screened by PCR using primers oSQT34 (5′-GACGGTGGCTGTCAAATACCAAGATATG-3′) and oSQT35 (5′-TCTCCTCCAGTTCACTTTTGACTAGTTGGG-3′). PCR products were analyzed on a QIAxcel capillary electrophoresis system (Qiagen). Miniprep DNA from clones that contained correctly sized PCR products were sent for DNA sequence confirmation with primers oSQT1 (5′-AGTAACAGCGGTAGAGGCAG-3′), oSQT3 (5′-ATTGGGCTACGATGGACTCC-3′), and oJS2980 (5′-TTAATTCAATATATTCATGAGGCAC-3′); oSQT1 anneals at the 5′ end of the TALE repeat array coding sequence and enables sequencing of the amino-terminal half of the assembled array, oSQT3 anneals at the 3′ end of the TALE repeat array coding sequence and enables sequencing of the carboxy-terminal half of the assembled array, and oJS2980 primes within the coding sequence of the FokI domain (downstream of oSQT3) and enables sequencing and verification of the carboxy-terminal 0.5 TALE repeat domain.. An example of DNA sequence for a FLASH-assembled TALEN is shown in Supplementary Figure 7.
We typically screened six colonies for each assembly as described above, followed by six additional colonies if necessary. With this approach, we have routinely identified one or more sequence-verified clones for >90% of assembly reactions. These percentages are derived primarily from experiments designed to construct DNA fragments encoding 16.5 TALE repeats. We have observed slightly lower frequencies of success when screening for clones encoding more than 16.5 TALE repeats.
EGFP TALEN activity and toxicity assays
EGFP reporter assays were performed in a clonal U2OS human cell line bearing an integrated construct that constitutively expresses an EGFP-PEST fusion protein. This clonal line was derived from a polyclonal U2OS EGFP-PEST reporter line (a gift from Dr. Toni Cathomen, Hannover Medical School). Clonal U2OS EGFP-PEST cells were cultured in Advanced DMEM (Life Technologies) supplemented with 10% FBS, 2 mM GlutaMax (Life Technologies), penicillin/streptomycin, and 400 μg/ml G418. Cells were transfected in triplicate with 500 ng of each TALEN plasmid DNA and 50 ng ptdTomato-N1 plasmid DNA using a Lonza 4D-Nucleofector System, Solution SE, and program DN-100 according to manufacturer’s instructions. 1 μg of ptdTomato-N1 plasmid alone was transfected in triplicate as a negative control. Cells were assayed for EGFP and tdTomato expression at 2 and 5 days post-transfection using a BD FACSAriaII flow cytometer.
A one-tailed independent two-sample t-test was used to test for increased percentages of EGFP disruption in cells transfected with nucleases (TALENs or ZFNs) compared with control transfected cells (Figure 2a). A one-tailed independent two-sample t-test was used to test for differences in the percentage of EGFP-disrupted cells transfected with TALENs or ZFNs at day 2 and day 5 post-transfection (Figure 2a). Analysis of variance (ANOVA) was performed on the ratio of day 2/day 5 EGFP-disrupted cells as a function of TALE array length (Figure 2c). ANOVA was performed on the ratio of day 2/day 5 tdTomato+ cells as a function of TALE array length (Figure 2d).
PCR amplification and sequence verification of endogenous human genes
PCR reactions to amplify targeted loci were performed using the primers shown in Supplementary Table 5. For most loci, we were able to use standard PCR conditions with Phusion Hot Start II high-fidelity DNA polymerase (Thermo-Fisher) performed according to manufacturer’s instructions for 35 cycles (98°C, 10 s denaturation; 68°C, 15 s annealing; 72°C, 30 s extension). For loci that did not amplify under standard conditions we used one of the following modifications: 1) the addition of betaine to a final concentration of 1.8M, 2) touchdown PCR ([98°C, 10 s; 72–62°C, −1°C/cycle, 15s; 72°C, 30s]10 cycles, [98°C, 10 s; 62°C, −1°C/cycle, 15s; 72°C, 30s]25 cycles) with 1.8M betaine, and 3) the addition of 3% or 5% DMSO and an annealing temperature of 65°C. PCR products were analyzed for correct size on a QIAxcel capillary electrophoresis system. Correctly sized products were treated with ExoSap-IT (Affymetrix) to remove unincorporated nucleotides or primers and sent for DNA sequencing to confirm the endogenous gene sequence.
T7 Endonuclease I assay for quantifying NHEJ-mediated mutation of endogenous human genes
U2OS-EGFP cells were cultured and transfected in duplicate as described above. Genomic DNA was isolated from cells transfected with TALEN-encoding or control plasmids using a high-throughput magnetic-bead based purification system (Agencourt DNAdvance) according to the manufacturer’s instructions. PCR to amplify endogenous loci was performed for 35 cycles as described above and fragments were purified with Ampure XP (Agencourt) according to manufacturer’s instructions. 200 ng of purified PCR product was denatured and reannealed in NEBuffer 2 (New England Biolabs) using a thermocycler with the following protocol (95°C, 5 min; 95-85°C at −2°C/s; 85-25°C at −0.1°C/s; hold at 4°C).33 Hybridized PCR products were treated with 10 U of T7 Endonuclease I at 37°C for 15 minutes in a reaction volume of 20 μl. Reactions were stopped by the addition of 2 μl 0.5 M EDTA, purified with Ampure XP, and quantified on a QIAxcel capillary electrophoresis system using method OM500. The sum of the area beneath TALEN-specific cleavage peaks (expressed as a percentage of the parent amplicon peak, denoted fraction cleaved) is used to estimate gene modification levels using the following equation as previously described33:
Sequence confirmation of endogenous gene mutations
11 genes that showed evidence of mutations in the T7 Endonuclease I assay were chosen for independent confirmation via Sanger sequencing. PCR products corresponding to these 11 sites were cloned into PCR-4-Blunt-Topo using a Topo cloning kit (Life Technologies) and transformed into XL-1 Blue cells. Plasmid DNA was isolated for multiple colonies from each transformation and then sent for DNA sequencing using either the T7 (5′-TAATACGACTCACTATAGGG-3′) and T3 (5′-ATTAACCCTCACTAAAGGGA-3′) primers or the M13F (5′-GTAAAACGACGGCCAG-3′) and T3 primers.
Supplementary Material
Acknowledgments
We thank Toni Cathomen for providing polyclonal U2OS-EGFP reporter cells and a T7EI protocol, Yanfang Fu for deriving the clonal U2OS-EGFP cell line, Drena Dobbs for support and encouragement, and Morgan Maeder, James Angstman, and Cherie Ramirez for helpful comments. This work was supported by a National Institutes of Health (NIH) Director’s Pioneer Award DP1 OD006862 (J.K.J.), NIH P50HG005550 (J.K.J.), the Jim and Ann Orr MGH Research Scholar Award (J.K.J.), NIH T32 CA009216 (J.D.S.), and NSF DBI-0923827 (D.R.).
Footnotes
Author contributions
J.D.S. and J.K.J. conceived of the FLASH method. D.R., S.T., C.K., J.F., and J.D.S. performed experiments. D.R., S.T., C.K., J.F., J.D.S., and J.K.J. wrote the manuscript.
Conflicts of interest
J.D.S. and J.K.J. are inventors on a patent application describing the FLASH method.
J.K.J. is a member of the Scientific Advisory Board of Transposagen Biopharmaceuticals, Inc.
References
- 1.Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 2.Bogdanove AJ, Schornack S, Lahaye T. TAL effectors: finding plant genes for disease and defense. Curr Opin Plant Biol. 2010;13:394–401. doi: 10.1016/j.pbi.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 3.Scholze H, Boch J. TAL effectors are remote controls for gene activation. Curr Opin Microbiol. 2011 doi: 10.1016/j.mib.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Boch J, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 5.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 6.Miller JC, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 7.Morbitzer R, Romer P, Boch J, Lahaye T. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc Natl Acad Sci U S A. 2010;107:21617–21622. doi: 10.1073/pnas.1013133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morbitzer R, Elsaesser J, Hausner J, Lahaye T. Assembly of custom TALE-type DNA binding domains by modular cloning. Nucleic Acids Res. 2011;39:5790–5799. doi: 10.1093/nar/gkr151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang F, et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geissler R, et al. Transcriptional activators of human genes with programmable DNA-specificity. PLoS ONE. 2011;6:e19509. doi: 10.1371/journal.pone.0019509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weber E, Gruetzner R, Werner S, Engler C, Marillonnet S. Assembly of designer TAL effectors by Golden Gate cloning. PLoS ONE. 2011;6:e19722. doi: 10.1371/journal.pone.0019722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christian M, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li T, et al. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2011;39:359–372. doi: 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahfouz MM, et al. De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc Natl Acad Sci U S A. 2011;108:2623–2628. doi: 10.1073/pnas.1019533108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mussolino C, et al. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li T, et al. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011;39:6315–6325. doi: 10.1093/nar/gkr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cermak T, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wood AJ, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hockemeyer D, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tesson L, et al. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol. 2011;29:695–696. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- 21.Sander JD, et al. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang P, et al. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- 23.Zhang F, et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maeder ML, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 26.Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19:1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gabriel R, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- 29.Yusa K, et al. Targeted gene correction of alpha1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478:391–394. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller JC, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 31.Doyon Y, et al. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods. 2011;8:74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- 32.Szczepek M, et al. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 33.Guschin DY, et al. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.