Abstract
Bacillus timonensis strain MM10403188T sp. nov. is the type strain of a proposed new species within the genus Bacillus. This strain, whose genome is described here, was isolated from the fecal flora of a healthy patient. B. timonensis is an aerobic Gram-negative rod shaped bacterium. Here we describe the features of this organism, together with the complete genome sequence and annotation. The 4,632,049 bp long genome (1 chromosome but no plasmid) contains 4,610 protein-coding and 74 RNA genes, including 5 rRNA genes.
Key words: Bacillus timonensis, genome
Introduction
Bacillus timonensis strain MM10403188T (= CSUR P162 = DSM 25372) is designated as the type strain of B. timonensis, a new Gram-negative aerobic, indole-positive bacillus that was isolated from the stool of a healthy Senegalese patient as part of a “culturomics” study aiming at cultivating individually all species within human feces.
To date, DNA-DNA hybridization and G+C content determination [1] remain the gold standard methods for the definition of bacterial species, despite the development of 16S rRNA PCR and sequencing which have deeply changed bacterial taxonomy [2]. Over recent years, high throughput genome sequencing provided a wealth of genetic information [3]. In an effort to include genomic data in bacterial taxonomy we recently used a polyphasic approach [4] that includes genomic data, MALDI-TOF spectrum and main phenotypic characteristics to describe new bacterial species [5,6] .
Here we present a summary classification and a set of features for B. timonensis sp. nov. strain MM10403188T together with the description of the complete genomic sequencing and annotation. These characteristics support the circumscription of the species B. timonensis.
The genus Bacillus (Cohn 1872) was created in 1872 [6]. To date, this genus, mostly comprised of Gram-positive, motile, and spore-forming bacteria, is made of 256 species and 7 subspecies with validly published names [7]. Members of the genus Bacillus are ubiquitous bacteria isolated from various environments including soil, fresh and sea water, food, and occasionally from humans in whom they are either pathogens, such as B. anthracis and B. cereus, or opportunists in immunocompromised patients [7]. Apart from anthrax, caused by B. anthracis [8], and toxi-infections caused by B. cereus, Bacillus species may be involved in a variety of aspecific human infections, including cutaneous, ocular, central nervous system or bone infections, pneumonia, endocarditis and bacteremia [9].
Classification and features
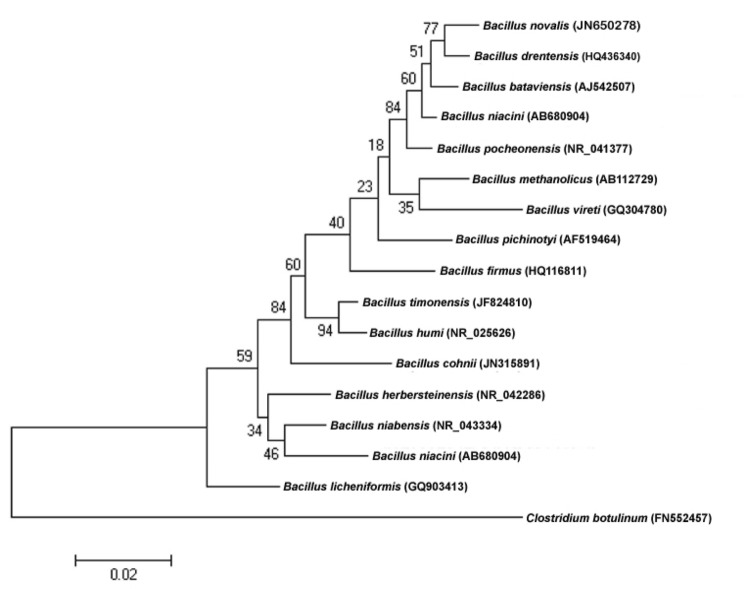
A stool sample was collected from a healthy 16-year-old male Senegalese volunteer patient living in Dielmo (a rural village in the Guinean-Sudanian zone in Senegal), who was included in a research protocol. The patient gave an informed and signed consent, and the agreement of the National Ethics Committee of Senegal and the local ethics committee of the IFR48 (Marseille, France) was obtained under agreements 09-022 and 11-017). The fecal specimen was preserved at -80°C after collection and sent to Marseille. Strain MM10403188 (Table 1) was isolated in June 2011 by cultivation on 5% sheep blood-enriched Brain Heart Infusion agar with (Becton Dickinson, Heidelberg, Germany). This strain exhibited a 98.2% nucleotide sequence similarity with Bacillus humi, the phylogenetically closest validated Bacillus species (Figure 1). This value was lower than the 98.7% 16S rRNA gene sequence threshold recommended by Stackebrandt and Ebers to delineate a new species without carrying out DNA-DNA hybridization [2].
Table 1. Classification and general features of Bacillus timonensis strain MM10403188T.
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS [10] | |
| Phylum Firmicutes | TAS [11-13] | ||
| Class Bacilli | TAS [14,15] | ||
| Order Bacillales | TAS [16,17] | ||
| Family Bacillaceae | TAS [16,18] | ||
| Genus Bacillus | TAS [16,19,20] | ||
| Species Bacillus timonensis | IDA | ||
| Type strain MM10403188T | IDA | ||
| Gram stain | negative | IDA | |
| Cell shape | rod | IDA | |
| Motility | motile | IDA | |
| Sporulation | sporulating | IDA | |
| Temperature range | mesophile | IDA | |
| Optimum temperature | 37°C | IDA | |
| MIGS-6.3 | Salinity | growth in BHI medium + 5% NaCl | IDA |
| MIGS-22 | Oxygen requirement | aerobic | IDA |
| Carbon source | unknown | NAS | |
| Energy source | unknown | NAS | |
| MIGS-6 | Habitat | human gut | IDA |
| MIGS-15 | Biotic relationship | Free living | IDA |
| MIGS-14 | Pathogenicity Biosafety level Isolation |
unknown 2 human feces |
NAS |
| MIGS-4 | Geographic location | Senegal | IDA |
| MIGS-5 | Sample collection time | September 2010 | IDA |
| MIGS-4.1 | Latitude | 13.7167 | IDA |
| MIGS-4.1 | Longitude | -16.4167 | IDA |
| MIGS-4.3 | Depth | Surface | IDA |
| MIGS-4.4 | Altitude | 51 m above sea level | IDA |
Evidence codes - IDA: Inferred from Direct Assay; TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [21]. If the evidence is IDA, then the property was directly observed for a live isolate by one of the authors or an expert mentioned in the acknowledgements.
Figure 1.
Phylogenetic tree highlighting the position of Bacillus timonensis strain MM10403188T relative to other type strains within the Bacillus genus. GenBank accession numbers are indicated in parentheses. Sequences were aligned using CLUSTALW, and phylogenetic inferences obtained using the maximum-likelihood method within the MEGA software. Numbers at the nodes are percentages of bootstrap values obtained by repeating the analysis 500 times to generate a majority consensus tree. Clostridium botulinum was used as an outgroup. The scale bar represents a 2% nucleotide sequence divergence.
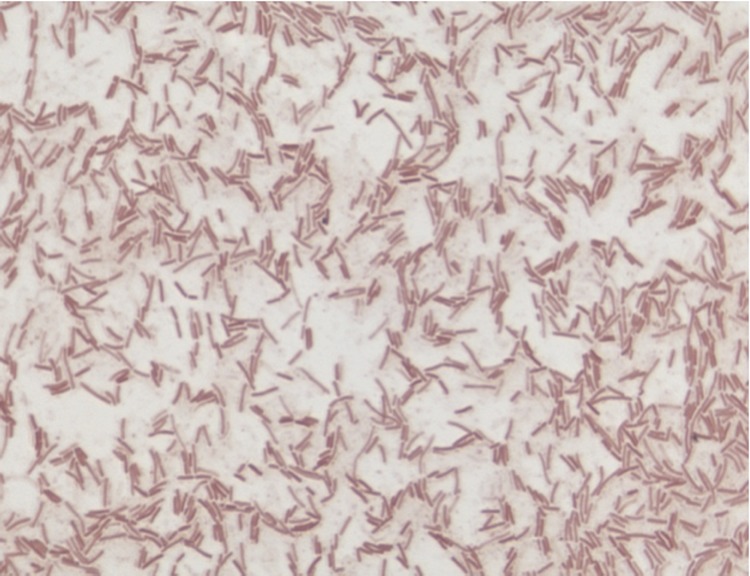
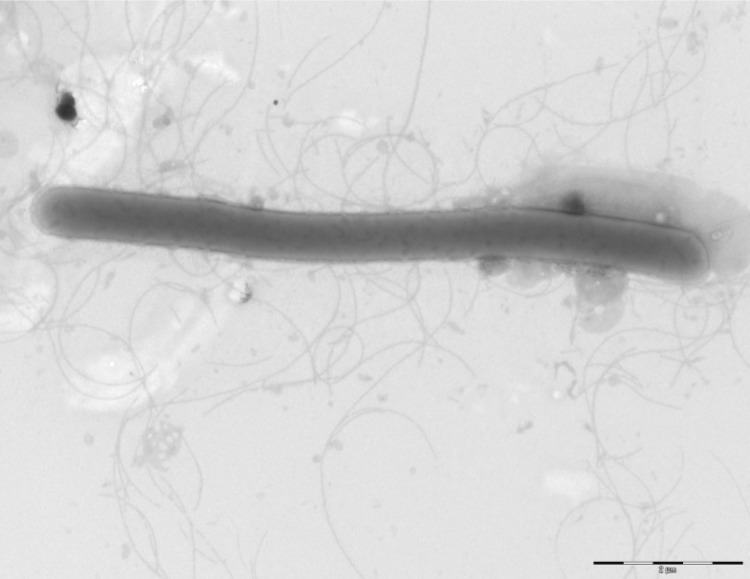
Different growth temperatures (25, 30, 37, 45°C) were tested. Growth occurred at all tested temperatures, but optimal growth occurred between 30 and 37°C. Colonies were 3 mm in diameter on blood-enriched BHI agar. Growth of the strain was tested under anaerobic and microaerophilic conditions using GENbag anaer and GENbag microaer systems, respectively (BioMérieux), and in aerobic conditions, with or without 5% CO2. Growth was achieved in aerobic (with and without CO2) and microaerophilic conditions. No growth was observed in anaerobic conditions. Gram staining showed Gram negative bacilli (Figure 2). A motility test was positive. Cells grown on agar are sporulated and have a mean diameter of 0.66 µm (Figure 3).
Figure 2.
Gram staining of B. timonensis strain MM10403188T
Figure 3.
Transmission electron microscopy of B. timonensis strain MM10403188T, using a Morgani 268D (Philips) at an operating voltage of 60kV. The scale bar represents 900 nm.
Strain MM10403188T exhibited oxidase activity but not catalase activity, and was positive for indole. Using API 50CH, a positive reaction was obtained for L-arabinose, D-lactose, D-melibiose, D-trehalose, D-saccharose, and D-turanose fermentation. A weak reaction was obtained for aesculin. Other tests were negative. Using API-ZYM, positive reactions were obtained for esterase, α-chimotrypsine, β-glucorinidase, and α- and β-glucosinidase. B. timonensis was susceptible to penicillin G, amoxicillin, vancomycin, gentamicin, erythromycin, doxycyclin, rifampicin, and ciprofloxacin but resistant to trimethoprim/sulfamethoxazole.
By comparison with B. humi, B. timonensis differed in Gram staining, in culture atmosphere, as B. humi was able to grow anaerobically, in catalase activity, in spore forming capacity, in indole production, and in carbohydrate metabolism, notably for arbutin, salicin, L-arabinose, melibiose, turanose, and trehalose [22].
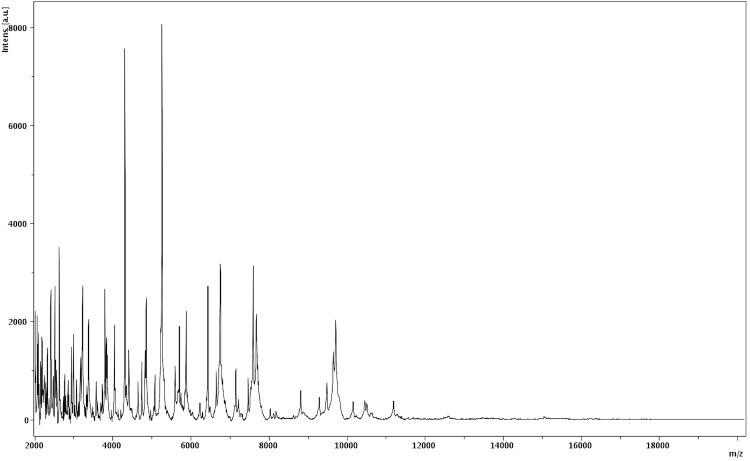
Matrix-assisted laser-desorption/ionization time-of-flight (MALDI-TOF) MS protein analysis was carried out as previously described [23]. Briefly, a pipette tip was used to pick one isolated bacterial colony from a culture agar plate, and to spread it as a thin film on a MTP 384 MALDI-TOF target plate (Bruker Daltonics, Leipzig, Germany). Four distinct deposits were done for strain MM10403188 from four isolated colonies. Each smear was overlaid with 2µL of matrix solution (saturated solution of alpha-cyano-4-hydroxycinnamic acid) in 50% acetonitrile, 2.5% tri-fluoracetic-acid, and allowed to dry for five minutes. Measurements were performed with a Microflex spectrometer (Bruker). Spectra were recorded in the positive linear mode for the mass range of 2,000 to 20,000 Da (parameter settings: ion source 1 (IS1), 20 kV; IS2, 18.5 kV; lens, 7 kV). A spectrum was obtained after 675 shots at a variable laser power. The time of acquisition was between 30 seconds and 1 minute per spot. The four MM10403188 spectra were imported into the MALDI BioTyper software (version 2.0, Bruker) and analyzed by standard pattern matching (with default parameter settings) against the main spectra of 3,769 bacteria including 129 spectra from 98 Bacillus species, notably B. humi, used as reference data, in the BioTyper database. The method of identification included the m/z from 3,000 to 15,000 Da. For every spectrum, 100 peaks at most were taken into account and compared with spectra in the database. A score enabled the presumptive identification and discrimination of the tested species from those in the database: a score > 2 with a validated species enabled the identification at the species level, a score > 1.7 but < 2 enabled the identification at the genus level; and a score < 1.7 did not enable any identification. For strain MM10403188T, the obtained score was 1.2, thus suggesting that our isolate was not a member of a known species. We incremented our database with the spectrum from strain MM10403188 (Figure 4). The spectrum was made available online in our free-access URMS database [24].
Figure 4.
Reference mass spectrum from B. timonensis strain MM10403188T. Spectra from 12 individual colonies were compared and a reference spectrum was generated.
Genome sequencing information
Genome project history
The organism was selected for sequencing on the basis of its phylogenetic position and 16S rRNA similarity to other members of the genus Bacillus, and is part of a “culturomics” study of the human digestive flora aiming at isolating all bacterial species within human feces. It was the 60th genome of a Bacillus species and the first genome of Bacillus timonensis sp. nov. A summary of the project information is shown in Table 2. The Genbank accession number is CAET00000000 and consists of 146 contigs.
Table 2. Project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | High-quality draft |
| MIGS-28 | Libraries used | 454 GS shotgun and paired-end 3- kb libraries |
| MIGS-29 | Sequencing platform | 454 GS FLX Titanium |
| MIGS-31.2 | Sequencing coverage | 19× |
| MIGS-30 | Assemblers | Newbler version 2.5.3 |
| MIGS-32 | Gene calling method | PRODIGAL |
| INSDC ID | 112529 | |
| Genbank Date of Release | February 28th, 2012 | |
| Gold ID | Gi13534 | |
| NCBI project ID | CAET00000000 | |
| MIGS-13 | Project relevance | Study of the human gut microbiome |
Growth conditions and DNA isolation
B. timonensis sp. nov. strain MM10403188T, CSUR P162, DSM 25372, was grown aerobically on 5% sheep blood-enriched BHI agar at 37°. Four petri dishes were spread and growth from the plates was resuspended in 3x500µl of TE buffer and stored at 80°C. Then, 500µl of this suspension were thawed, centrifuged 3 minutes at 10,000 rpm and resuspended in 3x100µL of G2 buffer (EZ1 DNA Tissue kit, Qiagen). A first mechanical lysis was performed by glass powder on the Fastprep-24 device (Sample Preparation system, MP Biomedicals, USA) using 2x20 seconds cycles. DNA was then treated with 2.5µg/µL lysozyme (30 minutes at 37°C) and extracted using the BioRobot EZ1 Advanced XL (Qiagen). The DNA was then concentrated and purified using the Qiamp kit (Qiagen). The yield and the concentration was measured by the Quant-it Picogreen kit (Invitrogen) on the Genios Tecan fluorometer at 50ng/µl.
Genome sequencing and assembly
DNA (5 µg) was mechanically fragmented on a Hydroshear device (Digilab, Holliston, MA,USA) with an enrichment size at 3-4kb. The DNA fragmentation was visualized through the Agilent 2100 BioAnalyzer on a DNA labchip 7500 with an optimal size of 3.345kb. The library was constructed according to the 454 GS FLX Titanium paired-end protocol. Circularization and nebulization were performed and generated a pattern with an optimum at 492 bp. After PCR amplification through 15 cycles followed by double size selection, the single stranded paired end library was then quantified on the Quant-it Ribogreen kit (Invitrogen) on the Genios Tecan fluorometer at 339 pg/µL. The library concentration equivalence was calculated as 12,6E+08 molecules/µL. The library was stored at -20°C until further use.
The shotgun library was clonally amplified with 3cpb and the paired-end library was amplified with lower cpb (1 cpb) in 4 emPCR reactions with the GS Titanium SV emPCR Kit (Lib-L) v2 (Roche). The yields of the emPCR was 5.97% for the shotgun and 15.92% for the paired end as expected by the range of 5 to 20% from the Roche procedure.
Approximately 790,000 beads for a 1/4 region and 340,000 beads for a 1/8 region were loaded on the GS Titanium PicoTiterPlate PTP Kit 70×75 and sequenced with the GS FLX Titanium Sequencing Kit XLR70 (Roche). The run was performed overnight and then analyzed on the cluster through the gsRunBrowser and Newbler assembler (Roche). For the shotgun sequencing, 112,962 passed filter wells were obtained and generated 34.48Mb with a length average of 322 bp. For the shotgun sequencing, 213,882 passed filter wells were obtained and generated 50.6 Mb with a length average of 236 bp. The passed filter sequences were assembled Using Newbler with 90% identity and 40bp as overlap. The final assembly identified 11 scaffolds and 89 contigs (>1500bp) generating a genome size of 4.6 Mb.
Genome annotation
Open Reading Frames (ORFs) were predicted using Prodigal [25] with default parameters but the predicted ORFs were excluded if they were spanning a sequencing gap region. The predicted bacterial protein sequences were searched against the GenBank database [26] and the Clusters of Orthologous Groups (COG) databases using BLASTP. The tRNAScanSE tool [27] was used to find tRNA genes, whereas ribosomal RNAs were found by using RNAmmer [28] and BLASTn against the GenBank database. ORFans were identified if their BLASTP E-value was lower than 1e-03 for alignment length greater than 80 amino acids. If alignment lengths were smaller than 80 amino acids, we used an E-value of 1e-05. Such parameter thresholds have already been used in previous works to define ORFans.
To estimate the mean level of nucleotide sequence similarity at the genome level between Bacillus species, we compared the ORFs only using BLASTN and the following parameters: a query coverage of ≥ 70% and a minimum nucleotide length of 100 bp.
Genome properties
The genome is 4,632,049 bp long (1 chromosome, but no plasmid) with a 37.30% GC content (Figure 5 and Table 3). Of the 4,684 predicted genes, 4,610 were protein-coding genes and 74 were RNAs. A total of 3,399 genes (75.56%) were assigned a putative function. Three hundred forty genes were identified as ORFans (7.4%). The remaining genes were annotated as hypothetical proteins. The properties and the statistics of the genome are summarized in Tables 3. The distribution of genes into COGs functional categories is presented in Table 4.
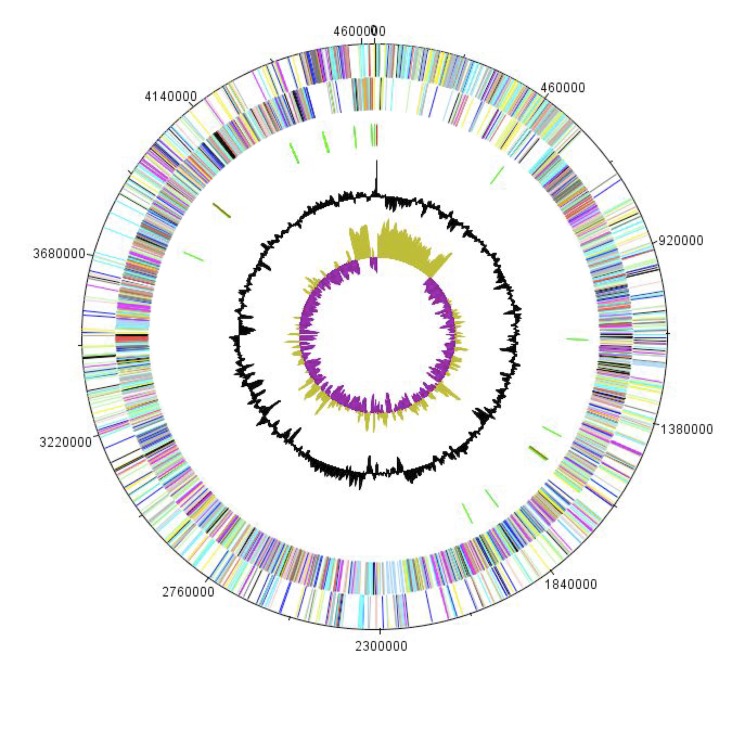
Figure 5.
Graphical circular map of the chromosome. From outside to the center: Genes on the forward strand (colored by COG categories), genes on the reverse strand (colored by COG categories), RNA genes (tRNAs green, rRNAs red), GC content, and GC skew.
Table 3. Nucleotide content and gene count levels of the genome.
| Attribute | Value | % of totala |
|---|---|---|
| Genome size (bp) | 4,632,049 | |
| DNA Coding region (bp) | 3,959,694 | 85.48 |
| DNA G+C content (bp) | 1,727,754 | 37.3 |
| Total genes | 4,684 | 100 |
| RNA genes | 74 | 1.58 |
| Protein-coding genes | 4,610 | 98.42 |
| Genes with function prediction | 3,643 | 77.75 |
| Genes assigned to COGs | 3,399 | 75.56 |
| Genes with peptide signals | 189 | 4.03 |
| Genes with transmembrane helices | 1,261 | 26.92 |
a The total is based on either the size of the genome in base pairs or the total number of protein coding genes in the annotated genome
Table 4. Number of genes associated with the 25 general COG functional categories.
| Code | Value | % agea | Description |
|---|---|---|---|
| J | 181 | 3.93 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0 | RNA processing and modification |
| K | 310 | 6.72 | Transcription |
| L | 169 | 3.67 | Replication, recombination and repair |
| B | 1 | 0.02 | Chromatin structure and dynamics |
| D | 39 | 0.85 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0 | Nuclear structure |
| V | 71 | 1.54 | Defense mechanisms |
| T | 193 | 4.19 | Signal transduction mechanisms |
| M | 197 | 4.27 | Cell wall/membrane biogenesis |
| N | 67 | 1.45 | Cell motility |
| Z | 0 | 0 | Cytoskeleton |
| W | 0 | 0 | Extracellular structures |
| U | 49 | 1.06 | Intracellular trafficking and secretion |
| O | 114 | 2.47 | Posttranslational modification, protein turnover, chaperones |
| C | 184 | 3.99 | Energy production and conversion |
| G | 349 | 7.57 | Carbohydrate transport and metabolism |
| E | 412 | 8.94 | Amino acid transport and metabolism |
| F | 97 | 2.10 | Nucleotide transport and metabolism |
| H | 121 | 2.62 | Coenzyme transport and metabolism |
| I | 150 | 3.25 | Lipid transport and metabolism |
| P | 245 | 5.31 | Inorganic ion transport and metabolism |
| Q | 100 | 2.17 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 594 | 12.89 | General function prediction only |
| S | 361 | 7.83 | Function unknown |
| - | 606 | 13.15 | Not in COGs |
a The total is based on the total number of protein coding genes in the annotated genome
Comparison with the genomes from other Bacillus species
Genome sequences are currently available for more than 25 validly named Bacillus species. Here we compared the genome sequence of B. timonensis strain MM10403188T with that of B. licheniformis strain ATCC 14580, the most closely related phylogenetic neighbor for which the genome sequence is available. The draft genome sequence of B. timonensis is larger than B. licheniformis (4.6 Mb and 4.2 Mb, respectively) but its G+C content is lower (37.30 and 46.19%, respectively). B. timonensis has more predicted genes than B. licheniformis (4,684 and 4,356, respectively), and more genes assigned to COGs (3,399 and 3,130, respectively). However, the distribution of genes into COG categories (Table 4) was highly similar in both genomes. In addition, B. timonensis shared a mean 86.10% (range 76.4-93%) sequence similarity with B. licheniformis at the genome level.
Although the degree of 16S rRNA similarity was elevated (98.2%) between strain MM10403188 and B. humi strain DSM 16318, both strains exhibited several phenotypic and genomic differences, and we formally propose the creation of Bacillus timonensis sp. nov. that contains the strain MM10403188T. This strain has been found in Senegal.
Description of Bacillus timonensis sp. nov.
Bacillus timonensis (tim.on.en′sis. L. gen. masc. n. timonensis, of Timone, the name of the hospital where strain MM10403188T was cultivated.) Isolated from stool from an asymptomatic Senegalese patient. B. timonensis is an aerobic Gram-negative bacterium. Grows on axenic medium at 37°C in an aerobic atmosphere. Colonies were 3 mm in diameter on blood-enriched BHI agar. Cells grown on agar are sporulated and have a mean diameter of 0.66 µm. A positive reaction was obtained for L-arabinose, D-lactose, D-melibiose, D-trehalose, D-saccharose, and D-turanose fermentation. Positive reactions were obtained for oxidase, esterase, α-chimotrypsine, β-glucorinidase, and α- and β-glucosinidase activity. No catalase activity was exhibited. Positive for indole. By comparison with B. humi, B. timonensis differs in Gram staining, in culture atmosphere, as B. humi grows anaerobically, in catalase activity, in spore forming capacity, in indole production, and in carbohydrate metabolism, notably for arbutin, salicin, L-arabinose, melibiose, turanose, and trehalose. B. timonensis is susceptible to penicillin G, amoxicillin, vancomycin, gentamicin, erythromycin, doxycyclin, rifampicin, and ciprofloxacin but resistant to trimethoprim/sulfamethoxazole. Motile. The G+C content of the genome is 37.30%. The 16S rRNA and genome sequences are deposited in GenBank under accession numbers JF824810 and CAET00000000, respectively.
The type strain MM10403188T (= CSUR P162 = DSM 253720) was isolated from the fecal flora of a healthy patient from Senegal.
References
- 1.Rossello-Mora R. DNA-DNA Reassociation Methods Applied to Microbial Taxonomy and Their Critical Evaluation. In: Stackebrandt E (ed), Molecular Identification, Systematics, and population Structure of Prokaryotes. Springer, Berlin, 2006, p. 23-50. [Google Scholar]
- 2.Stackebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 2006; 33:152-155 [Google Scholar]
- 3.Welker M, Moore ER. Applications of whole-cell matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry in systematic microbiology. Syst Appl Microbiol 2011; 34:2-11 10.1016/j.syapm.2010.11.013 [DOI] [PubMed] [Google Scholar]
- 4.Tindall BJ, Rosselló-Móra R, Busse HJ, Ludwig W, Kämpfer P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol 2010; 60:249-266 10.1099/ijs.0.016949-0 [DOI] [PubMed] [Google Scholar]
- 5.Lagier JC, El Karkouri K, Nguyen TT, Armougom F, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Anaerococcus senegalensis sp. nov. Stand Genomic Sci 2012; 6:116-125 10.4056/sigs.2415480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mishra AK, Lagier JC, Robert C, Raoult D, Fournier PE. 2012. Non-contiguous finished genome sequence and description of Clostridium senegalense sp. nov. Stand.Genomic.Sci. In press</jrn>5. Cohn F. Untersuchungen über Bakterien. Beitrage zur Biologie der Pflanzen Heft 1872; 1:127-224 [Google Scholar]
- 7.Jernigan JA, Stephens DS, Ashford DA, Omenaca C, Topiel MS, Galbraith M, Tapper M, Fisk TL, Zaki S, Popovic T, et al. Bioterrorism-related inhalational anthrax: the first 10 cases reported in the United States. Emerg Infect Dis 2001; 7:933-944 10.3201/eid0706.010604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bottone EJ. Bacillus cereus, a volatile human pathogen. Clin Microbiol Rev 2010; 23:382-398 10.1128/CMR.00073-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archae, Bacteria, and Eukarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibbons NE, Murray RGE. Proposals Concerning the Higher Taxa of Bacteria. Int J Syst Bacteriol 1978; 28:1-6 10.1099/00207713-28-1-1 [DOI] [Google Scholar]
- 11.Garrity GM, Holt JG. The Road Map to the Manual. In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 1, Springer, New York, 2001, p. 119-169. [Google Scholar]
- 12.Murray RGE. The Higher Taxa, or, a Place for Everything...? In: Holt JG (ed), Bergey's Manual of Systematic Bacteriology, First Edition, Volume 1, The Williams and Wilkins Co., Baltimore, 1984, p. 31-34. [Google Scholar]
- 13.List Editor List of new names and new combinations previously effectively, but not validly, published. List no. 132. Int J Syst Evol Microbiol 2010; 60:469-472 10.1099/ijs.0.022855-0 [DOI] [PubMed] [Google Scholar]
- 14.Ludwig W, Schleifer KH, Whitman WB. Class I. Bacilli class nov. In: De Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 3, Springer-Verlag, New York, 2009, p. 19-20. [Google Scholar]
- 15.Skerman VBD, Sneath PHA. Approved list of bacterial names. Int J Syst Bact 1980; 30:225-420 10.1099/00207713-30-1-225 [DOI] [Google Scholar]
- 16.Prevot AR. Dictionnaire des bactéries pathogens. In: Hauduroy P, Ehringer G, Guillot G, Magrou J, Prevot AR, Rosset, Urbain A (eds). Paris, Masson, 1953, p.1-692. [Google Scholar]
- 17.Fischer A. Untersuchungen über bakterien. Jahrbücher für Wissenschaftliche Botanik 1895; 27:1-163 [Google Scholar]
- 18.Gibson T, Gordon RE. Genus I. Bacillus Cohn 1872, 174; Nom. gen. cons. Nomencl. Comm. Intern. Soc. Microbiol. 1937, 28; Opin. A. Jud. Comm. 1955, 39. In: Buchanan RE, Gibbons NE (eds), Bergey's Manual of Determinative Bacteriology, Eighth Edition, The Williams and Wilkins Co., Baltimore, 1974, p. 529-550. [Google Scholar]
- 19.Mathews WC, Caperna J, Toerner JG, Barber RE, Morgenstern H. Neutropenia is a risk factor for Gram-negative Bacillus bacteremia in human immunodeficiency virus-infected patients: results of a nested case-control study. Am J Epidemiol 1998; 148:1175-1183 10.1093/oxfordjournals.aje.a009606 [DOI] [PubMed] [Google Scholar]
- 20.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stackebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 2006; 33:152-155 [Google Scholar]
- 22.Heyrman J, Rodriguez-Diaz M, Devos J, Felske A, Logan NA, De Vos P. Bacillus arenosi sp. nov., Bacillus arvi sp. nov., Bacillus humi sp. nov., isolated from soil. Int J Syst Evol Microbiol 2005; 55:111-117 10.1099/ijs.0.63240-0 [DOI] [PubMed] [Google Scholar]
- 23.Seng P, Drancourt M, Gouriet F, La SB, Fournier PE, Rolain JM, Raoult D. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis 2009; 49:543-551 10.1086/600885 [DOI] [PubMed] [Google Scholar]
- 24.URMS database. http://ifr48.timone.univ-mrs.fr/portail2/index.php?option=com_content&task=view
- 25.Prodigal. http://prodigal.ornl.gov
- 26.Benson DA, Karsch-Mizrachi I, Clark K, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res 2012; 40:D48-D53 10.1093/nar/gkr1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lowe TM, Eddy SR. t-RNAscan-SE: a program for imroved detection of transfer RNA gene in genomic sequence. Nucleic Acids Res 1997; 25:955-964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 2007; 35:3100-3108 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]