Abstract
Epithelia are sheets of connected cells that are essential across the animal kingdom. Experimental observations suggest that the dynamical behavior of many single-layered epithelial tissues has strong analogies with that of specific mechanical systems, namely large networks consisting of point masses connected through spring-damper elements and undergoing the influence of active and dissipating forces. Based on this analogy, this work develops a modeling framework to enable the study of the mechanical properties and of the dynamic behavior of large epithelial cellular networks. The model is built first by creating a network topology that is extracted from the actual cellular geometry as obtained from experiments, then by associating a mechanical structure and dynamics to the network via spring-damper elements. This scalable approach enables running simulations of large network dynamics: the derived modeling framework in particular is predisposed to be tailored to study general dynamics (for example, morphogenesis) of various classes of single-layered epithelial cellular networks. In this contribution we test the model on a case study of the dorsal epithelium of the Drosophila melanogaster embryo during early dorsal closure (and, less conspicuously, germband retraction).
Index Terms: Epithelium, Cellular network, Nonlinear dynamical model, Spring-damper system, Discrete element method, Early dorsal closure, Morphogenesis
1 Introduction
Quantitatively understanding the mechanical structure and dynamical properties of epithelial cellular networks is a compelling but complex task. Three main factors contribute to the difficulty of this goal.
Firstly, single cells are made up of a number of distinct components, each contributing to their mechanical structure [1]. The mechanical characteristics of networks of cells thus hinge on the characteristics of single cells, each with their complex structural features [2]. Encompassing these components easily leads to high-dimensional, nonlinear, spatially distributed models of cellular networks that are not likely to be prone to mathematical analysis or to simulation on a computer over large cellular networks.
Secondly, at the cellular network level, dynamics are often influenced by a combination of different forces (both internal and external to the network) acting simultaneously [3], [4], [5] – isolating individual force mechanism contributions in the lab can be a daunting task – we contend that a synthetic model can be an alternative solution.
Thirdly, the influence of genetic processes on the mechanical properties of cellular network is currently an open field of investigation [6]: conducting manipulations in the expression of certain genes can lead to significantly different mechanical properties. Encompassing such a dependence at the modeling level can be very difficult.
This work focuses on the first two of the three issues described above. With the general goal of developing a quantitative model comes a tradeoff between model descriptiveness and precision on the one hand, and size, computability, and ease of analysis on the other. Taking up this latter perspective, the main objective of this work is to develop a quantitative mathematical model for the study of the dynamics of large and heterogeneous epithelial networks. The model furthermore enables the investigation of the role played by forces acting on the epithelium, as well as the study of the influence of the non-uniformity of its mechanical properties on its dynamics. The model is developed with the main intention of enabling the simulation of complex, large-dimensional networks of cells.
After benchmarking two different modeling approaches over their ability to encompass the complexity of epithelial networks and the capacity of modeling subtleties of single cellular mechanics, this work presents a model that strikes a balance between computable high-level abstractions [7], [8], [9], [10] and specific low-level refined characterizations [11], [12], [13]. We claim that the main features of the proposed modeling framework are that 1. it is biologically well-grounded; 2. it is tractable for analysis and large-scale simulations; and 3. it is promising in a number of diverse applications focusing on classes of single-layered epithelial cellular networks.
Structure of the article
Section 2 recapitulates biological knowledge that is at the basis of cellular mechanics. Based on this insight, Section 3 proposes important modeling criteria and distills them towards the derivation of a novel modeling framework for single-layered epithelial cellular networks. This part motivates the use of the discrete-element approach as a principal constituent of the model. This Section also elaborates on the mathematical formulation: first, the underlying graphical structure is described; then, the dynamics are associated to the structure by introducing spring-damper elements. Section 4 discusses the potentials and the limitations of the proposed modeling framework. While this work does not claim to lead to any new biological insight, with the goal of validating the approach Section 5 proposes a experimental study, which focuses on simulating the dorsal epithelium of D. melanogaster embryo during early dorsal closure (and in part germband retraction). Section 5.1 presents how experimental data is utilized as a basis for the modeling framework (definition of the graphical structure). Section 5.2 discusses how the dynamics are added to the model, as well as how external forces and constraints are included. Section 5.3 presents the outcomes of the simulations: it focuses on both qualitative and quantitative assessments of modeling properties and of simulation assumptions. Finally, Section 6 concludes the work.
2 Biological Background
This Section recalls key empirical evidence on cellular mechanics. In order to gain understanding on its mechanical and dynamical characteristics, a single epithelial cell is dissected in terms of its own structural components and of its connections to adjacent components.
2.1 Cellular Mechanics
Cells are highly dynamic: they stretch, crawl, change shape and divide [14]. Their mechanical properties depend on their internal structure [15]. In many critical biological processes, cells both exert and respond to forces toward and from their surroundings [16]. The mechanical properties of a cell are thus intimately related to its physical nature, as well as to its position within a network of similar cells.
Cellular mechanics are determined by three key aspects: the internal structure of the cells, which hinges on the presence of the cytoskeleton (Sec. 2.1.1); cell-cell connections within the epithelium, as well as the connection of epithelial cells to other tissue layers (Sec. 2.1.2); and two additional important mechanical characteristics [16], [17]: nonlinear elasticity and anisotropy (Sec. 2.1.3).
2.1.1 Cellular structure
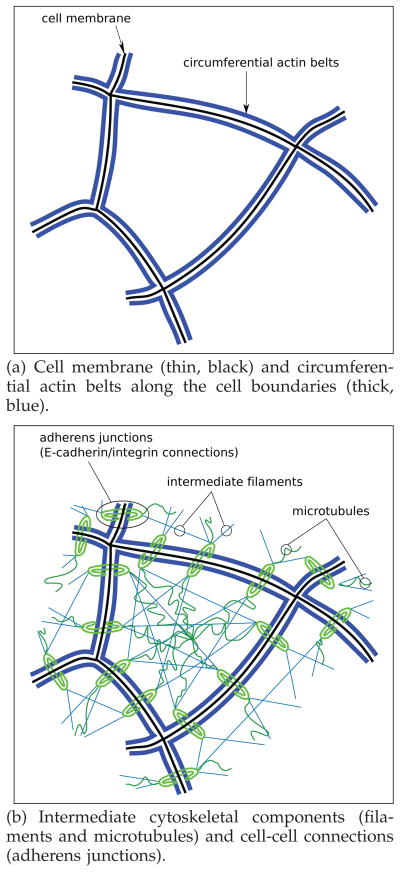
The cytoskeleton is a complex, heterogeneous and dynamic structure, which affects both elastic and viscous characteristics of cells [8], [18], [19], [20] (for possible modeling frameworks to encompass these properties see [10], [21], [22], [23]). The cytoskeleton is a biopolymer network consisting of three major components (Figure 1): microfilaments (made of actin), intermediate filaments, and microtubules. In addition to these major components, a myriad of filament cross-linker, motor and regulatory proteins also play a role.
Fig. 1.
Pictorial top-view of a single cell in an epithelial network.
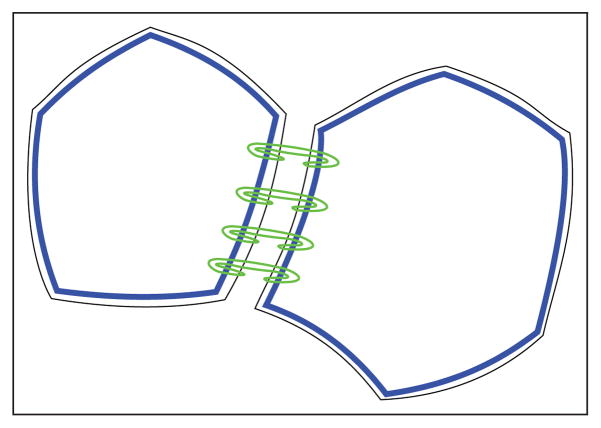
Cell tensional forces depend on the three components mentioned above [24]. With regards to the first filaments, actin accumulates as a circumferential belt along the cell membrane, at a specific apical-basal depth. Circumferential actin belts of adjacent cells are connected to each other through adherens junctions, see Figures 1(b) and 2, which thus contribute to cytoskeletal structure and dynamics by connecting cytoskeleta of adjacent cells. An adherens junction is a cell junction whose cytoplasmic face is linked to the cytoskeleton. The connection of circumferential actin belts of adjacent cells creates a two-dimensional network of actin, within the epithelial cellular network.
Fig. 2.
Two-dimensional pictorial top-view of two adjacent epithelial cells: actin accumulation (blue) along cell boundaries (black outer lines). Adherens junctions (green) connect different circumferential actin bundles between the cytoskeleta of adjacent cells along their boundaries.
As for intermediate filaments, they operate as ropes connecting two points on the cell membrane. They form a pattern of intersecting lines over the cell, as depicted in Figure 1(b). The tension sustaining actin filaments is balanced by interconnected structural elements that bear compression, called microtubules [25], with The flexible protein structure [9]. They act as struts pushing the cell membrane outwards, as depicted in Figure 1(b). Pushing microtubules and pulling intermediate actin filaments meet at the cell membrane, around adherens junctions, contributing together with actin belts to cell shape stability.
While the discussion has focused on the two-dimensional surface of circumferential actin belts and the elements in between, there also exist cytoskeletal components connecting the apical and basal cell membranes, thus developing along the third dimension of the cell (the apical-basal axis, which crosses the cell surface). These filaments govern the stability of the thickness of the cell under stretching and compressing loads.
While recent studies have addressed the issue of modeling the thickness of epithelial cells [2], [26], since in epithelia much of the mechanical properties of cell networks is dictated by the cell apical surface, a two-dimensional simplification is a good compromise that captures many of the properties of interest. Also in the present study, we shall focus on two-dimensional cellular properties and argue that, under careful working assumptions, the effect of this third dimension can be left out.
2.1.2 Cell-cell connections and connection to the extra-cellular matrix
The different cytoskeletal components of adjacent cells are connected by junctions (Figures 1(b) and 2), which are composed of (among others) integrin and E-cadherin proteins. These proteins are cell adhesion molecules nestled in the cell membrane and are connected to cell adhesion molecules of neighboring cells [27] (see Section 2.1.2), creating a cellular aggregate. The cell adhesion molecules can float through the cell membrane plane, which suggests that the bilipid cell membrane does not play a significant contribution to the structural balance of the cell. Most of the force is sustained by cytoskeletal components that cross cell membranes. In developing epithelia, cell junctions often rearrange at a high frequency, which facilitates cell motility and cell proliferation. In many other cases – with a much lower frequency – rearrangements help stabilizing the epithelium. Because epithelia serve primarily as a structural layer to protect underlying organs and processes, stable adherens junctions are needed to seal cells together into an aggregate. In this work we shall focus on cellular networks where rearrangements have very low occurrence and thus can be disregarded.
The extracellular matrix, and more specifically the basement membrane, is situated underneath the epithelium. Epithelial cellular networks are connected to the extracellular matrix [18], particularly to the basement membrane via focal adhesions, where integrins serve as anchors [27]. The number of adhesions affects the dynamical characteristic of this viscous interaction.
2.1.3 Additional mechanical characteristics
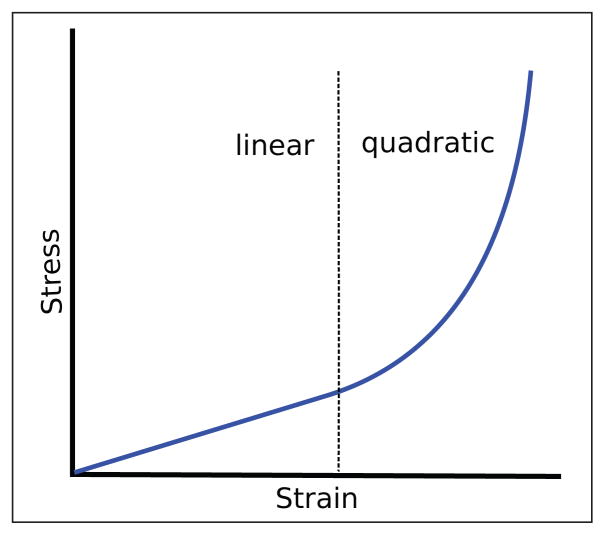
The overall elasticity of cells can be attributed to a number of different components, including the cell boundary, the cytoskeleton, and cell-cell connections. It can be verified experimentally that this elasticity is nonlinear [16]. The elastic modulus [28] of a cell depends on the degree of externally applied forces and of internal stress, as well as on the mechanical properties of its environment [16]. Unlike in materials that display an elastic constant that is independent of the applied stress (at least approximately, within a large stress regime) in networks of semi-flexible polymers the elastic modulus increases under increasing applied stress. This phenomenon is called stress stiffening: a typical nonlinear (quadratic) elasticity characteristic of a cell is represented in Figure 3. The cell elasticity thus cannot be modeled by simple Hookean springs. Furthermore, recent experiments have unveiled the prestress characteristics (namely, presence of tension at equilibrium) of different cytoskeletal components and of the cell as a whole [20], [25], [29]. Evidence (both at the cellular [15], [29] and cytoskeletal level [19], [25]) has shown a linear relation between pre-stress and the stiffness coefficient. Following the analogy with spring elements exerting forces proportional to stiffness and strain, this results in a quadratic elasticity characteristic. Furthermore, by severing individual actin filaments and microtubules and analyzing the dynamical response of the cell, it was shown that the dynamical characteristics of cytoskeletal components are viscoelastic. These responses can be encompassed by leveraging the Voigt element [30], which employs a spring and a damper in parallel (see also Sec. 3.4 for the mathematical details and Sec. 4.1 for alternatives or extensions).
Fig. 3.
Nonlinearity in cell elasticity (stress-stiffening).
The spatial organization of cytoskeletal components creates cellular structures which are inherently anisotropic. For instance, microtubules often determine the direction of elongation of a cell [17]. A feature of stretched cells is the alignment of their cell boundaries. Hence, the circumferential actin filaments that organize along the cell membrane, tend to sustain and propagate most of the load in the network. This property of the cell boundaries, in addition to the discussed nonlinear elasticity behavior, contribute to the cell stability.
3 Modeling Framework
Section 3.1 distills the details on cellular mechanics discussed in the previous section into a few essential modeling criteria for single-layer epithelial cellular networks. Among the many cellular modeling alternatives in the literature [2], Section 3.2 motivates the modeling choices by comparison against one alternative known modeling framework. Section 3.3 describes the underlying structure of the model. This leads to the formal introduction of the dynamical modeling framework in Section 3.4.
3.1 Modeling principles
We synthesize the biological knowledge presented in Section 2 into the following key principles, which inspire the development of a model for epithelial cellular networks:
-
The cellular architecture is discrete:
Single epithelial cells behave mechanistically as discrete entities composed of different interconnected cytoskeletal constituents, and are able to sustain both tensional and compressional loads. They do not behave as mechanical (viscous or viscoelastic) continua. The discrete nature of the actin network and of intermediate filaments should be incorporated in the model. A large proportion of the actin architecture is organized over a two-dimensional surface, governing most of the observed cell dynamics [31], [32].
-
Anisotropy depends both on cell geometry and on network topology:
Single cells have highly anisotropic mechanical structures, determined by their physical characteristics as well as by their geometry. Along with local anisotropy, the network topology is often necessary to explain the existence of global properties or certain global dynamics, such as the alignment of patches of cells or the propagation of forces through the cellular network. Taking into account both single cell geometry and global topology of the epithelial network is therefore necessary to study anisotropy and to explain properties at the network level.
-
Nonlinear and temporal mechanical characteristics of cell appear due to different structural components:
Cell components display characteristics of nonlinear elasticity, which thus emerges at the cellular level. The cytoskeletal prestress contributes to the nonlinearity in cell deformation [19], [20] and should be integrated with ideas from the tensegrity model [24]. Relatively simple nonlinear elasticity characteristics (namely, quadratic relations) have been observed at a cellular level as well as over a larger tissue level [16], [25], [29]. Furthermore, more complex properties (hysteresis, memory) can play a role in specific instances, thus the model should be extensible to accommodate them.
-
Modeling volume preservation can be complex:
Volume preservation is a feature of epithelial cells deforming in a network. However, since there is no clear relation between the stress in the surface directions (planar dimension) and the thickness (height) of the cellular network [26], volume preservation of cellular components should be imposed under controlled conditions and over specific spatial models.
3.2 Comparison between FEM and DEM models
Finite and discrete element methods (FEM and DEM) are numerical techniques developed to compute approximate solutions of partial differential equations (PDE) and of integral equations [30], [33]. These methods are in particular used for solving a set of nonlinear PDE over time-varying domains whenever the desired precision varies over the entire domain, or in case the solution lacks smoothness. The common feature of these two techniques is the application of a mesh discretization of a continuous domain into a set of discrete sub-domains, called elements. We draw a qualitative comparison between FEM and DEM techniques. This comparison is attuned to the modeling principles explained above and, with focus on the problem under study, is intended to motivate the choice of DEM as the basis of the model.
FEM represent continuous objects by meshing them into volumetric elements. Within each of these elements the mechanical properties are defined as constant or continuous functions. When spatial properties such as incompressibility, osmotic pressure, or density are important, then using these elements is indispensable. The FEM approach assumes that strain and stress vary continuously over the introduced volumetric elements, which is a delicate assumption when modeling systems endowed with a discrete mechanical structure.
DEM on the other hand consider a nodal mesh of a given object, in which nodes are associated to point masses and connected via discrete elements (discrete elements can be specific mechanical components). Internal forces can be exerted in any direction between nodes. The DEM approach is useful when the presence and organization of distinct elements resembles the physical structure of the system.
Hybrid combinations with both element types are also possible [22], for instance on structures consisting of beams (modeled with volumetric finite elements) and rods (modeled with discrete elements).
The four criteria derived in Section 3.1 are used to compare the two modeling methods in Table 1. For each of the criteria, the table includes a simple assessment (positive vs. negative sign) of the two modeling frameworks. The DEM approach is selected as the basis of the model that will be developed in Section 3 for the following reasons. The DEM method is suited to easily represent the discrete tensegrity structure of cellular mechanics. The use of DEM also enables the study of anisotropy due to cellular geometry and structural organization (network topology). Cellular volume preservation is an elusive task (though FEM techniques may enable encompassing it), and therefore is not considered in the early stage of modeling framework development. The DEM has shown interesting results in other modeling studies, where nonlinear mechanical characteristics were incorporated. These studies showed qualitative resemblance of tissue deformation [34], [35], [36] and displayed stability properties over high-dimensional models [37]. Time-dependent properties (hysteresis, memory effects) can be incorporated, albeit at increased computational costs. In conclusion, DEM promises to encompass the features of interest for the problem at hand.
TABLE 1.
Comparison of FEM and DEM modeling frameworks.
| 1) Cellular architecture is discrete | |||
|
| |||
| FEM | DEM | ||
| ± | Representation of discrete tensegrity structure unclear | + | Can represent discrete tensegrity structure |
| + | Can be combined with DEM as a hybrid model | − | To keep computations tractable, number of components per cell is limited to a lumped version of the real cell layout |
|
| |||
| 2) Anisotropy also depends on cell geometry and network topology | |||
|
| |||
| FEM | DEM | ||
| + | Finite elements can be adapted to cellular geometry | + | Discrete elements can be adapted to cellular geometry |
| ± | Incorporation of anisotropy of actin accumulation bundles to be understood | + | Anisotropy of actin accumulation bundles can easily be incorporated |
| − | Computational issues related to modeling large scale cellular networks | + | Can scale to model large cellular networks and thus encompass their topological features |
|
| |||
| 3) Nonlinear and temporal mechanical characteristics | |||
|
| |||
| FEM | DEM | ||
| + | Can be incorporated in finite elements | + | Can be incorporated in discrete elements |
| − | May lead to heavy computations, even for average-sized cellular network | + | Nonlinear elasticity qualitatively resembles tissue deformation |
| + | Nonlinear elasticity contributes to model stability, prevents collapse | ||
| − | Temporal characteristics can be computationally demanding | ||
|
| |||
| 4) Volume preservation | |||
|
| |||
| FEM | DEM | ||
| + | Can be imposed through volumetric properties | ± | Its effect can be approximated but not formally embedded |
| ± | May result in artifacts and computational issues (stiffness), due to limited knowledge on properties of cell height | − | Approximation likely related to introduction of dynamical artifacts, requires evaluation of resulting mechanical properties |
3.3 Graphical representation of underlying cellular structure
Consider a general graph-theoretical structure. This structure will be generated from experimental data, as discussed in Section 5.1. The graph consists of three different features: vertices, edges, and faces. Vertices are contained in a set
 of cardinality N, which stores the index i ∈ {1, 2, …, N} of each vertex vi. In Figure 4 vertices are indicated by dots and their indices are denoted by the adjacent numbers. An edge (circled numbers in Figure 4) ei,j = {vi, vj } is defined as a pair of adjacent vertices vi and vj, where j ≠ i and i, j ∈ {1, 2, …, N}. Edges are stored as pairs in a Ne × 2 set
of cardinality N, which stores the index i ∈ {1, 2, …, N} of each vertex vi. In Figure 4 vertices are indicated by dots and their indices are denoted by the adjacent numbers. An edge (circled numbers in Figure 4) ei,j = {vi, vj } is defined as a pair of adjacent vertices vi and vj, where j ≠ i and i, j ∈ {1, 2, …, N}. Edges are stored as pairs in a Ne × 2 set
 , where Ne is the cardinality of set
, where Ne is the cardinality of set
 . The edge index k ∈ {1, 2, …, Ne} is defined by the row number in
. The edge index k ∈ {1, 2, …, Ne} is defined by the row number in
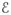 . The graph
. The graph
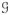 = (
= (
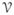 ,
,
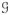 ) describes the full topology of a network. Consider the N × Ne incidence matrix H of a graph. Its entries are formulated as
) describes the full topology of a network. Consider the N × Ne incidence matrix H of a graph. Its entries are formulated as
Fig. 4.
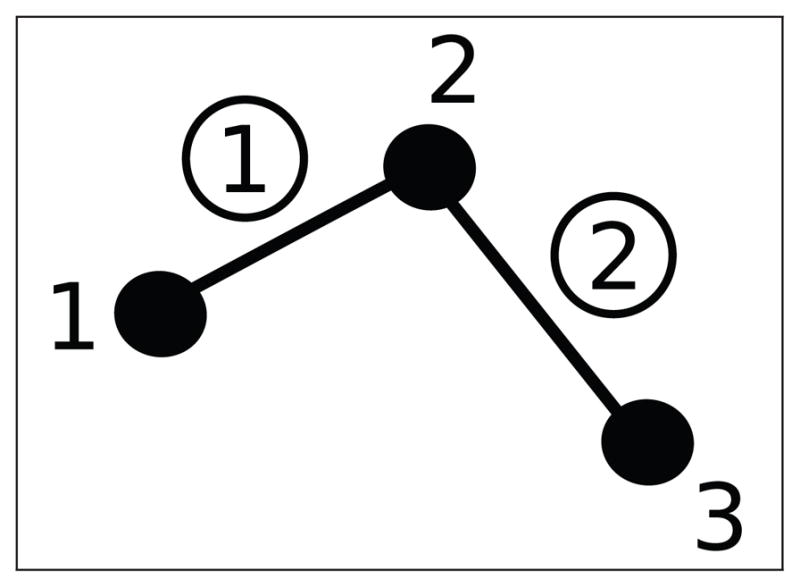
A small graph
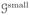 . Loose numbers denote vertices, whereas circled numbers denote edges.
. Loose numbers denote vertices, whereas circled numbers denote edges.
As an example, consider a small graph
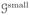 consisting of three vertices and two edges, as depicted in Figure 4. The incidence matrix follows:
consisting of three vertices and two edges, as depicted in Figure 4. The incidence matrix follows:
The N × N Laplacian matrix L describes which vertices are adjacent to each other and is calculated using the incidence matrix [38], as given by L = HH⊤. For
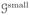 this yields
this yields
More specifically, the Laplacian L = L⊤ indicates whether a vertex vi is connected to another vertex vj (lij = lji = −1) or not (lij = lji = 0). The diagonal represents the degree
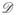 (vi) = lii, which is the number of edges that are adjacent to vertex vi.
(vi) = lii, which is the number of edges that are adjacent to vertex vi.
The third important feature in the network is represented by its cell faces. In a cellular network a face f represents (the apical side of) a cell and is defined as the set of its vertices. The degree of a face f is denoted by
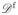 (f) and denotes the number of vertices (or edges) it consists of, e.g.
(f) and denotes the number of vertices (or edges) it consists of, e.g.
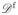 (f1) = 4. Since in this project the data is delivered as graphs of vertices and edges (
(f1) = 4. Since in this project the data is delivered as graphs of vertices and edges (
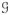 = (
= (
 ,
,
 )), an algorithm has been implemented in order to retrieve the information of the faces.
)), an algorithm has been implemented in order to retrieve the information of the faces.
3.4 Dynamical Model: the Spring-Damper element
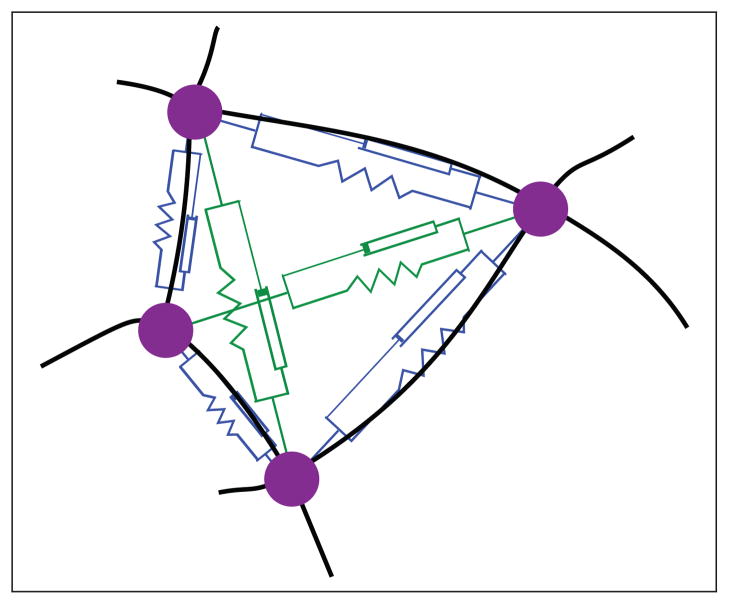
Assume that the underlying cellular geometry (that is, the underlying network topology) is known. A mesh is overlaid to the cell junctions, that is to the points where cell boundaries intersect (the magenta dots in Figure 5). A model is created as a two-dimensional, tiled surface and is represented using the graph theoretical network notation given in Subsection 3.3. Within this data structure three different modeling elements (discussed below) are discussed and associated to different structural features in epithelial cellular networks.
Fig. 5.
A spring-damper model of a single cell. Discrete points (large magenta dots) are adapted to the actual cell shape extracted from experiments (see Section 5.1). Black lines denote cell boundaries. Blue spring-damper elements are associated to boundaries, whereas green ones to intermediate pairs of non-adjacent points.
First, a magenta dot in Figure 5 represents a point mass element mi associated to vertex vi, i ∈ {1, 2, …, N}. This models the inertia of the network via a constant mass value. However, let us importantly remark that the modeling approach is not intended to imply that inertial forces are reflecting the underlying biology: the actual effect of the mass in the model dynamics can be eliminated by simple rescaling (as discussed later, we have in practice used it to normalize the simulations time span). The second law of Newton is applied to each point mass element at vi by considering the global contribution all the forces F⃗i acting upon the point:
where is the second time derivative of the position x⃗i of point mass mi, and x⃗ denotes a vector collecting all the points x⃗i. We will shortly express explicitly the force contribution in the equation above. The point masses are placed at vertices, where different cell boundaries come together. It should be noted that in reality cell junctions are not fixed, but they may rearrange with respect to the adjacent cell boundaries: the present modeling framework neglects these rearrangements. The implications of this assumption are further elaborated in Section 4.2.
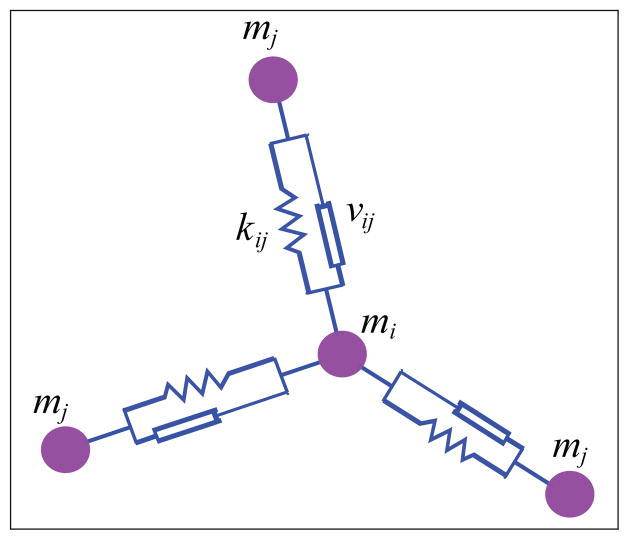
The second element models the circumferential actin belts, which organize along cell boundaries and connect adjacent cell vertices. Since these belts propagate their elastic energy to adjacent components through these vertices, a mechanistic equivalent will be used in the model. The mechanical behavior of actin belts is viscoelastic and modeled by the presence of spring-damper elements (Voigt elements) between two adjacent cell vertices, as depicted in Figure 5 [37], [34]. Section 4.1 discusses possible alternatives to the Voigt elements. In the following, these modules are called boundary elements. A boundary element encompasses the belts of two adjacent cells, since these two cells are connected through adherens junctions. Consider a point mass element mi connected to three other point mass elements mj, j ∈
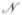 (vi), where
(vi), where
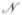 (vi) denotes the set of vertices adjacent to vertex vi, as depicted in Figure 6. The point masses are connected through spring-damper elements, characterized respectively by stiffness coefficients ki,j and damping coefficients νi,j. The mathematical equation for point mass element mi connected to the point masses mj via a spring-damper element, is given by:
(vi) denotes the set of vertices adjacent to vertex vi, as depicted in Figure 6. The point masses are connected through spring-damper elements, characterized respectively by stiffness coefficients ki,j and damping coefficients νi,j. The mathematical equation for point mass element mi connected to the point masses mj via a spring-damper element, is given by:
where li,j is the length of boundary element ei,j, is its time derivative, di,j is the resting length related to the spring at boundary element ei,j, and is the normalized vector accounting for orientation and direction of the element ei,j.
Fig. 6.
A single point mass element mi connected to three point mass elements (all denoted as mj ) through spring-damper elements.
As a third modeling element, the cytoskeleton (consisting of intermediate filaments and microtubules, see Figure 1) is modeled similarly to the circumferential actin belts using spring-damper elements, which in the following will be denoted as intermediate elements. With reference to the nodal characteristic of each single cell, elements are placed between all non-adjacent mass elements within a cell, as depicted in green on Figure 5. The collection of intermediate elements represent a discrete abstraction of the cytoskeleton. Section 4.2 explains the potential as well as the limitations of this approach. The mathematical equations for the elements are identical to those of the external elements, that is
where
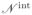 (vi) denotes the set of vertices laying within a face to which vertex vi belongs, but which are not directly adjacent to vi.
(vi) denotes the set of vertices laying within a face to which vertex vi belongs, but which are not directly adjacent to vi.
As anticipated in Section 2.1.3, the model considers nonlinear elasticity in cellular mechanics. The relationship between applied prestress and stiffness has a linear characteristic, both on the cell [29] and on the cytoskeletal component level [25]. To illustrate this relation, consider a spring, as depicted in Figure 7. The classical constitutive equation given by the Hooke law for the magnitude of the elastic force exerted by a spring is
Fig. 7.
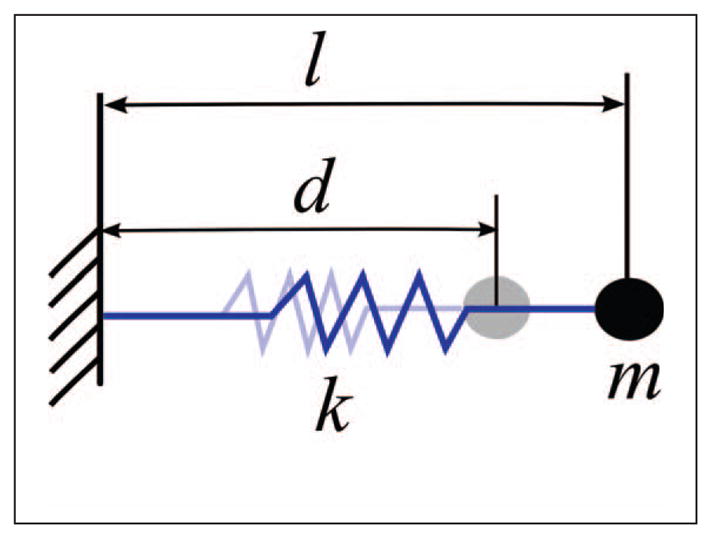
One-dimensional spring model used to explain the applied prestress.
| (1) |
where k denotes the stiffness coefficient, l is the current length and d is the resting length of the spring. Prestress is defined as the fraction of the deviation from the resting length, which is . Now consider an affine relation between stiffness and applied prestress, as observed in experiments [29]. This yields a coefficient
| (2) |
where k0 is the nominal (linear) stiffness coefficient at resting length and k1 accounts for the stiffness due to applied prestress. Substituting the affine relation in (2) into equation (1) for the elastic force results in a quadratic (and thus nonlinear) elastic force-deformation (stress-strain) relation:
The stiffness coefficients ki,j of the spring-damper elements in the model are characterized according to this nonlinear relation. This means that a pair of parameters ( ) is introduced for each spring-damper element. Section 4.1 discusses possible extensions of the expressions considered in (1)–(2).
In addition to the relations developed for the three main modeling elements, the framework assumes that the connections between cells and the extracellular matrix are viscous. A friction force is added, which acts on each point mass element mi. is inversely proportional to the velocity of point mass mi,
where λi is the positive friction coefficient related to point mass mi.
All the described components can be mathematically encompassed within a system of second-order ordinary differential equations (ODE). Formally, consider a point mass mi at vertex vi representing 6 state variables, i.e. 3 position variables x⃗i and 3 velocity variables
. The dynamical ODE for mi is presented in (3) on page 8, where ki,j (·) and νi,j denote the stiffness and damping coefficients of an element between masses mi and mj; li,j is the distance between the two masses;
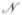 (vi) denotes the adjacent vertices of vi, and
(vi) denotes the adjacent vertices of vi, and
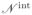 (vi) denotes the non-adjacent vertices within all the neighboring faces of vi. The stiffness and damping forces can only act in the direction of the element between the two masses, which is
. The parameter λi denotes the friction coefficient at mass mi, whereas
denotes the sum of all possible external, time-dependent forces μ acting on mass mi. (In Section 5.2 we shall consider a few examples for external forces and show that they also enable the embedding of spatial constraints in the model.)
(vi) denotes the non-adjacent vertices within all the neighboring faces of vi. The stiffness and damping forces can only act in the direction of the element between the two masses, which is
. The parameter λi denotes the friction coefficient at mass mi, whereas
denotes the sum of all possible external, time-dependent forces μ acting on mass mi. (In Section 5.2 we shall consider a few examples for external forces and show that they also enable the embedding of spatial constraints in the model.)
| (3) |
The model in (3) is nonlinear, due to the presence of the lengths li,j and their time derivatives, and because of the varying stiffness coefficients ki,j (li,j ). A matrix formulation can be derived using the Laplacian L defined above. First introduce a matrix
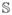 based on (3), which structure is related to that of L: notice that ||x⃗j − x⃗i|| = li,j, and define the non-diagonal entries (i ≠ j) of
based on (3), which structure is related to that of L: notice that ||x⃗j − x⃗i|| = li,j, and define the non-diagonal entries (i ≠ j) of
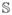 as
as
and similarly the diagonal entries as
Subsequently,
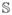 can be exploited to rewrite the dynamical equations in (3) for the whole network of point masses in matrix notation as follows:
can be exploited to rewrite the dynamical equations in (3) for the whole network of point masses in matrix notation as follows:
| (4) |
where M is a 3N × 3N diagonal matrix composed of the masses mi, and Λ is a 3N × 3N diagonal matrix with the friction coefficients λi.
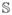 is time-dependent and contains all the nonlinearities of the equations. The values of ki,j (·), li,j, and dli,j/dt need to be updated in time for all spring-damper elements: the Laplacian L can be used to assign the updated variables to the corresponding entries in
is time-dependent and contains all the nonlinearities of the equations. The values of ki,j (·), li,j, and dli,j/dt need to be updated in time for all spring-damper elements: the Laplacian L can be used to assign the updated variables to the corresponding entries in
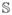 . Using smart vectorial implementations in MATLAB, the simulation algorithm can integrate the dynamics in (4) of a mass spring-damper network with dimensions in the order of hundreds of vertices in a computationally rapid way. The simulation can also benefit from techniques developed in the computer graphics community, for the development of simulators of soft-body dynamics [39]. We shall further discuss computational issues of the model in Section 5.2.
. Using smart vectorial implementations in MATLAB, the simulation algorithm can integrate the dynamics in (4) of a mass spring-damper network with dimensions in the order of hundreds of vertices in a computationally rapid way. The simulation can also benefit from techniques developed in the computer graphics community, for the development of simulators of soft-body dynamics [39]. We shall further discuss computational issues of the model in Section 5.2.
4 Discussion on the Model
The presented modeling framework incorporates both biological insight and engineering principles. Based on this modeling framework, the dynamical equations can be extended by introducing terms that account for new knowledge gained from both experiments and simulations. On the other hand, the choices made on the model architecture also pose limitations over its general application. We discuss next both potential and the limitations of the model.
4.1 Potential: model extensions and applications
The dynamical model allows to embed the effect of general, time-varying, non-linear, external forces. With reference to Figure 3 and the related discussion on nonlinearity of cellular elasticity, it is possible to include hysteresis and memory effects in the stress/strain characteristic by direct modification of Equation (2): the first feature can be useful to prevent dynamical high-frequency oscillations that are usually not observed experimentally, whereas the second can lead to the modeling of permanent cellular deformations. However, this can lead to higher computational costs. The choice of viscoelastic Voigt modules as interconnections between cellular junctions has been motivated by literature evidence and model testing, but can be as well substituted by Maxwell elements or – at the expense of an increase in complexity – by standard-linear or generalized-Maxwell elements [30]. The model also allows for the introduction of physical and spatial constraints, as will be discussed in the case study of Section 5.
The presence of an underlying dynamical model allows generating simulation outputs that can be matched to time-lapse experiments. This may help to explain mechanical properties of the network, for instance by identifying its stiffness parameters or friction coefficients. There are a number of advanced parameter identification techniques that could be used with this goal [40], [41], [42]. The model can furthermore help quantitatively studying certain morphogenetic effects such as germband retraction, groove formation, or dorsal closure. At the cell level, the study of elastic properties via laser ablation experiments can also represent an interesting potential to test the model on, where generated data would be matched to the model simulations.
4.2 Limitations
The cytoskeleton, scattered throughout the whole cell (cf. Subsection 2.1.1), influences most of the mechanical behavior of cells. Besides, other factors contribute to the internal mechanics of cells, such as osmotic pressure, the (apical-basal) height of the cell, the presence of microtubules. The internal spring-damper elements represent a lumped, two-dimensional abstraction of the cytoskeleton. This representation is not able to model the highly scattered cytoskeleton to its full extent, neither can it distinguish between different contributors to internal mechanics (e.g., microtubules vs. intermediate filaments).
The model describes a two-dimensional surface structure. It neglects the depth of the cell and only considers the mechanics and dynamics at the surface level. This assumption is meaningful since there is limited insight in the three dimensional deformation of epithelial structures, and because it represents an abstraction that makes the model computable over large networks of cells. Hence, this framework can help explaining behaviors of the surface-level properties of an epithelium, even with regards to its spatial dynamics (when for example used for full embryo studies), rather than giving insight on the volumetric deformations and dynamics (including volume preservation) of single epithelial cells or small clusters of cells.
The model assumes that cell junctions maintain their fixed configuration. In other words, adherens junction rearrangements are neglected. This means the framework cannot model cell division, cell death, or cell motility. Depending on the morphogenetic events under study (as it is the instance in the case study of Section 5), cell rearrangement may not play a significant role. It should be stressed that the modeling framework has the potential to incorporate these characteristics, albeit with a non-trivial extension of the underlying graphical structure and with an expected increase in computations (required to update the data structure and to integrate the dynamics over time). This extension could enable the study of higher-level phenotypes such as cell motility, cell proliferation and apical constriction, and to understand their relations to the dynamics and mechanics of epithelial cellular networks.
5 Case Study: from Experimental Data to Model and to Simulations
The dorsal epidermis of the D. melanogaster embryo during dorsal closure provides a solid basis for a case study. Firstly, at these stages, cells of different shapes get stretched along their dorsoventral axis. Secondly, at each segment boundary a specific group of cells, called the groove (as we will discuss later, in our data this corresponds to the central vertical “column” of cells in Figure 8(b)), cells get organized in order to form a single column of rectangular cells [6]. Thirdly, this row of cells deepens in the tissue in order to shape the segmental grooves [3], [5], [8]. Altogether these stereotypical features in a developing embryo provide a good comparison benchmark.
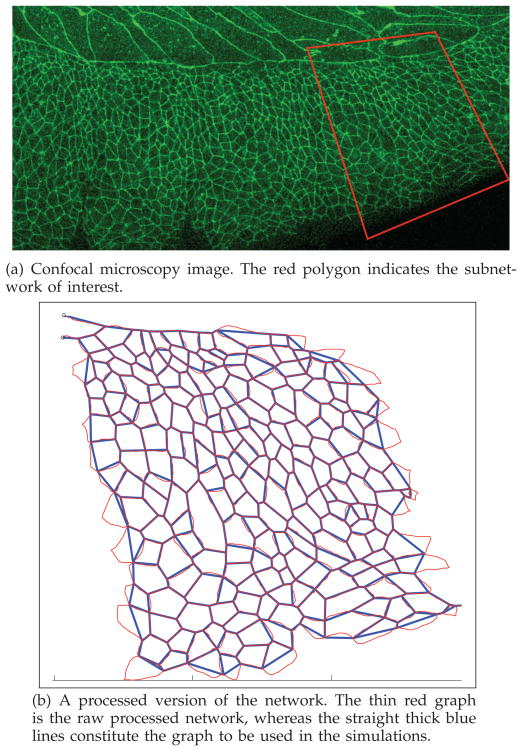
Fig. 8.
Isolated cellular network at starting time, used for model building.
We test the presented dynamical model on a cellular network extracted from experimental data covering dorsal closure and, less conspicuously, germband retraction. We build a dynamical model first by extracting a cellular network from data (Sec. 5.1), then by associating dynamics to the elements of the network (Sec. 5.2). We then perform simulations by exciting the model with external forces and introducing physical constraints (Sec. 5.2), to produce simulations resembling dorsal closure. While the goal of this simulation is solely to provide an interesting computational validation for the model, rather than to present new biological knowledge, it also suggests that the use of this model can lead to qualitative and quantitative conclusions on the mechanical properties of the epithelium of the embryo by comparing the simulation outcomes to the experimental data (Sec. 5.3).
5.1 From Experiments to Model
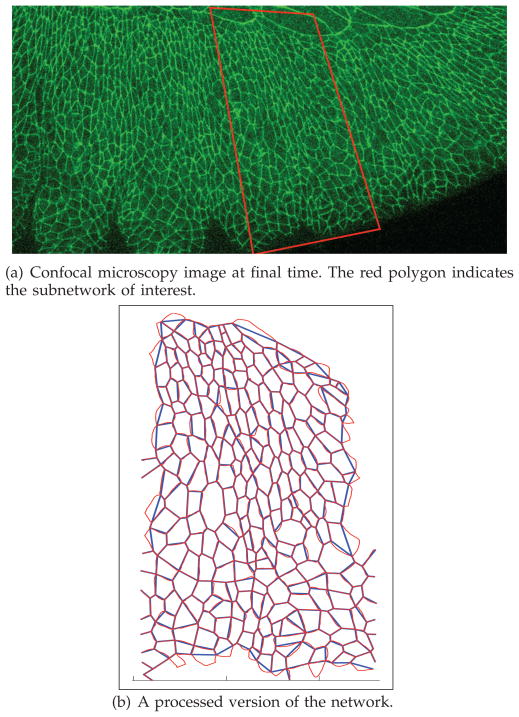
We consider a planar confocal microscopy image of a rectangular section of the lateral epithelium before dorsal closure occurs, as in Figure 8(a). An image processing software [43], developed in-house and implemented in MATLAB, is used to segment a rectangular section of the frame into a cellular network, namely a graph. The software employs a randomized search algorithm that scans the entire image and originates the graph. The output of the segmentation algorithm results in the thin red cellular network, depicted in Figure 8(b). The cellular sides of the network are subsequently straightened (see overlaid blue network) to obtain a graph. This graph contains the relevant information on boundaries (edges) and vertices of the patch of cells under study. The graph is represented and stored via the data structure that has been detailed in Section 3.3 (recall that the data structure accounts for the cell vertices, the cell boundaries (edges), and the cell areas (faces)).
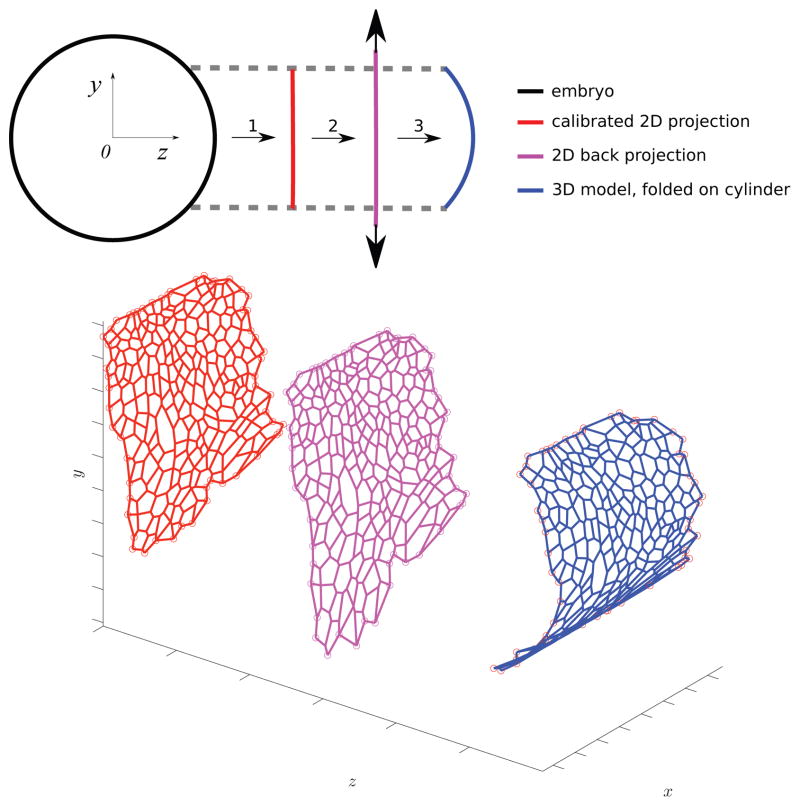
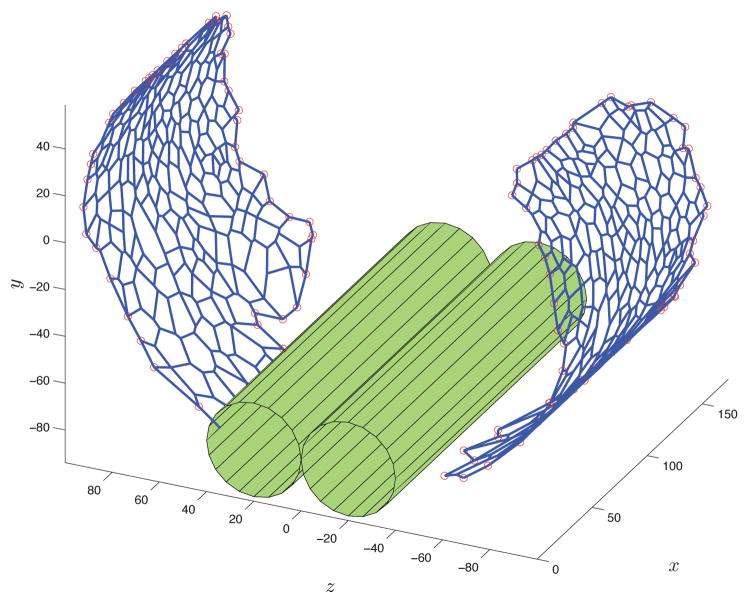
Figure 9 gives a pictorial representation of an embryo, with the position of the segment of the epithelium under study. The two dimensional cellular network that has been extracted from the data is morphed to assume a three dimensional embryo-like shape. The obtained three dimensional network matches its two-dimensional projection. The morphing procedure consists of three steps, as detailed in Figure 10. First, based on the microscope settings (e.g., its orientation w.r.t the embryo), the processed network is calibrated (rotated and translated) with respect to a cylindrical body resembling the embryo. Subsequently, the network is stretched out and finally, it is folded three dimensionally to adapt to the model of the epithelium, as indicated in blue color in Figure 10. Based on our experimental data, the radius of the embryo has been set to be Rbody = 100μm.
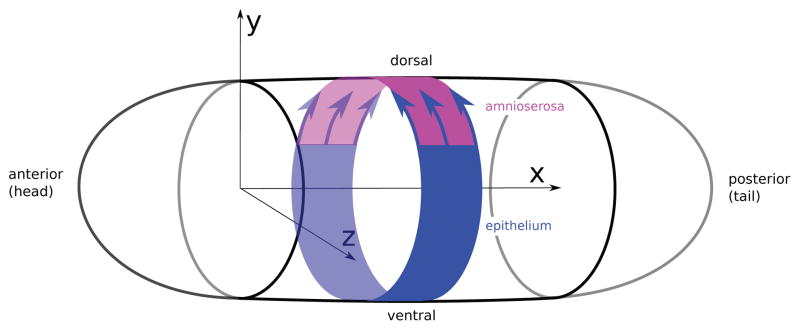
Fig. 9.
Pictorial model of the embryo, depicting a segment of the epithelium (blue). The dorsal closure forces acting on the epithelium over the amnioserosa (pink region) are indicated by the blue arrows.
Fig. 10.
Top: A pictorial longitudinal view of the embryo with indication of calibration, back projection, and folding steps, which together yield the folded, three-dimensional model of the epithelium. Bottom: A three-dimensional view of the morphing procedure.
5.2 Adding Dynamics to the Model
Given a cellular network extracted from data, we overlay the spring-damper elements on it. The dynamics in equation (3) are thus associated to each vertex of the stored cellular network and allow for the inclusion of external forces. The model in equation (3) depends on a set of parameters, which characterize spring and damper elements (both boundary and internal ones, both for stiffness and resting prestress), friction in the model, as well as external forces and constraints (the latter will be discussed shortly). These parameters need to be instantiated according to the data and to the goal of the simulation. In this study, their value is selected from a set of simulation tests, each of which is driven by a specific external force. The goal of these tests is, given a specific cellular network and a set of forces, to produce outputs that are qualitatively acceptable, namely that do not present unstable, unrealistic, on non-physical dynamics. Notice from Equation (3) that there is a linear relationship between the time horizon of a simulation and the value of the mass. We have thus decided to rescale both in order to normalize the first to span the unit interval.
One important set of forces that acts on epidermal cells comes from the developing central nervous system (CNS). The ventral epithelium closely wraps around the CNS, thus the epithelium experiences the CNS as a physical constraint. While the precise mechanical characteristics of the CNS have not been investigated, it is observed that folds rarely appear in the epidermis juxtaposed to the CNS. Only strong grooves, generated in genetic over-expression experiments, partially pinch into the CNS. This observation suggests that in general the CNS is able to resist pressure exerted by the overlaying epidermis. In our model the CNS is abstracted as two adjacent and parallel cylinders, as in Figure 11. The radius of each cylinder of the CNS, RCNS is estimated from cross section experiments to be one fifth of the radius of the embryo Rbody. Ideally the CNS only acts on mass elements that are in contact with it. However, modeling the CNS as a hard physical constraint may be both biologically unrealistic (organs are not infinitely rigid) and practically undesired (it may lead to stiff dynamics). Inspired by an approach developed in the computer graphics community for the development of simulators of soft-body dynamics [39], the interaction between embryo and CNS is implemented as an external force acting on the epithelium along a direction that is normal to the CNS cylinder. Let us denote the distance vector from the axis of the CNS to a vertex i by r⃗i. Our implementation employs a polynomial formulation that depends on a parameter u ∈ ℕ:
Fig. 11.
Three-dimensional model of the epithelium of the embryo (symmetric representation). The green cylinders denote the central nervous system. Notice that part of the ventral epithelium has not been captured by the microscope and hence has not been modeled.
The interaction of the epithelium with the CNS can be tuned and stiff dynamics can be avoided.
The ventral vertices of the cellular network ideally would lie under the ventral line of the embryo, as in Figure 9. Unfortunately, a limited section of the network close to the ventral line is not included in the model from the data, due to the limited range of the microscope – see Figure 11. Thanks to the limited dynamics of the ventral cells this drawback is not too severe: assuming symmetry between two vertical halves of the embryo and because of their adjacency to the ventral line, the ventral vertices are constrained to move exclusively along the ventral line, that is in x-direction. This means that the dynamics in the y- and z-direction are set to zero.
How to prevent the global cellular network from collapsing over itself? The inner body of the embryo can be imagined as a fluid mass exerting a pressure force on the ectoderm, whenever there is a pressure difference between the interior and the exterior of the embryo. This phenomenon can be modeled using the Laplace-Young law [44]. Assume that the embryo is a cylindrical vessel, as depicted in Figure 12. The larger the cylindrical radius Rbody, the larger the boundary tension T (dashed red arrows in Figure 12) required to withstand a given pressure difference Δp (solid cyan arrows in Figure 12) over the boundary. This property can be derived from the Laplace-Young equation, which relates the pressure difference to the shape of the surface: . This effect is implemented over the whole cellular network, assuming a constant pressure difference Δp.
Fig. 12.
The Laplace-Young Law relates the pressure difference Δp over a surface to the surface tension T.
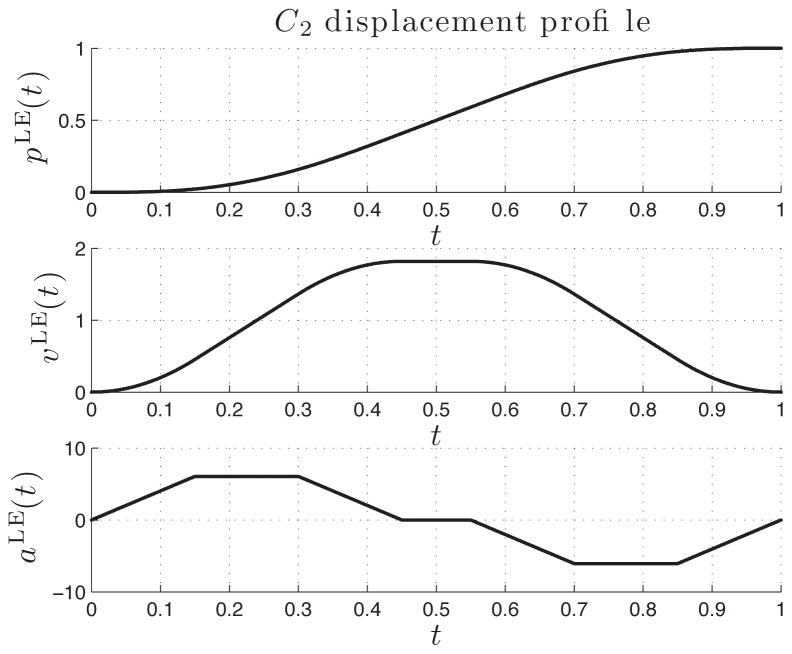
Dorsal closure is the preponderant morphogenetic movement showing in the experimental data for the case study, and is driven by the leading edge cells, namely the row of cells lying most dorsally within the epithelium. The leading edge is pulled locally, and the corresponding cells move over the embryo surface up toward the dorsal line, while propagating the pull over the rest of the epithelium. We assume to have knowledge only of the final position of the leading edge vertices (aligned along the dorsal line). We simulate DC forces by imposing a
 continuous trajectory (that is, a trajectory represented with a twice-continuous function of time) on all leading edge vertices, between their initial and final configuration. The trajectory is thus only imposed on the dynamics in the y- and z-directions, the x-direction being left undisturbed. A
continuous trajectory (that is, a trajectory represented with a twice-continuous function of time) on all leading edge vertices, between their initial and final configuration. The trajectory is thus only imposed on the dynamics in the y- and z-directions, the x-direction being left undisturbed. A
 trajectory provides complete flexibility in order to obtain smooth motion and to eliminate jerk from the mechanical components [45]. The input trajectory, along with velocity and acceleration profiles, are depicted in Figure 13.
trajectory provides complete flexibility in order to obtain smooth motion and to eliminate jerk from the mechanical components [45]. The input trajectory, along with velocity and acceleration profiles, are depicted in Figure 13.
Fig. 13.
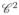 trajectory imposed on the leading edge vertices. Top down displacement profile pLE(t), velocity profile vLE(t), and acceleration profile aLE(t). The time axis has been normalized.
trajectory imposed on the leading edge vertices. Top down displacement profile pLE(t), velocity profile vLE(t), and acceleration profile aLE(t). The time axis has been normalized.
Simulations are run in MATLAB, on a laptop with a Pentium 2.66 GHz Intel Core 2 Duo processor and 4 GB of memory at 1067 MHz DDR3. The Runge-Kutta 4,5 method [46] is used to numerically integrate the dynamics in (4). The cellular network is made up of 253 vertices and thus the dynamical model has 1518 ODEs. The average integration time for a simulation taks less than three minutes. The total time spent on the post-processing of the data and the graphical analysis amounts to about two minutes.
The outputs of the integration procedure, namely the position and the velocities of all the vertices of the network in time, are saved as Visualization ToolKit (VTK) files and exported to the ParaView software environment [47]. Paraview is an open-source software that imports the generated VTK files and compiles them allowing for spatial computer graphics and visualization.
5.3 Simulations
We select a stiffness coefficient k0 for the central vertical column of cells in Figure 8(b) that is twice as high as that of the remaining cells. These cells display an accumulation of cytoskeletal proteins compared to their non-groove neighbors [6]. This is the case for instance for Enabled, which is implicated in the organization of actin filaments [48], as well as the beta-catenin Armadillo, which links the adherens junctions to the cytoskeleton [49]. Higher stiffness of these specific cells is related to the emergence of grooves, a phenomenon we are not going to further focus on.
We expect the cells to stretch vertically and show alignment, particularly around the mid column of Figure 8(b). We are thus interested in quantitatively assessing the quality of the simulations with respect
-
2
to the alignment of cells, and
-
3
to their elongation.
This outcome may indicate that the accumulation of cytoskeletal components in the groove cells is consistent with a different behavior of these cells at the mechanical level and may be responsible for their rectangular shape. This possibility will need to be addressed with genetic tools in vivo. Therefore, this contribution has no explicit goal to shed new light over the biological data under consideration: we are instead interested to qualitatively show that simulations of the model, which has been initialized to fit the data, can reproduce the behavior observed in experiments. This indicates the possible general use of the proposed modeling framework in similar studies.
5.3.1 Qualitative analysis of the simulation scenarios
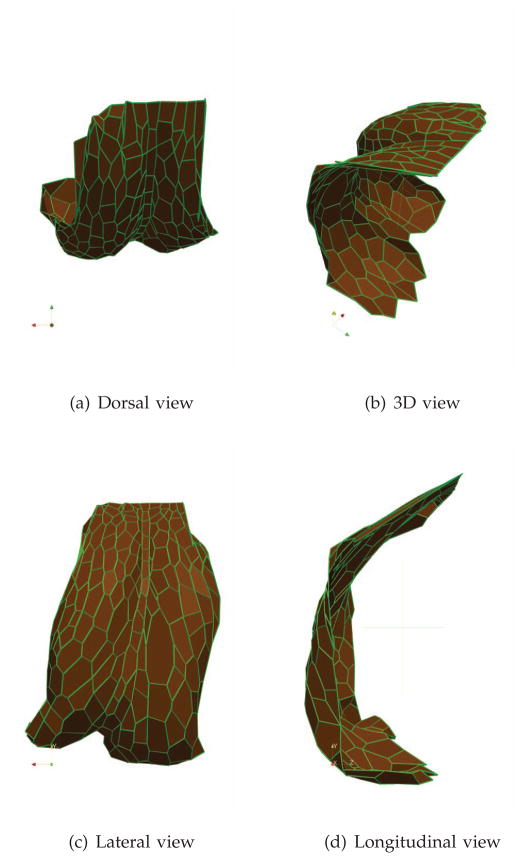
Figure 14 displays the rendered outputs of a single simulation. They are to be compared with the image in Figure 15(a), representing the network of cells in Figure 8(a) at a later stage, after dorsal closure has ensued.
Fig. 14.
Three-dimensional simulation outcomes. The cartesian axes are x (red), y (yellow), and z (green).
Fig. 15.
Experimental cellular network at final time, used as a benchmark for the simulation outcomes.
Notice that the indenting central column approaches the CNS cylinder and interacts with it, which calls for the use of the corresponding external forces discussed earlier. The dorsal and lateral view clearly indicate that cell boundaries line up along columns in dorsoventral direction. In particular, the central column lines up in an almost perfect ladder configuration. It is also quite clear that cells stretch in the vertical direction. A limitation of the simulations is visible on the leading edge which, unlike in the experimental image, is nicely aligned: this artificial effect is due to the imposed simulation dynamics (input trajectory) over the leading edge and can be possibly eliminated by feeding to the simulation a more realistic trajectory for those specific cells.
5.3.2 Quantitative analysis of the simulation scenarios
We now consider the processed cellular network in Figure 15(b), which is extracted from the experimental data in Figure 15(a). It clearly relates to the network in Figure 8(b), except for a few extra cells that have been caught by the microscope and appear close to the ventral and lateral boundaries. We exclude these extra cells from the statistics that are going to be elaborated below. No noticeable junction rearrangement is observed in the data.
As discussed earlier, we are interested in quantitatively comparing alignment and elongation of the cells from the simulated network with those of the cells from the experimental data. We introduce the following metrics, for each cell:
the ratio κ = xL/xl between the two axes of the cell, where xL denotes the longer axis and xl the shorter one. This quantity is a number greater than one that denotes the elongation of the cell;
the angle of orientation of the principal (longer) axis of the cell. This is a quantity α ∈ [−180, 180] degrees, computed from the vertical line, namely α = 0 along the vertical axis and α growing clockwise.
For both κ and α we compute mean m and the standard deviation σ, over the whole cohort of cells. The value of σ is particularly important for the orientation angle α, which is a quantity that spans [−180, 180].
Such metrics can be easily computed over the cellular networks present in Figures 8(b) and 15(b), which are extracted from the experimental data. Furthermore, we compute these metrics on the two-dimensional network that is obtained by projecting the lateral-view output of the simulation (see Figure 14(c)): this network is reported in Figure 16. (Notice that this two dimensional projection inevitably suffers from a few cells crossing each other: we shall eliminate these cells from the statistics developed below.)
Fig. 16.
Two dimensional projection of the lateral-view simulation output in Figure 14(c).
We set up two statistical comparisons. The first is based on the whole network (see Table 2, rows denoted by “W”), whereas the second is based on the group of cells adjacent to the central vertical column (see Table 2, rows denoted by “C”). For consistency, we selected the cells in this region by matching the experimental outcomes with the simulations. We discussed above how we expect a more ordinate alignment of cells within this region.
TABLE 2.
Metrics on cells - κ denotes the elongation of the cell, whereas α the angle of orientation of the cell; m represents the mean value and σ the standard deviation; W indicates statistics over the whole network, whereas C over the cells adjacent to the central vertical column.
| m(κ) | σ(κ) | m(α) | σ(α) | |
|---|---|---|---|---|
| Initial exp. data, Fig. 8(b), W | 2.48 | 1.93 | −23.68 | 48.66 |
| Initial exp. data, Fig. 8(b), C | 2.29 | 1.18 | −33.63 | 27.78 |
| Final exp. data, Fig. 15(b), W | 3.40 | 1.71 | −11.01 | 25.48 |
| Simulation data, Fig. 16, W | 4.09 | 4.36 | −4.55 | 32.05 |
| Final exp. data, Fig. 15(b), C | 4.04 | 1.31 | −6.15 | 9.01 |
| Simulation data, Fig. 16, C | 4.44 | 2.37 | −8.63 | 16.45 |
The first two rows of Table 2 display statistics based on the experimental data at the initial time, as per Fig. 8(b), and serve as reference. (Notice in particular that the statistics on the orientation display a high variance, as expected.) The second pair of rows focus on statistics at the final time, for the whole network. They compare the experimental data with the outcomes of the simulations. Similarly, the last two rows draw statistics at the final time, for the cells belonging to the central vertical region. Notice how the simulation outputs are consistent with the observed experimental data, particularly when the statistics are focused on the central region of the network. As expected, we observe a general increase in elongation of the cells (cfr. m(κ) values, for similar values of variance), as well as an alignment toward the vertical axis especially in the cells belonging to the central region (cfr. small values of m(α) and decreasing values of σ(α) for rows denoted with C).
Let us conclude by adding that of course, the selection of more complex metrics is possible (in order to enable a comparison of strain maps, for example), as well as the extension of the correspondence over the whole set of images taken from the time-lapse movie (this would account for detailed matching of the motion).
6 Conclusions
The main goal of this work has been that of developing and implementing an adaptable and scalable modeling framework for single-layered epithelial cellular structures. The modeling architecture has been motivated by a set of criteria distilled from biological knowledge on cellular mechanics, and grounded on comparisons with other modeling options from the literature. After discussing the data structure underlying the implementation of the model, the dynamical model has been the main focus of the exposition.
As a case study, the work has considered the process of early dorsal closure on the embryo of Drosophila melanogaster. The work leveraged developed software to extract the cellular geometry from a two-dimensional microscopy image, which has enabled realistic simulations that have been compared to the experimental data.
While this work has no explicit goal of biological relevance over the considered case study, we expect that the outcomes of the simulations can be meaningful to derive conclusions on the elasticity and stiffness properties of the epithelium, as well as on the force distributions and profiles playing a role in early dorsal closure. More generally, the proposed modeling framework promises to be a general platform to investigate structure and dynamics of single-layered epithelial cellular networks in a number of diverse applications.
Supplementary Material
Acknowledgments
This research is in part funded by the European Commission under the NoE FP7-ICT-2009-5 257462, by the European Commission under Marie Curie grant MANTRAS PIRG-GA-2009-249295, by NWO under VENI grant 016.103.020, by grants R01 GM097081 and R01 GM097081, and by NCI through the PSOC grant.
Biographies
 Alessandro Abate received a Laurea in Electrical Engineering in October 2002 from the University of Padova (Italy), an MS in May 2004 and a PhD in December 2007, both in Electrical Engineering and Computer Sciences, at UC Berkeley. He has been an International Fellow in the CS Lab at SRI International in Menlo Park (CA), and a PostDoctoral Researcher at Stanford University, in the Department of Aeronautics and Astronautics. Since June 2009, he has been an Assistant Professor at the Delft Center for Systems and Control (DCSC), TU Delft - Delft University of Technology. His research interests are in the analysis, verification, and control of probabilistic and hybrid systems, and in their general application over a number of domains, particularly in systems biology.
Alessandro Abate received a Laurea in Electrical Engineering in October 2002 from the University of Padova (Italy), an MS in May 2004 and a PhD in December 2007, both in Electrical Engineering and Computer Sciences, at UC Berkeley. He has been an International Fellow in the CS Lab at SRI International in Menlo Park (CA), and a PostDoctoral Researcher at Stanford University, in the Department of Aeronautics and Astronautics. Since June 2009, he has been an Assistant Professor at the Delft Center for Systems and Control (DCSC), TU Delft - Delft University of Technology. His research interests are in the analysis, verification, and control of probabilistic and hybrid systems, and in their general application over a number of domains, particularly in systems biology.
 Stephane Vincent is a lecturer at the Ecole Normale Superieure de Lyon in Lyon, France. He conducted his thesis research in Markus Affolter’s laboratory at the Biozentrum, Basel Switzerland, where he studied the roles of FGF and DPP on cell migration and identified DOF as a key adaptor specific to the FGF pathway. He received his post-doctoral training in the laboratory of Norbert Perrimon at the department of Genetics, Harvard Medical School and in the laboratory of Jeff Axelrod at the department of Pathology, Stanford School of Medicine. During this training, he focused first on the specification of the segmental groove cells in the Drosophila embryo and then on groove morphogenesis.
Stephane Vincent is a lecturer at the Ecole Normale Superieure de Lyon in Lyon, France. He conducted his thesis research in Markus Affolter’s laboratory at the Biozentrum, Basel Switzerland, where he studied the roles of FGF and DPP on cell migration and identified DOF as a key adaptor specific to the FGF pathway. He received his post-doctoral training in the laboratory of Norbert Perrimon at the department of Genetics, Harvard Medical School and in the laboratory of Jeff Axelrod at the department of Pathology, Stanford School of Medicine. During this training, he focused first on the specification of the segmental groove cells in the Drosophila embryo and then on groove morphogenesis.
 Roel Dobbe received his Bachelor degree cum laude in Mechanical Eng. at Delft University of Technology in 2007. In 2010 he finished his Master in Systems & Control cum laude at Delft Center for Systems & Control at Delft University of Technology. He graduated on a project in collaboration with and executed at EECS at University of California Berkeley, as a Huygens Scholar. His Msc research focused on Hybrid Systems and Control, with application in Systems Biology.
Roel Dobbe received his Bachelor degree cum laude in Mechanical Eng. at Delft University of Technology in 2007. In 2010 he finished his Master in Systems & Control cum laude at Delft Center for Systems & Control at Delft University of Technology. He graduated on a project in collaboration with and executed at EECS at University of California Berkeley, as a Huygens Scholar. His Msc research focused on Hybrid Systems and Control, with application in Systems Biology.
 Alberto Silletti got the Master degree cum laude in Computer Eng. at University of Padua in 2006. In 2010 he got the PhD degree in Science and Information Tecnology. He currently works as Postdoctoral Researcher at the Dep. of Information Engineering, University of Padua. His research areas and international publications range over computer vision algorithmics, image analysis and decisional systems.
Alberto Silletti got the Master degree cum laude in Computer Eng. at University of Padua in 2006. In 2010 he got the PhD degree in Science and Information Tecnology. He currently works as Postdoctoral Researcher at the Dep. of Information Engineering, University of Padua. His research areas and international publications range over computer vision algorithmics, image analysis and decisional systems.
 Neal Master graduated with Highest Honors from the University of California at Berkeley in May 2012 with a Bachelors of Science in Electrical Engineering and Computer Sciences. He was an undergraduate research assistant with Professor Claire Tomlin and has previously worked at Lawrence Berkeley National Laboratory in the National Energy Research Scientific Computing Center (NERSC) as well as at Oracle Corporation in Burlington, MA. He is currently a first year graduate student in the Department of Electrical Engineering at Stanford University. He has been awarded a Stanford Graduate Fellowship (SGF) as well as a National Defense Science and Engineering Graduate (NDSEG) fellowship. His research interests focus on the analysis and control of large scale distributed dynamical systems.
Neal Master graduated with Highest Honors from the University of California at Berkeley in May 2012 with a Bachelors of Science in Electrical Engineering and Computer Sciences. He was an undergraduate research assistant with Professor Claire Tomlin and has previously worked at Lawrence Berkeley National Laboratory in the National Energy Research Scientific Computing Center (NERSC) as well as at Oracle Corporation in Burlington, MA. He is currently a first year graduate student in the Department of Electrical Engineering at Stanford University. He has been awarded a Stanford Graduate Fellowship (SGF) as well as a National Defense Science and Engineering Graduate (NDSEG) fellowship. His research interests focus on the analysis and control of large scale distributed dynamical systems.
 Jeffrey D. Axelrod MD PhD joined the faculty at Stanford in 1998, and is currently Associate Professor of Pathology. The primary focus of his research program is the acquisition of a mechanistic understanding of the intra- and inter-cellular signaling events that regulate Planar Cell Polarity (PCP), a fundamental biological process that polarizes epithelial cells within the plane of the epithelium, to produce parallel arrays oriented with respect to the tissue axes. Disturbances of PCP signaling result in numerous developmental defects, and PCP is implicated in several disease states.
Jeffrey D. Axelrod MD PhD joined the faculty at Stanford in 1998, and is currently Associate Professor of Pathology. The primary focus of his research program is the acquisition of a mechanistic understanding of the intra- and inter-cellular signaling events that regulate Planar Cell Polarity (PCP), a fundamental biological process that polarizes epithelial cells within the plane of the epithelium, to produce parallel arrays oriented with respect to the tissue axes. Disturbances of PCP signaling result in numerous developmental defects, and PCP is implicated in several disease states.
 Claire J. Tomlin is a Professor of Electrical Engineering and Computer Sciences at Berkeley, where she holds the Charles A. Desoer Chair in Engineering. She held the positions of Assistant, Associate, and Full Professor at Stanford from 1998–2007, and in 2005 joined Berkeley. She received the Erlander Professorship of the Swedish Research Council in 2009, a MacArthur Fellowship in 2006, and the Eckman Award of the American Automatic Control Council in 2003. She works in hybrid systems and control, with applications to biology, robotics, and air traffic systems.
Claire J. Tomlin is a Professor of Electrical Engineering and Computer Sciences at Berkeley, where she holds the Charles A. Desoer Chair in Engineering. She held the positions of Assistant, Associate, and Full Professor at Stanford from 1998–2007, and in 2005 joined Berkeley. She received the Erlander Professorship of the Swedish Research Council in 2009, a MacArthur Fellowship in 2006, and the Eckman Award of the American Automatic Control Council in 2003. She works in hybrid systems and control, with applications to biology, robotics, and air traffic systems.
Footnotes
Additional on-line Material
Videos of the simulation outputs can be found at: http://www.dcsc.tudelft.nl/~aabate/tcbb
Contributor Information
Alessandro Abate, Email: a.abate@tudelft.nl, Delft Center for Systems and Control, TU Delft, Delft, The Netherlands.
Stéphane Vincent, Email: stephane.vincent11@ens-lyon.fr, Ecole Normale Supérieure de Lyon, Lyon, France.
Roel Dobbe, Delft Center for Systems and Control, TU Delft, Delft, The Netherlands.
Alberto Silletti, Email: alberto.silletti@gmail.com, Department of Information Engineering, University of Padova, Padova, Italy.
Neal Master, Email: neal.m.master@EECS.berkeley.EDU, Department of Electrical Engineering and Computer Sciences, University of California at Berkeley, Berkeley, CA, USA.
Jeffrey D. Axelrod, Email: jaxelrod@stanford.edu, Department of Pathology, Stanford University School of Medicine, Stanford, CA, USA
Claire J. Tomlin, Email: tomlin@EECS.berkeley.EDU, Department of Electrical Engineering and Computer Sciences, University of California at Berkeley, Berkeley, CA, USA.
References
- 1.Boal D. Mechanics of the Cell. Cambridge University Press; 2011. [Google Scholar]
- 2.Alexander R, Anderson M, Chaplain, KR, editors. Single-cell-based Models in Biology and Medicine. Springer Verlag; 2007. [Google Scholar]
- 3.Peralta X, Toyama Y, Hutson M, Montague R, Venakides S, Kiehart D, Edwards G. Upregulation of forces and morphogenic asymmetries in dorsal closure during Drosophila development. Biophysical Journal. 2007;92(7):2583–2596. doi: 10.1529/biophysj.106.094110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solon J, Kaya-Copur A, Colombelli J, Brunner D. Pulsed forces timed by a ratchet-like mechanism drive directed tissue movement during dorsal closure. Cell. 2009;137(7):1331–1342. doi: 10.1016/j.cell.2009.03.050. [DOI] [PubMed] [Google Scholar]
- 5.Layton A, YTY, Yang G, Edwards G, Kiehart D, Venakides S. Drosophila morphogenesis: tissue force laws and the modeling of dorsal closure. HFSP Journal. 2009;3(6):441–460. doi: 10.2976/1.3266062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vincent S, Perrimon N, Axelrod J. Hedgehog and Wingless stabilize but do not induce cell fate during Drosophila dorsal embryonic epidermal patterning. Development. 2008;135:2767–2775. doi: 10.1242/dev.017814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibson M, Nagpal R, Perrimon N. The emergence of geometric order in proliferating metazoan epithelia. Nature. 2006:1038–1041. doi: 10.1038/nature05014. [DOI] [PubMed] [Google Scholar]
- 8.Blanchard G, Murugesu S, Adams R, Martinez-Arias A, Gorfinkiel N. Cytoskeletal dynamics and supracellular organisation of cell shape fluctuations during dorsal closure. Development. 2010;137(16):2743–2752. doi: 10.1242/dev.045872. [DOI] [PubMed] [Google Scholar]
- 9.Stamenovic D. Models of cytoskeletal mechanics based on tensegrity. In: Kazempur-Mofrad MR, Kamm RD, editors. Cytoskeletal Mechanics. Cambridge University Press; 2006. pp. 103–128. [Google Scholar]
- 10.Cickovski T, Huang C, Chaturvedi R, Glimm T, Hentschel H, Alber M, Glazier J, Newman S, Izaguirre J. A framework for three-dimensional simulation of morphogenesis. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2005 Oct;2:273–288. doi: 10.1109/TCBB.2005.46. [DOI] [PubMed] [Google Scholar]
- 11.Chen H, Brodland G. Cell-level finite element studies of viscous cells in planar aggregates. Journal of Biomechanical Engineering. 2000;122:394–401. doi: 10.1115/1.1286563. [DOI] [PubMed] [Google Scholar]
- 12.Brodland G, Viens D, Veldhuis J. A new cell-based fe model for the mechanics of embryonic epithelia. Computer Methods in Biomechanics and Biomedical Engineering. 2007 Apr;10(2):121–128. doi: 10.1080/10255840601124704. [DOI] [PubMed] [Google Scholar]
- 13.Brodland G. Computational modeling of cell sorting, tissue engulfment, and related phenomena: A review. Applied Mechanics Review. 2004;57(1):47–76. [Google Scholar]
- 14.Karp G. Cell and Molecular Biology: Concepts and Experiments. Wiley; 2004. [Google Scholar]
- 15.Kasza K, Rowat A, Liu J, Angelini T, Brangwynne C, Koenderink G, Weitz D. The cell as a material. Current Opinion in Cell Biology. 2007;19:101–107. doi: 10.1016/j.ceb.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Discher D, Janmey P, Wang Y. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 17.Pesen D, Hoh J. Micromechanical architecture of the endothelial cell cortex. Biophysical Journal. 2005;88:670–679. doi: 10.1529/biophysj.104.049965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Potard U, Butler J, Wang N. Cytoskeletal mechanics in confluent epithelial cells probed through integrins and e-cadherins. American Journal of Physiology. 1997;272(41):1654–1663. doi: 10.1152/ajpcell.1997.272.5.C1654. [DOI] [PubMed] [Google Scholar]
- 19.Hu S, Chen J, Fabry B, Numaguchi Y, Gouldstone A, Ingber D, Fredberg J, Butler J, Wang N. Intracellular stress tomography reveals stress focusing and structural anisotropy in cytoskeleton of living cells. American Journal of Physiology Cell Physiology. 2003;285:C1082–C1090. doi: 10.1152/ajpcell.00159.2003. [DOI] [PubMed] [Google Scholar]
- 20.Kumar S, Maxwell I, Heisterkamp A, Polte T, Lele T, Salanga M, Mazur E, Ingber D. Viscoelastic retraction of single living stress fibers and its impact on cell shape, cytoskeletal organization, and extracellular matrix mechanics. Biophysical Journal. 2006 May;90:3762–3773. doi: 10.1529/biophysj.105.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiebe C, Brodland G. Tensile properties of embryonic epithelia measured using a novel instrument. Journal of Biomechanics. 2005;38:2087–2094. doi: 10.1016/j.jbiomech.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 22.Brodland G, Chen D, Veldhuis J. A cell-based constitutive model for embryonic epithelia and other planar aggregates of biological cells. International Journal of Plasticity. 2006;22:965–995. [Google Scholar]
- 23.Kamm R, Mofrad M. Cytoskeletal Mechanics: Models and Measurements. 1. Cambridge University Press; 2006. [Google Scholar]
- 24.Ingber D. Tensegrity I. cell structure and hierarchical systems biology. Journal of Cell Science. 2003;116:1157–1173. doi: 10.1242/jcs.00359. [DOI] [PubMed] [Google Scholar]
- 25.Stamenovic D, Mijailovich M, Tolic-Nørrelykke I, Chen J, Wang N. Cell prestress. II. Contribution of microtubules. American Journal of Physiology. 2002;282:617–624. doi: 10.1152/ajpcell.00271.2001. [DOI] [PubMed] [Google Scholar]
- 26.Chen X, Brodland G. Mechanical determinants of epithelium thickness in early-stage embryos. Journal of Mechanical Behavior of Biomedical Materials. 2009;2:494–501. doi: 10.1016/j.jmbbm.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 27.Zaidel-Bar R, Cohen M, Addadi L, Geiger B. Hierarchical assembly of cell–matrix adhesion complexes. Biochemical Society Transactions. 2004;32(3):416–420. doi: 10.1042/BST0320416. [DOI] [PubMed] [Google Scholar]
- 28.Beer F, Johnston E, Dewolf J, Mazurek D. Mechanics of the Cell. McGraw Hil; 2009. [Google Scholar]
- 29.Wang N, Tolic-Nørrelykke I, Chen J, Mijailovich S, Butler J, Fredberg J, Stamenovic D. Cell prestress. I. Stiffness and prestress are closely associated in adherent contractile cells. American Journal of Physiology. 2002;282:606–616. doi: 10.1152/ajpcell.00269.2001. [DOI] [PubMed] [Google Scholar]
- 30.Fung Y. Biomechanics: Mechanical Properties of Living Tissues. 2. Springer-Verlag; 1993. [Google Scholar]
- 31.Vasioukhin V, Fuchs E. Actin dynamics and cell-cell adhesion in epithelia. Current Opinion in Cell Biology. 2001;13:76–84. doi: 10.1016/s0955-0674(00)00177-0. [DOI] [PubMed] [Google Scholar]
- 32.Luo Y, Xu X, Lele T, Kumar S, Ingber D. A multi-modular tensegrity model of an actin stress fiber. Journal of Biomechanics. 2008;41:2379–2387. doi: 10.1016/j.jbiomech.2008.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Munjiza A. The Combined Finite-Discrete Element Method. Wiley; 2004. [Google Scholar]
- 34.Mollemans W, Schutyser F, Van Cleynenbreugel J, Suetens P. Tetrahedral mass spring model for fast soft tissue deformation. In: Ayache N, Delingette H, editors. Surgery Simulation and Soft Tissue Modeling. Vol. 2673. Springer-Verlag; 2003. pp. 145–154. [Google Scholar]
- 35.Odell G, Oster G, Burnside B, Alberch P. A mechanical model for epithelial morphogenesis. Journal of Mathematical Biology. 1980;9:291–295. doi: 10.1007/BF00276030. [DOI] [PubMed] [Google Scholar]
- 36.Terzopoulos D, Waters K. Physically-based facial modeling, analysis, and animation. Journal of Visualization and Computer Animation. 1990;1(2):73–80. [Google Scholar]
- 37.Cooper L, Maddock S. Preventing collapse within mass-spring-damper models of deformable objects. Proceedings of the Fifth International Conference in Central Europe on Computer Graphics and Visualization; Plzn, Czech Republic. 1997. pp. 196–204. [Google Scholar]
- 38.Arcak M, Sontag E. A passivity-based stability criterion for a class of biochemical reaction networks. Journal of Mathematical Biosciences and Engineering. 2008;5(1):1–19. doi: 10.3934/mbe.2008.5.1. [DOI] [PubMed] [Google Scholar]
- 39.Nealen A, Müller M, Keiser R, Boxerman E, Carlson M. Physically based deformable models in computer graphics. Computer Graphics Forum. 2005;25(4):809–836. [Google Scholar]
- 40.Chen D, Paden B. Stable inversion of nonlinear non-minimum phase systems. International Journal of Control. 1996;64(1):81–97. [Google Scholar]
- 41.Devasia S, Paden B. Stable inversion for nonlinear nonminimum-phase time-varying systems. IEEE Transactions on Automatic Control. 1998;43(2):283–288. [Google Scholar]
- 42.Raffard R, Amondlirdviman K, Axelrod J, Tomlin C. An adjoint-based parameter identification algorithm applied to planar cell polarity signaling. IEEE Transactions on Automatic Control. 2008;53:109–121. [Google Scholar]
- 43.Silletti A, Cenedese A, Abate A. The emergent structure of the Drosophila Wing - a dynamic model generator. Proceedings of the International Conference on Computer Vision, Theory and Applications (VISAPP 09); Lisboa, Portugal. February 2009; pp. 406–410. [Google Scholar]
- 44.Phillips R, Kondev J, Theriot J. Physical Biology of the Cell. 1. Garland Science; 2009. [Google Scholar]
- 45.Tsai L. Robot analysis: the mechanics of serial and parallel manipulators. Wiley & Sons; 1999. [Google Scholar]
- 46.Dormand J, Prince P. A family of embedded Runge-Kutta formulae. Journal of Computational Applied Mathematics. 1980;6:19–26. [Google Scholar]
- 47.Ahrens J, Geveci B, Law C. ParaView: An end-user tool for large data visualization. In: Hansen C, Johnson C, editors. The Visualization Handbook. Elsevier; 2005. [Google Scholar]
- 48.Kiger A, Baum B, Jones S, Jones M, Coulson A, Echeverri C, Perrimon N. A functional genomic analysis of cell morphology using RNA interference. Journal of Biology. 2003;2(4):27. doi: 10.1186/1475-4924-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peifer M. The product of the Drosophila segment polarity gene armadillo is part of a multi-protein complex resembling the vertebrate adherens junction. Journal of cell science. 1993;105(4):993–1000. doi: 10.1242/jcs.105.4.993. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.