Summary
Bacteria establish stable communities, known as biofilms, that are resistant to antimicrobials. Biofilm robustness is due to the presence of an extracellular matrix, which for several species - among them Bacillus subtilis - includes amyloid-like protein fibers. In this work, we show that B. subtilis biofilms can be a simple and reliable tool for screening of molecules with anti-amyloid activity. We identified two molecules, AA-861 and parthenolide, which efficiently inhibited biofilms by preventing the formation of amyloid-like fibers. We found that parthenolide also disrupted pre-established biofilms. These molecules also impeded the formation of biofilms of other bacterial species that secrete amyloid proteins, such as Bacillus cereus and Escherichia coli. Furthermore, the identified molecules decreased the conversion of the yeast protein New1 to the prion state in a heterologous host, indicating the broad range of activity of the molecules.
Introduction
Bacteria have the propensity to aggregate on any surface in ordered communities termed biofilms (O’Toole et al., 2000). To form biofilms bacteria secrete a matrix made up of a variety of molecules, sometimes including amyloid-like fibers (Blanco et al., 2012). Amyloid proteins were initially diagnosed as the etiological agent of important human diseases such as Alzheimer’s or Parkinson’s (Murphy, 2002). Based on their pathogenicity, amyloid proteins were originally thought to be exclusively non-functional misfolded proteins. The amyloid fold results in proteolysis-resistant fibers with characteristic morphological and biochemical properties. However, the ability to fold into an amyloid state is a property shared by many proteins and peptides and it is not necessarily related to pathogenesis (Blanco et al., 2012; Dobson, 2004; Fowler et al., 2007; Maury, 2009). Indeed, functional proteins that fold into an amyloid state have been found in different microorganisms (Blanco et al., 2012; Chapman et al., 2002; Claessen et al., 2002; Dueholm, et al., 2010; Gebbink et al., 2005; Romero et al., 2010; Wosten and de Vocht, 2000). A role for these proteins in cell-cell interactions, cell-surface interactions and biofilm formation has been demonstrated and they have thus been designated functional amyloid proteins (Badtke et al., 2009; Maury, 2009). Efforts to understand the nature of amyloid folding have led to screens for molecules that might inhibit the aggregation of these proteins. To date, a variety of molecules have been found to affect the polymerization of amyloid proteins responsible for important human disorders or involved in biofilm formation and virulence of the uropathogenic bacterium Escherichia coli (Amijee et al., 2009; Cegelski et al., 2009; Nie et al., 2011).
Bacillus subtilis forms robust floating biofilms (pellicles) in standing liquid cultures. The most striking visual feature of such pellicles is their extensive degree of wrinkling. The formation of biofilms is dependent on the secretion of the protein TasA and its assembly into amyloid-like fibers (Branda et al., 2006; Romero et al., 2010). The disassembly of biofilms relies on the detachment of these fibers from cell surfaces (Kolodkin-Gal et al., 2010; Romero and Kolter, 2011). In this study, we showed that B. subtilis biofilms can be used as a simple and reliable biological system to screen for molecules with anti-biofilm and/or anti-amyloid activity. Using this system we found two molecules, AA-861 and parthenolide, that arrested biofilm formation by B. subtilis, E. coli and Bacillus cereus. We also showed that both molecules interfere with the polymerization of TasA into amyloid-like fibers. Further, these two molecules inhibited conversion of the yeast New1 protein to the prion state.
Results
Screening for anti-biofilm molecules
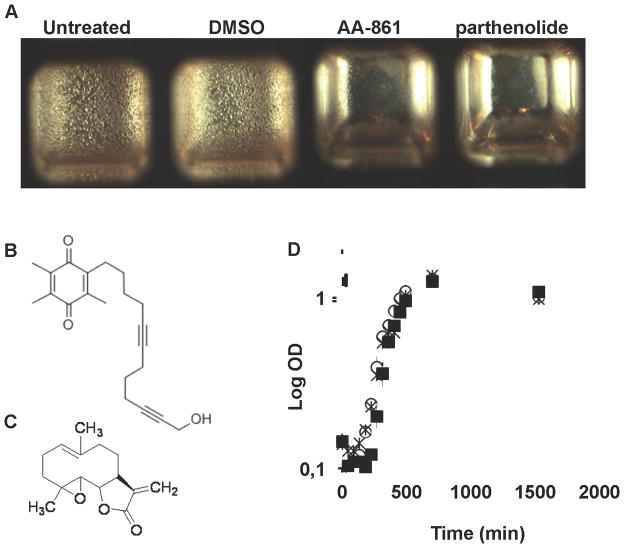
At the air-liquid interface of standing liquid cultures B. subtilis forms biofilms with wrinkles as a key distinguishable feature. Alterations of this phenotype have been used to screen collections of mutants and define regulatory genes and genes responsible for the synthesis of structural components of the extracellular matrix (Branda et al., 2004). We used the simplicity of this experimental set-up as a principle to screen for molecules with anti-biofilm activity. We obtained a small collection of known bioactive molecules from the BIOMOL–ICCB Known Bioactives collection from the ICCB Longwood Screening Facility (Harvard Medical School, Boston, MA, US). The collection originated from BIOMOL International, LP, Plymouth Meeting, PA, USA. The complete list of molecules in the known bioactives collection can be found at the following URL: http://iccb.med.harvard.edu/screening/compound_libraries/bioactives_biomol_med.htm. The collection was screened using a 384-well plate and positive hits were selected based on the absence of wrinkled pellicles (Figure 1A). This collection contains 480 small molecules whose mammalian cellular targets and/or biological activities have been well characterized. Two molecules, AA-861, a benzoquinone derivative (Figure 1B) and parthenolide, a sesquiterpene lactone (Figure 1C) inhibited the formation of B. subtilis biofilms (Figure 1A). A growth curve of cells grown in the presence or absence of these molecules showed that the concentration employed in the biofilm assay did not affect bacterial growth (Figure 1D).
Figure 1. Screening of molecules with anti-biofilm activity.
384 well microplates filled with MSgg medium were inoculated with B. subitlis 3610 cells and aliquots of a collection of small molecules at a final concentration of 12.5 μg/ml were added. After 24 h of incubation, plates were assessed for presence or absence of pellicles. (A) A close view of one of the plates showing the inhibition of pellicle provoked by two different molecules, (B) Structure of AA-861, a benzoquinone derivative, and (C) parthenolide, a sesquiterpene lactone. (D) A growth curve of B. subtilis 3610 in MSgg liquid medium showed no variation in bacterial growth in the absence (○) or presence of 50 μM of AA-861 (■), or parthenolide (×).
The anti-biofilm molecules act on the TasA amyloid protein
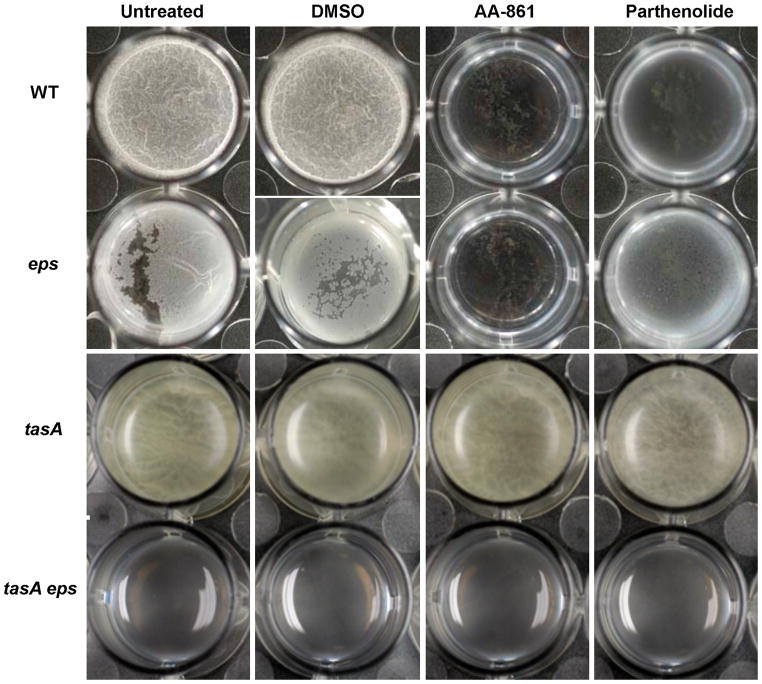
The B. subtilis extracellular matrix is made up of two main components: an exopolysaccharide (EPS) and the amyloid-like fibers formed by the TasA protein (Branda et al., 2006; Romero et al., 2010). We hypothesized that the anti-biofilm compounds could function to target one of the components of the extracellular matrix. Both EPS and TasA contribute to biofilm formation and only a mutant lacking both of these components is completely defective in pellicle formation (Branda et al., 2006). Thus, we could distinguish which component is affected by analyzing the effect of the compounds on mutants lacking either TasA or EPS. To test this, we analyzed the effect of the compounds on wild-type cells, individual eps or tasA mutants and a double mutant lacking both components of the extracellular matrix, in 24-well microtiter dishes. As observed in our primary screen, both molecules prevented the formation of wrinkly pellicles when added at a concentration of 50 μM, whereas the DMSO control looked similar to the untreated sample (Figure 2). The eps mutant grew as previously seen, forming a fragile broken pellicle, but this pellicle was completely inhibited in the presence of AA-861 and partially inhibited with parthenolide. In contrast, a tasA mutant was refractory to the activity of both molecules and produced the same thin and easily disrupted pellicles as the untreated controls (Figure 2). This suggested that both compounds could specifically target the protein component of the matrix. As expected, a tasA eps double mutant lacking both components of the extracellular matrix produced no pellicles under all conditions.
Figure 2. TasA is the main target of the anti-biofilm molecules.
Biofilm assays were performed to determine the main target of AA-861 and parthenolide. MSgg medium was inoculated with cells of wild-type, eps or tasA single mutants or double tasA, eps mutant. The molecules were added at 50 μM. DMSO was used as a negative control. Images of pellicles were taken after 24 hr of incubation at 30°C with no agitation (see also Figure S1 and Figure S2).
The anti-biofilm activity is not related to inhibition of tasA gene expression
The identified molecules are not toxic to B. subtilis cells (Figure 1D) and they appear to target TasA specifically (Figure 2). We therefore wanted to evaluate whether the anti-biofilm activity was due to a reduction in the expression level of the tasA gene. To test this we used the transcriptional fusion of the promotor of the tasA operon to the yfp protein (PtapA-yfp) and analyzed expression of this reporter using flow cytometry (Vlamakis et al., 2008). As previously observed, after 24 hr of incubation most untreated wild-type cells expressed PtapA-yfp (Figure S1A). The addition of either AA-861 or parthenolide did not alter the expression pattern significantly. In addition, TasA protein could be detected at similar levels in the matrix and in the medium before and after treatment with these two compounds (Figure S1B). We noted a slight decrease in TasA protein levels in the cell-associated fraction of treated cells relative to untreated control, however since this did not correspond to a decrease in the protein outside of cells (matrix or medium fraction) we do not believe this could account for the biofilm inhibition. These results indicated that the molecules do not appear to alter tasA transcription or TasA localization. Therefore, we hypothesized that the molecules might inhibit biofilm formation by physically interfering with the assembly of the TasA amyloid fibers.
The anti-biofilm activity is due to inhibition of TasA polymerization
Next, we tested if AA-861 and parthenolide might inhibit the polymerization of TasA amyloid-like fibers on the cell surface. We used transmission electron microscopy and immuno-gold labeling to study the localization of TasA in cells of biofilms after 24 hr of growth. In untreated controls, TasA formed long fibers that emanated from cells and were decorated with gold-labeled anti-TasA antibodies (Figure 3A, E). DMSO-treated cells looked similar to the untreated cells (Figure 3B, F). In contrast, gold-labelled antibodies remained associated to the cell surface and no fibers were apparent in cells treated with 50 μM of either AA-861 (Figure 3C, G) or parthenolide (Figure 3D, H).
Figure 3. The molecules inhibit fibers formation in cell surfaces.
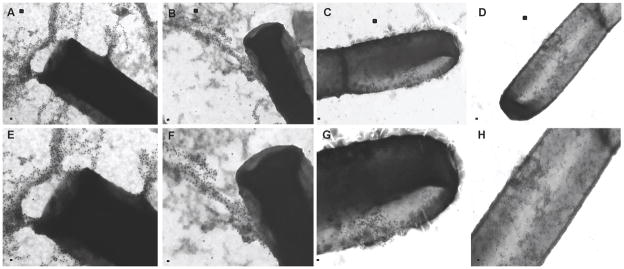
Transmission electron micrographs of negatively stained, anti-TasA immunogold labeled samples from pellicles grown for 24 hr in MSgg medium. (A) Untreated cells or (B) cells treated with DMSO as a negative control show wild-type fibers. Cells treated with 50 μM of (C) AA-861 and (D) parthenolide did not show fibers although the gold-labeled antibodies remained associated to cell surfaces. Panels E–H represent a magnified view of areas marked in panels A–D. Scale bars, 100 nm in panels A–D and 200 nm in panels E–H (see also Figure S2).
These findings favored the hypothesis that the anti-biofilm activity of these molecules is due to the fact that they prevent the assembly of TasA into functional amyloid-like fibers. We have previously demonstrated that the formation of TasA fibers requires at least two proteins, TasA and TapA (Romero et al., 2010; Romero et al., 2011). TasA is the major subunit of the fibers and TapA is a minor constituent that is necessary for the initiation of fiber polymerization and the anchoring of the fibers to the cell surface (Romero et al., 2011). Therefore, we asked whether the inhibitory effect of these molecules could be overcome by increasing the amount of TasA or TapA protein. To test this, we performed biofilm assays with two strains, DR6 (Phypespank-TasA) and DR8 (Phyperspank-TapA), expressing either TasA or TapA under the control of an IPTG-inducible promoter. The strain overproducing TasA formed robust pellicle in the presence of parthenolide similar to the untreated or DMSO-treated cells and an improved pellicle in the presence of AA-861 relative to the wild-type cells treated with this molecule. In contrast, the strain overproducing TapA showed the same sensitivity to the molecules as the wild-type strain (Figure S2). Thus, we conclude that the two molecules inhibited biofilm formation by interfering with the assembly of TasA into amyloid-like fibers.
The molecules obstruct the formation of TasA amyloid-like fibers in vitro
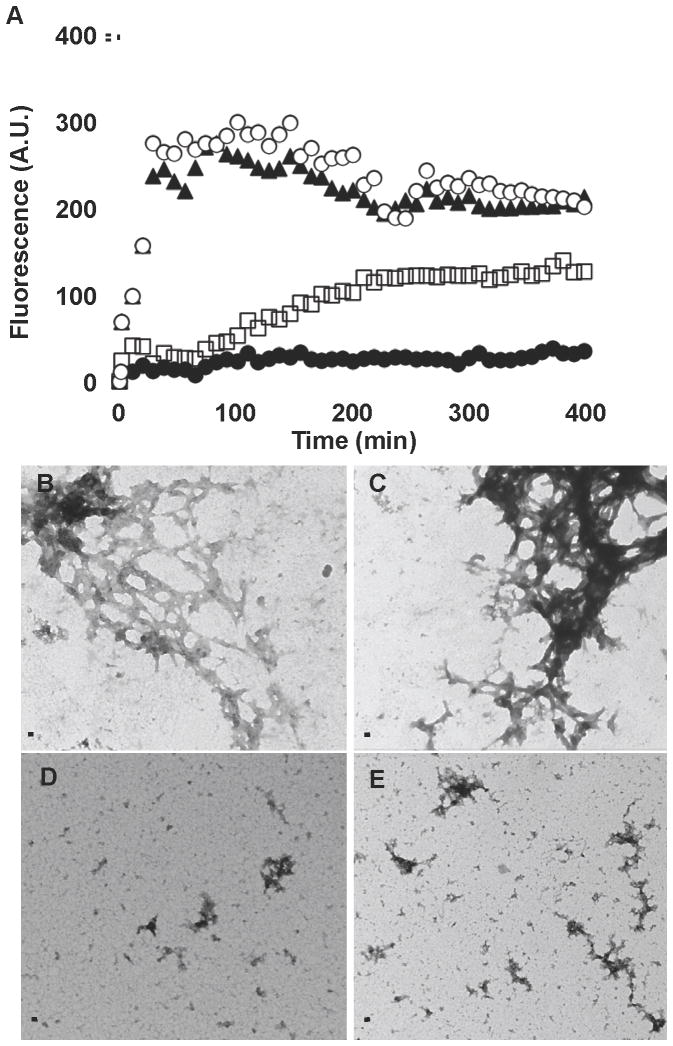
To further determine if both molecules inhibit TasA polymerization we studied the kinetics of TasA polymerization using an in vitro assay with thioflavin T. Thioflavin T displays increased fluorescence at 495 nm when it is bound to the beta sheet-rich fold found in amyloids. For this analysis purified TasA was first treated with formic acid. After vacuum drying, the protein was resuspended in physiological buffer with thioflavin T, with or without the addition of 50 μM of AA-861 or parthenolide. For TasA resuspended in buffer (○) or with DMSO (▲) the fluorescence signal increased rapidly and reached a maximum after 100 min (Figure 4A). Both AA-861 and parthenolide reduced the increase in fluorescence, albeit to different levels. First, a delay in the initiation of the increase in fluorescence was observed in the presence of both molecules. After 100 min, the parthenolide treated sample (□) displayed a small and slow increase in fluorescence. In contrast, no substantial increase in fluorescence was observed in the presence of AA-861 (●). Neither molecule displayed background fluorescence in these conditions when assayed without protein. To confirm a loss of fiber formation, after 24 hr of incubation the samples were analyzed by electron microscopy. In the untreated or DMSO-treated controls, fibers could be observed (Figure 4B, C). In contrast, AA-861 prevented fiber formation and only small aggregates were observed (Figure 4D). Parthenolide allowed the formation of small fibers and disorganized aggregates (Figure 4E). It thus appears that these molecules do indeed inhibit TasA polymerization into fibers.
Figure 4. The molecules inhibit the polymerization of TasA in vitro.
(A) Thioflavin T and fluorescence were used to study the kinetics of polymerization of TasA. Purified TasA was treated with formic acid, dried under vacuum and mixed with physiological buffer (○) or DMSO (▲), plus 50 μM of AA-861 (●) or parthenolide (□). Transmission electron micrographs of samples taken after 24 hr of incubation of (B) TasA in buffer, (C) TasA treated with DMSO, (D) TasA treated with AA-861 and (E) TasA treated with parthenolide. Scale bars, 100 nm.
We then wanted to test whether AA-861 and parthenolide could destroy pre-formed biofilms in vivo. To test this, we analyzed pellicles of wild-type and eps mutant strains. We used the eps mutant to generate weaker pellicles that were totally dependent on TasA fibers as our previous data indicate that TasA was the primary target of the molecules. This increased the sensitivity of the assay in order to better visualize the effect of the molecules. Pellicles were grown for 12 hr and then the molecules were added at different concentrations, prior to visualization 12 hr later (Figure 5). Concentrations of AA-861 of 100 μM to 200 μM were necessary to observe any significant inhibition of biofilm, with a more dramatic effect observed in the eps mutant. Parthenolide was more effective, and a concentration of 100 μM was sufficient to promote the destruction of 12-hour-old pellicles.
Figure 5. The molecules destroy preformed biofilms.
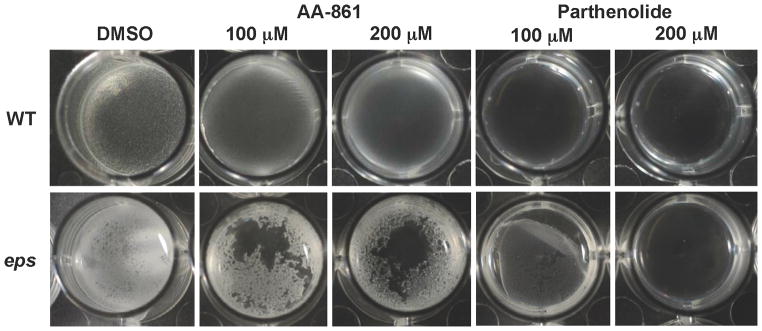
Biofilms of wild-type and eps mutants were grown in MSgg medium for 12 hr at 30°C. Then, DMSO, AA-861 or parthenolide were added at 100 μM or 200 μM, and pictures of the pellicles were taken 12 hr later.
The molecules work additively to inhibit biofilm formation
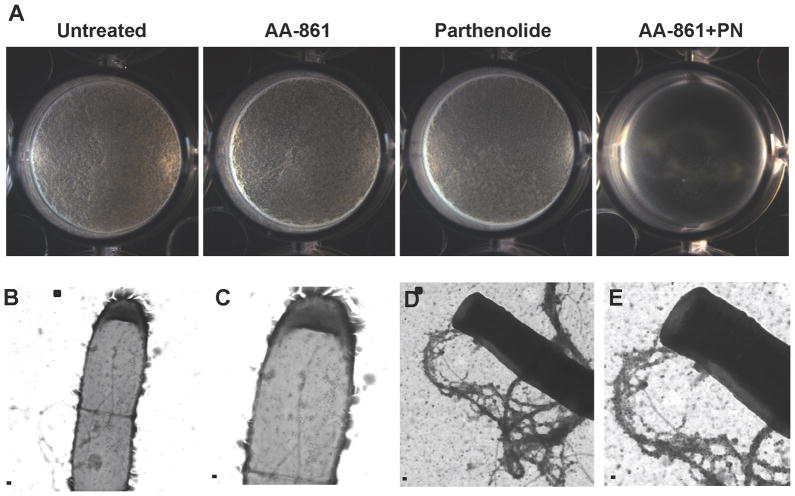
All of our results indicated that AA-861 and parthenolide might antagonize TasA in different ways. While AA-861 was more efficient at inhibiting polymerization of the fibers (Figure 4) parthenolide was more potent at destroying pre-formed biofilms (Figure 5). Therefore we asked whether these molecules might work additively to inhibit biofilm formation. In order to test this, we performed pellicle assays of wild-type strains using concentrations of each molecule that failed to inhibit biofilm formation. 20 μM of either AA861 or parthenolide separately had no effect on biofilm formation relative to the untreated samples (Figure 6A). However a combination of both molecules at a final concentration of 40 μM (20 μM each) completely inhibited the formation of pellicles. No inhibitory activity was observed when the concentration of each molecule was reduced to 10 μM (Figure S3). Transmission electron microscopy coupled with immuno-gold labeling proved that TasA was associated to cell surfaces of the wild-type strain treated with the combination of both molecules as indicated by the presence of gold particles, but TasA was no longer observed in the form of fibers (Figure 6B, C). Untreated cells produced dense and highly labeled fibers attached to the cells (Figure 6D, E).
Figure 6. The molecules inhibit biofilm formation with additive effects.
(A) The formation of biofilm by the wild-type strain was evaluated in MSgg medium alone or with the addition 20 μM of each molecule added separately or in combination (20 μM of each). Pictures of the wells were taken after 24 hr of incubation at 30°C. Transmission electron microscopy and immunogold labeling were used to determine the presence or absence of TasA fibers. (B–C) Cells treated with both molecules appeared decorated with gold particles but did not show any TasA fibers. (D–E) Untreated cells showed long fibers labeled with gold particles. Panels C and E are magnified views of the areas marked in panels B and D. Scale bars, 500 nm in panels B & D and 200 nm in panels C & E (see also Figure S3).
The molecules affect other amyloid proteins
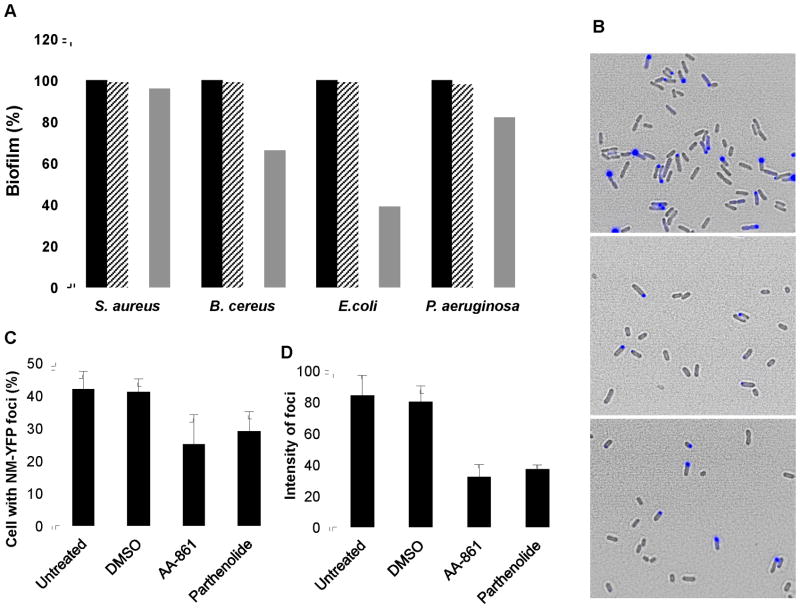
We have shown that AA-861 and parthenolide efficiently inhibit the formation of B. subtilis biofilms by inhibiting polymerization of the amyloid-like fibers of TasA. In order to determine the anti-biofilm potency of these molecules and their specificity towards amyloid proteins, we tested them against a subset of bacterial pathogens, two Gram-negative species, Escherichia coli and Pseudomonas aeruginosa, and two Gram-positive species, Bacillus cereus and Staphylococcus aureus. Of these bacteria, E. coli is known to produce the amyloid fiber Curli, and B. cereus, though not reported to produce amyloid proteins, contains homologs of the tasA gene. All of these organisms form surface-attached biofilms (unlike the floating pellicle of B. subtilis) and these biofilms can be quantified using a standard crystal violet assay (O’Toole and Kolter, 1998). The untreated controls were normalized to 100% of biofilm formation and the treatments with DMSO, AA-861 or parthenolide were expressed as percentages of the untreated value (Figure 7A). The activity of these molecules varied depending on the bacterial species. Both were effective at preventing the formation of biofilms by E. coli and B. cereus. However, they did not show any detectable anti-biofilm activity against S. aureus and P. aeruginosa, organisms not thought to produce an amyloid component during biofilm formation. These findings support the specific anti-amyloid activity of AA-861 and parthenolide.
Figure 7. The molecules affect other amyloid proteins.
(A) Anti-biofilm activity against other bacterial species that form surface-adhered biofilms. The crystal violet method was used to quantify biofilm formation. The absorbance at 590 nm was measured and values were represented relative to untreated controls which were normalized to 100%. The molecules were used at a final concentration of 50 μM. Untreated cells (■), treated with DMSO (
 ), AA-861 (□) or parthenolide (
), AA-861 (□) or parthenolide (
 ). (B–D) The molecules reduce the aggregation of New1 yeast prion expressed in E. coli cells. (B) Fluorescence and phase contrast microscopy images were taken of E. coli cells expressing the yeast prion domain New1 fused to CFP and induced with 5 μM IPTG (upper panel). A decrease in the number of cells accumulating New1-CFP foci was observed after treatment with 120 μM of (middle panel) AA-861 and (lower panel) parthenolide. (C) The number of cells accumulating New1-CFP foci was quantified and expressed as percentage of cells with foci, and (D) the average intensity of these foci expressed as arbitrary units.
). (B–D) The molecules reduce the aggregation of New1 yeast prion expressed in E. coli cells. (B) Fluorescence and phase contrast microscopy images were taken of E. coli cells expressing the yeast prion domain New1 fused to CFP and induced with 5 μM IPTG (upper panel). A decrease in the number of cells accumulating New1-CFP foci was observed after treatment with 120 μM of (middle panel) AA-861 and (lower panel) parthenolide. (C) The number of cells accumulating New1-CFP foci was quantified and expressed as percentage of cells with foci, and (D) the average intensity of these foci expressed as arbitrary units.
Since both AA-861 and parthenolide showed activity against bacterial amyloid proteins, we assessed their effect on a non-bacterial amyloid protein, the New1 yeast prion (Garrity et al., 2010). In order to do this, we used the E. coli strain SG811, expressing the N-terminal amyloidogenic domain of New1 fused to CFP and under control of an IPTG inducible promotor. New1 has the ability to convert to the prion state upon IPTG induction in this heterologous system (Garrity et al., 2010). Cultures of SG811 were induced with 5 μM IPTG and incubated with 120 μM of DMSO (negative control), AA-861 or parthenolide, at 30°C for 24 hr. We increased the concentration of molecules from the 50 μM that was used in B. subtilis to 120 μM for this experiment because the lower concentration was ineffective in this assay. 120 μM is similar to the concentration used in other reports detect activity of amyloid inhibitors (Cegelski et al., 2009; Roberts et al., 2009). The aggregation of the prion domain was confirmed using fluorescence microscopy by the presence of a bright focus in the cells (Figure 7B upper panel). Interestingly, a reduction in the number of cells accumulating a focus was observed after treatment with AA-861 (Figure 7B middle panel) or parthenolide (Figure 7B lower panel). To quantify these results, we estimated the number of cells with a focus (Figure 7C) and the average intensity of the focus (Figure 7D). In comparison to untreated or DMSO controls, a decrease in the percentages of cells with a focus was obtained with AA-861 (40%) and parthenolide (31%). These differences were more striking when the average intensity of each focus was compared (Figure 7D). Indeed, reductions between 55–60% were observed in the treatment with parthenolide or AA-861, respectively. These results indicate that both molecules target the aggregation of amyloid proteins of different origins.
Discussion
In this study we show that B. subtilis biofilms are effective for screening for molecules with anti-biofilm and/or anti-amyloid properties. Among a collection of hundreds of known bioactive molecules, we found two that efficiently inhibited biofilm formation by specifically targeting amyloids. One of them, AA-861 is a benzoquinone derivative with anti-inflammatory activity (Yoshimoto et al., 1982). The other, parthenolide is a sesquiterpene lactone with anti-inflammation and anti-cancer activities (Mathema et al., 2012). It has been shown for certain molecules that anti-amyloid activity results from molecule aggregates and not monomers (Feng et al., 2008). Whether the inhibitory activity of these compounds is due to monomers or aggregates is unclear at this time.
The anti-inflammatory activity of AA-861 is exerted by competing with the enzyme 5-lipoxigenase, which provokes a reduction of the levels of leukotrienes, and as a consequence, a decrease in the aggressiveness of inflammation associated with diverse human diseases (Titos, et al., 2003). Besides this main biological activity, AA-861 was later shown to slightly reduce amyloid formation of amylin, the amyloid polypeptide found in human pancreatic islet (Tomiyama, et al., 1997). These findings support our conclusion that the anti-biofilm effect of AA-861 could result from preventing the TasA protein from forming functional amyloid-like fibers. Indeed, our in vitro experiments supported this hypothesis as AA-861 arrested the aggregation of TasA into fibers. The mechanism by which AA-861 and other quinone derivatives reduce amyloidogenesis still remains obscure. However, it is likely that a direct interaction of AA-861 with different forms of the amyloid proteins could impede the polymerization of the fiber as is observed with rifampicin, which has anti-amyloid activity against amylin and α-synuclein, the peptide responsible for Parkinson’s disease (Li et al., 2004).
Parthenolide is a natural product found in the plant Tanacetum parthenium (feverfew) which has been used as a herbal treatment to reduce fever and pain. There has been some scientific interest in parthenolide because it has been shown to have anti-cancer and anti-inflammatory activities (Mathema et al., 2012). However, there are no published studies of the effects of parthenolide in humans. Here we have demonstrated that parthenolide also has an anti-biofilm activity by reducing amyloid fiber formation and destroying pre-formed biofilms. A dual activity of being anti-inflammatory and anti-amyloid has been previously reported for another group of molecules, the non-steroidal anti-inflammatory drugs (NSAIDs) (Gasparini et al., 2004; Weggen et al., 2001). Epidemiological studies found that patients treated with NSAIDs had a low propensity of suffering Alzheimer’s disease (Szekely and Zandi, 2010). Whether parthenolide could exhibit similar effects remains to be determined.
We have additionally shown that the anti-biofilm activity of AA-861 and parthenolide is effective against E. coli and B. cereus. E. coli produces the amyloid fiber Curli, and it was recently shown that two ring-fused 2-pyridones with pilicide activity inhibited Curli polymerization. This inhibition of both curli and type I pili was correlated with reduced virulence and a decreased ability of E. coli to colonize host tissues and form biofilms (Cegelski et al., 2009). B. cereus has not been reported to form amyloid-like fibers. However, its genome harbors homologs of the B. subtilis TasA protein. Therefore, it is reasonable to think this homolog might polymerize into amyloid fibers that are involved in biofilm formation. Though not covered in the study, we speculate that the molecules could antagonize the formation of amyloid fibers in E. coli and B. cereus similar to what we have found in B. subtilis. In contrast, P. aeruginosa and S. aureus are not reported to have amyloid proteins, and biofilms of these species were immune to the action of AA-861 and parthenolide, an observation that pointed toward the specificity of both molecules for amyloid proteins. Finally, these molecules were not only active against bacterial amyloids, both molecules arrested the conversion of the yeast New1 protein to its prion state. The mechanism by which this inhibition happens remains to be elucidated. It is important to note that before any molecule can be described as a general anti-amyloid compound, studies must be performed on a variety of amyloids. Indeed, FN075, a ring-fused 2-pyridone that inhibits Curli fibrillation in E. coli actually stimulates amyloid formation of α-synuclein (Horvath et al., 2012). Our analysis of AA-861 and parthenolide thus far suggests these molecules may be general inhibitors as we have observed inhibition of TasA and New1 aggregation and inhibition of biofilm formation in E. coli and B. cereus which have amyloid proteins as major matrix components.
Significance
In order to form a biofilm, bacterial cells have to produce, secrete and assemble different macromolecules into the network that is the extracellular matrix (Branda et al., 2005). Amyloid proteins represent key components of the biofilm matrix of several microorganisms (Maury, 2009; Romero et al., 2010). These microbial amyloid fibers are structurally and biochemically similar to the pathogenic variants found in humans. Therefore, they are good candidates to be used as targets in screens for molecules that may serve two purposes: anti-biofilm and anti-amyloid agents (Cegelski et al., 2009; Nie et al., 2011; Roberts et al., 2009). In order to screen large libraries of molecules it is important to have simple and robust assay. In this study we show that B. subtilis biofilm inhibition can provide such an assay. We found two molecules that efficiently arrested biofilm formation. One of them, AA-861 is a benzoquinone derivative (Yoshimoto et al., 1982), and the other, parthenolide, a sesquiterpene lactone with anti-cancer and anti-inflammatory activities (Mathema et al., 2012). We also showed that these molecules have the ability to arrest the aggregation of New1, a yeast prion protein. These findings open the door for large-scale screens to identify additional molecules with similar activities and for structure-activity relationship studies with the molecules identified.
Experimental procedures
Regents and growth conditions
AA-861 (A3711), a benzoquinone derivative and parthenolide (P0667) were obtained from Sigma and dissolved in DMSO at a concentration of 5 mg/ml. The molecules were used at final concentration of 50 μM when added from the beginning to the cultures, 100 μM to 200 μM when added after 12 h of growth, and 120 μM in the experiments with New1.
LB broth: 1% tryptone (Difco), 0.5% yeast extract (Difco), 0.5% NaCl. MSgg broth: 100 mM morpholinepropane sulphonic acid (MOPS) (pH 7), 0.5% glycerol, 0.5% glutamate, 5 mM potassium phosphate (pH 7), 50 μg/ml tryptophan, 50 μg/ml phenylalanine, 2 mM MgCl2, 700 μM CaCl2, 50 μM FeCl3, 50 μM MnCl2, 2 μM thiamine, 1 μM ZnCl2 (Branda, et al., 2001). TSB (Sigma Aldrich) supplemented with 0.5% glucose and 3% NaCl. YESCA: 1% Casamino Acids, 0.1% yeast extract. M63 minimal medium: 1.2% KH2PO4, 2.8% K2HPO4, 0.8 g (NH4)2SO4 supplemented with 1 mmol l−1 MgSO4, 0.2% glucose and 0.5% Casamino Acids.
Strains
The strains used in this study were B. subitlis NCIB3610, and derivative mutants SSB488 (epsA-O::tet), CA017 (tasA::kan), SSB572 (epsA-O::tet tasA::spc) (Romero et al., 2010), DR6 (tasA::km, lacA:: Phyperspank-tasA, mls) (Romero et al., 2011), B. cereus ATCC14579, E. coli MC4100, E. coli GS811 (BW27785 attB::ahp lacIq Ptac new150–100-cfp-3xha; produces New1 (residues 50–100) fused to CFP and 3 HA tags from a chromosomal construct integrated at lambda attachment site) (Garrity et al., 2010), S. aureus and P. aeruginosa.
To construct the strain DR-8 ((tapA-sipW-tasA)::spc, amyE::(tapA(13-234)-sipW-tasA) (cm), lacA::Phyperspank-tapA), the mutant strain lacking the tapA-sipW-tasA operon (SSB149) was transformed by natural competence with the plasmid pDFR8 and positive clones were used as donor strains for transferring the constructs into B. subtilis strain mutant (FC268) by means of SPP1-mediated generalized transduction (Yasbin and Young, 1974). To generate the plasmid pDFR8 (lacA::Phyperspank-tapA), the open reading frame of tapA was amplified with the primers TapA-F (5′-ggc cat GTC GAC ttt tac agg agg taa gat atg ttt cga ttg-3′) and TapA-R (5′-ggc cat GCA TGC tta ctg atc agc ttc att gct ttt ttc-3′). The PCR product was digested with SalI and SphI and cloned into the plasmid pDR111 digested with the same enzymes. Then, a digest of pDFR-7 with XhoI and BamHI containing the fragment Phyperspank-tapA was cloned into pDR183 digested with the same enzymes resulting in pDFR8 (lacA::Phyperspank-tapA).
Biofilm assays
Pellicle formation assay in standing liquid cultures was used to test biofilm development of B. subtilis. The different strains were grown in MSgg broth in a 24-well microtiter dish and incubated without agitation at 30°C for 48 h (Romero et al., 2011). For the screen, library compounds were pin-transferred to 384-well plates into 30μL of MSgg, followed by addition of 10μL of an overnight culture suspension of B. subtilis NCIB3610 in MSgg to a final OD600 of 0.05. The final concentration of each compound was 12.5 μg/ml. Plates were incubated at 30°C for 24h, and then sco red visually or by measuring OD600. For screening by OD600, a Z′ score of 0.71 was determined by comparing the average and standard deviation of the OD600 obtained on untreated wells or wells harboring 3mM of D-Tyrosine, a known inhibitor of biofilm formation in B. subtilis (Kolodkin-Gal et al., 2010; Romero and Kolter, 2011). The crystal violet assay was used to test the formation of biofilm for the rest of the bacterial species, as previously reported (O’Toole and Kolter, 1998). Briefly, S. aureus was grown in TSB medium, B. cereus was grown in LB, E. coli was grown in YESCA and P. aeruginosa in M63 glucose medium. All of them where incubated without agitation at 30°C for 24–36 h. A 1% crystal violet solution was added and after 15 min of incubation the amount of biomass attached to the well surfaces was quantified after solubilizing the dye with 70% ethanol and measuring the absorbance at 590 nm.
SDS page and immunoblot assays
For localization of TasA within B. subtilis biofilms, pellicles of 24h produced in MSgg broth were fractionated in three biochemical compartments: matrix, cell and medium, as previously described (Branda et al., 2006). Protein preparations were developed in SDS-PAGE with 10% polyacrylamide and blotted onto PVDF membrane using standard procedures. For detection of TasA, blots were probed with anti-TasA antibodies (raised in rabbits) at dilution 1:20,000. A secondary anti-rabbit IgG antibody conjugated to horseradish peroxidase (BioRad) was used at a dilution of 1:20,000. Blots were developed using the Pierce super signal detection system (Pierce, Thermo Scientific).
Flow cytometry
Biofilms of cells harboring PtapA-yfp fusions (ZK3755 and DR7) were harvested at 24 and dispersed as previously described (Vlamakis et al., 2008). After fixation in 4% paraformaldehyde solution for 7 min, washes with PBS and re-suspension in GTE buffer, single cells were obtained by mild sonication (Branda et al., 2006). For flow cytometric analysis, cell suspensions were measured on a BD LSR II flow cytometer (BD Biosciences) using a solid state laser and a laser excitation of 488 nm coupled with 530/30 and 505LP sequential filters. The photomultiplier voltage was set at 400–500 V. Every sample was analyzed by counting 50,000 events using FACS Diva (BD Biosciences) software to capture the data (Lopez et al., 2009). Further analysis was performed using FlowJo 8.7.2 software (http://www.flowjo.com).
In vitro assays for study assembly of TasA amyloid-like fibers
The effect of the different molecules on TasA polymerization was analyzed using the Thioflavin-T (ThT) assay in 96 micro-well plates, as previously reported (Romero et al., 2010). Purified TasA was depolymerized by treatment with 10% formic acid solution for two minutes. After drying under vacuum in a Speed-vac at 30°C the remaining product containing TasA was resuspended in 50mM NaCl, 20 mM Tris, (pH 7) buffer or buffer containing the different molecules at 50 μM and incubated at room temperature. The wells were previously blocked with 2.5 mg/ml bovine serum albumin in PBS for 2h at 30 °C and shaking. After blocking each well was filled with the following mixture: 10 μM protein samples, 20 μM ThT solution, PBS buffer and distilled water to completing a final volume of 200 μL. The fluorescence was measure in a spectrophotometer (Spectra Max M2, Molecular Devices, Sunnyvale, CA) fluorescence plate reader set up at 438 excitation and 495 nm emission, at 30 °C and shaking for a total of 24 h In this assay, the final concentration of the molecules was 50 μM.
Transmission electron microscopy and immunolabeling
For negative staining analysis, protein solutions were spotted onto a carbon or formvar/carbon coated grid, for a few minutes. The excess of liquid was discharged in a filter paper, and the grids stained with a 1% aqueous uranyl acetate solution for 2 minutes.
Immunolocalization of TasA was performed on intact cells from 24 hr biofilms as previously described (Romero et al., 2011). Briefly, intact cells were absorbed in hydrophilic carbon coated grids. Then the samples were blocked in 1% non-fat dry milk in PBS with 0.1% Tween 20 for 30 min and on anti-TasA antibodies diluted 1:150 in the same buffer blocking buffer for 2 h, rinsed in PBST and exposed to goat-anti-rabbit 20 nm gold secondary antibody diluted 1:50 (TedPella) for 1 h, and rinsed. All grids were contrasted with uranyl acetate as described above for visualization. The samples were examined in a Tecnai G2 Spirit BioTWIN microscope at an accelerating voltage of 80 KV. Images were taken with an AMT 2k CCD camera (Romero et al., 2011).
Fluorescence microscopy
E. coli cells of the strain SG811 (BW27785 attB::ahp lacIq Ptac new150–100-cfp-3xha; produces New1 (residues 50–100) fused to CFP) were grown in microtiter plates induced with 5 μM of IPTG and incubated with 120 μM of AA-861 or parthenolide. DMSO was used as a negative control. After 24 hr of incubation at 30°C, cells were pelleted and fixed with a 4% paraformaldehyde solution for 7 minutes, washed in PBS and resuspended in GTE buffer (Romero et al., 2011). 10 mL drops were deposited in slides pre-treated with a thin layer of agarose (1%). Images were taken in a Nikon Eclipse TE2000-U microscope with X-cite 120 illumination system and a Hamamatsu digital camera model ORCA-ER. CFP fluorescence signal was taken using the filter set (Ex436/500). The exposure times were 800 msec. For quantification of cells accumulating foci and the intensity of the foci, approximately 1,000 cells were analyzed per treatment. Image processing was done using MetaMorph Software, ImageJ and Photoshop.
Supplementary Material
Highlights.
B. subtilis biofilm as tool for screening of anti-biofilm and anti-amyloid molecules.
AA-861 and parthenolide are anti-biofilm agents.
Amyloid protein fibers are the main targets of AA-861 and parthenolide.
Inhibition of amyloid fiber formation as main mode of action.
Acknowledgments
We would like to thank Richard Losick and members of the Kolter laboratory for helpful discussions. We thank Adam Driks (Loyola University Medical Center, Maywood, IL) for kindly providing antibodies against TasA. We thank Viknesh Sivanathan of Ann Hochschild’s lab for providing the E. coli strain for anti-prion assays and experimental suggestions. We thank M. Ericsson, for help and guidance in the electron microscope. We thank the ICCB-Longwood Screening Facility for the BIOMOL–ICCB Known Bioactives collection and for technical advise. This work was funded by National Institutes of Health grants to R.K. (GM58213 and GM82137) as well as grants to R.K. by the Harvard Accelerator Fund. D.R. is the recipient of a MEC/Fulbright post-doctoral fellowship from Secretaría General de Estado de Universidades e Investigación del Ministerio de Educación y Ciencia (Spain). The small-molecule screen was funded in part through grant U54 AI057159 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amijee H, Madine J, Middleton DA, Doig AJ. Inhibitors of protein aggregation and toxicity. Biochem Soc Trans. 2009;37:692–696. doi: 10.1042/BST0370692. [DOI] [PubMed] [Google Scholar]
- Badtke MP, Hammer ND, Chapman MR. Functional amyloids signal their arrival. Sci Signal. 2009;2:pe43. doi: 10.1126/scisignal.280pe43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco LP, Evans ML, Smith DR, Badtke MP, Chapman MR. Diversity, biogenesis and function of microbial amyloids. Trends Microbiol. 2012;20:66–73. doi: 10.1016/j.tim.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Chu F, Kearns DB, Losick R, Kolter R. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol. 2006;59:1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Gonzalez-Pastor JE, Dervyn E, Ehrlich SD, Losick R, Kolter R. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J Bacteriol. 2004;186:3970–3979. doi: 10.1128/JB.186.12.3970-3979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Vik S, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Cegelski L, Pinkner JS, Hammer ND, Cusumano CK, Hung CS, Chorell E, Aberg V, Walker JN, Seed PC, Almqvist F, et al. Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat Chem Biol. 2009;5:913–919. doi: 10.1038/nchembio.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen D, Wosten HA, van Keulen G, Faber OG, Alves AM, Meijer WG, Dijkhuizen L. Two novel homologous proteins of Streptomyces coelicolor and Streptomyces lividans are involved in the formation of the rodlet layer and mediate attachment to a hydrophobic surface. Mol Microbiol. 2002;44:1483–1492. doi: 10.1046/j.1365-2958.2002.02980.x. [DOI] [PubMed] [Google Scholar]
- Dobson CM. Principles of protein folding, misfolding and aggregation. Semin Cell Dev Biol. 2004;15:3–16. doi: 10.1016/j.semcdb.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Dueholm MS, Petersen SV, Sonderkaer M, Larsen P, Christiansen G, Hein KL, Enghild JJ, Nielsen JL, Nielsen KL, Nielsen PH, et al. Functional amyloid in Pseudomonas. Mol Microbiol. 2010;77:1009–1020. doi: 10.1111/j.1365-2958.2010.07269.x. [DOI] [PubMed] [Google Scholar]
- Feng BY, Toyama BH, Wille H, Colby DW, Collins SR, May BC, Prusiner SB, Weissman J, Shoichet BK. Small-molecule aggregates inhibit amyloid polymerization. Nat Chem Biol. 2008;4:197–199. doi: 10.1038/nchembio.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid--from bacteria to humans. Trends Biochem Sci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Garrity SJ, Sivanathan V, Dong J, Lindquist S, Hochschild A. Conversion of a yeast prion protein to an infectious form in bacteria. Proc Natl Acad Sci U S A. 2010;107:10596–10601. doi: 10.1073/pnas.0913280107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparini L, Rusconi L, Xu H, del Soldato P, Ongini E. Modulation of beta-amyloid metabolism by non-steroidal anti-inflammatory drugs in neuronal cell cultures. J Neurochem. 2004;88:337–348. doi: 10.1111/j.1471-4159.2004.02154.x. [DOI] [PubMed] [Google Scholar]
- Gebbink MF, Claessen D, Bouma B, Dijkhuizen L, Wosten HA. Amyloids-a functional coat for microorganisms. Nat Rev Microbiol. 2005;3:333–341. doi: 10.1038/nrmicro1127. [DOI] [PubMed] [Google Scholar]
- Horvath I, Weise CF, Andersson EK, Chorell E, Sellstedt M, Bengtsson C, Olofsson A, Hultgren SJ, Chapman M, Wolf-Watz M, et al. Mechanisms of protein oligomerization: inhibitor of functional amyloids templates alpha-synuclein fibrillation. J Am Chem Soc. 2012;134:3439–3444. doi: 10.1021/ja209829m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin-Gal I, Romero D, Cao S, Clardy J, Kolter R, Losick R. D-amino acids trigger biofilm disassembly. Science. 2010;328:627–629. doi: 10.1126/science.1188628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhu M, Rajamani S, Uversky VN, Fink AL. Rifampicin inhibits alpha-synuclein fibrillation and disaggregates fibrils. Chem Biol. 2004;11:1513–1521. doi: 10.1016/j.chembiol.2004.08.025. [DOI] [PubMed] [Google Scholar]
- Lopez D, Vlamakis H, Losick R, Kolter R. Cannibalism enhances biofilm development in Bacillus subtilis. Mol Microbiol. 2009;74:609–618. doi: 10.1111/j.1365-2958.2009.06882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathema VB, Koh YS, Thakuri BC, Sillanpaa M. Parthenolide, a sesquiterpene lactone, expresses multiple anti-cancer and anti-inflammatory activities. Inflammation. 2012;35:560–565. doi: 10.1007/s10753-011-9346-0. [DOI] [PubMed] [Google Scholar]
- Maury CP. The emerging concept of functional amyloid. J Intern Med. 2009;265:329–334. doi: 10.1111/j.1365-2796.2008.02068.x. [DOI] [PubMed] [Google Scholar]
- Murphy RM. Peptide aggregation in neurodegenerative disease. Annu Rev Biomed Eng. 2002;4:155–174. doi: 10.1146/annurev.bioeng.4.092801.094202. [DOI] [PubMed] [Google Scholar]
- Nie Q, Du XG, Geng MY. Small molecule inhibitors of amyloid beta peptide aggregation as a potential therapeutic strategy for Alzheimer’s disease. Acta Pharmacol Sin. 2011;32:545–551. doi: 10.1038/aps.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- O’Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- Roberts BE, Duennwald ML, Wang H, Chung C, Lopreiato NP, Sweeny EA, Knight MN, Shorter J. A synergistic small-molecule combination directly eradicates diverse prion strain structures. Nat Chem Biol. 2009;5:936–946. doi: 10.1038/nchembio.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero D, Aguilar C, Losick R, Kolter R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci U S A. 2010;107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero D, Kolter R. Will biofilm disassembly agents make it to market? Trends Microbiol. 2011;19:304–306. doi: 10.1016/j.tim.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero D, Vlamakis H, Losick R, Kolter R. An accessory protein required for anchoring and assembly of amyloid fibres in B. subtilis biofilms. Mol Microbiol. 2011;80:1155–1168. doi: 10.1111/j.1365-2958.2011.07653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekely CA, Zandi PP. Non-steroidal anti-inflammatory drugs and Alzheimer’s disease: the epidemiological evidence. CNS Neurol Disord Drug Targets. 2010;9:132–139. doi: 10.2174/187152710791012026. [DOI] [PubMed] [Google Scholar]
- Titos E, Claria J, Planaguma A, Lopez-Parra M, Villamor N, Parrizas M, Carrio A, Miquel R, Jimenez W, Arroyo V, et al. Inhibition of 5-lipoxygenase induces cell growth arrest and apoptosis in rat Kupffer cells: implications for liver fibrosis. FASEB J. 2003;17:1745–1747. doi: 10.1096/fj.02-1157fje. [DOI] [PubMed] [Google Scholar]
- Tomiyama T, Kaneko H, Kataoka K, Asano S, Endo N. Rifampicin inhibits the toxicity of pre-aggregated amyloid peptides by binding to peptide fibrils and preventing amyloid-cell interaction. Biochem J. 1997;322 ( Pt 3):859–865. doi: 10.1042/bj3220859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlamakis H, Aguilar C, Losick R, Kolter R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 2008;22:945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weggen S, Eriksen JL, Das P, Sagi SA, Wang R, Pietrzik CU, Findlay KA, Smith TE, Murphy MP, Bulter T, et al. A subset of NSAIDs lower amyloidogenic Abeta42 independently of cyclooxygenase activity. Nature. 2001;414:212–216. doi: 10.1038/35102591. [DOI] [PubMed] [Google Scholar]
- Wosten HA, de Vocht ML. Hydrophobins, the fungal coat unravelled. Biochim Biophys Acta. 2000;1469:79–86. doi: 10.1016/s0304-4157(00)00002-2. [DOI] [PubMed] [Google Scholar]
- Yasbin RE, Young FE. Transduction in Bacillus subtilis by bacteriophage SPP1. J Virol. 1974;14:1343–1348. doi: 10.1128/jvi.14.6.1343-1348.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto T, Yokoyama C, Ochi K, Yamamoto S, Maki Y, Ashida Y, Terao S, Shiraishi M. 2,3,5-Trimethyl-6-(12-hydroxy-5,10-dodecadiynyl)-1,4-benzoquinone (AA861), a selective inhibitor of the 5-lipoxygenase reaction and the biosynthesis of slow-reacting substance of anaphylaxis. Biochim Biophys Acta. 1982;713:470–473. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.