Abstract
This review provides a translational and unifying summary of metabolic syndrome genetics and highlights evidence that genetic studies are starting to unravel and untangle origins of the complex and challenging cluster of disease phenotypes. The associated genes effectively express in the brain, liver, kidney, arterial endothelium, adipocytes, myocytes and β cells. Progression of syndrome traits has been associated with ectopic lipid accumulation in the arterial wall, visceral adipocytes, myocytes, and liver. Thus it follows that the genetics of dyslipidemia, obesity, and non-alcoholic fatty liver (NAFLD) disease are central in triggering progression of the syndrome to overt expression of disease traits, and have become a key focus of interest for early detection and for designing prevention and treatments. To support the “birds’ eye view” approach we provide a road-map depicting commonality and interrelationships between the traits and their genetic and environmental determinants based on known risk factors, metabolic pathways, pharmacological targets, treatment responses, gene networks, pleiotropy, and association with circadian rhythm. Although only a small portion of the known heritability is accounted for and there is insufficient support for clinical application of gene-based prediction models, there is direction and encouraging progress in a rapidly moving field that is beginning to show clinical relevance.
Introduction
There is accumulating evidence that insulin resistance and associated biochemical derangements precede atherogenesis and beta cell failure by several years1, indicating that there is a window of time during which prediction would be useful. The window extends further, since many of the traits have been identified in childhood and adolescence suggesting that early recognition of genotypes may precede disease progression and enable institution of preventive measures before the traits develop into overt disease. Even when the syndrome presents at an early age, it is more usual for more than one trait to be present, so it is realistic to approach the problem by recognizing the cluster in childhood and adolescence 2. Since gene-gene and gene-environment interaction occurs with time, study of young age groups is less likely to have confounding effects and a popular strategy has been to search for novel loci in pediatric cohorts and to attain replication of the findings3.
The cluster of three or more out of five criteria of the metabolic syndrome as defined by the National Cholesterol Education Program (NCEP), is predictive of both cardiovascular disease and type 2 diabetes and has been recommended for clinical use 4. However, it is uncertain whether the syndrome is best represented by dichotomization of the variables or whether they should be assessed as continuous variables which have provided better prediction when used with the Framingham Risk Equation 5, 6. It is also proposed that the syndrome contains four clusters with latent underlying linking factors, but it remains uncertain whether clinical identification of the syndrome has any advantages over separate evaluation of each component 7. Blood pressure and hyperglycemia have been linked separately from the remaining factors such as waist circumference, triglyceride, and HDL-C 8. The presence of hypertriglyceridemia with increased waist circumference has been identified as a strong predictor of coronary artery disease (CAD)9 and has been recommended as a screening phenotype 10. However, it has been debated whether obesity is a stronger underlying factor than insulin resistance since obese individuals can escape the metabolic syndrome and remain metabolically healthy, whereas lean individuals can be insulin resistant with increased cardio-metabolic risk, particularly if they have a first degree relative with type 2 diabetes 11. Also the hypothesis that insulin resistance is the main underlying factor has been challenged, since many cases with the syndrome have insulin resistance measures below the first quartile 12.
To account for the rapid and variable increase in obesity and metabolic syndrome prevalence, the argument for gene-environment interaction has gained momentum. It was originally proposed that phenotype expression may occur when conditions of nutritional excess prevail, supporting the concept that the metabolic syndrome results from an array of “thrifty” genes that are latent in the normal state but manifest after prolonged nutritional excess often associated with obesity 13. It is possible that efficient storage of nutrients had a selective advantage but the subsequent effects such as obesity and ectopic fat accumulation are deleterious. In some areas of metabolic syndrome research the concept is viable and supports lifestyle intervention. The mechanism that promotes accumulation of fat and lipid metabolites in liver and muscle resulting in insulin resistance has been defined by Shulman et al and has been recently reviewed 14 and the process can potentially be reversed with exercise 15. These observations support the role of excessive organ fat storage in the progression of insulin resistance to diabetes and fatty liver disease. However, each of the criteria have been shown to be associated with several genes and SNPs within each gene resulting in complex polygenic inheritance that, as a whole would have been less likely to have provided selective advantage. Consequently alternative hypotheses favoring more direct gene-environment interaction have been proposed 16, but current approaches have involved gene association methods, particularly genome-wide association scanning (GWAS), requiring large populations with replication, since the traits are variable and interactive resulting in variable association. Because of the complexity of the cardio-metabolic phenotype, most reviews and studies, including GWAS, have preferred to use single traits and not the cluster of traits as a syndrome or score as the phenotype. However, to obtain a clinical perspective of the syndrome’s complex genetic inheritance, the traits will be reviewed in a sequence corresponding to three main anatomic locations of their expression and modulation of cardio-metabolic effects; hypothalamic genes modulating obesity, hepatic genes with effects on dyslipidemia and excessive hepatic fat deposition, hepatic, renal and possibly endothelial genes on blood pressure and the beta cell genes modulating the impairment of insulin secretion. Genes that express in more than one location such as the FTO in the hypothalamus and adipocyte are discussed under the organ where the effect appears most prominent. However there are examples of pleiotropic effects and effects acting via gene networks and interacting metabolic pathways that can have significant effects on more than one trait. Circulating hormones, cytokines and lipoproteins that change with obesity and insulin resistance also can modulate traits associated with gene interaction in the target organ. In this regard recent evidence for the effects of lipoproteins on the β-cell is included. Effects of age and interrelationships between the traits and their genetic determinants are discussed, and a model of how interaction may occur is presented in Figures 1–2.
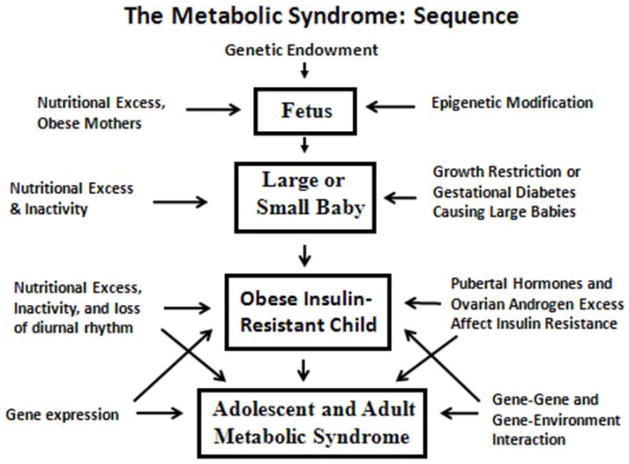
Figure 1.
The fetus, endowed with a genotype, becomes exposed to the maternal environment coinciding with susceptibility to metabolic programming by hormones, nutrients and stresses (see text). Programming continues during childhood leading to expression of metabolic syndrome traits.
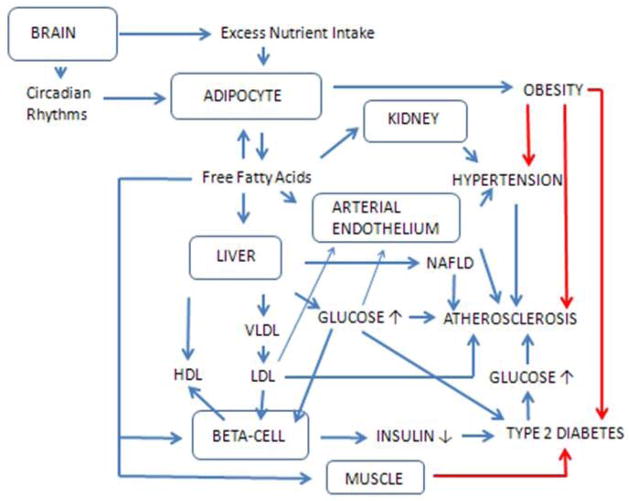
Figure 2.
The genes express in six main locations: in the brain, adipocyte, kidney, liver, arterial endothelium and β-cell. Perturbations in metabolic pathways programmed by the respective genes result in alterations in plasma metabolites (lipids carried in lipoproteins, glucose and fatty acids) and insulin, resulting in progression of metabolic syndrome traits leading to disease expression. Effects of insulin resistance are shown by the red lines.
Hypothalamic Genes
Obesity measures such as waist or body mass index (BMI) are components of most childhood and adult definitions of the metabolic syndrome and both have strong association with insulin resistance 17 and are highly correlated, however BMI has been used in most genetic studies because of its availability and widespread acceptance.
Rare monogenic forms of obesity have clearly provided insight on possible mechanisms for the development of severe obesity 18 and has led to the question whether polymorphisms within these known genes are involved in polygenic inheritance of obesity in the general population, since 60–90% of the BMI variance within a population is accounted for by inheritance 19. The discoveries lead to definition of energy homeostasis pathways in animal models; in particular the leptin-melanocortin pathway responsible for satiation 20. MC4R deficiency, the commonest of the clinically occurring monogenic forms, has been described in association with severe obesity, increased lean mass, increased linear growth, hyperphagia beginning in childhood, and severe hyperinsulinemia in heterozygous carriers but with greater severity in homozygotes 21. The increased linear growth has been associated with incomplete growth hormone suppression consistent with possible interference with somatostatin suppression of growth hormone pulsatility 22. Several functional polymorphisms in the gene have been detected and some have protected against obesity, but larger scale GWAS have identified positive association. BMI association with 2.8 million single nucleotide polymorphisms (SNPs) in 123,865 individuals with follow-up in a significant number revealed 14 known obesity susceptibility loci and identified 18 new loci. Some of the loci such as MC4R, POMC, SH2B1 and BDNF mapped near key hypothalamic regulators of energy balance. One of the loci was near GIPR, an incretin receptor 23 supporting a predisposition to type 2 diabetes with pleiotropic effects in the hypothalamus and β-cell.
A single common variant in intron 1 of the FTO (fat mass and obesity-associated) gene (rs9939609) was identified in association with type 2 diabetes, and further analysis revealed that the higher BMI in individuals with diabetes accounted for the association 24. The analysis included 7477 UK children from the Avon Longitudinal Study of Parents and Children (ALSPAC) cohort who had anthropometric measures at birth and at 7, 8, 9, 10, and 11 years of age and 4320 children from the Northern Finland 1966 birth cohort, leading to the conclusion that the FTO allele is not associated with changes in fetal growth as reflected by birth weight, but is associated with changes in BMI in children by the age of 7, and persisting to puberty 24. The association with obesity has been confirmed in other longitudinal studies in childhood 25, 26 including a Dutch study showing association with higher BMI, fat mass index, and leptin concentrations during puberty but declining at ages 13–14 years, a finding thought to be consistent with hormonal effects of puberty 27. The association of severe obesity with FTO has been studied using a haplotype approach. The investigators examined the linkage disequilibrium (LD) block structure of a region surrounding the candidate FTO rs9939609 SNP and determined the best haplotype composed of a three SNP combination associated with severe obesity. The calculation of a risk score based on the haplotype yielded an attributable risk of 34% for severe obesity suggesting that the approach has clinical use for examining risk in predisposed families 28.
The finding that FTO mRNA is abundant in mouse hypothalamic nuclei and codes for 2-oxoglutarate-dependent nucleic-acid demethylase, supports a regulatory role in energy balance, appetite and sympathetic outflow to the circulatory system 29. The FTO variants have early effects on obesity since they affect the rate of weight gain in African and European American youth 26. These observations are consistent with the finding that high fat intake and low physical activity modify the association between genetic variation in the FTO genotype and obesity 30. Since the common intron 1 FTO variant was initially associated with diabetes, and phenotypic interactions appear to be diabetogenic, the association with diabetes appears likely. The question has been explored in a meta-analysis of South Asian populations in whom BMI and waist association with FTO is similar to that seen in Europeans but a strong association with diabetes is only partly accounted for by BMI 31. A recent large scale meta-analyses study conducted on 96,551 individuals from East and South Asia confirmed the association of rs9939609 with type 2 diabetes independent of obesity 32. Also the FTO gene has been associated with hypertension and obesity in adolescents within a French Canadian founder population supporting pleiotropy 33.
Hepatic Genes
a. Dyslipidemia
Since, not all obese individuals have elevated triglycerides, and non-obese cases can present with elevated levels 34, 35, there is support for genetic predisposition for explaining abnormal levels and for gene-environment interactions with obesity and dietary intake as the main modifiers. Four classic Frederickson phenotypes (IIb, III, IV and V) originally described at the National Institutes of Health have been characterized as having an elevated triglyceride. With the exception of Type III hyperlipidemia, which has a distinct monogenic association with APOE polymorphism coding for homozygous apolipoprotein E2 and expressing when the individual becomes obese, the remaining three phenotypes were found to have overlapping genotypes 36. Interestingly, the genotypes had previously been identified in GWAS performed on subjects with mild triglyceride elevations. Thus clinically relevant dyslipidemia with high triglyceride as a component, can often be associated with triglyceride-associated polymorphisms. SNPs in genes such as APOAV and APOE, can result in severely increased triglyceride 37 and many cases were found to be carriers of APOAV variants (S19W or −1131 T>C). Studies on APOAV polymorphisms have consistently shown association with triglyceride elevation following the discovery that homozygous apoA-V deficiency results in severe hypertriglyceridemia. Also the −455 T>C polymorphism in the APOCIII gene promoter region is associated with increased triglyceride levels. The −455C and −482T alleles fail to respond to insulin-mediated down-regulation so that transcription remains active and plasma apoC-III is increased 38. The activated state for apoC-III transcription occurs in insulin resistance, which accounts for increases in plasma apoC-III and triglyceride in obesity and in the metabolic syndrome 39. In a multi-ethnic population sample, the triglyceride was 20% higher for −455C carriers, particularly in females who were also shown to have low HDL-C40. Findings in GWAS and meta-analysis studies have reported a strong association of common variants near APOAV-AIV-CIII-AI gene cluster with serum triglycerides 41 suggesting influence of polymorphisms on expression of apoC-III and apoA-V.
Although cultural, environmental and hormonal factors determine HDL-C, a genetic component accounts for up to 76% of the variation in HDL-C 42. High heritability of HDL-C and HDL-associated traits provide a strong rationale for identifying loci that may uncover pathways crucial for HDL regulation and treatment design. Regulatory genes involved in HDL metabolism mediated by apoA-I, LCAT, endothelial lipase and ABCA1 have been associated with severe HDL deficiencies 43. However, the population frequencies of the major gene abnormalities are small and association with disease has been ambivalent 44, supporting a case for functional assays to represent the HDL phenotype, such as apoA-I and measures of cholesterol efflux 45. Candidate gene association studies provide evidence that variation in HDL-regulatory genes have an effect on HDL which is dependent on environment 46 and as many as 20% of cases with a low HDL-C have known mutations.
GWAS using apoA-I and HDL-C as phenotypic markers, done in a predominantly American Indian population, located quantitative trait loci in regions of the genome that contain known candidate genes located in 6p, 9q and 15q regions 47, findings which could lead to further investigation to identify association with single nucleotide polymorphisms. The 15q region has been recognized to have a significant interaction with diabetes, BMI, smoking, alcohol intake and gender 48. After serial adjustments, the LOD score increased from 1.75 to 4.52, supporting multiple endogenous and environmental influences including obesity. The region contains the gene for hepatic lipase suggesting that it has HDL-determining polymorphisms. The 9q locus contains the ABCA1 gene, coding for the cholesterol transporter regulating efflux from cells to HDL, and located at 9q31.1. Moreover, the ABCA1-C230 allele was associated with low HDL-C in exclusively American Indian populations 49. This is important since carriers of loss of function mutations in ABCA1 display pancreatic beta-cell dysfunction supporting a role for ABCA1 in removing cholesterol from beta cells 50.
Susceptibility to changes in HDL composition and function occur in obesity in part due to triglyceride elevation, triglyceride-enrichment of HDL mediated by cholesterol ester transfer protein (CETP) is followed by degradation of HDL by hepatic triglyceride lipase, dissociation of apoA-I and subsequent renal catabolism51. It follows that in hypertriglyceridemic conditions CETP activity has an HDL-reducing role. Conversely, CETP deficiency secondary to a gene defect results in extreme elevations in HDL-C 52, but there has been controversy on whether the large HDL particles formed in CETP deficiency perform an adequate protective function. Nevertheless, CETP inhibition is the basis for a series of pharmaceutical agents designed to raise HDL-C, although the initial ILLUMINATE trial was abruptly terminated when a disproportionate number of deaths occurred in the treatment arm. Hypertension, attributed to an off-target effect of Torcetrapib on increasing aldosterone, was a recognized problem which has been eliminated in newer compounds.
Genetic variation in the CETP gene has been extensively studied for association with variation in HDL-C in different populations 53, 54. A meta-analysis reported CETP genotypes to be associated with moderate inhibition of CETP activity and inverse association with CAD or no increased risk 55. Other studies have reported greater risk associated with low CETP activity secondary to severe genetic deficiency56. A recent prospective study from the community-based Framingham Heart Study also reported greater risk with low CETP activity57. More recently it has been shown that polymorphisms in the CETP promoter region determine activity. GWAS in Caucasians has revealed association of the variant -2568 C/A (rs3764261) with HDL-C variation and the finding has been replicated in different ethnic groups 58, 59.
The role of three SNPs in the promoter region (−2568 C/A, −1700 C/T), −998 A/G) and the well-known non-coding SNP (397 A/G) identified as a restriction fragment (Taq1b) in the first intron, were studied in the unique Sikh population of Northern India who are known to have a high prevalence of type 2 diabetes and CAD despite much lower obesity rates 60. The −2568 C/A allele showed a strong association with increased HDL-C and decreased blood pressure. Although none of the SNPs were individually associated with CETP activity, low activity was associated with greater CAD risk and there was significant interaction between the CETP SNPs studied as haplotypes and CETP activity for affecting HDL-C 61. These results suggest that more complete genotyping could serve to define individual risk and response to therapies designed to raise HDL-C by inhibiting CETP.
b. Hepatic Fat and NAFLD
The NAFLD begins as simple steatosis and progresses to inflammation with risk for cirrhosis and liver cancer and is independently associated with increased risk of CAD 62. It has been proposed as a pre-diabetes phenotype and as a component of the metabolic syndrome 63. When fatty acids are mobilized from peripheral adipocytes in obese individuals, they are delivered to the liver and serve as a source for triglyceride which can either be stored or become incorporated into VLDL 64. It has been proposed that adipocytes become abnormal in obesity and that fatty acid release is excessive, particularly in Asian Indians who tend to be insulin resistant despite being relatively non-obese 65. Adipocyte dysfunction results in part from ectonucleotide pyrophosphate phosphodiesterase (ENPP1) over-expression, which may account for excessive mobilization of fatty acids leading to ectopic fat deposits as well as increases in VLDL triglyceride formation 66. In the multi-ethnic Dallas Heart Study, logistic regression analysis revealed significant interactions between the ENPP1 genotype, age, and body mass index (BMI) within each ethnic group and an ENPP1 allele predicted diabetes when a recessive model was tested. Consequently it was speculated that ethnic differences in the allele frequency could contribute to susceptibility to type 2 diabetes in African Americans and Hispanics 67.
Defective maturation of the VLDL particle in the golgi at the stage when triglyceride is transferred to apoB by microsomal triglyceride transfer protein coded for by MTTP 68, could account for excess liver fat storage leading to non-alcoholic fatty liver disease. This concept is supported by observations that MTTP polymorphism may impact non-alcoholic steatohepatitis by modulating lipoprotein metabolism and post-prandial lipemia. Carriers of the −493 G/T allele have a more atherogenic lipid profile 69, which also has a deleterious effect on beta cell function 70. Genetic determinants of VLDL formation as a cause of both atherosclerosis and fatty liver disease are supported by association of apoC-III polymorphisms with NAFLD. Asian Indian carriers of the APOCIII variant alleles (C-482T, T-455C, or both) had a 30% increase in apoC-III levels and a 60% increase in triglyceride, as compared with the wild-type homozygotes. The prevalence of NAFLD was 38% among variant-allele carriers compared to 0% among wild-type homozygotes, and association with insulin resistance was significant 71 Furthermore, apo-CIII overexpressing mice are predisposed to diet-induced hepatic steatosis and hepatic insulin resistance 72. These observations are explained by the dual role of apoC-III in VLDL assembly in the liver and in inhibiting VLDL lipolysis 73. Missense mutation in APOCIII within the C-terminal lipid binding domain of human apoC-III results in impaired assembly and secretion of VLDL providing evidence that apoC-III plays a role in the formation of lipoproteins 74, whereas apoC-III non-competitively inhibits activity by direct interaction with lipoprotein lipase 75. It is possible that a combination of polymorphisms could result in large sized VLDL as has been observed in NAFLD in an adolescent population independent of adiposity and insulin resistance, and interestingly the NMR lipid profile was characterized as having increased small dense LDL and a decrease in the number of large HDL particles 76, supporting the association of NAFLD with increased risk for atherosclerosois in adults 77 and with increased IMT in adolescents 78. These findings support the concept that NAFLD genotypes have pleiotropic effects, or alternatively the effects arise from a biochemical cascade leading to excessive hepatic fat storage.
GWAS of 2111 participants of the Dallas Heart Study revealed a robust association of liver fat defined by magnetic spectroscopy with the I148M allele of the PNPLA3 gene 79 and the association also occurs in children and adolescents80. Furthermore a meta-analysis of 16 studies showed association with disease severity. PNPLA3 has a strong effect on susceptibility to more aggressive disease with higher necroinflammatory scores and progression to fibrosis 81. The gene PNPLA3 codes for patatin-like phospholipase domain-containing a protein known as adiponutrin which plays a role in hepatic triglyceride hydrolysis, but the specific function is being investigated. It has been associated with increased alanine transaminase level, a marker of fatty liver disease, in Hispanics, Europeans, and Asian Indians 82, 83. Interestingly the S453I allele was associated with lower hepatic fat content and was more frequent in African Americans who had the lowest hepatic fat content, suggesting a protective effect 79. Although hepatic fat accumulation has been associated with insulin resistance there has been no association of the PNPLA3 allele with glucose intolerance, however associated obesity and alcohol consumption act independently with the PNPLA3 allele to increase serum transaminases 84.
Hypertension
Insight into the field of hypertension genetics has been provided by previous reviews85, 86. As with the other syndrome traits, the heritability of blood pressure is high ranging from 30–40% 87. Furthermore, systolic blood pressure has been associated with greater risk of mortality from CAD and stroke than diastolic with strong relationships to dietary salt intake 88 and ingestion of sugars and sugar-sweetened beverages 89 supporting gene-environment interactions. An increase in systolic blood pressure precedes diastolic and both are associated with obesity 90 with an abundance of evidence to support the associated effect of insulin resistance. Rare monogenic forms of hypertension have provided evidence for a regulatory role of key metabolic pathways and have been the basis for candidate gene population studies. Using such an approach, 24-hour ambulatory blood pressure has been associated with five polymorphisms in the KCNJ1 gene coding for an inward-rectifying apical potassium channel expressed in the thick ascending limb of Henle and throughout the distal nephron of the kidney. It has the potential to cause expression of antenatal Bartter Syndrome Type 2 when the abnormal allele is inherited 91. Also ambulatory blood pressure is associated with common variations in the WNK1 gene known to cause pseudohypoaldosteronism type 2 or Gordon syndrome92. Furthermore, association of WNK1 with blood pressure in childhood underscores its possible association with evolving hypertension at young ages93. Additional association with variants in CASR, NR3C2, SCNN1, and SCNN1B, all of which are known to have had mutations causing rare Mendelian defects in blood pressure regulation, provide support for the hypothesis that relevant polymorphisms influence conventional pathways involved in blood pressure regulation 91. However, GWAS has shown that only some of the associations are in or near genes involved in known hypertension-related metabolic pathways. The International Consortium for Blood Pressure GWAS studied 200,000 individuals of European descent and identified sixteen loci of which only six contained genes that are known or suspected to regulate blood pressure, which include NPR3, GUCY1A3- GUCY1B3, ADM, GNAS-EDN3, NPPA-NPPB, and CYP17A1 and their known metabolic roles have been have been comprehensively reviewed 94. Interestingly CYP17A1 achieved the most GWAS significance and is the site for a known Mendelian-inherited mutation causing hypertension by increasing mineralocorticoids in the adrenal steroid pathway and causing a rare form of congenital adrenal hyperplasia.
Data from the National Health and Nutrition Examination Survey showed the prevalence of hypertension to be 40% in African Americans compared to 27% in European Americans 95 leading to the hypothesis that part of the excess burden in African Americans is due to genetic susceptibility 96. Genome-wide and candidate gene associations have been examined in the Candidate Gene Association Resource Consortium consisting of 8591 African Americans. Novel associations were detected for diastolic blood pressure on chromosome 5 near GPR98 and ARRDC3 and for systolic blood pressure on chromosome 21 in C21orf91. Two of the top SNPs were not replicated in previously studied independent African American cohorts. However, several European American SNPs in SH2B3, TBX3-TBX5 and CSK-ULK3 did replicate supporting similarities in inheritance and associated complexities due to environmental and cultural factors 96.
The Beta Cell
a) Lipoproteins and the Beta Cell
Epidemiological observations supporting a role for HDL in the pathogenesis of diabetes have been supported by in vitro studies showing that addition of LDL to isolated human and rat islets decreases glucose stimulated insulin secretion and is attributed to cholesterol uptake by islet LDL receptors 97. Furthermore, the effect of intracellular accumulation of cholesterol is strongly influenced by HDL-mediated cholesterol efflux via the ATP-binding cassette transporter A1 (ABCA1), since mice lacking the LDL receptor and the ABCA1 transporter were not protected from effects of added LDL on decreasing beta cell insulin secretion, suggesting that HDL-mediated efflux plays a critical protective role 98. Further studies have revealed that high cholesterol content in the beta cell membrane down-regulates insulin secretion by influencing membrane depolarization, the signal for calcium influx and calcium-mediated insulin secretion 99. These studies provide a plausible explanation for the role of HDL in protecting the beta cell from cholesterol-induced toxicity.
b) Intrinsic Beta Cell Genes
The majority of gene variants associated with type 2 diabetes such as TCF7L2, CDKAL1, CDKN2A/B, HHEX-IDE, IGF2BP2, SLC30A8, KCNJ11, WFS1, JAZF1, TSPAN8, CD123/CAMK1D and MTNR1B, are implicated in β-cell functions such as glucose-stimulated insulin secretion, incretin effects on β-cell stimulation, and proinsulin to insulin conversion 100, 101. However the variants associated with fasting glucose levels in the normoglycemic population such as GCK, GCKR, G6PC2 and MTNR1B 102, do not always influence risk for type 2 diabetes but may only influence fasting glucose homeostasis both individually and when combined 103, 104.
Since fatty acids, LDL and HDL interact with the beta cell, it is possible that the levels, function and corresponding lipoprotein metabolism-determining genotypes interact with known SNPs which determine beta cell function and survival, and have a compounding effect on the beta cell. If so, those populations that have very high diabetes incidence may be collectively predisposed by influx of cholesterol, fatty acids and genes coding for beta cell metabolism. For example the Khatri Sikhs in Northern India are very susceptible to both type 2 diabetes and cardiovascular disease. Four of six SNPs for the TCF7L2 gene and two variants within the KCNQ1 gene were associated with type 2 diabetes 105, 106. Three of the four TCF7L2 SNPs were associated with LDL-C levels 105. In separate studies the CDK5 gene contained an allele associated with decreased HDL-C 107. In addition a GWAS performed in the same population has identified significant linkage signals for HDL-C at 10q21.2 and for LDL-C at 10p11.23 108.
Effects of Age
The increasing presentation of the metabolic syndrome in children and adolescents observed over the past two decades has coincided with increasing prevalence in adults and descending age of onset for both obesity and type 2 diabetes 109. Since the likelihood of risk factor appearance increases with age, most GWAS are adjusted for age. Although risk begins at birth most GWAS are done on large populations over age 18 years, however expression of risk factors in childhood and adolescence may represent more significant lifetime effects than if they presented later. Longitudinal studies indicate association of risk with insulin resistance and obesity in youth with gender and ethnic differences with tracking of BMI, blood pressure, and lipids to middle age adulthood 110. Analyses from 4 longitudinal cohorts beginning in childhood showed that the strength of the associations between baseline risk factors and adult carotid intima-media thickness is dependent on childhood age 111. For the most part these studies indicate that phenotypes in adolescence are similar to those in adults, and GWAS have shown replication. The PNPLA3 association with NAFLD is an example of replication of adult GWAS findings in youth, answering the question whether this association begins at early ages when preventive measures would be appropriate80.
Thrifty Phenotype
Significantly, over the past two decades there has been accumulation of epidemiological data showing a relationship of low birth weight associated with relatively less nutrient supply for the fetus leading to metabolic syndrome traits in adulthood including obesity, hypertension and progression to type 2 diabetes. Based on their own and accumulating evidence, Hales and Barker proposed the thrifty phenotype hypothesis stating that “epidemiological associations between poor fetal and infant growth and the subsequent development of type 2 diabetes and the metabolic syndrome result from the effects of poor nutrition in early life, which produces permanent changes in glucose-insulin metabolism”112 (Figure 1).
It is also clearly evident that both nutritional excess and exposure to high maternal glucose during gestation can result in large babies and prediction of similar metabolic syndrome traits giving rise to the observation that the association with birth-weight is often u-shaped113. However, the relative roles of genes and environment to these relationships remains a focus of further study. A review of 11 animal models investigating glycemic control in offspring of mothers exposed to a high fat diet during gestation has identified risk for type 2 diabetes and obesity in the offspring especially in males, and the loss of glucose tolerance is independent of maternal obesity, birth weight, or post-weaning macronutrient intake114. The experiments elucidating mechanisms whereby fetal systems are modulated by hormones (cortisol, insulin and leptin) changes in blood supply, oxidative stress, transcription, DNA methylation and histone acetylation has been reviewed and could not only serve as a foundation for therapeutic strategy but also could lead to studies on interaction with genotypes115.
Metabolic Pathways
Although investigations on genetic determinants of cardio-metabolic risk have progressed over the past decade following increased characterization of the genome and use of GWAS, a large portion of the heritability is unaccounted for and many of the genes commonly found in GWAS have small effects 81, 84, 116, 117. Also the SNPs cannot yet be used to predict disease onset or response to treatment as can be done with monogenic forms of diabetes, dyslipidemia or hypertension 117. Furthermore metabolic pathways that have been responsive to treatment with pharmaceutical agents or exercise, or have been associated with disease have rarely contained genes that have been identified by large scale studies such as GWAS. Therefore common pathways that may be responsive to treatments with pharmaceutical agents or exercise, may contain enzymes that are encoded by gene candidates for drug targets.
Sookoian et al have reviewed the question whether SNPs identified by GWAS are related to metabolic pathways 81. They used GWAS data and gene enrichment analysis based on neighboring genes and protein interaction networks. Results of the analysis support evidence for a network regulated by nuclear receptor proteins including retinoid X receptor (RXR) and farnesoid x receptor (FXR). They may be driving the expression of metabolic syndrome traits by their interaction with genes associated with metabolic pathways, cell differentiation and oxidative stress. KLF14 which encodes the transcription factor Kruppel-like factor 14, is an example of a gene located by GWAS, which has a central role in a regulatory network involving ten genes identified by gene expression profiling 118. The studies identified a single locus associated with a variety of metabolic syndrome traits including obesity, dyslipidemia and measures of insulin resistance. The method provides a way to identify variability in gene expression and to distinguish whether factors regulate the transcript level of the gene itself, known as cis-regulated expression quantitative trait loci (cis eQTLs) or transcripts of other genes (trans eQTLs). Spector et al have identified ten genes that have trans association with the same SNP that regulates KLF14 expression in the cis mode. One of the genes, SLC7A10, is associated with mediating neutral amino acid transport and has been associated with HDL and obesity 118. Further studies identifying a gene network of variants related to obesity and the metabolic syndrome has been conducted by Monda et al 119 who extracted and compiled data from GWAS and gene networks using the National Center for Biotechnology Information (NCBI). Based on the reported phenotypes, the results were grouped into six domains (obesity, dyslipidemia, type 2 diabetes, glucose, blood pressure and inflammation) that had been reported in GWAS. There were no apparent network drivers, but APOE, APOC1, CETP, GCKR, LPL, FADS1, FTO, MADD, HNF1A, SLC30A8 and TCF7L2 had inter-connections.
Maury et al 120 have proposed that mechanisms regulating sleep play a central role in the pathogenesis of the metabolic syndrome and have reviewed the influence of genotype and environment on sleep and circadian rhythms and the effects of sleep rhythm disruption. Based on evidence that both environmental effects or social factors resulting in sleep disruption and abnormal metabolic regulation of the internal clock system have been associated with obesity, diabetes, cardiovascular disease, thrombosis and inflammation, it is feasible that studies of sleep-regulated effects could lead to effective treatments. CLOCK, BMAL1, PER2, NNAS2 and MTNR1B encode proteins that function to regulate the mammalian clock and have been linked to features of the metabolic syndrome such as obesity, diabetes and hypertension 120. Furthermore differences in circadian gene expression in adipose tissue may influence the rate of fatty acid overflow resulting in deposition in liver, muscle and islets leading to NAFLD and diabetes.
Resistance to insulin action occurring in the liver fat cells and muscle is associated with many of the metabolic syndrome characteristics. This observation has led to the hypothesis that insulin resistance regulates the metabolic syndrome 121 which has been used to test for associations among components by factor analysis using insulin resistance as the central component 122, but the theory has often been questioned 123–125. Alternatively, it is possible that obesity is the factor driving the appearance of the syndrome and its progression, particularly blood pressure 124. However the opposing views may not be incompatible since both mechanisms may strain metabolic pathways and trigger disruption, and polymorphisms that involve pathways regulating expression of metabolic syndrome traits may accelerate the disruption. Since the metabolic syndrome is a strong predictor of type 2 diabetes 4, 126, it follows that predictive gene polymorphisms overlap; however, many of the mutations predicting monogenic diabetes have largely involved the β-cell and polymorphisms that have predicted the type 2 diabetes phenotype have often been β-cell-specific. Based on these observations Doria et al have concluded in an insightful review that type 2 diabetes has a progressive pathogenesis beginning with insulin resistance and progressing to β-cell failure 117. Furthermore, the authors emphasize that genes interact with one another and with the environment. The same group headed by Ronald Kahn have shown that a compound effect of gene interaction in a rodent model with separate and combined knockouts of the insulin receptor and IRS-1 genes results in a compounding effect of the two genes. However, neither gene defect alone could produce diabetes in more than 10% of the mice but when combined more than 50% developed diabetes at young ages 127; a phenomenon known as epistasis. It has been established that environmental effects modify the expression of insulin resistance via effects on post-receptor metabolic pathways 117 including epigenetic modifications such as DNA methylation. This effect can occur in the fetus 128, in peripheral white blood cells in adolescents 129, and involves the peroxisome proliferator-activated receptor gamma coactivator 1alpha (PGC-1α or PPARGC1A gene) promoter in NAFLD in association with insulin resistance 130, which is also an important coactivator and major regulator of exercise-induced adaptation within physiological ranges 131.
Drug Targets and Genes that Predict Treatment Response
Since the enzyme, 11β-hydroxysteroid dehydrogenase type 1 (11β-HSD1) catalyses intracellular conversion of inert 11-ketosteroids to cortisol and corticosterone and has been identified as being increased in obese human and rodent adipose tissue, it has been a focus of investigation 132. Transgenic mice selectively overexpressing the enzyme in adipose tissue show many of the metabolic syndrome traits 133. Conversely knock-out 11β-HSD1 mice showed cardio-protective traits including an improved lipid and lipoprotein profile 134. Human studies have shown increased activity in subcutaneous fat tissues 135, suggesting that the enzyme might be a suitable selective target for pharmaceutical intervention that not only influences activity in the fat cell by down-regulating harmful glucocorticoid effects but also has beneficial pleiotropic effects on syndrome traits 136. Emodin, an active ingredient of Chinese herbs has been show to selectively inhibit 11β-HSD1 in mice suggesting that analogues might be developed for therapeutic use 136. Studies in identical twins have shown association enzyme activity with environmental factors but not genotype 137, however, polymorphisms of 11β-HSD1 have been associated with diabetes, hypertension and apolipoprotein levels 138–140 and the rs3753519 polymorphism has been associated with pediatric-onset obesity in Spanish children 141.
Adenosine-monophosphate-activated kinase (AMPK) is a key regulator of metabolism involving pathways central to regulation of obesity and the metabolic syndrome 142, consequently it has emerged as a drug target 143. It is a large heterotrimeric enzyme composed of a catalytic and two regulatory subunits encoded by separate genes. AMPK serves as a central metabolic switch sensing cellular energy status through modulation via its phosphorylation and activation. Liver AMPK controls hepatic glucose production by inhibiting expression of the gluconeogenic enzymes, phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase. The AICAR (5-aminoimidazole-4-carboxamide-1-β-D-ribofuranoside) and metformin down-regulate both enzymes. Furthermore AMPK is activated by several stimuli, including exercise, hypoxia, hypoglycemia, calcium, hormones (adiponectin, leptin, ciliary neurotrophic factor, ghrelin), interleukin-6, α-lipoic acid and resveratrol 143. It follows that genes encoding enzymes, cofactors and transcription factors involved in AMPK-associated pathways could represent significant candidate genes for prospective studies with both predictive and therapeutic implications. AREBP (AICAR response element binding protein) is an example of a transcription factor that binds to the PEPCK promoter and represses translation in a phosphorylation-dependent manner. When overexpressed in mice, investigators were able to show that AICAR could reduce fasting-induced upregulation of PEPCK 144 supporting PEPCK activation by AMPK as a target.
With the hypothesis that polymorphisms in metabolic pathways involving metformin’s action may determine treatment responses, investigators in the United Kingdom conducted a meta-analysis using the glycemic response to metformin as the phenotype. A SNP at a locus containing ATM, the ataxia telangiectasia mutated gene (rs11212617) was associated with the response 145. In a rat hepatoma cell line, specific inhibition of ATM attenuated the phosphorylation and activation of AMPK in response to metformin 145. The data suggest that ATM, a gene known to be involved in DNA repair and cell cycle control, plays a role in the effect of metformin upstream of AMP-activated protein kinase. Although the effect attributed to the polymorphism is small, the study identified an important upstream AMPK regulator adding information to how metformin works, and also identifies a possible mechanistic link with DNA repair and cancer prevention. Shu et al have investigated polymorphisms in SLC22A1 which encodes organic cation transporter 1(OCT-1) functions to facilitate absorption of metformin into hepatocytes and identified association with reduced responsiveness to metformin’s effect on glucose levels during an oral glucose tolerance test 146. Also association with HbA1c was shown with a variant in SLC47A1 encoding the multidrug and toxin extrusion protein 1 involved in the excretion of metformin into bile and urine 147. Furthermore, the two genes both acting to increase intracellular metformin, have an interactive effect 148. The roles of both genes were supported by the Diabetes Prevention Program investigators 149 who confirmed association of the glucose-lowering response to metformin and variants in SLC47A1, and interactions with SLC22A1 and the AMPK genes STK11, PRKAA1 and PRKAA2.
The pharmacogenetics of other anti-diabetes drugs has been well reviewed 150. Besides metformin, it is likely that genes encoding the receptor and pathways for PPARG agonists constituting the thiozolidinediones, are most likely to be associated with the metabolic syndrome because of their role in insulin sensitivity. Of 133 PPARG SNPs tested in the TRIPOD study, eight showed evidence for association with response to Troglitazone therapy defined as change in tolerance to intravenous glucose (IVGTT) 151. However there was no association with fasting glucose suggesting that response in confined to measures of insulin resistance such is the minimal model IVGTT whereas fasting glucose is more likely to be a measure of β-cell failure. In studies where overt type 2 diabetes has been the phenotype the majority of associated polymorphisms have encoded proteins known to be involved in β-cell metabolism; for example TCF7L2, KCNJ11 and HHEX have shown robust association 152, 153. The HHEX gene was shown to be associated with increased risk for type 2 diabetes in mainly Asians and Caucasians by meta-analysis 154, and is involved in β-cell development and function via interaction with hepatic nuclear factor 1α (HNF1α) 155. Similarly the c-allele of rs13266634 located in SLC30A8 (ZNT8) has been associated with insulin and glucagon levels and type 2 diabetes in East Asians and Europeans in a meta-analysis 156. Interestingly the glucokinase gene (GCK) has activating mutations causing hypoglycemia that might provide structural and functional models leading to drug targets for treating type 2 diabetes 157. In GoDARTs study, investigators examined the medication response of metformin and sulphonylurea based on the TCF7L2 genotypes. The carriers of the at risk ‘T’ allele responded less well to sulphonylurea therapy than metformin158. In the Diabetes Prevention Program (DPP), the lifestyle modifications were shown to reduce the risk of diabetes conferred by risk variants of TCF7L2 at rs7093146. In placebo participants who carried the homozygous risk genotype (TT), had 80% higher risk for developing diabetes compared to the lifestyle intervention group carrying the same risk genotypes 159.
Statins and fibrates alone or in combination are frequent choices for treatment of the metabolic syndrome dyslipidemia indicating a need for genetic prediction of treatment response so that effective lipid lowering can be attained by tailoring treatment to individual requirements. Brautbar et al have recently identified lipoprotein lipase gene variants that affect apoC-III lowering by paradoxically increasing apoC-III levels 160. The study offers a feasible explanation for disappointing clinical outcomes in trials that evaluated the efficacy of fibrates such as FIELD and ACCORD 161–163. The importance of apoC-III is underscored by association of polymorphisms in the promoter with coronary heart disease, particularly in the insulin response element 164. There is strong association of apoC-III bound to apoB-containing lipoproteins with the number of metabolic syndrome criteria 165, coronary heart disease events in the CARE trial 166, whereas apoC-III bound to apoA-I-containing particles was a predictor of angiographic change in the Cholesterol Lowering Atherosclerosis Study in response to colestipol and niacin 167. It is unknown how apoC-III, an LPL inhibitor, may interact with LPL to determine the response to fibrates. ApoB and the LDL receptor may also determine response to therapy such as statins in familial hypercholesterolemia 168 and anti-hypertensive medications 169.
Proposed Model for Phenotype Interaction
Monogenic models such as the lipodystrophy syndromes could serve as a model 170. but there has been accumulating evidence for multigenic origin, and changes in the traits throughout life. Therefore the serial nature of the syndrome should be taken into account since the traits are susceptible to interaction both at the gene level as shown by epigenetic modifications and at the pathway level as shown by modifications coinciding with the development of insulin resistance and obesity (Figure 1). Based on known metabolic pathways, genotype and phenotype associations and epidemiological studies, we propose an outline for genetic modulation of clinical cardio-metabolic phenotypes such as obesity, hypertension, dyslipidemia, NAFLD, and glucose intolerance leading to atherosclerosis and type 2 diabetes (Figure 2).
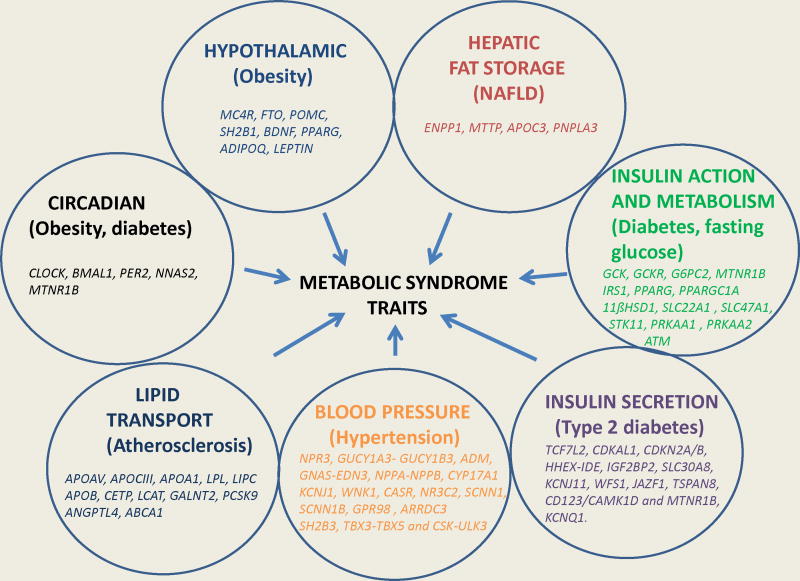
In addition to the standard five criteria, NAFLD, a newly recognized addition to the metabolic syndrome, appears to have a central predisposing role for cardiovascular disease and type 2 diabetes. There is evidence, cited in the text, to support inter-relationship in regard to progression to atherosclerosis and diabetes phenotypes. The classic lipid derangement observed in insulin resistance consisting of elevated triglyceride, small LDL particles in increased numbers and low HDL-C has significant association with genotype and risk prediction. Cross-sectional and sequential clinical investigations beginning at early phases of the pathogenesis are needed to determine more precise inter-relationship of phenotypes to each other and to the respective genotypes while contributing to improved characterization. In addition improved phenotypic characterization and relationship to genotypes is needed to uncover new pathways and targets for intervention 171. To achieve this goal it will be necessary to understand overlapping relationships of polymorphisms with traits, their expression during the lifespan and interrelationships either by pleiotropism or common pathways. We propose beginning this process by sorting the genes by their respective metabolic functions (Table 1, Figure 3).
Table 1.
List of Genetic loci in Metabolic Syndrome Pathway
| Category | Gene Name | Chromosome | Entrez Gene ID | Role |
|---|---|---|---|---|
| Hypothalamic Genes | FTO† | 16q12.2 | 79068 | Severe obesity/insulin resistance |
| MC4R† | 18q21.32 | 4160 | Member of G-protein coupled receptor family, signaling hormone involved in energy homoeostasis | |
| PPARG† | 3p25.2 | 5468 | Transcription factor involved in adipogenesis and type 2 diabetes risk | |
| ADIPOQ | 3q27.3 | 9370 | Adipose tissue specific protein involved in insulin sensitizing and anti-atherosclerotic properties | |
| LEPTIN | 7q31.3 | 3952 | Signaling hormone affects directly or indirectly on the central nervous system to inhibit food intake and/or regulate energy expenditure as part of a homeostatic mechanism | |
| Hepatic Genes | ||||
| Dyslipidemia | APOE-CI-CII-CIV† | 19q13.32 | 2282 | Cluster of triglyceride-rich lipoprotein receptor ligands for LDL receptor –related proteins |
| APOB† | 2p24.1 | 338 | Main apolipoprotein of chylomicrons and low density lipoproteins, functions as a recognition signal for the cellular binding and internalization of LDL particles | |
| APOAV-AIV-CIII-AI† | 11q23.3 | 117536 | Cluster of apolipoproteins plays an important role in regulating the plasma triglyceride levels | |
| GALNT2 | 1q42.13 | 2590 | Catalyzes the initial reaction in O-linked oligosaccharide biosynthesis | |
| PCSK9 | 1p32.3 | 255738 | Decreases plasma cholesterol and LDL cholesterol and provides protection from coronary artery disease | |
| CETP† | 16q13 | 1071 | Exchanges cholesterol esters for triglycerides from HDL and triglyceride rich lipoproteins | |
| LCAT† | 16q22.1 | 3931 | Required for remodeling HDL particles into their spherical forms | |
| ABCA1† | 2p23.3 | 2646 | Functions as a cholesteral efflux pump in the cellular lipid removal pathway. Mutations in this gene cause Tangier’ disease and familial HDL deficiency. | |
| LPL† | 8p21.3 | 4023 | Catalyzes the hydrolysis of triglycerides to release free fatty acids into the circulation | |
| LIPC† | 15q21.3 | 3990 | Encodes hepatic triglyceride lipase in liver and hydrolyses triglycerides | |
| ANGPTL4† | 19p13.2 | 51129 | Plasma hormone directly involved in regulating glucose homeostasis, lipid metabolism, and insulin sensitivity and also acts as an apoptosis factor for vascular endothelial cells | |
| NAFLD | MTTP | 4q23 | 4547 | Catalyzes the transport of triglyceride, cholesterol ester, and phospholipid between phospholipid surfaces |
| ENPP1 | 6q23.2 | 5167 | Involved primarily in ATP hydrolysis at the plasma membrane. Appears to modulate insulin sensitivity | |
| APOCIII† | 11q23.3 | 345 | Inhibits lipoprotein lipase; it delays catabolism of triglyceride-rich particles, induces the development of hypertriglyceridemia | |
| PNPLA3† | 22q13.31 | 80339 | Triacylglycerol lipase that mediates triacylglycerol hydrolysis in adipocytes | |
| Hypertension | WNK1 | 12p13.33 | 65125 | A key regulator of blood pressure by controlling the transport of sodium and chloride ions |
| KCNJ1 | 11q24.3 | 3758 | Mutations in this gene have been associated with Bartter syndrome, which is characterized by salt wasting, hypercalciuria, and low blood pressure | |
| NPR3 | 5p13.3 | 4883 | Encodes natriuretic peptides which regulate blood volume and pressure, pulmonary hypertension, and cardiac function | |
| GUCY1A3 | 4q32.1 | 2982 | guanylyl cyclases are groups of enzymes that mediate important communication between the heart, intestine and kidney to regulate blood volume and Na+ balance. | |
| GNAS | 20q13.32 | 4686 | Guanine nucleotide-binding proteins (G proteins) are involved as modulators or transducers in various transmembrane signaling systems | |
| NPPA-NPPB | 1q36.22 | 9757 | Natriuretic peptide receptors are associated with intracellular guanylyl cyclase activity and involved in homeostasis of body fluid volume | |
| CYP17A1† | 10q24.32 | 1586 | Mono-oxygenases which catalyze many reactions involved in drug metabolism and synthesis of cholesterol, steroids and lipids; gene variants associated with hypertension | |
| C21orf91† | 21q21.1 | 54149 | Gene variants associated with systolic blood pressure | |
| GPR98† | 5q14.3 | 84059 | Associated with Usher syndrome 2 and familial febrile seizures, gene variants associated with diastolic blood pressure | |
| ARRDC3 | 5q14.3 | 57561 | Gene variants associated with diastolic blood pressure | |
| β-cell function, insulin secretion and insulin resistance, and type 2 diabetes genes | GCKR† | 2p23.3 | 2646 | Enzyme regulators, controls activity of glucokinase in liver and brain |
| G6PC2† | 2q24.3 | 57818 | Enzyme, transport channel, key role in glucose homeostasis | |
| CDKN2A-B† | 9p21.3 | 1029 | Enzyme, anti-oncogene involved in pancreatic carcinomas, type 2 diabetes | |
| GLUT4 | 17 p13.1 | 6517 | Solute carrier family 2, mediates insulin-stimulation glucose uptake in adipocytes & muscles | |
| INSR | 19 p13.3 | 3643 | Signaling hormone receptor tyrosine kinase | |
| HNF4A† | 20q12 | 3172 | Transcription factor regulates genes required for glucose transport and metabolism | |
| ADAM 30† | 1p12-p11 | 11085 | Disintegrin and metalloproteinase domain-containing protein 30 has been implicated in a variety of biological processes and associated with type 2 diabetes risk | |
| NOTCH2† | 1p13-p11 | 4853 | Transcription regulator, type 2 diabetes | |
| THADA† | 2p21 | 63892 | Death receptor membrane protein, gene variants associated with type 2 diabetes | |
| ADAMTS9† | 3p14.3 | 56999 | Enzyme, anti-oncogene, associated with type 2 diabetes | |
| JAZF1† | 7p15.2 | 221895 | Transcription factor, increases risk for prostate cancer, type 2 diabetes | |
| TSPAN8† | 12q14.1 | 7103 | Regulatory protein involved in cell development, growth and motility, type 2 diabetes | |
| IGF2BP2† | 3q27.2 | 10644 | Regulatory enzyme influences insulin secretion | |
| CDKAL1† | 6p22.2 | 54901 | Variant confers risk through reduced insulin secretion | |
| GCK† | 7p14 | 2645 | Modulates insulin secretion, glucolysis, energy pathways | |
| SLC30A8† | 8q24.11 | 169026 | Facilitates transportation of zinc from cytoplasm into insulin containing vesicles | |
| TCF7L2† | 10 q25.2 | 6934 | Transcription regulator influences insulin secretion | |
| INS | 11 p15.5 | 3630 | Signaling hormone, increases cell permeability to monosaccharides, amino acids and fatty acids | |
| CDC123† | 10p13 | 8872 | Involved in transcription regulation, insulin secretion | |
| HHEX | 10q23.33 | 3087 | Transcription factor involved in hematopoietic differentiation, pancreatic development, insulin secretion | |
| KCNJ11† | 11 p15.1 | 3767 | Ion channel transporter | |
Gene association detected in GWAS
Figure 3.
Interrelationship between obesity, hepatic fat, insulin action, insulin secretion, blood pressure, dyslipidemia, and circadian clock with metabolic syndrome. Seven groups of genes that affect metabolic syndrome traits are summarized (see table for details).
Conclusions
This review summarizes the rapidly moving field of metabolic syndrome genetics by covering advances in the commonly encountered clinical traits as opposed to a more specialized focus on one trait. The evidence supports progress in unraveling the origins of a complex and interrelated cluster, and provides insight on how each of the traits may relate to one another, either through common genes, common and overlapping pathways or by shared end-points. We propose commonality and interrelationships between the traits and their genetic and environmental determinants, which includes sequential development from conception through gestation, childhood and adolescence. Progress in the field is encouraging and is beginning to show some potential for clinical prediction and identification of drug targets. Reviewing progress in genetics of the syndrome as a whole does not argue against coning down on individual components in studies that provide insight and have made significant discoveries of genes for which key functions can be determined. Since understanding biochemical mechanisms and interactions between pathways is central to unveiling the cluster, key questions relate to the small effect size of multiple common mutations when assessed in a multi-genic background, making it difficult to support successful pathway-based pharmaceutical interventions. However, discovery of missing heritability from rare variants with larger effects and genes coding for novel drug targets together with gene-gene and gene-environmental interactions and effects of lifestyle interventions are important considerations in actively investigated approaches.
Acknowledgments
This work was partly supported by NIH grants (K01TW006087 and R01DK082766) funded by the Fogarty International Center (FIC) and National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), and a seed grant from University of Oklahoma Health Sciences Center, Oklahoma City, OK, USA. Technical assistance provided by Latonya Been in manuscript preparation is duly acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mottillo S, Filion KB, Genest J, Joseph L, Pilote L, Poirier P, Rinfret S, Schiffrin EL, Eisenberg MJ. The metabolic syndrome and cardiovascular risk a systematic review and meta-analysis. Journal of the American College of Cardiology. 2010;56:1113–32. doi: 10.1016/j.jacc.2010.05.034. [DOI] [PubMed] [Google Scholar]
- 2.Morrison JA, Ford ES, Steinberger J. The pediatric metabolic syndrome. Minerva medica. 2008;99:269–87. [PubMed] [Google Scholar]
- 3.Wang YZ, Huang YN, Sun KY, Qi JH, Xiang L. Leptin gene transfer regulates fibromuscular development and lipid deposition in muscles via SIRT1, FOXO3a and PGC-1alpha in mice in vivo. International journal of molecular medicine. 2011;28:617–23. doi: 10.3892/ijmm.2011.711. [DOI] [PubMed] [Google Scholar]
- 4.Ford ES, Li C, Sattar N. Metabolic syndrome and incident diabetes: current state of the evidence. Diabetes Care. 2008;31:1898–904. doi: 10.2337/dc08-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reaven GM. The metabolic syndrome: time to get off the merry-go-round? Journal of internal medicine. 2011;269:127–36. doi: 10.1111/j.1365-2796.2010.02325.x. [DOI] [PubMed] [Google Scholar]
- 6.Stern MP, Williams K, Gonzalez-Villalpando C, Hunt KJ, Haffner SM. Does the metabolic syndrome improve identification of individuals at risk of type 2 diabetes and/or cardiovascular disease? Diabetes Care. 2004;27:2676–81. doi: 10.2337/diacare.27.11.2676. [DOI] [PubMed] [Google Scholar]
- 7.Kahn R, Buse J, Ferrannini E, Stern M. The metabolic syndrome: time for a critical appraisal: joint statement from the American Diabetes Association and the European Association for the Study of Diabetes. Diabetes Care. 2005;28:2289–304. doi: 10.2337/diacare.28.9.2289. [DOI] [PubMed] [Google Scholar]
- 8.Boyko EJ, Doheny RA, McNeely MJ, Kahn SE, Leonetti DL, Fujimoto WY. Latent class analysis of the metabolic syndrome. Diabetes research and clinical practice. 2010;89:88–93. doi: 10.1016/j.diabres.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemieux I, Pascot A, Couillard C, Lamarche B, Tchernof A, Almeras N, Bergeron J, Gaudet D, Tremblay G, Prud’homme D, Nadeau A, Despres JP. Hypertriglyceridemic waist: A marker of the atherogenic metabolic triad (hyperinsulinemia; hyperapolipoprotein B; small, dense LDL) in men? Circulation. 2000;102:179–84. doi: 10.1161/01.cir.102.2.179. [DOI] [PubMed] [Google Scholar]
- 10.Lemieux I, Poirier P, Bergeron J, Almeras N, Lamarche B, Cantin B, Dagenais GR, Despres JP. Hypertriglyceridemic waist: a useful screening phenotype in preventive cardiology? The Canadian journal of cardiology. 2007;23 (Suppl B):23B–31B. doi: 10.1016/s0828-282x(07)71007-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Utzschneider KM, Van de Lagemaat A, Faulenbach MV, Goedecke JH, Carr DB, Boyko EJ, Fujimoto WY, Kahn SE. Insulin resistance is the best predictor of the metabolic syndrome in subjects with a first-degree relative with type 2 diabetes. Obesity. 2010;18:1781–7. doi: 10.1038/oby.2010.77. [DOI] [PubMed] [Google Scholar]
- 12.Meigs JB, Rutter MK, Sullivan LM, Fox CS, D’Agostino RB, Sr, Wilson PW. Impact of insulin resistance on risk of type 2 diabetes and cardiovascular disease in people with metabolic syndrome. Diabetes Care. 2007;30:1219–25. doi: 10.2337/dc06-2484. [DOI] [PubMed] [Google Scholar]
- 13.Neel JV. The “thrifty genotype” in 1998. Nutrition reviews. 1999;57:S2–9. doi: 10.1111/j.1753-4887.1999.tb01782.x. [DOI] [PubMed] [Google Scholar]
- 14.Samuel VT, Shulman GI. Mechanisms for insulin resistance: common threads and missing links. Cell. 148:852–71. doi: 10.1016/j.cell.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rabol R, Petersen KF, Dufour S, Flannery C, Shulman GI. Reversal of muscle insulin resistance with exercise reduces postprandial hepatic de novo lipogenesis in insulin resistant individuals. Proc Natl Acad Sci U S A. 108:13705–9. doi: 10.1073/pnas.1110105108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.WSG Gene-diet interactions in childhood obesity. Curr Genomics. 2011;12:180–9. doi: 10.2174/138920211795677903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farin HM, Abbasi F, Reaven GM. Body mass index and waist circumference both contribute to differences in insulin-mediated glucose disposal in nondiabetic adults. The American journal of clinical nutrition. 2006;83:47–51. doi: 10.1093/ajcn/83.1.47. [DOI] [PubMed] [Google Scholar]
- 18.Farooqi IS, O’Rahilly S. Genetic factors in human obesity. Obesity reviews: an official journal of the International Association for the Study of Obesity. 2007;8 (Suppl 1):37–40. doi: 10.1111/j.1467-789X.2007.00315.x. [DOI] [PubMed] [Google Scholar]
- 19.Stunkard AJ, Harris JR, Pedersen NL, McClearn GE. The body-mass index of twins who have been reared apart. N Engl J Med. 1990;322:1483–7. doi: 10.1056/NEJM199005243222102. [DOI] [PubMed] [Google Scholar]
- 20.Farooqi IS, O’Rahilly S. New advances in the genetics of early onset obesity. Int J Obes (Lond) 2005;29:1149–52. doi: 10.1038/sj.ijo.0803056. [DOI] [PubMed] [Google Scholar]
- 21.Farooqi IS, Keogh JM, Yeo GS, Lank EJ, Cheetham T, O’Rahilly S. Clinical spectrum of obesity and mutations in the melanocortin 4 receptor gene. N Engl J Med. 2003;348:1085–95. doi: 10.1056/NEJMoa022050. [DOI] [PubMed] [Google Scholar]
- 22.Martinelli CE, Keogh JM, Greenfield JR, Henning E, van der Klaauw AA, Blackwood A, O’Rahilly S, Roelfsema F, Camacho-Hubner C, Pijl H, Farooqi IS. Obesity due to melanocortin 4 receptor (MC4R) deficiency is associated with increased linear growth and final height, fasting hyperinsulinemia, and incompletely suppressed growth hormone secretion. J Clin Endocrinol Metab. 2011;96:E181–8. doi: 10.1210/jc.2010-1369. [DOI] [PubMed] [Google Scholar]
- 23.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Allen HL, Lindgren CM, Luan J, Magi R, Randall JC, Vedantam S, Winkler TW, Qi L, Workalemahu T, Heid IM, Steinthorsdottir V, Stringham HM, Weedon MN, Wheeler E, Wood AR, Ferreira T, Weyant RJ, Segre AV, Estrada K, Liang L, Nemesh J, Park JH, Gustafsson S, Kilpelainen TO, Yang J, Bouatia-Naji N, Esko T, Feitosa MF, Kutalik Z, Mangino M, Raychaudhuri S, Scherag A, Smith AV, Welch R, Zhao JH, Aben KK, Absher DM, Amin N, Dixon AL, Fisher E, Glazer NL, Goddard ME, Heard-Costa NL, Hoesel V, Hottenga JJ, Johansson A, Johnson T, Ketkar S, Lamina C, Li S, Moffatt MF, Myers RH, Narisu N, Perry JR, Peters MJ, Preuss M, Ripatti S, Rivadeneira F, Sandholt C, Scott LJ, Timpson NJ, Tyrer JP, van Wingerden S, Watanabe RM, White CC, Wiklund F, Barlassina C, Chasman DI, Cooper MN, Jansson JO, Lawrence RW, Pellikka N, Prokopenko I, Shi J, Thiering E, Alavere H, Alibrandi MT, Almgren P, Arnold AM, Aspelund T, Atwood LD, Balkau B, Balmforth AJ, Bennett AJ, Ben-Shlomo Y, Bergman RN, Bergmann S, Biebermann H, Blakemore AI, Boes T, Bonnycastle LL, Bornstein SR, Brown MJ, Buchanan TA, Busonero F, Campbell H, Cappuccio FP, Cavalcanti-Proenca C, Chen YD, Chen CM, Chines PS, Clarke R, Coin L, Connell J, Day IN, den Heijer M, Duan J, Ebrahim S, Elliott P, Elosua R, Eiriksdottir G, Erdos MR, Eriksson JG, Facheris MF, Felix SB, Fischer-Posovszky P, Folsom AR, Friedrich N, Freimer NB, Fu M, Gaget S, Gejman PV, Geus EJ, Gieger C, Gjesing AP, Goel A, Goyette P, Grallert H, Grassler J, Greenawalt DM, Groves CJ, Gudnason V, Guiducci C, Hartikainen AL, Hassanali N, Hall AS, Havulinna AS, Hayward C, Heath AC, Hengstenberg C, Hicks AA, Hinney A, Hofman A, Homuth G, Hui J, Igl W, Iribarren C, Isomaa B, Jacobs KB, Jarick I, Jewell E, John U, Jorgensen T, Jousilahti P, Jula A, Kaakinen M, Kajantie E, Kaplan LM, Kathiresan S, Kettunen J, Kinnunen L, Knowles JW, Kolcic I, Konig IR, Koskinen S, Kovacs P, Kuusisto J, Kraft P, Kvaloy K, Laitinen J, Lantieri O, Lanzani C, Launer LJ, Lecoeur C, Lehtimaki T, Lettre G, Liu J, Lokki ML, Lorentzon M, Luben RN, Ludwig B, Manunta P, Marek D, Marre M, Martin NG, McArdle WL, McCarthy A, McKnight B, Meitinger T, Melander O, Meyre D, Midthjell K, Montgomery GW, Morken MA, Morris AP, Mulic R, Ngwa JS, Nelis M, Neville MJ, Nyholt DR, O’Donnell CJ, O’Rahilly S, Ong KK, Oostra B, Pare G, Parker AN, Perola M, Pichler I, Pietilainen KH, Platou CG, Polasek O, Pouta A, Rafelt S, Raitakari O, Rayner NW, Ridderstrale M, Rief W, Ruokonen A, Robertson NR, Rzehak P, Salomaa V, Sanders AR, Sandhu MS, Sanna S, Saramies J, Savolainen MJ, Scherag S, Schipf S, Schreiber S, Schunkert H, Silander K, Sinisalo J, Siscovick DS, Smit JH, Soranzo N, Sovio U, Stephens J, Surakka I, Swift AJ, Tammesoo ML, Tardif JC, Teder-Laving M, Teslovich TM, Thompson JR, Thomson B, Tonjes A, Tuomi T, van Meurs JB, van Ommen GJ, Vatin V, Viikari J, Visvikis-Siest S, Vitart V, Vogel CI, Voight BF, Waite LL, Wallaschofski H, Walters GB, Widen E, Wiegand S, Wild SH, Willemsen G, Witte DR, Witteman JC, Xu J, Zhang Q, Zgaga L, Ziegler A, Zitting P, Beilby JP, Farooqi IS, Hebebrand J, Huikuri HV, James AL, Kahonen M, Levinson DF, Macciardi F, Nieminen MS, Ohlsson C, Palmer LJ, Ridker PM, Stumvoll M, Beckmann JS, Boeing H, Boerwinkle E, Boomsma DI, Caulfield MJ, Chanock SJ, Collins FS, Cupples LA, Smith GD, Erdmann J, Froguel P, Gronberg H, Gyllensten U, Hall P, Hansen T, Harris TB, Hattersley AT, Hayes RB, Heinrich J, Hu FB, Hveem K, Illig T, Jarvelin MR, Kaprio J, Karpe F, Khaw KT, Kiemeney LA, Krude H, Laakso M, Lawlor DA, Metspalu A, Munroe PB, Ouwehand WH, Pedersen O, Penninx BW, Peters A, Pramstaller PP, Quertermous T, Reinehr T, Rissanen A, Rudan I, Samani NJ, Schwarz PE, Shuldiner AR, Spector TD, Tuomilehto J, Uda M, Uitterlinden A, Valle TT, Wabitsch M, Waeber G, Wareham NJ, Watkins H, Wilson JF, Wright AF, Zillikens MC, Chatterjee N, McCarroll SA, Purcell S, Schadt EE, Visscher PM, Assimes TL, Borecki IB, Deloukas P, Fox CS, Groop LC, Haritunians T, Hunter DJ, Kaplan RC, Mohlke KL, O’Connell JR, Peltonen L, Schlessinger D, Strachan DP, van Duijn CM, Wichmann HE, Frayling TM, Thorsteinsdottir U, Abecasis GR, Barroso I, Boehnke M, Stefansson K, North KE, McCarthy MI, Hirschhorn JN, Ingelsson E, Loos RJ. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–48. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, Shields B, Harries LW, Barrett JC, Ellard S, Groves CJ, Knight B, Patch AM, Ness AR, Ebrahim S, Lawlor DA, Ring SM, Ben-Shlomo Y, Jarvelin MR, Sovio U, Bennett AJ, Melzer D, Ferrucci L, Loos RJ, Barroso I, Wareham NJ, Karpe F, Owen KR, Cardon LR, Walker M, Hitman GA, Palmer CN, Doney AS, Morris AD, Smith GD, Hattersley AT, McCarthy MI. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–94. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hallman DM, Friedel VC, Eissa MA, Boerwinkle E, Huber JC, Jr, Harrist RB, Srinivasan SR, Chen W, Dai S, Labarthe DR, Berenson GS. The association of variants in the FTO gene with longitudinal body mass index profiles in non-Hispanic white children and adolescents. Int J Obes. 2011 doi: 10.1038/ijo.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu G, Zhu H, Dong Y, Podolsky RH, Treiber FA, Snieder H. Influence of common variants in FTO and near INSIG2 and MC4R on growth curves for adiposity in African- and European-American youth. Eur J Epidemiol. 2011;26:463–73. doi: 10.1007/s10654-011-9583-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rutters F, Nieuwenhuizen AG, Bouwman F, Mariman E, Westerterp-Plantenga MS. Associations between a single nucleotide polymorphism of the FTO Gene (rs9939609) and obesity-related characteristics over time during puberty in a Dutch children cohort. The Journal of clinical endocrinology and metabolism. 2011;96:E939–42. doi: 10.1210/jc.2010-2413. [DOI] [PubMed] [Google Scholar]
- 28.Gonzalez JR, Gonzalez-Carpio M, Hernandez-Saez R, Serrano Vargas V, Torres Hidalgo G, Rubio-Rodrigo M, Garcia-Nogales A, Nunez Estevez M, Luengo Perez LM, Rodriguez-Lopez R. FTO Risk Haplotype Among Early Onset and Severe Obesity Cases in a Population of Western Spain. Obesity. 2011 doi: 10.1038/oby.2011.325. [DOI] [PubMed] [Google Scholar]
- 29.Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, Yeo GS, McDonough MA, Cunliffe S, McNeill LA, Galvanovskis J, Rorsman P, Robins P, Prieur X, Coll AP, Ma M, Jovanovic Z, Farooqi IS, Sedgwick B, Barroso I, Lindahl T, Ponting CP, Ashcroft FM, O’Rahilly S, Schofield CJ. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–72. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sonestedt E, Roos C, Gullberg B, Ericson U, Wirfalt E, Orho-Melander M. Fat and carbohydrate intake modify the association between genetic variation in the FTO genotype and obesity. The American journal of clinical nutrition. 2009;90:1418–25. doi: 10.3945/ajcn.2009.27958. [DOI] [PubMed] [Google Scholar]
- 31.Rees SD, Islam M, Hydrie MZ, Chaudhary B, Bellary S, Hashmi S, O’Hare JP, Kumar S, Sanghera DK, Chaturvedi N, Barnett AH, Shera AS, Weedon MN, Basit A, Frayling TM, Kelly MA, Jafar TH. An FTO variant is associated with Type 2 diabetes in South Asian populations after accounting for body mass index and waist circumference. Diabet Med. 2011;28:673–80. doi: 10.1111/j.1464-5491.2011.03257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Kilpelainen TO, Liu C, Zhu J, Liu Y, Hu C, Yang Z, Zhang W, Bao W, Cha S, Wu Y, Yang T, Sekine A, Choi BY, Yajnik CS, Zhou D, Takeuchi F, Yamamoto K, Chan JC, Mani KR, Been LF, Imamura M, Nakashima E, Lee N, Fujisawa T, Karasawa S, Wen W, Joglekar CV, Lu W, Chang Y, Xiang Y, Gao Y, Liu S, Song Y, Kwak SH, Shin HD, Park KS, Fall CH, Kim JY, Sham PC, Lam KS, Zheng W, Shu X, Deng H, Ikegami H, Krishnaveni GV, Sanghera DK, Chuang L, Liu L, Hu R, Kim Y, Daimon M, Hotta K, Jia W, Kooner JS, Chambers JC, Chandak GR, Ma RC, Maeda S, Dorajoo R, Yokota M, Takayanagi R, Kato N, Lin X, Loos RJ. Association of genetic variation in FTO with risk of obesity and type 2 diabetes with data from 96,551 East and South Asians. Diabetologia. doi: 10.1007/s00125-011-2370-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pausova Z, Syme C, Abrahamowicz M, Xiao Y, Leonard GT, Perron M, Richer L, Veillette S, Smith GD, Seda O, Tremblay J, Hamet P, Gaudet D, Paus T. A common variant of the FTO gene is associated with not only increased adiposity but also elevated blood pressure in French Canadians. Circulation Cardiovascular genetics. 2009;2:260–9. doi: 10.1161/CIRCGENETICS.109.857359. [DOI] [PubMed] [Google Scholar]
- 34.Lim SY, Ha HS, Kwon HS, Lee JH, Yim HW, Yoon KH, Lee WC, Son HY, Park YM. Factors Associated with Insulin Resistance in a Middle-Aged Non-Obese Rural Population: The Chungju Metabolic Disease Cohort (CMC) Study. Epidemiology and health. 2011;33:e2011009. doi: 10.4178/epih/e2011009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Musso C, Graffigna M, Soutelo J, Honfi M, Ledesma L, Miksztowicz V, Pazos M, Migliano M, Schreier LE, Berg GA. Cardiometabolic risk factors as apolipoprotein B, triglyceride/HDL-cholesterol ratio and C-reactive protein, in adolescents with and without obesity: cross-sectional study in middle class suburban children. Pediatric diabetes. 2011;12:229–34. doi: 10.1111/j.1399-5448.2010.00710.x. [DOI] [PubMed] [Google Scholar]
- 36.Alvarez Caro F, Diaz Martin JJ, Riano Galan I, Perez Solis D, Venta Obaya R, Malaga Guerrero S. [Classic and emergent cardiovascular risk factors in schoolchildren in Asturias] Anales de pediatria. 2011;74:388–95. doi: 10.1016/j.anpedi.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 37.Hunter SM, Frerichs RR, Webber LS, Berenson GS. Social status and cardiovascular disease risk factor variables in children: the Bogalusa Heart Study. Journal of chronic diseases. 1979;32:441–9. doi: 10.1016/0021-9681(79)90104-8. [DOI] [PubMed] [Google Scholar]
- 38.Chen M, Breslow JL, Li W, Leff T. Transcriptional regulation of the apoC-III gene by insulin in diabetic mice: correlation with changes in plasma triglyceride levels. J Lipid Res. 1994;35:1918–24. [PubMed] [Google Scholar]
- 39.Li WW, Dammerman MM, Smith JD, Metzger S, Breslow JL, Leff T. Common genetic variation in the promoter of the human apo CIII gene abolishes regulation by insulin and may contribute to hypertriglyceridemia. J Clin Invest. 1995;96:2601–5. doi: 10.1172/JCI118324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dallongeville J, Meirhaeghe A, Cottel D, Fruchart JC, Amouyel P, Helbecque N. Polymorphisms in the insulin response element of APOC-III gene promoter influence the correlation between insulin and triglycerides or triglyceride-rich lipoproteins in humans. Int J Obes Relat Metab Disord. 2001;25:1012–7. doi: 10.1038/sj.ijo.0801658. [DOI] [PubMed] [Google Scholar]
- 41.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, Wahlstrand B, Hedner T, Corella D, Tai ES, Ordovas JM, Berglund G, Vartiainen E, Jousilahti P, Hedblad B, Taskinen MR, Newton-Cheh C, Salomaa V, Peltonen L, Groop L, Altshuler DM, Orho-Melander M. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–97. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snieder H, van Doornen LJ, Boomsma DI. Dissecting the genetic architecture of lipids, lipoproteins, and apolipoproteins: lessons from twin studies. Arteriosclerosis, thrombosis, and vascular biology. 1999;19:2826–34. doi: 10.1161/01.atv.19.12.2826. [DOI] [PubMed] [Google Scholar]
- 43.Qin S, Kawano K, Bruce C, Lin M, Bisgaier C, Tall AR, Jiang X. Phospholipid transfer protein gene knock-out mice have low high density lipoprotein levels, due to hypercatabolism, and accumulate apoA-IV-rich lamellar lipoproteins. Journal of lipid research. 2000;41:269–76. [PubMed] [Google Scholar]
- 44.Larsen TM, Toubro S, van Baak MA, Gottesdiener KM, Larson P, Saris WH, Astrup A. Effect of a 28-d treatment with L-796568, a novel beta(3)-adrenergic receptor agonist, on energy expenditure and body composition in obese men. The American journal of clinical nutrition. 2002;76:780–8. doi: 10.1093/ajcn/76.4.780. [DOI] [PubMed] [Google Scholar]
- 45.Posadas Romero C. [Some physiopathologic features of metabolic syndrome] Archivos de cardiologia de Mexico. 2007;77 (Suppl 4):S4-42-7. [PubMed] [Google Scholar]
- 46.Vaziri ND. Dyslipidemia of chronic renal failure: the nature, mechanisms, and potential consequences. American journal of physiology Renal physiology. 2006;290:F262–72. doi: 10.1152/ajprenal.00099.2005. [DOI] [PubMed] [Google Scholar]
- 47.Le Goff W, Guerin M, Chapman MJ. Pharmacological modulation of cholesteryl ester transfer protein, a new therapeutic target in atherogenic dyslipidemia. Pharmacology & therapeutics. 2004;101:17–38. doi: 10.1016/j.pharmthera.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 48.Sakai N, Yamashita S, Hirano K, Ishigami M, Arai T, Kobayashi K, Funahashi T, Matsuzawa Y. Decreased affinity of low density lipoprotein (LDL) particles for LDL receptors in patients with cholesteryl ester transfer protein deficiency. European journal of clinical investigation. 1995;25:332–9. doi: 10.1111/j.1365-2362.1995.tb01710.x. [DOI] [PubMed] [Google Scholar]
- 49.Joy T, Hegele RA. The end of the road for CETP inhibitors after torcetrapib? Curr Opin Cardiol. 2009;24:364–71. doi: 10.1097/hco.0b013e32832ac166. [DOI] [PubMed] [Google Scholar]
- 50.Kruit JK, Brunham LR, Verchere CB, Hayden MR. HDL and LDL cholesterol significantly influence beta-cell function in type 2 diabetes mellitus. Current opinion in lipidology. 2010;21:178–85. doi: 10.1097/MOL.0b013e328339387b. [DOI] [PubMed] [Google Scholar]
- 51.Thompson A, Di Angelantonio E, Sarwar N, Erqou S, Saleheen D, Dullaart RP, Keavney B, Ye Z, Danesh J. Association of cholesteryl ester transfer protein genotypes with CETP mass and activity, lipid levels, and coronary risk. JAMA. 2008;299:2777–88. doi: 10.1001/jama.299.23.2777. [DOI] [PubMed] [Google Scholar]
- 52.Nagano M, Yamashita S, Hirano K, Kujiraoka T, Ito M, Sagehashi Y, Hattori H, Nakajima N, Maruyama T, Sakai N, Egashira T, Matsuzawa Y. Point mutation (−69 G-->A) in the promoter region of cholesteryl ester transfer protein gene in Japanese hyperalphalipoproteinemic subjects. Arteriosclerosis, thrombosis, and vascular biology. 2001;21:985–90. doi: 10.1161/01.atv.21.6.985. [DOI] [PubMed] [Google Scholar]
- 53.Yilmaz H, Isbir T, Agachan B, Karaali ZE. Effects of cholesterol ester transfer protein Taq1B gene polymorphism on serum lipoprotein levels in Turkish coronary artery disease patients. Cell biochemistry and function. 2005;23:23–8. doi: 10.1002/cbf.1124. [DOI] [PubMed] [Google Scholar]
- 54.Padmaja N, Ravindra Kumar M, Soya SS, Adithan C. Common variants of Cholesteryl ester transfer protein gene and their association with lipid parameters in healthy volunteers of Tamilian population. Clinica chimica acta; international journal of clinical chemistry. 2007;375:140–6. doi: 10.1016/j.cca.2006.06.031. [DOI] [PubMed] [Google Scholar]
- 55.Rhyne J, Ryan MJ, White C, Chimonas T, Miller M. The two novel CETP mutations Gln87X and Gln165X in a compound heterozygous state are associated with marked hyperalphalipoproteinemia and absence of significant coronary artery disease. Journal of molecular medicine. 2006;84:647–50. doi: 10.1007/s00109-006-0070-4. [DOI] [PubMed] [Google Scholar]
- 56.Bruce CJ, Nishimura RA. Newer advances in the diagnosis and treatment of mitral stenosis. Current problems in cardiology. 1998;23:125–92. doi: 10.1016/s0146-2806(98)80004-2. [DOI] [PubMed] [Google Scholar]
- 57.Vasan RS, Pencina MJ, Robins SJ, Zachariah JP, Kaur G, D’Agostino RB, Ordovas JM. Association of circulating cholesteryl ester transfer protein activity with incidence of cardiovascular disease in the community. Circulation. 2009;120:2414–20. doi: 10.1161/CIRCULATIONAHA.109.872705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nature genetics. 2008;40:161–9. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hegele RA. Plasma lipoproteins: genetic influences and clinical implications. Nature reviews Genetics. 2009;10:109–21. doi: 10.1038/nrg2481. [DOI] [PubMed] [Google Scholar]
- 60.Sanghera DK, Bhatti JS, Bhatti GK, Ralhan SK, Wander GS, Singh JR, Bunker CH, Weeks DE, Kamboh MI, Ferrell RE. The Khatri Sikh Diabetes Study (SDS): study design, methodology, sample collection, and initial results. Human biology. 2006;78:43–63. doi: 10.1353/hub.2006.0027. [DOI] [PubMed] [Google Scholar]
- 61.Schierer A, Been L, Ralhan S, Wander G, Aston C, Sanghera D. Genetic variation in cholesterol ester transfer protein (CETP), serum CETP activity, and coronary artery disease risk in Asian Indian diabetic cohort. Pharmacogenetics and Genomics. 2011 doi: 10.1097/FPC.0b013e32834dc9ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Targher G, Day CP, Bonora E. Risk of cardiovascular disease in patients with nonalcoholic fatty liver disease. N Engl J Med. 2010;363:1341–50. doi: 10.1056/NEJMra0912063. [DOI] [PubMed] [Google Scholar]
- 63.Kotronen A, Yki-Jarvinen H. Fatty liver: a novel component of the metabolic syndrome. Arterioscler Thromb Vasc Biol. 2008;28:27–38. doi: 10.1161/ATVBAHA.107.147538. [DOI] [PubMed] [Google Scholar]
- 64.Brunzell JD, Chait A, Bierman EL. Pathophysiology of lipoprotein transport. Metabolism: clinical and experimental. 1978;27:1109–27. doi: 10.1016/0026-0495(78)90157-9. [DOI] [PubMed] [Google Scholar]
- 65.Chandalia M, Grundy SM, Adams-Huet B, Abate N. Ethnic differences in the frequency of ENPP1/PC1 121Q genetic variant in the Dallas Heart Study cohort. J Diabetes Complications. 2007;21:143–8. doi: 10.1016/j.jdiacomp.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 66.Pan W, Ciociola E, Saraf M, Tumurbaatar B, Tuvdendorj D, Prasad S, Chandalia M, Abate N. Metabolic consequences of ENPP1 overexpression in adipose tissue. Am J Physiol Endocrinol Metab. 2011;301:E901–11. doi: 10.1152/ajpendo.00087.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chandalia M, Grundy SM, Adams-Huet B, Abate N. Ethnic differences in the frequency of ENPP1/PC1 121Q genetic variant in the Dallas Heart Study cohort. Journal of diabetes and its complications. 2007;21:143–8. doi: 10.1016/j.jdiacomp.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 68.Sparks JD, Sparks CE. Overindulgence and metabolic syndrome: is FoxO1 a missing link? The Journal of clinical investigation. 2008;118:2012–5. doi: 10.1172/JCI35693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gambino R, Cassader M, Pagano G, Durazzo M, Musso G. Polymorphism in microsomal triglyceride transfer protein: a link between liver disease and atherogenic postprandial lipid profile in NASH? Hepatology. 2007;45:1097–107. doi: 10.1002/hep.21631. [DOI] [PubMed] [Google Scholar]
- 70.Musso G, Gambino R, Cassader M. Lipoprotein metabolism mediates the association of MTP polymorphism with beta-cell dysfunction in healthy subjects and in nondiabetic normolipidemic patients with nonalcoholic steatohepatitis. The Journal of nutritional biochemistry. 2010;21:834–40. doi: 10.1016/j.jnutbio.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 71.Petersen KF, Dufour S, Hariri A, Nelson-Williams C, Foo JN, Zhang XM, Dziura J, Lifton RP, Shulman GI. Apolipoprotein C3 gene variants in nonalcoholic fatty liver disease. N Engl J Med. 2010;362:1082–9. doi: 10.1056/NEJMoa0907295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee HY, Birkenfeld AL, Jornayvaz FR, Jurczak MJ, Kanda S, Popov V, Frederick DW, Zhang D, Guigni B, Bharadwaj KG, Choi CS, Goldberg IJ, Park JH, Petersen KF, Samuel VT, Shulman GI. Apolipoprotein CIII overexpressing mice are predisposed to diet-induced hepatic steatosis and hepatic insulin resistance. Hepatology. 2011;54:1650–60. doi: 10.1002/hep.24571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taskinen MR, Adiels M, Westerbacka J, Soderlund S, Kahri J, Lundbom N, Lundbom J, Hakkarainen A, Olofsson SO, Orho-Melander M, Boren J. Dual metabolic defects are required to produce hypertriglyceridemia in obese subjects. Arteriosclerosis, thrombosis, and vascular biology. 2011;31:2144–50. doi: 10.1161/ATVBAHA.111.224808. [DOI] [PubMed] [Google Scholar]
- 74.Qin W, Sundaram M, Wang Y, Zhou H, Zhong S, Chang CC, Manhas S, Yao EF, Parks RJ, McFie PJ, Stone SJ, Jiang ZG, Wang C, Figeys D, Jia W, Yao Z. Missense mutation in APOC3 within the C-terminal lipid binding domain of human ApoC-III results in impaired assembly and secretion of triacylglycerol-rich very low density lipoproteins: evidence that ApoC-III plays a major role in the formation of lipid precursors within the microsomal lumen. The Journal of biological chemistry. 2011;286:27769–80. doi: 10.1074/jbc.M110.203679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang CS, McConathy WJ, Kloer HU, Alaupovic P. Modulation of lipoprotein lipase activity by apolipoproteins. Effect of apolipoprotein C-III. The Journal of clinical investigation. 1985;75:384–90. doi: 10.1172/JCI111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cali AM, Zern TL, Taksali SE, de Oliveira AM, Dufour S, Otvos JD, Caprio S. Intrahepatic fat accumulation and alterations in lipoprotein composition in obese adolescents: a perfect proatherogenic state. Diabetes Care. 2007;30:3093–8. doi: 10.2337/dc07-1088. [DOI] [PubMed] [Google Scholar]
- 77.Targher G, Arcaro G. Non-alcoholic fatty liver disease and increased risk of cardiovascular disease. Atherosclerosis. 2007;191:235–40. doi: 10.1016/j.atherosclerosis.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 78.Pacifico L, Cantisani V, Ricci P, Osborn JF, Schiavo E, Anania C, Ferrara E, Dvisic G, Chiesa C. Nonalcoholic fatty liver disease and carotid atherosclerosis in children. Pediatric research. 2008;63:423–7. doi: 10.1203/PDR.0b013e318165b8e7. [DOI] [PubMed] [Google Scholar]
- 79.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, Hobbs HH. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–5. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Romeo S, Sentinelli F, Cambuli VM, Incani M, Congiu T, Matta V, Pilia S, Huang-Doran I, Cossu E, Loche S, Baroni MG. The 148M allele of the PNPLA3 gene is associated with indices of liver damage early in life. J Hepatol. 2010;53:335–8. doi: 10.1016/j.jhep.2010.02.034. [DOI] [PubMed] [Google Scholar]
- 81.Sookoian S, Pirola CJ. Metabolic syndrome: from the genetics to the pathophysiology. Current hypertension reports. 2011;13:149–57. doi: 10.1007/s11906-010-0164-9. [DOI] [PubMed] [Google Scholar]
- 82.Romeo S, Sentinelli F, Dash S, Yeo GS, Savage DB, Leonetti F, Capoccia D, Incani M, Maglio C, Iacovino M, O’Rahilly S, Baroni MG. Morbid obesity exposes the association between PNPLA3 I148M (rs738409) and indices of hepatic injury in individuals of European descent. Int J Obes. 2010;34:190–4. doi: 10.1038/ijo.2009.216. [DOI] [PubMed] [Google Scholar]
- 83.Yuan X, Waterworth D, Perry JR, Lim N, Song K, Chambers JC, Zhang W, Vollenweider P, Stirnadel H, Johnson T, Bergmann S, Beckmann ND, Li Y, Ferrucci L, Melzer D, Hernandez D, Singleton A, Scott J, Elliott P, Waeber G, Cardon L, Frayling TM, Kooner JS, Mooser V. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. American journal of human genetics. 2008;83:520–8. doi: 10.1016/j.ajhg.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sookoian S, Pirola CJ. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. 2011;53:1883–94. doi: 10.1002/hep.24283. [DOI] [PubMed] [Google Scholar]
- 85.Pravenec M, Petretto E. Insight into the genetics of hypertension, a core component of the metabolic syndrome. Curr Opin Clin Nutr Metab Care. 2008;11:393–7. doi: 10.1097/MCO.0b013e32830366f6. [DOI] [PubMed] [Google Scholar]
- 86.Basson J, Simino J, Rao DC. Between candidate genes and whole genomes: time for alternative approaches in blood pressure genetics. Curr Hypertens Rep. 14:46–61. doi: 10.1007/s11906-011-0241-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hopkins PN, Hunt SC. Genetics of hypertension. Genetics in medicine: official journal of the American College of Medical Genetics. 2003;5:413–29. doi: 10.1097/01.gim.0000096375.88710.a6. [DOI] [PubMed] [Google Scholar]
- 88.Elliott P, Stamler J. Evidence on salt and blood pressure is consistent and persuasive. International journal of epidemiology. 2002;31:316–9. discussion 31–2. [PubMed] [Google Scholar]
- 89.Brown IJ, Stamler J, Van Horn L, Robertson CE, Chan Q, Dyer AR, Huang CC, Rodriguez BL, Zhao L, Daviglus ML, Ueshima H, Elliott P. Sugar-sweetened beverage, sugar intake of individuals, and their blood pressure: international study of macro/micronutrients and blood pressure. Hypertension. 2011;57:695–701. doi: 10.1161/HYPERTENSIONAHA.110.165456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu A, Abbasi F, Reaven GM. Adiposity indices in the prediction of metabolic abnormalities associated with cardiovascular disease in non-diabetic adults. Nutrition, metabolism, and cardiovascular diseases: NMCD. 2011;21:553–60. doi: 10.1016/j.numecd.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tobin MD, Tomaszewski M, Braund PS, Hajat C, Raleigh SM, Palmer TM, Caulfield M, Burton PR, Samani NJ. Common variants in genes underlying monogenic hypertension and hypotension and blood pressure in the general population. Hypertension. 2008;51:1658–64. doi: 10.1161/HYPERTENSIONAHA.108.112664. [DOI] [PubMed] [Google Scholar]
- 92.Tobin MD, Raleigh SM, Newhouse S, Braund P, Bodycote C, Ogleby J, Cross D, Gracey J, Hayes S, Smith T, Ridge C, Caulfield M, Sheehan NA, Munroe PB, Burton PR, Samani NJ. Association of WNK1 gene polymorphisms and haplotypes with ambulatory blood pressure in the general population. Circulation. 2005;112:3423–9. doi: 10.1161/CIRCULATIONAHA.105.555474. [DOI] [PubMed] [Google Scholar]
- 93.Tobin MD, Timpson NJ, Wain LV, Ring S, Jones LR, Emmett PM, Palmer TM, Ness AR, Samani NJ, Smith GD, Burton PR. Common variation in the WNK1 gene nd blood pressure in childhood: the Avon Longitudinal Study of Parents and Children. Hypertension. 2008;52:974–9. doi: 10.1161/HYPERTENSIONAHA.108.118414. [DOI] [PubMed] [Google Scholar]
- 94.Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, Pihur V, Vollenweider P, O’Reilly PF, Amin N, Bragg-Gresham JL, Teumer A, Glazer NL, Launer L, Zhao JH, Aulchenko Y, Heath S, Sober S, Parsa A, Luan J, Arora P, Dehghan A, Zhang F, Lucas G, Hicks AA, Jackson AU, Peden JF, Tanaka T, Wild SH, Rudan I, Igl W, Milaneschi Y, Parker AN, Fava C, Chambers JC, Fox ER, Kumari M, Go MJ, van der Harst P, Kao WH, Sjogren M, Vinay DG, Alexander M, Tabara Y, Shaw-Hawkins S, Whincup PH, Liu Y, Shi G, Kuusisto J, Tayo B, Seielstad M, Sim X, Nguyen KD, Lehtimaki T, Matullo G, Wu Y, Gaunt TR, Onland-Moret NC, Cooper MN, Platou CG, Org E, Hardy R, Dahgam S, Palmen J, Vitart V, Braund PS, Kuznetsova T, Uiterwaal CS, Adeyemo A, Palmas W, Campbell H, Ludwig B, Tomaszewski M, Tzoulaki I, Palmer ND, Aspelund T, Garcia M, Chang YP, O’Connell JR, Steinle NI, Grobbee DE, Arking DE, Kardia SL, Morrison AC, Hernandez D, Najjar S, McArdle WL, Hadley D, Brown MJ, Connell JM, Hingorani AD, Day IN, Lawlor DA, Beilby JP, Lawrence RW, Clarke R, Hopewell JC, Ongen H, Dreisbach AW, Li Y, Young JH, Bis JC, Kahonen M, Viikari J, Adair LS, Lee NR, Chen MH, Olden M, Pattaro C, Bolton JA, Kottgen A, Bergmann S, Mooser V, Chaturvedi N, Frayling TM, Islam M, Jafar TH, Erdmann J, Kulkarni SR, Bornstein SR, Grassler J, Groop L, Voight BF, Kettunen J, Howard P, Taylor A, Guarrera S, Ricceri F, Emilsson V, Plump A, Barroso I, Khaw KT, Weder AB, Hunt SC, Sun YV, Bergman RN, Collins FS, Bonnycastle LL, Scott LJ, Stringham HM, Peltonen L, Perola M, Vartiainen E, Brand SM, Staessen JA, Wang TJ, Burton PR, Artigas MS, Dong Y, Snieder H, Wang X, Zhu H, Lohman KK, Rudock ME, Heckbert SR, Smith NL, Wiggins KL, Doumatey A, Shriner D, Veldre G, Viigimaa M, Kinra S, Prabhakaran D, Tripathy V, Langefeld CD, Rosengren A, Thelle DS, Corsi AM, Singleton A, Forrester T, Hilton G, McKenzie CA, Salako T, Iwai N, Kita Y, Ogihara T, Ohkubo T, Okamura T, Ueshima H, Umemura S, Eyheramendy S, Meitinger T, Wichmann HE, Cho YS, Kim HL, Lee JY, Scott J, Sehmi JS, Zhang W, Hedblad B, Nilsson P, Smith GD, Wong A, Narisu N, Stancakova A, Raffel LJ, Yao J, Kathiresan S, O’Donnell CJ, Schwartz SM, Ikram MA, Longstreth WT, Jr, Mosley TH, Seshadri S, Shrine NR, Wain LV, Morken MA, Swift AJ, Laitinen J, Prokopenko I, Zitting P, Cooper JA, Humphries SE, Danesh J, Rasheed A, Goel A, Hamsten A, Watkins H, Bakker SJ, van Gilst WH, Janipalli CS, Mani KR, Yajnik CS, Hofman A, Mattace-Raso FU, Oostra BA, Demirkan A, Isaacs A, Rivadeneira F, Lakatta EG, Orru M, Scuteri A, Ala-Korpela M, Kangas AJ, Lyytikainen LP, Soininen P, Tukiainen T, Wurtz P, Ong RT, Dorr M, Kroemer HK, Volker U, Volzke H, Galan P, Hercberg S, Lathrop M, Zelenika D, Deloukas P, Mangino M, Spector TD, Zhai G, Meschia JF, Nalls MA, Sharma P, Terzic J, Kumar MV, Denniff M, Zukowska-Szczechowska E, Wagenknecht LE, Fowkes FG, Charchar FJ, Schwarz PE, Hayward C, Guo X, Rotimi C, Bots ML, Brand E, Samani NJ, Polasek O, Talmud PJ, Nyberg F, Kuh D, Laan M, Hveem K, Palmer LJ, van der Schouw YT, Casas JP, Mohlke KL, Vineis P, Raitakari O, Ganesh SK, Wong TY, Tai ES, Cooper RS, Laakso M, Rao DC, Harris TB, Morris RW, Dominiczak AF, Kivimaki M, Marmot MG, Miki T, Saleheen D, Chandak GR, Coresh J, Navis G, Salomaa V, Han BG, Zhu X, Kooner JS, Melander O, Ridker PM, Bandinelli S, Gyllensten UB, Wright AF, Wilson JF, Ferrucci L, Farrall M, Tuomilehto J, Pramstaller PP, Elosua R, Soranzo N, Sijbrands EJ, Altshuler D, Loos RJ, Shuldiner AR, Gieger C, Meneton P, Uitterlinden AG, Wareham NJ, Gudnason V, Rotter JI, Rettig R, Uda M, Strachan DP, Witteman JC, Hartikainen AL, Beckmann JS, Boerwinkle E, Vasan RS, Boehnke M, Larson MG, Jarvelin MR, Psaty BM, Abecasis GR, Chakravarti A, Elliott P, van Duijn CM, Newton-Cheh C, Levy D, Caulfield MJ, Johnson T, Tang H, Knowles J, Hlatky M, Fortmann S, Assimes TL, Quertermous T, Go A, Iribarren C, Absher D, Risch N, Myers R, Sidney S, Ziegler A, Schillert A, Bickel C, Sinning C, Rupprecht HJ, Lackner K, Wild P, Schnabel R, Blankenberg S, Zeller T, Munzel T, Perret C, Cambien F, Tiret L, Nicaud V, Proust C, Uitterlinden A, van Duijn C, Whitteman J, Cupples LA, Demissie-Banjaw S, Ramachandran V, Smith A, Folsom A, Morrison A, Chen IY, Bis J, Volcik K, Rice K, Taylor KD, Marciante K, Smith N, Glazer N, Heckbert S, Harris T, Lumley T, Kong A, Thorleifsson G, Thorgeirsson G, Holm H, Gulcher JR, Stefansson K, Andersen K, Gretarsdottir S, Thorsteinsdottir U, Preuss M, Schreiber S, Konig IR, Lieb W, Hengstenberg C, Schunkert H, Fischer M, Grosshennig A, Medack A, Stark K, Linsel-Nitschke P, Bruse P, Aherrahrou Z, Peters A, Loley C, Willenborg C, Nahrstedt J, Freyer J, Gulde S, Doering A, Meisinger C, Klopp N, Illig T, Meinitzer A, Tomaschitz A, Halperin E, Dobnig H, Scharnagl H, Kleber M, Laaksonen R, Pilz S, Grammer TB, Stojakovic T, Renner W, Marz W, Bohm BO, Winkelmann BR, Winkler K, Hoffmann MM, Siscovick DS, Musunuru K, Barbalic M, Guiducci C, Burtt N, Gabriel SB, Stewart AF, Wells GA, Chen L, Jarinova O, Roberts R, McPherson R, Dandona S, Pichard AD, Rader DJ, Devaney J, Lindsay JM, Kent KM, Qu L, Satler L, Burnett MS, Li M, Reilly MP, Wilensky R, Waksman R, Epstein S, Matthai W, Knouff CW, Waterworth DM, Hakonarson HH, Walker MC, Hall AS, Balmforth AJ, Wright BJ, Nelson C, Thompson JR, Ball SG, Felix JF, Demissie S, Loehr LR, Rosamond WD, Folsom AR, Benjamin E, Aulchenko YS, Haritunians T, Couper D, Murabito J, Wang YA, Stricker BH, Gottdiener JS, Chang PP, Willerson JT, Boger CA, Fuchsberger C, Gao X, Yang Q, Schmidt H, Ketkar S, Pare G, Atkinson EJ, Lohman K, Cornelis MC, Probst-Hensch NM, Kronenberg F, Tonjes A, Eiriksdottir G, Launer LJ, Rampersaud E, Mitchell BD, Struchalin M, Cavalieri M, Giallauria F, Metter J, de Boer J, Siscovick D, Zillikens MC, Feitosa M, Province M, de Andrade M, Turner ST, Wild PS, Schnabel RB, Wilde S, Munzel TF, Leak TS, Koenig W, Zgaga L, Zemunik T, Kolcic I, Minelli C, Hu FB, Johansson A, Zaboli G, Ellinghaus D, Imboden M, Nitsch D, Brandstatter A, Kollerits B, Kedenko L, Magi R, Stumvoll M, Kovacs P, Boban M, Campbell S, Endlich K, Nauck M, Badola S, Curhan GC, Franke A, Rochat T, Paulweber B, Wang W, Schmidt R, Shlipak MG, Borecki I, Kramer BK, Gyllensten U, Hastie N, Heid IM, Fox CS, Felix SB, Watzinger N, Homuth G, Aragam J, Zweiker R, Lind L, Rodeheffer RJ, Greiser KH, Deckers JW, Stritzke J, Lackner KJ, Ingelsson E, Kullo I, Haerting J, Reffelmann T, Redfield MM, Werdan K, Mitchell GF, Arnett DK, Blettner M, Friedrich N, Benjamin EJ, Lord GM, Gale DP, Wass MN, Ahmadi KR, Beckmann J, Bilo HJ, Cook HT, Cotlarciuc I, Davey Smith G, de Silva R, Deng G, Devuyst O, Dikkeschei LD, Dimkovic N, Dockrell M, Dominiczak A, Ebrahim S, Eggermann T, Floege J, Forouhi NG, Gansevoort RT, Han X, Homan van der Heide JJ, Hepkema BG, Hernandez-Fuentes M, Hypponen E, de Jong PE, Kleefstra N, Lagou V, Lapsley M, Luttropp K, Marechal C, Nordfors L, Penninx BW, Perucha E, Pouta A, Roderick PJ, Ruokonen A, Sanna S, Schalling M, Schlessinger D, Schlieper G, Seelen MA, Smit JH, Stenvinkel P, Sternberg MJ, Swaminathan R, Ubink-Veltmaat LJ, Wallace C, Waterworth D, Zerres K, Waeber G, Maxwell PH, McCarthy MI, Lightstone L. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–9. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cutler JA, Sorlie PD, Wolz M, Thom T, Fields LE, Roccella EJ. Trends in hypertension prevalence, awareness, treatment, and control rates in United States adults between 1988–1994 and 1999–2004. Hypertension. 2008;52:818–27. doi: 10.1161/HYPERTENSIONAHA.108.113357. [DOI] [PubMed] [Google Scholar]
- 96.Fox ER, Young JH, Li Y, Dreisbach AW, Keating BJ, Musani SK, Liu K, Morrison AC, Ganesh S, Kutlar A, Ramachandran VS, Polak JF, Fabsitz RR, Dries DL, Farlow DN, Redline S, Adeyemo A, Hirschorn JN, Sun YV, Wyatt SB, Penman AD, Palmas W, Rotter JI, Townsend RR, Doumatey AP, Tayo BO, Mosley TH, Jr, Lyon HN, Kang SJ, Rotimi CN, Cooper RS, Franceschini N, Curb JD, Martin LW, Eaton CB, Kardia SL, Taylor HA, Caulfield MJ, Ehret GB, Johnson T, Chakravarti A, Zhu X, Levy D, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, Pihur V, Vollenweider P, O’Reilly PF, Amin N, Bragg-Gresham JL, Teumer A, Glazer NL, Launer L, Zhao JH, Aulchenko Y, Heath S, Sober S, Parsa A, Luan J, Arora P, Dehghan A, Zhang F, Lucas G, Hicks AA, Jackson AU, Peden JF, Tanaka T, Wild SH, Rudan I, Igl W, Milaneschi Y, Parker AN, Fava C, Chambers JC, Kumari M, Go MJ, van der Harst P, Kao WH, Sjogren M, Vinay DG, Alexander M, Tabara Y, Shaw-Hawkins S, Whincup PH, Liu Y, Shi G, Kuusisto J, Seielstad M, Sim X, Nguyen KD, Lehtimaki T, Matullo G, Wu Y, Gaunt TR, Onland-Moret NC, Cooper MN, Platou CG, Org E, Hardy R, Dahgam S, Palmen J, Vitart V, Braund PS, Kuznetsova T, Uiterwaal CS, Campbell H, Ludwig B, Tomaszewski M, Tzoulaki I, Palmer ND, Aspelund T, Garcia M, Chang YP, O’Connell JR, Steinle NI, Grobbee DE, Arking DE, Hernandez D, Najjar S, McArdle WL, Hadley D, Brown MJ, Connell JM, Hingorani AD, Day IN, Lawlor DA, Beilby JP, Lawrence RW, Clarke R, Collins R, Hopewell JC, Ongen H, Bis JC, Kahonen M, Viikari J, Adair LS, Lee NR, Chen MH, Olden M, Pattaro C, Hoffman Bolton JA, Kottgen A, Bergmann S, Mooser V, Chaturvedi N, Frayling TM, Islam M, Jafar TH, Erdmann J, Kulkarni SR, Bornstein SR, Grassler J, Groop L, Voight BF, Kettunen J, Howard P, Taylor A, Guarrera S, Ricceri F, Emilsson V, Plump A, Barroso I, Khaw KT, Weder AB, Hunt SC, Bergman RN, Collins FS, Bonnycastle LL, Scott LJ, Stringham HM, Peltonen L, Perola M, Vartiainen E, Brand SM, Staessen JA, Wang TJ, Burton PR, Soler Artigas M, Dong Y, Snieder H, Wang X, Zhu H, Lohman KK, Rudock ME, Heckbert SR, Smith NL, Wiggins KL, Shriner D, Veldre G, Viigimaa M, Kinra S, Prabhakaran D, Tripathy V, Langefeld CD, Rosengren A, Thelle DS, Corsi AM, Singleton A, Forrester T, Hilton G, McKenzie CA, Salako T, Iwai N, Kita Y, Ogihara T, Ohkubo T, Okamura T, Ueshima H, Umemura S, Eyheramendy S, Meitinger T, Wichmann HE, Cho YS, Kim HL, Lee JY, Scott J, Sehmi JS, Zhang W, Hedblad B, Nilsson P, Smith GD, Wong A, Narisu N, Stancakova A, Raffel LJ, Yao J, Kathiresan S, O’Donnell C, Schwartz SM, Ikram MA, Longstreth WT, Jr, Seshadri S, Shrine NR, Wain LV, Morken MA, Swift AJ, Laitinen J, Prokopenko I, Zitting P, Cooper JA, Humphries SE, Danesh J, Rasheed A, Goel A, Hamsten A, Watkins H, Bakker SJ, van Gilst WH, Janipalli C, Mani KR, Yajnik CS, Hofman A, Mattace-Raso FU, Oostra BA, Demirkan A, Isaacs A, Rivadeneira F, Lakatta EG, Orru M, Scuteri A, Ala-Korpela M, Kangas AJ, Lyytikainen LP, Soininen P, Tukiainen T, Wurz P, Ong RT, Dorr M, Kroemer HK, Volker U, Volzke H, Galan P, Hercberg S, Lathrop M, Zelenika D, Deloukas P, Mangino M, Spector TD, Zhai G, Meschia JF, Nalls MA, Sharma P, Terzic J, Kumar MJ, Denniff M, Zukowska-Szczechowska E, Wagenknecht LE, Fowkes FG, Charchar FJ, Schwarz PE, Hayward C, Guo X, Bots ML, Brand E, Samani N, Polasek O, Talmud PJ, Nyberg F, Kuh D, Laan M, Hveem K, Palmer LJ, van der Schouw YT, Casas JP, Mohlke KL, Vineis P, Raitakari O, Wong TY, Tai ES, Laakso M, Rao DC, Harris TB, Morris RW, Dominiczak AF, Kivimaki M, Marmot MG, Miki T, Saleheen D, Chandak GR, Coresh J, Navis G, Salomaa V, Han BG, Kooner JS, Melander O, Ridker PM, Bandinelli S, Gyllensten UB, Wright AF, Wilson JF, Ferrucci L, Farrall M, Tuomilehto J, Pramstaller PP, Elosua R, Soranzo N, Sijbrands EJ, Altshuler D, Loos RJ, Shuldiner AR, Gieger C, Meneton P, Uitterlinden AG, Wareham NJ, Gudnason V, Rettig R, Uda M, Strachan DP, Witteman JC, Hartikainen AL, Beckmann JS, Boerwinkle E, Boehnke M, Larson MG, Jarvelin MR, Psaty BM, Abecasis GR, Elliott P, van Duijn CM, Newton-Cheh C. Association of genetic variation with systolic and diastolic blood pressure among African Americans: the Candidate Gene Association Resource study. Hum Mol Genet. 2011;20:2273–84. doi: 10.1093/hmg/ddr092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rutti S, Ehses JA, Sibler RA, Prazak R, Rohrer L, Georgopoulos S, Meier DT, Niclauss N, Berney T, Donath MY, von Eckardstein A. Low- and high-density lipoproteins modulate function, apoptosis, and proliferation of primary human and murine pancreatic beta-cells. Endocrinology. 2009;150:4521–30. doi: 10.1210/en.2009-0252. [DOI] [PubMed] [Google Scholar]
- 98.Kruit JK, Kremer PH, Dai L, Tang R, Ruddle P, de Haan W, Brunham LR, Verchere CB, Hayden MR. Cholesterol efflux via ATP-binding cassette transporter A1 (ABCA1) and cholesterol uptake via the LDL receptor influences cholesterol-induced impairment of beta cell function in mice. Diabetologia. 2010;53:1110–9. doi: 10.1007/s00125-010-1691-2. [DOI] [PubMed] [Google Scholar]
- 99.Hao M, Bogan JS. Cholesterol regulates glucose-stimulated insulin secretion through phosphatidylinositol 4,5-bisphosphate. The Journal of biological chemistry. 2009;284:29489–98. doi: 10.1074/jbc.M109.038034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schafer SA, Machicao F, Fritsche A, Haring HU, Kantartzis K. New type 2 diabetes risk genes provide new insights in insulin secretion mechanisms. Diabetes Res Clin Pract. 93(Suppl 1):S9–24. doi: 10.1016/S0168-8227(11)70008-0. [DOI] [PubMed] [Google Scholar]
- 101.De Silva NM, Frayling TM. Novel biological insights emerging from genetic studies of type 2 diabetes and related metabolic traits. Curr Opin Lipidol. 2010;21:44–50. doi: 10.1097/MOL.0b013e328334fdb6. [DOI] [PubMed] [Google Scholar]
- 102.Takeuchi F, Katsuya T, Chakrewarthy S, Yamamoto K, Fujioka A, Serizawa M, Fujisawa T, Nakashima E, Ohnaka K, Ikegami H, Sugiyama T, Nabika T, Kasturiratne A, Yamaguchi S, Kono S, Takayanagi R, Yamori Y, Kobayashi S, Ogihara T, de Silva A, Wickremasinghe R, Kato N. Common variants at the GCK, GCKR, G6PC2-ABCB11 and MTNR1B loci are associated with fasting glucose in two Asian populations. Diabetologia. 53:299–308. doi: 10.1007/s00125-009-1595-1. [DOI] [PubMed] [Google Scholar]
- 103.Reiling E, van ’t Riet E, Groenewoud MJ, Welschen LM, van Hove EC, Nijpels G, Maassen JA, Dekker JM, t Hart LM. Combined effects of single-nucleotide polymorphisms in GCK, GCKR, G6PC2 and MTNR1B on fasting plasma glucose and type 2 diabetes risk. Diabetologia. 2009;52:1866–70. doi: 10.1007/s00125-009-1413-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen WM, Erdos MR, Jackson AU, Saxena R, Sanna S, Silver KD, Timpson NJ, Hansen T, Orru M, Grazia Piras M, Bonnycastle LL, Willer CJ, Lyssenko V, Shen H, Kuusisto J, Ebrahim S, Sestu N, Duren WL, Spada MC, Stringham HM, Scott LJ, Olla N, Swift AJ, Najjar S, Mitchell BD, Lawlor DA, Smith GD, Ben-Shlomo Y, Andersen G, Borch-Johnsen K, Jorgensen T, Saramies J, Valle TT, Buchanan TA, Shuldiner AR, Lakatta E, Bergman RN, Uda M, Tuomilehto J, Pedersen O, Cao A, Groop L, Mohlke KL, Laakso M, Schlessinger D, Collins FS, Altshuler D, Abecasis GR, Boehnke M, Scuteri A, Watanabe RM. Variations in the G6PC2/ABCB11 genomic region are associated with fasting glucose levels. J Clin Invest. 2008;118:2620–8. doi: 10.1172/JCI34566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sanghera DK, Nath SK, Ortega L, Gambarelli M, Kim-Howard X, Singh JR, Ralhan SK, Wander GS, Mehra NK, Mulvihill JJ, Kamboh MI. TCF7L2 polymorphisms are associated with type 2 diabetes in Khatri Sikhs from North India: genetic variation affects lipid levels. Ann Hum Genet. 2008;72:499–509. doi: 10.1111/j.1469-1809.2008.00443.x. [DOI] [PubMed] [Google Scholar]
- 106.Been LF, Ralhan S, Wander GS, Mehra NK, Singh J, Mulvihill JJ, Aston CE, Sanghera DK. Variants in KCNQ1 increase type II diabetes susceptibility in South Asians: a study of 3,310 subjects from India and the US. BMC Med Genet. 12:18. doi: 10.1186/1471-2350-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sanghera DK, Ortega L, Han S, Singh J, Ralhan SK, Wander GS, Mehra NK, Mulvihill JJ, Ferrell RE, Nath SK, Kamboh MI. Impact of nine common type 2 diabetes risk polymorphisms in Asian Indian Sikhs: PPARG2 (Pro12Ala), IGF2BP2, TCF7L2 and FTO variants confer a significant risk. BMC Med Genet. 2008;9:59. doi: 10.1186/1471-2350-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sanghera DK, Been LF, Ralhan S, Wander GS, Mehra NK, Singh JR, Ferrell RE, Kamboh MI, Aston CE. Genome-wide linkage scan to identify Loci associated with type 2 diabetes and blood lipid phenotypes in the sikh diabetes study. PLoS One. 6:e21188. doi: 10.1371/journal.pone.0021188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lampidonis AD, Rogdakis E, Voutsinas GE, Stravopodis DJ. The resurgence of Hormone-Sensitive Lipase (HSL) in mammalian lipolysis. Gene. 2011;477:1–11. doi: 10.1016/j.gene.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 110.Juhola J, Magnussen CG, Viikari JS, Kahonen M, Hutri-Kahonen N, Jula A, Lehtimaki T, Akerblom HK, Pietikainen M, Laitinen T, Jokinen E, Taittonen L, Raitakari OT, Juonala M. Tracking of serum lipid levels, blood pressure, and body mass index from childhood to adulthood: the Cardiovascular Risk in Young Finns Study. The Journal of pediatrics. 2011;159:584–90. doi: 10.1016/j.jpeds.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 111.Juonala M, Magnussen CG, Venn A, Dwyer T, Burns TL, Davis PH, Chen W, Srinivasan SR, Daniels SR, Kahonen M, Laitinen T, Taittonen L, Berenson GS, Viikari JS, Raitakari OT. Influence of age on associations between childhood risk factors and carotid intima-media thickness in adulthood: the Cardiovascular Risk in Young Finns Study, the Childhood Determinants of Adult Health Study, the Bogalusa Heart Study, and the Muscatine Study for the International Childhood Cardiovascular Cohort (i3C) Consortium. Circulation. 2010;122:2514–20. doi: 10.1161/CIRCULATIONAHA.110.966465. [DOI] [PubMed] [Google Scholar]
- 112.Hales CN, Barker DJ. Type 2 (non-insulin-dependent) diabetes mellitus: the thrifty phenotype hypothesis. Diabetologia. 1992;35:595–601. doi: 10.1007/BF00400248. [DOI] [PubMed] [Google Scholar]
- 113.Tamashiro KL, Moran TH. Perinatal environment and its influences on metabolic programming of offspring. Physiol Behav. 100:560–6. doi: 10.1016/j.physbeh.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ainge H, Thompson C, Ozanne SE, Rooney KB. A systematic review on animal models of maternal high fat feeding and offspring glycaemic control. Int J Obes (Lond) 35:325–35. doi: 10.1038/ijo.2010.149. [DOI] [PubMed] [Google Scholar]
- 115.Sebert S, Sharkey D, Budge H, Symonds ME. The early programming of metabolic health: is epigenetic setting the missing link? Am J Clin Nutr. 94:1953S–8S. doi: 10.3945/ajcn.110.001040. [DOI] [PubMed] [Google Scholar]
- 116.Florez JC. Genetic susceptibility to type 2 diabetes and implications for therapy. J Diabetes Sci Technol. 2009;3:690–6. doi: 10.1177/193229680900300413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Doria A, Patti ME, Kahn CR. The emerging genetic architecture of type 2 diabetes. Cell Metab. 2008;8:186–200. doi: 10.1016/j.cmet.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Small KS, Hedman AK, Grundberg E, Nica AC, Thorleifsson G, Kong A, Thorsteindottir U, Shin SY, Richards HB, Soranzo N, Ahmadi KR, Lindgren CM, Stefansson K, Dermitzakis ET, Deloukas P, Spector TD, McCarthy MI. Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat Genet. 2011;43:561–4. doi: 10.1038/ng.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Monda KL, North KE, Hunt SC, Rao DC, Province MA, Kraja AT. The genetics of obesity and the metabolic syndrome. Endocr Metab Immune Disord Drug Targets. 2010;10:86–108. doi: 10.2174/187153010791213100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Maury E, Ramsey KM, Bass J. Circadian rhythms and metabolic syndrome: from experimental genetics to human disease. Circ Res. 2010;106:447–62. doi: 10.1161/CIRCRESAHA.109.208355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Reaven GM. Banting lecture 1988. Role of insulin resistance in human disease. Diabetes. 1988;37:1595–607. doi: 10.2337/diab.37.12.1595. [DOI] [PubMed] [Google Scholar]
- 122.Hanley AJ, Karter AJ, Festa A, D’Agostino R, Jr, Wagenknecht LE, Savage P, Tracy RP, Saad MF, Haffner S. Factor analysis of metabolic syndrome using directly measured insulin sensitivity: The Insulin Resistance Atherosclerosis Study. Diabetes. 2002;51:2642–7. doi: 10.2337/diabetes.51.8.2642. [DOI] [PubMed] [Google Scholar]
- 123.Haffner SM, D’Agostino R, Jr, Festa A, Bergman RN, Mykkanen L, Karter A, Saad MF, Wagenknecht LE. Low insulin sensitivity (S(i) = 0) in diabetic and nondiabetic subjects in the insulin resistance atherosclerosis study: is it associated with components of the metabolic syndrome and nontraditional risk factors? Diabetes Care. 2003;26:2796–803. doi: 10.2337/diacare.26.10.2796. [DOI] [PubMed] [Google Scholar]
- 124.Salazar MR, Carbajal HA, Espeche WG, Dulbecco CA, Aizpurua M, Marillet AG, Echeverria RF, Reaven GM. Relationships among insulin resistance, obesity, diagnosis of the metabolic syndrome and cardio-metabolic risk. Diab Vasc Dis Res. 2011;8:109–16. doi: 10.1177/1479164111403170. [DOI] [PubMed] [Google Scholar]
- 125.Cheal KL, Abbasi F, Lamendola C, McLaughlin T, Reaven GM, Ford ES. Relationship to insulin resistance of the adult treatment panel III diagnostic criteria for identification of the metabolic syndrome. Diabetes. 2004;53:1195–200. doi: 10.2337/diabetes.53.5.1195. [DOI] [PubMed] [Google Scholar]
- 126.Ford ES, Schulze MB, Pischon T, Bergmann MM, Joost HG, Boeing H. Metabolic syndrome and risk of incident diabetes: findings from the European Prospective Investigation into Cancer and Nutrition-Potsdam Study. Cardiovasc Diabetol. 2008;7:35. doi: 10.1186/1475-2840-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bruning JC, Winnay J, Bonner-Weir S, Taylor SI, Accili D, Kahn CR. Development of a novel polygenic model of NIDDM in mice heterozygous for IR and IRS-1 null alleles. Cell. 1997;88:561–72. doi: 10.1016/s0092-8674(00)81896-6. [DOI] [PubMed] [Google Scholar]
- 128.Sinclair KD, Allegrucci C, Singh R, Gardner DS, Sebastian S, Bispham J, Thurston A, Huntley JF, Rees WD, Maloney CA, Lea RG, Craigon J, McEvoy TG, Young LE. DNA methylation, insulin resistance, and blood pressure in offspring determined by maternal periconceptional B vitamin and methionine status. Proc Natl Acad Sci U S A. 2007;104:19351–6. doi: 10.1073/pnas.0707258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gemma C, Sookoian S, Dieuzeide G, Garcia SI, Gianotti TF, Gonzalez CD, Pirola CJ. Methylation of TFAM gene promoter in peripheral white blood cells is associated with insulin resistance in adolescents. Mol Genet Metab. 2010;100:83–7. doi: 10.1016/j.ymgme.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 130.Sookoian S, Rosselli MS, Gemma C, Burgueno AL, Fernandez Gianotti T, Castano GO, Pirola CJ. Epigenetic regulation of insulin resistance in nonalcoholic fatty liver disease: impact of liver methylation of the peroxisome proliferator-activated receptor gamma coactivator 1alpha promoter. Hepatology. 2010;52:1992–2000. doi: 10.1002/hep.23927. [DOI] [PubMed] [Google Scholar]
- 131.Lira VA, Benton CR, Yan Z, Bonen A. PGC-1alpha regulation by exercise training and its influences on muscle function and insulin sensitivity. Am J Physiol Endocrinol Metab. 2010;299:E145–61. doi: 10.1152/ajpendo.00755.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wamil M, Seckl JR. Inhibition of 11beta-hydroxysteroid dehydrogenase type 1 as a promising therapeutic target. Drug Discov Today. 2007;12:504–20. doi: 10.1016/j.drudis.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 133.Masuzaki H, Paterson J, Shinyama H, Morton NM, Mullins JJ, Seckl JR, Flier JS. A transgenic model of visceral obesity and the metabolic syndrome. Science. 2001;294:2166–70. doi: 10.1126/science.1066285. [DOI] [PubMed] [Google Scholar]
- 134.Morton NM, Holmes MC, Fievet C, Staels B, Tailleux A, Mullins JJ, Seckl JR. Improved lipid and lipoprotein profile, hepatic insulin sensitivity, and glucose tolerance in 11beta-hydroxysteroid dehydrogenase type 1 null mice. J Biol Chem. 2001;276:41293–300. doi: 10.1074/jbc.M103676200. [DOI] [PubMed] [Google Scholar]
- 135.Lindsay RS, Wake DJ, Nair S, Bunt J, Livingstone DE, Permana PA, Tataranni PA, Walker BR. Subcutaneous adipose 11 beta-hydroxysteroid dehydrogenase type 1 activity and messenger ribonucleic acid levels are associated with adiposity and insulinemia in Pima Indians and Caucasians. J Clin Endocrinol Metab. 2003;88:2738–44. doi: 10.1210/jc.2002-030017. [DOI] [PubMed] [Google Scholar]
- 136.Feng Y, Huang SL, Dou W, Zhang S, Chen JH, Shen Y, Shen JH, Leng Y. Emodin, a natural product, selectively inhibits 11beta-hydroxysteroid dehydrogenase type 1 and ameliorates metabolic disorder in diet-induced obese mice. Br J Pharmacol. 2010;161:113–26. doi: 10.1111/j.1476-5381.2010.00826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kannisto K, Pietilainen KH, Ehrenborg E, Rissanen A, Kaprio J, Hamsten A, Yki-Jarvinen H. Overexpression of 11beta-hydroxysteroid dehydrogenase-1 in adipose tissue is associated with acquired obesity and features of insulin resistance: studies in young adult monozygotic twins. J Clin Endocrinol Metab. 2004;89:4414–21. doi: 10.1210/jc.2004-0153. [DOI] [PubMed] [Google Scholar]
- 138.Robitaille J, Brouillette C, Houde A, Despres JP, Tchernof A, Vohl MC. Molecular screening of the 11beta-HSD1 gene in men characterized by the metabolic syndrome. Obes Res. 2004;12:1570–5. doi: 10.1038/oby.2004.196. [DOI] [PubMed] [Google Scholar]
- 139.Nair S, Lee YH, Lindsay RS, Walker BR, Tataranni PA, Bogardus C, Baier LJ, Permana PA. 11beta-Hydroxysteroid dehydrogenase Type 1: genetic polymorphisms are associated with Type 2 diabetes in Pima Indians independently of obesity and expression in adipocyte and muscle. Diabetologia. 2004;47:1088–95. doi: 10.1007/s00125-004-1407-6. [DOI] [PubMed] [Google Scholar]
- 140.Franks PW, Knowler WC, Nair S, Koska J, Lee YH, Lindsay RS, Walker BR, Looker HC, Permana PA, Tataranni PA, Hanson RL. Interaction between an 11betaHSD1 gene variant and birth era modifies the risk of hypertension in Pima Indians. Hypertension. 2004;44:681–8. doi: 10.1161/01.HYP.0000144294.28985.d5. [DOI] [PubMed] [Google Scholar]
- 141.Olza J, Gil-Campos M, Leis R, Ruperez AI, Tojo R, Canete R, Gil A, Aguilera CM. A gene variant of 11beta-hydroxysteroid dehydrogenase type 1 is associated with obesity in children. Int J Obes (Lond) 2012 doi: 10.1038/ijo.2012.4. [DOI] [PubMed] [Google Scholar]
- 142.Hardie DG. AMPK: a key regulator of energy balance in the single cell and the whole organism. Int J Obes (Lond) 2008;32 (Suppl 4):S7–12. doi: 10.1038/ijo.2008.116. [DOI] [PubMed] [Google Scholar]
- 143.Zhang BB, Zhou G, Li C. AMPK: an emerging drug target for diabetes and the metabolic syndrome. Cell Metab. 2009;9:407–16. doi: 10.1016/j.cmet.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 144.Shirai T, Inoue E, Ishimi Y, Yamauchi J. AICAR response element binding protein (AREBP), a key modulator of hepatic glucose production regulated by AMPK in vivo. Biochem Biophys Res Commun. 2011;414:287–91. doi: 10.1016/j.bbrc.2011.08.120. [DOI] [PubMed] [Google Scholar]
- 145.Zhou K, Bellenguez C, Spencer CC, Bennett AJ, Coleman RL, Tavendale R, Hawley SA, Donnelly LA, Schofield C, Groves CJ, Burch L, Carr F, Strange A, Freeman C, Blackwell JM, Bramon E, Brown MA, Casas JP, Corvin A, Craddock N, Deloukas P, Dronov S, Duncanson A, Edkins S, Gray E, Hunt S, Jankowski J, Langford C, Markus HS, Mathew CG, Plomin R, Rautanen A, Sawcer SJ, Samani NJ, Trembath R, Viswanathan AC, Wood NW, Harries LW, Hattersley AT, Doney AS, Colhoun H, Morris AD, Sutherland C, Hardie DG, Peltonen L, McCarthy MI, Holman RR, Palmer CN, Donnelly P, Pearson ER. Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nat Genet. 2011;43:117–20. doi: 10.1038/ng.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shu Y, Sheardown SA, Brown C, Owen RP, Zhang S, Castro RA, Ianculescu AG, Yue L, Lo JC, Burchard EG, Brett CM, Giacomini KM. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J Clin Invest. 2007;117:1422–31. doi: 10.1172/JCI30558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Becker ML, Visser LE, van Schaik RH, Hofman A, Uitterlinden AG, Stricker BH. Genetic variation in the multidrug and toxin extrusion 1 transporter protein influences the glucose-lowering effect of metformin in patients with diabetes: a preliminary study. Diabetes. 2009;58:745–9. doi: 10.2337/db08-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Becker ML, Visser LE, van Schaik RH, Hofman A, Uitterlinden AG, Stricker BH. Interaction between polymorphisms in the OCT1 and MATE1 transporter and metformin response. Pharmacogenet Genomics. 2010;20:38–44. doi: 10.1097/FPC.0b013e328333bb11. [DOI] [PubMed] [Google Scholar]
- 149.Jablonski KA, McAteer JB, de Bakker PI, Franks PW, Pollin TI, Hanson RL, Saxena R, Fowler S, Shuldiner AR, Knowler WC, Altshuler D, Florez JC. Common variants in 40 genes assessed for diabetes incidence and response to metformin and lifestyle intervention in the diabetes prevention program. Diabetes. 2010;59:2672–81. doi: 10.2337/db10-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Distefano JK, Watanabe RM. Pharmacogenetics of Anti-Diabetes Drugs. Pharmaceuticals (Basel) 2010;3:2610–46. doi: 10.3390/ph3082610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wolford JK, Yeatts KA, Dhanjal SK, Black MH, Xiang AH, Buchanan TA, Watanabe RM. Sequence variation in PPARG may underlie differential response to troglitazone. Diabetes. 2005;54:3319–25. doi: 10.2337/diabetes.54.11.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gloyn AL, Weedon MN, Owen KR, Turner MJ, Knight BA, Hitman G, Walker M, Levy JC, Sampson M, Halford S, McCarthy MI, Hattersley AT, Frayling TM. Large-scale association studies of variants in genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) confirm that the KCNJ11 E23K variant is associated with type 2 diabetes. Diabetes. 2003;52:568–72. doi: 10.2337/diabetes.52.2.568. [DOI] [PubMed] [Google Scholar]
- 153.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, Styrkarsdottir U, Magnusson KP, Walters GB, Palsdottir E, Jonsdottir T, Gudmundsdottir T, Gylfason A, Saemundsdottir J, Wilensky RL, Reilly MP, Rader DJ, Bagger Y, Christiansen C, Gudnason V, Sigurdsson G, Thorsteinsdottir U, Gulcher JR, Kong A, Stefansson K. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–3. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 154.Cai Y, Yi J, Ma Y, Fu D. Meta-analysis of the effect of HHEX gene polymorphism on the risk of type 2 diabetes. Mutagenesis. 2011;26:309–14. doi: 10.1093/mutage/geq095. [DOI] [PubMed] [Google Scholar]
- 155.Tanaka H, Yamamoto T, Ban T, Satoh S, Tanaka T, Shimoda M, Miyazaki J, Noguchi T. Hex stimulates the hepatocyte nuclear factor 1alpha-mediated activation of transcription. Arch Biochem Biophys. 2005;442:117–24. doi: 10.1016/j.abb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 156.Cauchi S, Del Guerra S, Choquet H, D’Aleo V, Groves CJ, Lupi R, McCarthy MI, Froguel P, Marchetti P. Meta-analysis and functional effects of the SLC30A8 rs13266634 polymorphism on isolated human pancreatic islets. Mol Genet Metab. 2010;100:77–82. doi: 10.1016/j.ymgme.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 157.Gloyn AL, Noordam K, Willemsen MA, Ellard S, Lam WW, Campbell IW, Midgley P, Shiota C, Buettger C, Magnuson MA, Matschinsky FM, Hattersley AT. Insights into the biochemical and genetic basis of glucokinase activation from naturally occurring hypoglycemia mutations. Diabetes. 2003;52:2433–40. doi: 10.2337/diabetes.52.9.2433. [DOI] [PubMed] [Google Scholar]
- 158.Pearson ER, Donnelly LA, Kimber C, Whitley A, Doney AS, McCarthy MI, Hattersley AT, Morris AD, Palmer CN. Variation in TCF7L2 influences therapeutic response to sulfonylureas: a GoDARTs study. Diabetes. 2007;56:2178–82. doi: 10.2337/db07-0440. [DOI] [PubMed] [Google Scholar]
- 159.Knowler WC, Barrett-Connor E, Fowler SE, Hamman RF, Lachin JM, Walker EA, Nathan DM. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med. 2002;346:393–403. doi: 10.1056/NEJMoa012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Brautbar A, Virani SS, Belmont J, Nambi V, Jones PH, Ballantyne CM. LPL gene variants affect apoC-III response to combination therapy of statins and fenofibric acid in a randomized clinical trial of individuals with mixed dyslipidemia. J Lipid Res. 2012 doi: 10.1194/jlr.M020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Keech A, Simes RJ, Barter P, Best J, Scott R, Taskinen MR, Forder P, Pillai A, Davis T, Glasziou P, Drury P, Kesaniemi YA, Sullivan D, Hunt D, Colman P, d’Emden M, Whiting M, Ehnholm C, Laakso M. Effects of long-term fenofibrate therapy on cardiovascular events in 9795 people with type 2 diabetes mellitus (the FIELD study): randomised controlled trial. Lancet. 2005;366:1849–61. doi: 10.1016/S0140-6736(05)67667-2. [DOI] [PubMed] [Google Scholar]
- 162.Nilsson PM. ACCORD and Risk-Factor Control in Type 2 Diabetes. N Engl J Med. 2010;362:1628–30. doi: 10.1056/NEJMe1002498. [DOI] [PubMed] [Google Scholar]
- 163.Ginsberg HN, Elam MB, Lovato LC, Crouse JR, 3rd, Leiter LA, Linz P, Friedewald WT, Buse JB, Gerstein HC, Probstfield J, Grimm RH, Ismail-Beigi F, Bigger JT, Goff DC, Jr, Cushman WC, Simons-Morton DG, Byington RP. Effects of combination lipid therapy in type 2 diabetes mellitus. N Engl J Med. 2010;362:1563–74. doi: 10.1056/NEJMoa1001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Olivieri O, Stranieri C, Bassi A, Zaia B, Girelli D, Pizzolo F, Trabetti E, Cheng S, Grow MA, Pignatti PF, Corrocher R. ApoC-III gene polymorphisms and risk of coronary artery disease. J Lipid Res. 2002;43:1450–7. doi: 10.1194/jlr.m200145-jlr200. [DOI] [PubMed] [Google Scholar]
- 165.Alaupovic P, Blackett P, Wang W, Lee E. Characterization of the metabolic syndrome by apolipoproteins in the Oklahoma Cherokee. J Cardiometab Syndr. 2008;3:193–9. doi: 10.1111/j.1559-4572.2008.00022.x. [DOI] [PubMed] [Google Scholar]
- 166.Sacks FM, Alaupovic P, Moye LA, Cole TG, Sussex B, Stampfer MJ, Pfeffer MA, Braunwald E. VLDL, apolipoproteins B, CIII, and E, and risk of recurrent coronary events in the Cholesterol and Recurrent Events (CARE) trial. Circulation. 2000;102:1886–92. doi: 10.1161/01.cir.102.16.1886. [DOI] [PubMed] [Google Scholar]
- 167.Blankenhorn DH, Alaupovic P, Wickham E, Chin HP, Azen SP. Prediction of angiographic change in native human coronary arteries and aortocoronary bypass grafts. Lipid and nonlipid factors. Circulation. 1990;81:470–6. doi: 10.1161/01.cir.81.2.470. [DOI] [PubMed] [Google Scholar]
- 168.Chaves FJ, Real JT, Garcia-Garcia AB, Civera M, Armengod ME, Ascaso JF, Carmena R. Genetic diagnosis of familial hypercholesterolemia in a South European outbreed population: influence of low-density lipoprotein (LDL) receptor gene mutations on treatment response to simvastatin in total, LDL, and high-density lipoprotein cholesterol. J Clin Endocrinol Metab. 2001;86:4926–32. doi: 10.1210/jcem.86.10.7899. [DOI] [PubMed] [Google Scholar]
- 169.Liljedahl U, Lind L, Kurland L, Berglund L, Kahan T, Syvanen AC. Single nucleotide polymorphisms in the apolipoprotein B and low density lipoprotein receptor genes affect response to antihypertensive treatment. BMC Cardiovasc Disord. 2004;4:16. doi: 10.1186/1471-2261-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Joy T, Hegele RA. Genetics of metabolic syndrome: is there a role for phenomics? Curr Atheroscler Rep. 2008;10:201–8. doi: 10.1007/s11883-008-0032-0. [DOI] [PubMed] [Google Scholar]
- 171.Hegele RA. Phenomics, lipodystrophy, and the metabolic syndrome. Trends Cardiovasc Med. 2004;14:133–7. doi: 10.1016/j.tcm.2004.02.001. [DOI] [PubMed] [Google Scholar]