Abstract
Purpose.
We investigated the impact of PININ (PNN) and epithelial splicing regulatory protein 1 (ESRP1) on alternative pre-mRNA splicing in the corneal epithelial context.
Methods.
Isoform-specific RT-PCR assays were performed on wild-type and Pnn knockout mouse cornea. Protein interactions were examined by deconvolution microscopy and co-immunoprecipitation. For genome-wide alternative splicing study, immortalized human corneal epithelial cells (HCET) harboring doxycycline-inducible shRNA against PNN or ESRP1 were created. Total RNA was isolated from four biological replicates of control and knockdown HCET cells, and subjected to hGlue3_0 transcriptome array analysis.
Results.
Pnn depletion in developing mouse corneal epithelium led to disrupted alternative splicing of multiple ESRP-regulated epithelial-type exons. In HCET cells, ESRP1 and PNN displayed close localization in and around nuclear speckles, and their physical association in protein complexes was identified. Whole transcriptome array analysis on ESRP1 or PNN knockdown HCET cells revealed clear alterations in transcript profiles and splicing patterns of specific subsets of genes. Separate RT-PCR validation assays confirmed successfully specific changes in exon usage of several representative splice variants, including PAX6(5a), FOXJ3, ARHGEF11, and SLC37A2. Gene ontologic analyses on ESRP1- or PNN-regulated alternative exons suggested their roles in epithelial phenotypes, such as cell morphology and movement.
Conclusions.
Our data suggested that ESRP1 and PNN modulate alternative splicing of a specific subset of target genes, but not general splicing events, in HCET cells to maintain or enhance epithelial characteristics.
Genome-wide PNN- and ESRP1-regulated alternative pre-mRNA splicing events were identified in human corneal epithelial cells through whole transcriptome array analysis. The study suggests specific roles of PNN and ESRP1 in the regulation of epithelial-specific alternative splicing.
Introduction
The corneal epithelium provides the proper refractive surface for visual processing, and protects the eye against infections and damages through its unique differentiated characteristics, such as transparency, optimal curvature, and barrier capacity. Within this multilayered epithelium, cells residing at the different layers display distinctive morphology and gene expression profile, while undergoing orderly differentiation, apoptosis, and desquamation. Thus, the establishment of the specific corneal epithelial identity and execution of error-free maturation are crucial for healthy eye and ideal vision.
We reported recently PININ (PNN) as one of the proteins that have key roles during differentiation of corneal epithelium.1 In the study, conditional inactivation of Pnn in developing mouse corneal epithelium resulted in severe disruption in epithelial differentiation. Specifically, Pnn-depleted ocular surface ectoderm failed to commit toward corneal epithelium, and instead displayed epidermis-like qualities. Pnn's specific impact on the specification and maintenance of epithelial cell identity has been well demonstrated in other studies as well.2–5 Interestingly, an increasing number of recent studies by our group and others have revealed PNN's association with a large number of RNA splicing proteins, such as SNIP1, RNPS1, SRp75, and SRm300.6–10 PNN protein, indeed, contains two known domains that are necessary to interact with other RNA processing proteins, SR-like domain and RNPS1-SAP18-binding (RSB) motif. Consistently, PNN was shown to exert an influence on splice site selection of multiple splicing reporter minigenes, such as E-cadherin, E1A, and chimeric calcitonin/dhfr constructs,11,12 suggesting PNN's involvement in the modulation of alternative pre-mRNA splicing.
Alternative splicing of precursor mRNA is a fundamental process that allows the production of multiple transcripts from a single gene. In the past decade, extensive genome-wide studies revealed an astonishing complexity of eukaryotic transcriptomes largely due to alternative splicing.13,14 The complexity, which originates from the diverse exon/intron usage, exponentially expands the transcriptomic repertoire encoded by the limited metazoan genome.15,16 However, removing noncoding introns and joining coding exons together from primary transcripts of multi-exon genes require an exquisite level of precision and fidelity at the single-nucleotide level for constitutive and alternative exons. The consequences of defective splicing are well documented in countless reports of human diseases caused by mis-splicing.17–19 It is evident that this process requires far more information and direction than that contained within short splice donor and acceptor sequences at the intron–exon boundaries. The accuracy that prevents catastrophic disasters of reading frame error is accomplished in large part by the combinatorial contributions of additional genome-embedded cis-regulatory elements and transacting regulatory proteins.16,20
Interestingly, alternative splicing within the corneal epithelium has received little attention, and a relatively small number of genes have been reported so far to undergo alternative splicing. Yet, the list of alternatively-spliced genes includes some with integral ocular roles, such as p63 (ΔNp63α), PAX6(5a), plasma membrane calcium-ATPase, and MUC1.21–24 Although neither specific functions of the splice variants, nor molecular mechanisms dictating the specific timing and splicing patterns have been elucidated clearly, these studies all highlight the potential impact of alternative splicing on corneal epithelium. PAX6(5a) is an intriguing example of alternatively spliced products observed in corneal epithelium. PAX6(5a), one of two major products of PAX6 gene, contains additional exon (exon 5a) encoding 14 amino acids in its paired domain which confers unique DNA binding property.22,25 Indeed, a heterozygous missense mutation (V54D) located within exon 5a was linked to diverse ocular phenotypes, including corneal opacity in a group of human patients.26 Specifically, a splicing mutation of exon 5a, which leads to disruption in the ratio between PAX6 and PAX6(5a) altering net transcript level of each isoform without affecting the total PAX6 mRNA level, was connected to abnormal iris, corectopia, and corneal opacity in affected members of a family.22 Therefore, further identification and characterization of alternatively spliced genes and their associated elements in a context-specific manner, for example in health or disease, will greatly advance our knowledge on the onset and/or progression of those devastating vision-impairing diseases.
In addition to diseases, many other different conditions, such as cell or tissue type, signaling, and developmental state, have been linked closely to differential splicing, instating the concept of “RNA code” or “splicing code.”18,27,28 The establishment of RNA code is achieved, for the most part, through the variable expression/interaction of splicing proteins and/or context-specific regulatory factors. Examples of such factors would be the cell type-specific splicing proteins, such as Nova, nPTB, MBNL, and ESRPs.15,28,29 Epithelial splicing regulatory proteins (ESRP1 and ESRP2) are the first identified epithelial-restricted RNA binding proteins that modulate alternative splicing of a number of epithelial-type transcripts, including FGFR2, ENAH, CTNND1, and CD44.29,30 Exons that are regulated by ESRPs displayed high level of correlation with epithelial-to-mesenchymal transition (EMT), which leads to loss of cell adhesion, altered cell morphology, and acquisition of cellular motility/invasiveness.31–33 These observations are of our particular interest because, as mentioned earlier, many previous studies on PNN have revealed consistently PNN's specific impact on epithelial cell adhesion and motility,2,4,5,34 as well as its involvement in pre-mRNA splicing.6,7,9,10,35 Thus, considering their similar impact on epithelial phenotypes and alternative splicing, we hypothesize that PNN may function as a component of splicing network in epithelial cells through the functional connection with epithelial-specific splicing factors, such as ESRP proteins.
Here, we report specific regulatory roles of PNN and ESRP1 in alternative pre-mRNA splicing in a corneal epithelial context. To our knowledge, our study provides the first evidence of physical association between PNN and ESRP1. The whole transcriptome array analysis clearly demonstrates involvement of PNN and ESRP1 in the alternative splicing of a specific subset of genes, and identified novel PNN- and ESRP1-regulated alternative exons in human corneal epithelial cells (HCET).
Materials and Methods
Semi-Quantitative RT-PCR, Sequencing, and Quantitative RT-PCR (qRT-PCR)
Total RNA was isolated from corneas of embryonic day 17.5 (E17.5) mice or cultured HCET cells, and semi-quantitative RT-PCR was performed as described previously.3 Quantification of PCR products of alternatively spliced transcripts was performed by ChemiDoc XRS+ and Image Lab Software Version 4.0 (Bio-Rad, Hercules, CA) according to the manufacturer's suggestion. Inclusion of specific exons/introns was determined as percentage among all PCR products. For sequencing of PCR products, individual band was gel extracted with the QIAEX II Gel Extraction Kit (Qiagen, Inc., Valencia, CA), cloned into pCRII-TOPO vector (Invitrogen, Carlsbad, CA), and sequenced. Real-time qRT-PCR assays were performed by the relative standard curve method with RT2 SYBR Green ROX qPCR Mastermix (Qiagen, Inc.) on a 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA). Primer sequences and PCR conditions will be available upon request.
Epitope-Tagged Expression Plasmids
The detailed procedure to generate Flag-HA–tagged human PNN expression vector has been described previously.10 The full-length cDNA of human ESRP1 was generated by PCR with a primer pair: 5′-cccAAGCTTaccatgacggcctctccggattac-3′ and 5′-cgGGATCCCGaatacaaacccattctttggg-3′. Template cDNAs were prepared by reverse transcription of total RNA from HCET cells. Amplified 2065 base pair (bp) cDNA then was cloned into pEGFN1 vector (Clontech, Mountain View, CA) and confirmed by sequencing.
Cell Culture, Stable Cell Lines, and Experimental Animals
Immortalized HCET cells36 were maintained as reported previously.10 HCET cells that bicistronically express interleukin-2 receptor (IL-2R)-α and Flag-HA–PNN were generated with the methods described previously.10 To create a stable HCET cell line expressing ESRP1-GFP, HCET cells were transfected with GFP-tagged ESRP1 expression plasmids with FuGENE 6 transfection reagent (Roche, Indianapolis, IN) and selected in culture medium containing 400 μg/mL of G418 (Invitrogen). For doxycycline-inducible PNN or ESRP1 knockdown HCET cells, human TRIPZ shRNAmir clones (V2THS_170187 for PNN and V3THS_400802 for ESRP1) were purchased from Open Biosystems (Lafayette, CO). HCET cells transfected with either shRNAmir clone then were selected with 10 μg/mL of puromycin (Cellgro, Manassas, VA). For induction of shRNA expression, cells were treated with doxycycline at a concentration of 1 μg/mL with daily change of fresh doxycycline medium.
All mice used in the study were described previously.1 Animal procedures were adherent to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research, and approved by the Institutional Animal Care and Use Committee at University of Florida.
Immunofluorescence and Deconvolution Microscopy
Experimental HCET cells cultured on coverslips were fixed with methanol at −20°C for 2 minutes and processed as described previously.10 Primary antibodies were anti-PNN (mAb 2-2-143), anti-GFP (Abcam, Inc., Cambridge, MA), and anti-ESRP1 (Abgent, San Diego, CA). Deconvolution microscopy was performed with a DM IRBE Fluorescence microscope (Leica, Deerfield, IL) with Openlab and Volocity software (ImproVision, Lexington, MA) as reported previously.10
Co-Immunoprecipitation (Co-IP) Assay, GFP-Trap, and Immunoblotting
Co-IP assays were done with ANTI-FLAG M2 Agarose Affinity Gel (Sigma-Aldrich, St. Louis, MO) as described previously.11 GFP-Trap assays for ESRP1-GFP were performed with Chromotek-GFP-Trap-Agarose (Allele Biotech, San Diego, CA) as suggested by the manufacturer. Immunoblotting was performed as reported previously,2 with mouse monoclonal antibodies against PCNA (BD Transduction Labs, San Jose, CA), U2AF65 and α-tubulin (Sigma-Aldrich), and rabbit polyclonal antibodies against SF3b155 (Bethyl Lab, Inc., Montgomery, TX), SF3A1 (Novus Biologicals, Littleton, CO), and GFP (Abcam). Monoclonal antibody raised against PNN is mAb 2-2-143.
GG-H Human Transcriptome Array Analysis
Parental HCET, shRNA-PNN HCET, and shRNA-ESRP1 HCET cells were cultured for 3 days with/without doxycycline. Total RNA was isolated from four biological replicates of each sample group with RNesay Plus Mini Kit and treated with RNase-free DNase I (Qiagen). The RNA quality was verified with Agilent 2100 BioAnalyzer (Agilent Technologies, Inc., Santa Clara, CA). For GG-H array, samples were processed as previously described.37 Briefly, 105 ng total RNA was amplified into cRNA using WT Expression Kit (Applied Biosystems). Then, 15 μg of the resulting cRNA was used to generate biotin-labeled cDNA using The GeneChip Eukaryotic Double Strand Whole Transcript Protocol (Affymetrix, Santa Clara, CA). Then, 20 μg of cDNA was fragmented and labeled using Affymetrix GeneChip WT Terminal Labeling Kit (Affymetrix), and then hybridized onto arrays for 16 hours at 45°C. The arrays were scanned using Affymetrix Fluidics Station FS450 and GeneChip Scanner 3000 7G.
GG-H Array Data Processing
The raw data were preprocessed by Affymetrix Power Tools (APT) package for background correction, normalization, and summarizations with the Robust Multi-array Analysis (RMA) approach. The exon- and transcript-level intensities were determined by APT according to probe set definitions and annotations provided by Affymetrix, and expression levels of junction probes were calculated as expression of the corresponding probe sets. The detection above background (DABG) P value was used to filter the nonexpressing transcript and exons. A DABG P value below 0.05 was considered as expressed. All the statistical analyses were performed using R/Bioconductor statistical environment.38 The linear modeling approach and the empirical Bayes statistics as implemented in the LIMMA package39 were used for the differential transcript level analysis. The false discovery rate (FDR) for each P value was calculated based on Benjamini and Hochberg approach.40
To detect the alternatively spliced transcripts, the probe sets were filtered out if the signal was near the background noise (DABG <0.01). The Junction and Exon array Toolkits for Transcriptome Analysis (JETTA) software was used.41 The criteria used for selecting potential alternative candidates are shown in the Table. Our array data can be accessed at the NCBI Gene Expression Omnibus (GEO) data repository with the accession number GSE41996.
Table. .
hGlue 3_0 Exon Array Summary
|
|
Transcript Level* |
Alternative Splicing† |
||||
|
Upregulated |
Downregulated |
Total |
Included Probe Sets |
Excluded Probe Sets |
Total Probe Sets |
|
| Control HCET | 10 | 10 | 20 | 0 | 0 | 0 |
| shRNA-ESRP1 HCET | 31 | 16 | 47 | 47 | 15 | 62 (45 genes) |
| shRNA-PNN HCET | 84 | 36 | 120 | 226 | 97 | 323 (260 genes) |
Cut-off criteria for transcript level:fold change ≥1.5, P ≤ 0.05.
Cut-off criteria for alternative splicing: (1) The average transcript level expressions within a group should be higher than 40 in both conditions. (2) The differential expression at transcript level between the conditions should be <2. (3) The splicing index should be >1.5. (4) The Microarray Data Analysis System (MIDAS) P < 0.01. (5) The Microarray Analysis of Differential Splicing (MADS) P < 0.01. (6) Candidate exons should be supported by at least one junction probe satisfying the same criteria with the exon.
The selected statistically significant candidates were analyzed for functional enrichments using Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Inc., Redwood City, CA). Fisher's exact test was used to calculate the significant value, which determined the probability whether the association between the genes in the dataset and the functional pathway can be explained by chance alone. Functional groups with a P value less than 0.05 were considered to be statistically significant.
Results
Disruption in Alternative Pre-mRNA Splicing in Pnn Mutant Cornea
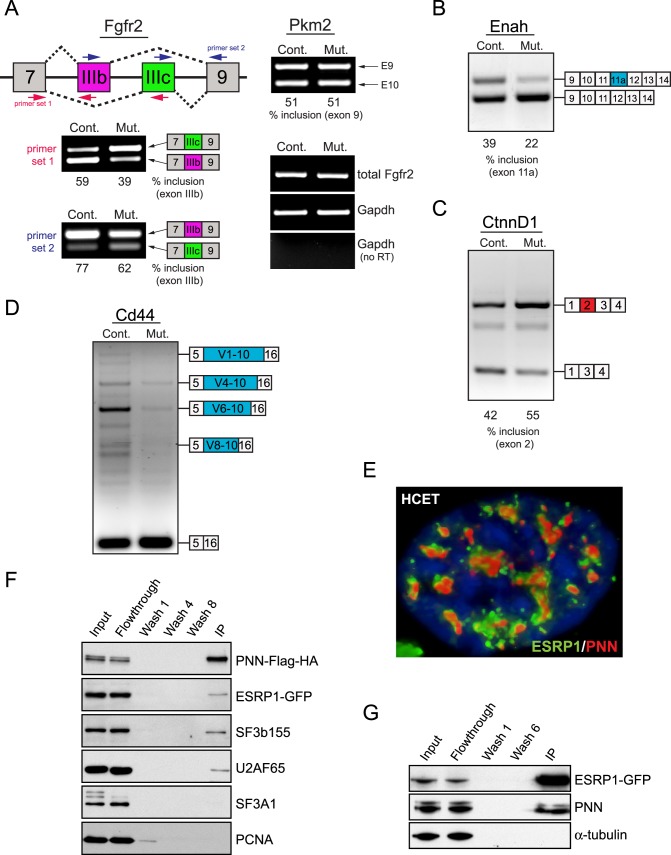
Evidence of PNN's role in alternative splicing is demonstrated through a series of alternative splicing-specific RT-PCR analyses on Pnn knockout mouse cornea. The results reveal clearly that loss of Pnn expression in developing corneal epithelial cells in Pax6-Cre;Pnn2f/1f mutant mice1 led to a pronounced decrease in the level of epithelial-type (IIIb) Fgfr2 transcript, but increased mesenchymal-type (IIIc) transcript (Fig. 1A), which is remarkably similar to those seen under knockdown of ESRPs.29 Importantly, Pnn depletion does not affect the splicing pattern of Pkm2 and total transcript level of Fgfr2 (Fig. 1A). Pnn mutant cornea also displayed specific changes in the splicing pattern of Enah, CtnnD1, and Cd44, other ESRP targets29 (Figs. 1B–D).
Figure 1. .
Disrupted alternative pre-mRNA splicing in PNN mutant cornea, and interaction between PNN and ESRP1. (A) A diagram shows alternatively spliced exons of Fgfr2 (IIIb in red and IIIc in green), and the location of primers for alternative splicing-specific RT-PCR assay. Pnn-deficiency in developing corneal epithelium of Pax6-Cre;Pnn2f/1f embryos led to increased exon IIIc usage with decreased exon IIIb inclusion. Inclusion percentage of exon IIIb is shown under each lane. No change was observed in alternative splicing of pyruvate kinase muscle-type (Pkm2) nor in total Fgfr2 expression (right panel). (B–D) Mutant corneas display differential exon usage in Enah (decreased inclusion of exon 11a), CtnnD1 (increased inclusion of exon 2), and Cd44 (decreased inclusion of variable exons). For simplification purposes, only a few major isoforms of Cd44 are indicated. All marked PCR products were cloned into the plasmid DNAs and sequenced. (E) Deconvolution microscopy reveals close localization of PNN (red) and ESRP1 (green) in a nucleus of a HCET cell. DNA is visualized as blue fluorescent signal with 4′,6-diamidino-2-phenylindole (DAPI). (F) Co-immunoprecipitation analysis using anti-Flag antibody indicates the presence of ESRP1 in PNN complex. Other splicing factors, such as SF3b155 and U2AF65, also are present in PNN complex, but not SF3A1 nor PCNA. (G) GFP-Trap IP for ESRP1-GFP demonstrates the presence of endogenous PNN (bottom band) as well as exogenous PNN-Flag-HA (top band) among copurified proteins. α-tubulin serves as a negative control for the assay.
Since these data strongly support not only PNN's role in alternative splicing, but also the potential of a functional connection between PNN and ESRPs, we next investigated possible interactions between PNN and ESRP1 proteins. Deconvolution microscopy analysis on ESRP1-GFP expressing HCET cells reveals a close localization of PNN and ESRP1 in nucleus with a distinct speckle-distribution pattern (Fig. 1E). Co-IP analysis of HCET cells expressing PNN-Flag-HA and ESRP1-GFP indicates that PNN is present in a complex with ESRP1 as well as with other splicing factors, such as SF3b155 and U2AF65 (Fig. 1F). Reciprocal IP analysis with GFP-Trap also reveals that PNN and ESRP1 are in the same protein complexes (Fig. 1G). Taken together, these observations suggest that PNN has a role in the regulation of alternative pre-mRNA splicing through its physical association with other RNA binding/processing proteins in a context-specific fashion.
Comparable Impact of ESRP1 and PNN on Transcript Profile of HCET Cells
The broader functional relevance of ESRP1 and PNN in the regulation of transcript level and alternative splicing in a corneal epithelial context was explored through a recently developed human transcriptome array (GG-H array).37 First, stable HCET cell lines were established for doxycycline-inducible expression of shRNA against ESRP1 or PNN, and successful knockdown of ESRP1 and PNN in each cell line was confirmed (see Supplementary Material and Supplementary Fig. S1, http://www.iovs.org/content/54/1/697/suppl/DC1). Since the earliest time point that shows sufficient knockdown of ESRP1 and PNN was day 3 after induction, all RNA samples were harvested at day 3 and subjected to array analysis. After rigorous quality control of the array data, subsets of specific candidate genes exhibiting differential transcript level or alternative splicing are successfully identified (see Table). Notably, none of 20 genes exhibiting differential transcript level in doxycycline-treated parental HCET cells was identified in shRNA-PNN or shRNA-ESRP1 HCET cells. In addition, doxycycline treatment alone does not affect alternative splicing of any genes. See Supplementary Material and Supplementary Table S1 (http://www.iovs.org/content/54/1/697/suppl/DC1) for the full list of identified genes. Interestingly, among the upregulated protein-coding genes upon either ESRP1 or PNN knockdown, 14 candidate transcripts are identified to be upregulated commonly in both cell lines (Fig. 2A, see Supplementary Material and Supplementary Table S2, http://www.iovs.org/content/54/1/697/suppl/DC1). Although the extent to which increased transcript levels are due to increased transcription or mRNA stability is yet to be determined, a significant percentage of overlapping transcripts, 14 of 24 (58%) for ESRP1 and 14 of 53 (26%) for PNN, may indicate the functional connection between ESRP1 and PNN. Separate RT-PCR validation assays for several representative transcripts from each group are shown in Figure 2B, where three commonly upregulated genes (TMPRSS11E, MMP13, and ADAM21) clearly are validated. GSDMA shows noticeable upregulation only in shRNA-ESRP1 HCET cells, whereas HAS2 exhibits increased transcript level only in shRNA-PNN HCET cells, as recognized by exon array. See Supplementary Material and Supplementary Figures S2 and S3 (http://www.iovs.org/content/54/1/697/suppl/DC1) for principal component analysis (PCA), hierarchical clustering heat map analysis, and exon expression profiles of several representative transcripts.
Figure 2. .
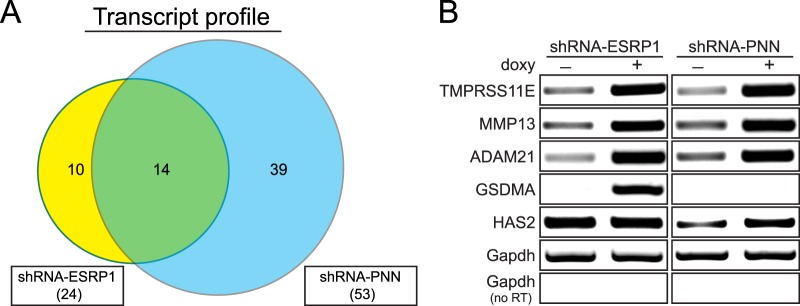
Similar impacts of PNN and ESRP1 on transcript profile. (A) A Venn diagram shows the number of upregulated protein-coding genes upon ESRP1 (yellow) or PNN (blue) knockdown. The total number of such genes for each cell line is shown in parentheses. Fold change in transcript level >1.5, P value < 0.05. (B) Validation of transcript levels of candidate genes from exon array by semiquantitative RT-PCR assays confirms specific alterations in transcript level of several representative genes. Three genes commonly upregulated in both cell lines and one unique gene from each cell line were chosen as examples of validation.
Identification and Validation of ESRP1- or PNN-Regulated Splicing Events
Data analysis of alternative splicing identified 45 genes (62 probe sets) and 260 genes (323 probe sets) from ESRP1 and PNN knockdown HCET cells, respectively (see Table, see Supplementary Material and Supplementary Fig. S4, http://www.iovs.org/content/54/1/697/suppl/DC1). Interestingly, only 7 of 11 overlapping genes between two cell lines displayed identical alterations in their splicing pattern, in which the inclusion of the same exons/introns was increased (see Supplementary Material and Supplementary Table S3, http://www.iovs.org/content/54/1/697/suppl/DC1). Although this observation might indicate that some of ESRP1-regulated splicing events also are under the control of PNN, the relatively small percentage of overlapping events suggests a narrow connection between PNN and ESRP1 in terms of the regulation of alternative pre-mRNA splicing.
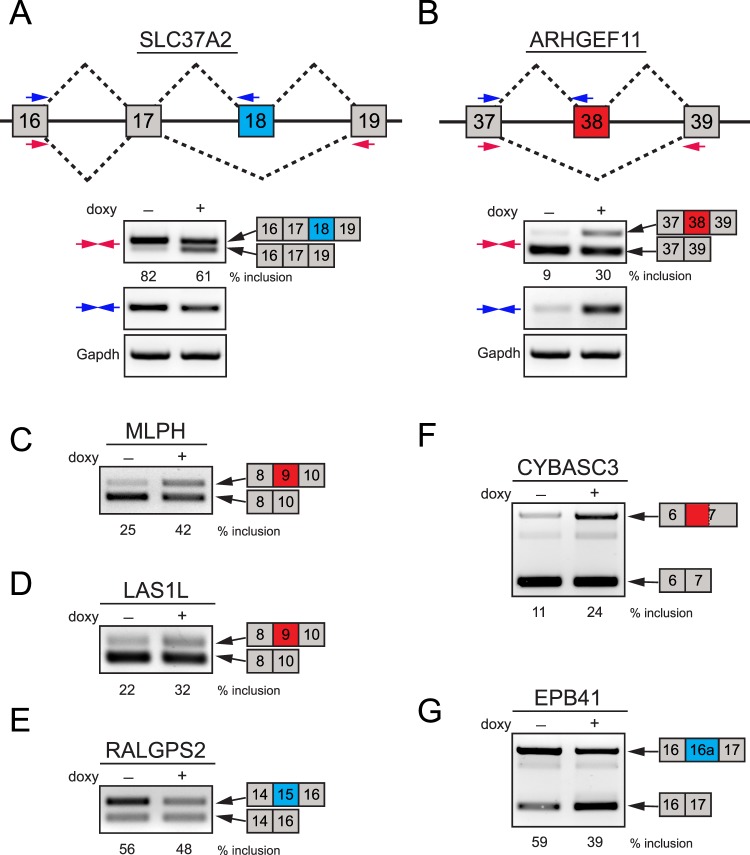
Alternative splicing of a large number of representative candidate genes was verified through RT-PCR assays. Several examples of validated genes upon ESRP1 knockdown in HCET cells are shown in Figure 3, and summarized in the Supplementary Material and Supplementary Table S4 (http://www.iovs.org/content/54/1/697/suppl/DC1). ARHGEF11 and SLC37A2, two known ESRP targets,33 have been shown to be alternatively spliced during EMT in mammary epithelial cells.42 Array and validation analyses successfully identified specific splicing changes in those ESRP cassette exons upon ESRP1 knockdown in HCET cells, demonstrating decreased inclusion of exon 18 for SLC37A2 and increased inclusion of exon 38 for ARHGEF11 (Figs. 3A, 3B). Other ESRP-regulated cassette exons, such as exon 9 of MLPH,31 exon 9 of LAS1L, and exon 15 of RALGPS2,33 also are confirmed clearly in our analysis as well (Figs. 3C–E). CYBASC3 also has been identified to exhibit increased usage of alternative 3′ splice site leading to the longer exon 7 upon simultaneous knockdown of ESRP1 and 2.30 As shown in Figure 3F, ESRP1 knockdown in HCET cells results in the substantial increase of alternative 3′ splice site usage as seen in a human prostate cell line,30 supporting the reliability of the array analysis and validation assays. Furthermore, our array also identified previously unidentified ESRP-regulated events in a corneal epithelial context. One such example is shown in Figure 3G, where knockdown of ESRP1 leads to the decreased inclusion of 450 base pair (bp) alternative cassette exon (16a) of EPB41.
Figure 3. .
Examples of validated ESRP1-regulated splicing events in HCET cells. (A, B) Two examples of alternatively spliced genes in ESRP1 knockdown HCET cells are presented. Diagrams show the structure of exons that are alternatively spliced in SLC37A2 and ARHGEF11. The location of two primer sets (colored arrows) used for RT-PCR validation for each gene is indicated. While SLC37A2 exhibited decreased inclusion of exon 18 (shown in blue), ARHGEF11 specifically displayed increased inclusion of exon 38 following ESRP1 knockdown. Inclusion percentages of alternative exons are shown for each condition. (C–E) Three examples of known ESRP-target events validated in ESRP1 knockdown HCET cells are shown. Alternative cassette exons of MLPH, LAS1L, and RALGPS2 showed specific changes in inclusion rate upon ESRP1 knockdown. (F) An example of increased alternative 3′ splice site usage is shown. Upon ESRP1 knockdown, CYBASC2 displayed enhanced usage of alternative 3′ splice site (779 bp upstream of major splice site of exon 7). (G) A previously unidentified ESRP1-regulated cassette exon in EPB41 is validated in HCET cells. Exon 16a of EPB41 showed decreased inclusion upon ESRP1 knockdown. All alternatively spliced PCR products shown in Figure 3 were verified through sequencing.
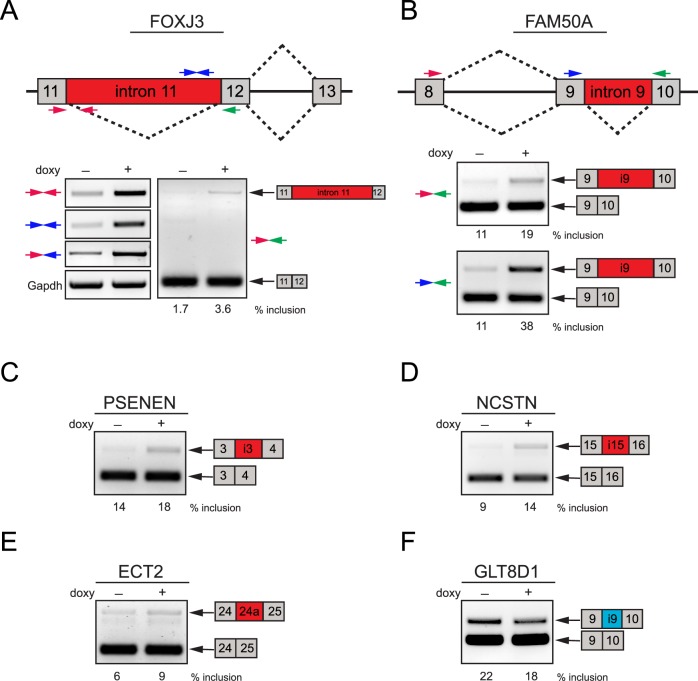
In PNN knockdown HCET cells, FOXJ3, a forkhead/winged helix transcription factor expressed in developing neuroectoderm, neural crest, and myotome,43 displays retained intron 11 as predicted in the exon array (Fig. 4A, see Supplementary Material and Supplementary Table S4, http://www.iovs.org/content/54/1/697/suppl/DC1). Inclusion of the entire intron 11, which is confirmed by separate RT-PCR assays with multiple primer sets, leads to premature translational termination and most likely nonsense-mediated decay (NMD) of alternatively spliced FOXJ3 transcripts. On the other hand, retained intron 9 of a transcription factor FAM50A, observed upon PNN knockdown (Fig. 4B), is not expected to cause a frame shift nor early termination, thus predicted to add 49 amino acids to FAM50A. These findings clearly highlight the complexity of splicing-dependent mRNA quality control mechanism and the importance of precise regulation of splicing events. Validation assays also indicate PNN's involvement in the regulation of alternative splicing of two essential components of the γ-secretase protein complex, which has a central role in Alzheimer's disease and Notch signaling pathway.44 Knockdown of PNN promotes inclusion of introns 3 and 15 in PSENEN (PEN-2) and NCSTN (nicastrin) transcripts, respectively (Figs. 4C, 4D). Finally, while an alternative cassette exon (24a) of a guanine nucleotide exchange factor, epithelial cell transforming sequence 2 oncogene (ECT2), also known as ARHGEF31, is found to be a PNN-silenced exon (Fig. 4E), inclusion of intron 9 in glycosyltransferase 8 domain containing 1 (GLT8D1) is determined to be enhanced by PNN (Fig. 4F).
Figure 4. .
Validation of PNN-regulated splicing events in HCET cells. (A, B) Validation of alternatively spliced genes upon PNN knockdown was achieved by semi-quantitative RT-PCR assays with multiple primer pairs for FOXJ3 and FAM50A. Diagrams show exon structure around alternative region and the location of primers (colored arrows) used for RT-PCR validation. Reduction of PNN led to the increased retention of introns 11 and 9 in their entire length on FOXJ3 and FAM50A, respectively. Inclusion percentage of retained introns is shown under the corresponding lanes. (C–F) Examples of validated PNN-regulated splicing events are presented. While alternative splicing changes of PSENEN, NCSTN, and ECT2 are PNN-silenced events, inclusion of intron 9 of GLT8D1 is a PNN-enhanced event. All PCR products shown were verified by sequencing.
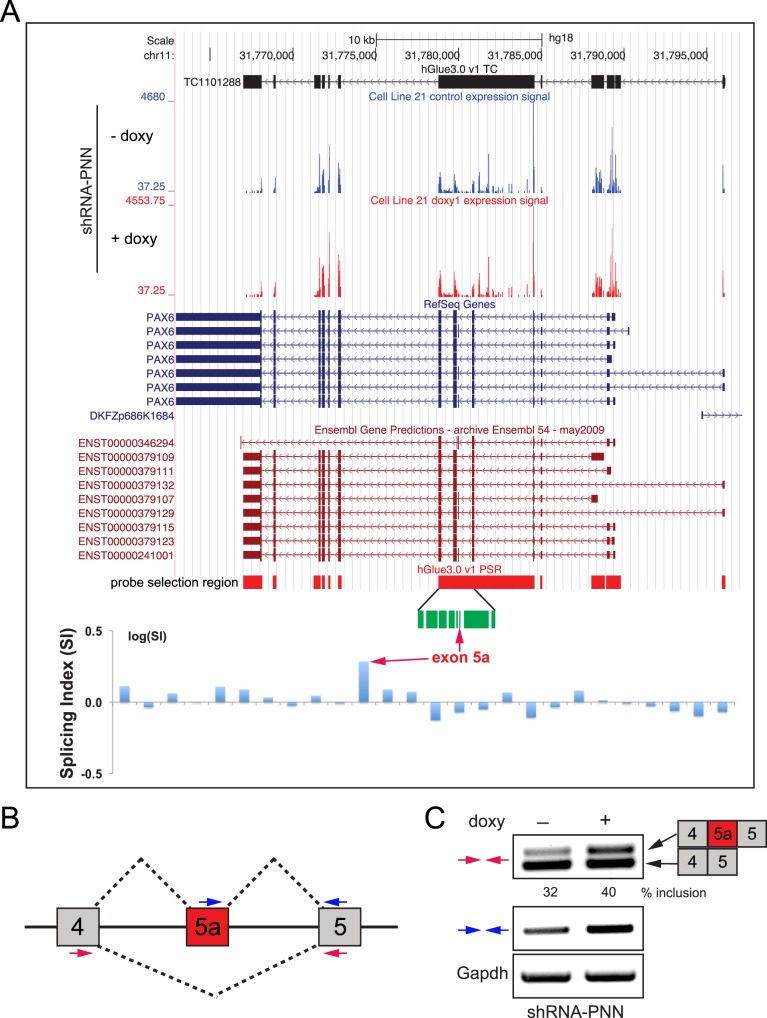
Most interestingly, the array data reveal that PNN knockdown HCET cells exhibit increased inclusion of exon 5a of PAX6 gene (splicing index = 1.9243, DABG P value < 0.001, MADS P value < 0.001, MIDAS P value < 0.01). While splicing indices for all other PAX6 exons failed to be considered significant, the inclusion event of exon 5a upon PNN knockdown was statistically significant (Fig. 5A). Separate RT-PCR analyses with isoform-specific primers confirmed increased exon 5a usage (Figs. 5B, 5C), indicating that PNN is integral to the regulation and execution of alternative splicing of PAX6(5a).
Figure 5. .
Impact of PNN on alternative splicing of PAX6. (A) UCSC genome browser view (human genome assembly hg18) of PAX6 locus is shown with exon array expression signal levels for control and doxycycline-treated shRNA-PNN HCET cells (blue, RefSeq entries; red, Ensembl transcripts). Probe selection region (PSR) of hGlue3_0 exon array is shown with an amplified view of exon 5a region (in green). Normalized splicing indices for each probe set are presented as bar graphs in logarithmic scale. Note the elevated splicing index for exon 5a (red arrows). (B) A schematic diagram shows the splicing pattern and exon structure of PAX6 around exon 5a. Two sets of primers used for RT-PCR validation are shown in colored arrows. (C) Separate RT-PCR validation assays confirm increased inclusion of exon 5a upon PNN knockdown in HCET cells.
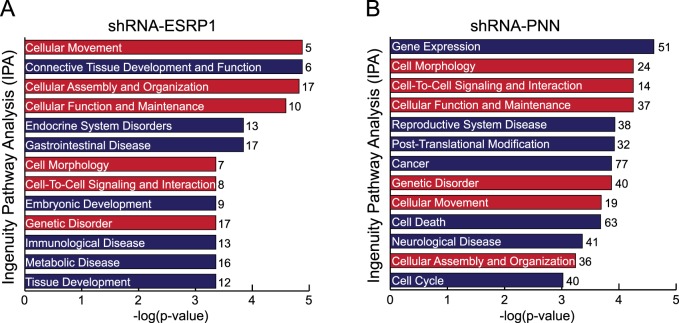
Analogous Functionality of ESRP1 and PNN Revealed by Gene Ontology Analysis
To gain insight into biological significance of ESRP1- and PNN-dependent changes in alternative splicing and to classify identified target genes into functional networks, ontologic analysis of genes showing alterations in splicing patterns was performed using the IPA. Consistent with the close connection of ESRP1 to EMT, the highest ranked biofunction identified by IPA on ESRP1-dependent exons is “Cellular Movement.” The IPA on PNN-dependent events indicates that 20% to 30% of alternatively spliced genes under PNN knockdown are connected functionally to “Gene Expression,” “Cancer,” and “Cell Death.” Interestingly, despite the limited connection in splicing between ESRP1 and PNN suggested by our current study, the analysis reveals noticeable overlap in affected biologic processes between two cell lines, including “Cell Morphology,” “Cellular Movement,” and “Cellular Assembly and Organization” (Figs. 6A, 6B), all of which are in great agreement with previously reported roles of ESRP1 and PNN in epithelial differentiation and maintenence.2,29,33 Therefore, although it is reasonable, at this point, to consider that ESRP1 and PNN may exert their impact on alternative splicing and epithelial phenotypes by distinct directions, considering their physical interaction and similar impact on gene expression profile, we believe that further studies involving deeper analysis of alternative splicing changes are needed to clarify their functional connection.
Figure 6. .
Functional overlap between ESRP1 and PNN indicated by a gene ontological analysis. (A, B) Biological processes that are significantly affected by alternatively spliced transcripts upon PNN or ESRP1 knockdown are identified by IPA. Categories whose –log(P value) is greater than 3 are shown. Numbers next to each bar show the number of alternatively spliced genes within each category. The categories overlapping in both cell lines are shown as red bars. Cut off criteria for analyzed genes: |SI| > 1.5 and MIDAS P value < 0.01.
Discussion
Our previous studies from Pnn hypomorphic mice45 and Pax6-Cre;Pnn2f/1f mutant mice1 demonstrated clearly indispensable roles of Pnn in corneal epithelial differentiation and morphogenesis. Reduced expression or conditional inactivation of Pnn in developing corneal epithelium resulted in the formation of persistent lens stalk resembling those seen in Peter's anomaly, and the squamous metaplasia of corneal epithelium. Since the etiology of these phenotypes found in Pnn mutant cornea presents immensely valuable opportunity, we focused on searching a mechanistic explanation at the molecular level. In the current study, as PNN has been linked closely to alternative pre-mRNA splicing,8,11,12 we investigated the dynamic genome-wide modulation of alternative pre-mRNA splicing by PNN and its interacting partner, ESRP1, in a corneal epithelial-specific context by using GG-H human transcriptome array. First of all, despite PNN's interaction with many basic splicing machinery proteins,6–10 our data suggested that PNN is not involved in the housekeeping roles of general splicing events, but rather it modulates alternative splicing in a specific manner. Although we do not rule out the possibility of false negative signals, our array and validation assays show clearly precise alteration at a specific splice junction even within a multi-exon gene, demonstrating a certain degree of directionality of PNN- or ESRP1-dependent splicing changes. Second, our genome-wide exon array identifies an exciting panel of alternatively spliced transcripts to be explored for their biologic significance in corneal epithelial development and maintenance. Third, despite their unique impacts on distinct subsets of target genes, our data indicated noticeable level of similarities in cellular response upon knockdown of PNN or ESRP1. Finally, the validation assays of many representative genes strongly support the integrity of our transcriptome array data and led us to the exciting confirmation of PNN's role in alternative splicing of PAX6, one of the master regulatory genes in the ocular system.
Our data revealed a negative correlation between the level of PNN and the inclusion level of PAX6 exon 5a, suggesting a role of PNN in the regulation of alternative splicing of PAX6(5a). A major splice variant of PAX6, PAX6(5a), has been shown to have a central effect on the development of various ocular tissues, such as mouse lens, iris, ciliary body, and chick neural retina.46–48 It is well known that two PAX6 products are expressed simultaneously in most ocular tissues and the ratio between them must be tightly controlled.49,50 A single nucleotide mutation at the splice acceptor of exon 5a, which promotes inclusion of exon 5a and consequently leads to a higher PAX6(5a)/PAX6 ratio, has been reported to cause distinct phenotypes in iris and cornea in human patients,22 underscoring the significance of tight regulation of alternative splicing. Due to its central impact on the global health of the ocular system, an extensive level of effort has been made to elucidate expression level, profile, or transcriptional activities of PAX6.25 However, a very limited amount of research has been performed so far on the regulatory mechanism of alternative splicing of PAX6(5a).51 To our knowledge, our study is the first report of a transacting molecule that influences splicing outcome of PAX6(5a). Further investigation of such transacting molecules in conjunction with associated cis-elements will assure our success in deciphering the complexity of PAX6-dependent developmental defects or pathologic conditions.
While a panel of candidate genes was discovered successfully through the array analysis, some of the alternatively spliced genes in Pnn knockout cornea were not selected through the current transcriptome array. Indeed, separate RT-PCR analyses indicated relatively marginal changes in ENAH and FGFR2 in PNN knockdown cells (data not shown). This simply might be due to the different cellular environment between developing mouse cornea and cultured human corneal epithelial cells. Unlike developing mouse cornea, HCET cells almost exclusively are expressing one specific isoform (IIIb form of FGFR2 and 11a-included isoform of ENAH), which would make detectable isoform switching difficult to occur. However, we suspect that the different extent of PNN downregulation between complete knockout and partial knockdown more likely accounts for the observed variations. Nevertheless, since Pnn clearly exhibited its dosage-dependent impact on multiple tissues of developing mouse embryos as described in our previous publication45 in which Pnnko/ko, Pnnko/hypo, and Pnnhypo/hypo mice showed Pnn dosage-dependent embryonic lethality around E3.5, E8.5, and E17.5, respectively, in this study, we specifically focused on PNN's impact on alternative splicing upon its knockdown in cultured cells of corneal epithelium, where we had observed previously Peter's Anomaly-like phenotypes in Pnnhypo/hypo mice (Joo JH, Sugrue SP, unpublished data, 2007).
Our study identifies ESRP1 as one of the components of PNN-containing protein complexes. Despite PNN's ubiquitous expression pattern in many cell or tissue types,52 our earlier studies have emphasized PNN's role in the establishment and maintenance of cellular identity of epithelial lineage, in which PNN promotes epithelial cell–cell adhesion, inhibits cell migration, and controls epithelial differentiation.1–3,5 Thus, the discovery of PNN's interaction with an epithelial cell-restricted RNA splicing protein, ESRP1, may provide valuable insight into the mechanism underlying PNN's established impact on epithelial cells. Reduction in ESRP proteins has been linked closely to compromised cell adhesion and enhanced cell mobility through their regulatory roles in the alternative splicing of epithelial-type transcripts of numerous genes.29–31,33 It is now becoming clear that the epithelial- or mesenchymal-specific splicing program is controlled tightly by a group of cell-type specific splicing factors.31,42,53 Thus, further identification of such factors and allied protein networks indisputably will advance our understandings on cell type specification and differentiation. However, our study reveals that only a limited number of splicing events are overlapping under knockdown of ESRP1 or PNN. While it is possible that their functional connection is restricted to the small number of splicing events, we suspect that relatively low knockdown efficiency of ESRP1 (60% reduction as determined by separate qRT-PCR assays) led to the incomplete detection of corneal-specific ESRP targets. Additionally, it also has been reported that while knockdown of ESRP1 alone is sufficient to affect ESRP target exons, knockdown of ESRP1 and ESRP2 is necessary to observe significant splicing changes in ESRP targets.30 Thus, single knockdown of ESRP1 in our experiments also might contribute to the identification of the small number of total or overlapping ESRP target genes. It will be of great relevance to analyze their functional relationship in more uniform experimental systems with enhanced detection depth.
PNN may have an effect on other aspects of alternative splicing events, such as histone modification and gene transcription. In fact, a large number of recent studies are demonstrating clearly the inseparable connection between histone modification and pre-mRNA splicing.54–56 Since PNN has been reported previously to be a component of transcriptional and histone remodeling complexes as well,7,11,34 it is tempting to speculate PNN functions as an epithelial-specific bridge molecule connecting those two fundamental elements of gene regulation.
In summary, our genome-wide approach demonstrates clearly specific roles of PNN and ESRP1 in the regulation of alternative pre-mRNA splicing of a subset of genes in a corneal epithelial context. Further studies are warranted to elucidate their independent or cooperative roles in the epithelial-specific splicing program. Finally, our current study provided us with an exciting list of alternatively spliced genes that demand further investigation for their biologic relevance in corneal epithelium.
Supplementary Material
Acknowledgments
Nicholas W. Dunn, Daniel M. Bryan, and Rick A. Swain provided technical assistance.
Footnotes
Supported by National Institutes of Health Grants R01 EY007883, P30 EY021721.
Disclosure: J.-H. Joo, None; G.P. Correia, None; J.-L. Li, None; M.-C. Lopez, H.V. Baker, None; S.P. Sugrue, None
References
- 1. Joo JH, Kim YH, Dunn NW, Sugrue SP. Disruption of mouse corneal epithelial differentiation by conditional inactivation of pnn. Invest Ophthalmol Vis Sci. 2010; 51: 1927–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Joo JH, Alpatov R, Munguba GC, Jackson MR, Hunt ME, Sugrue SP. Reduction of Pnn by RNAi induces loss of cell-cell adhesion between human corneal epithelial cells. Mol Vis. 2005; 11: 133–142 [PubMed] [Google Scholar]
- 3. Joo JH, Taxter TJ, Munguba GC, et al. Pinin modulates expression of an intestinal homeobox gene, Cdx2, and plays an essential role for small intestinal morphogenesis. Dev Biol. 2010; 345: 191–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shi Y, Ouyang P, Sugrue SP. Characterization of the gene encoding pinin/DRS/memA and evidence for its potential tumor suppressor function. Oncogene. 2000; 19: 289–297 [DOI] [PubMed] [Google Scholar]
- 5. Shi Y, Tabesh M, Sugrue SP. Role of cell adhesion-associated protein, pinin (DRS/memA), in corneal epithelial migration. Invest Ophthalmol Vis Sci. 2000; 41: 1337–1345 [PubMed] [Google Scholar]
- 6. Bracken CP, Wall SJ, Barre B, Panov KI, Ajuh PM, Perkins ND. Regulation of cyclin D1 RNA stability by SNIP1. Cancer Res. 2008; 68: 7621–7628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dellaire G, Makarov EM, Cowger JJ, et al. Mammalian PRP4 kinase copurifies and interacts with components of both the U5 snRNP and the N-CoR deacetylase complexes. Mol Cell Biol. 2002; 22: 5141–5156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Murachelli AG, Ebert J, Basquin C, Le Hir H, Conti E. The structure of the ASAP core complex reveals the existence of a Pinin-containing PSAP complex. Nat Struct Mol Biol. 2012; 19: 378–386 [DOI] [PubMed] [Google Scholar]
- 9. Sakashita E, Tatsumi S, Werner D, Endo H, Mayeda A. Human RNPS1 and its associated factors: a versatile alternative pre-mRNA splicing regulator in vivo. Mol Cell Biol. 2004; 24: 1174–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zimowska G, Shi J, Munguba G, et al. Pinin/DRS/memA interacts with SRp75, SRm300 and SRrp130 in corneal epithelial cells. Invest Ophthalmol Vis Sci. 2003; 44: 4715–4723 [DOI] [PubMed] [Google Scholar]
- 11. Alpatov R, Shi Y, Munguba GC, et al. Corepressor CtBP and nuclear speckle protein PNN/DRS differentially modulate transcription and splicing of the E-cadherin gene. Mol Cell Biol. 2008; 28: 1584–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang P, Lou PJ, Leu S, Ouyang P. Modulation of alternative pre-mRNA splicing in vivo by pinin. Biochem Biophys Res Commun. 2002; 294: 448–455 [DOI] [PubMed] [Google Scholar]
- 13. Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008; 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- 14. Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008; 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hallegger M, Llorian M, Smith CW. Alternative splicing: global insights. FEBS J. 2010; 277: 856–866 [DOI] [PubMed] [Google Scholar]
- 16. Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010; 463: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003; 17: 419–437 [DOI] [PubMed] [Google Scholar]
- 18. Spitali P, Aartsma-Rus A. Splice modulating therapies for human disease. Cell. 2012; 148: 1085–1088 [DOI] [PubMed] [Google Scholar]
- 19. Tazi J, Bakkour N, Stamm S. Alternative splicing and disease. Biochim Biophys Acta. 2009; 1792: 14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005; 6: 386–398 [DOI] [PubMed] [Google Scholar]
- 21. Di Iorio E, Barbaro V, Ruzza A, Ponzin D, Pellegrini G, De Luca M. Isoforms of DeltaNp63 and the migration of ocular limbal cells in human corneal regeneration. Proc Natl Acad Sci U S A. 2005; 102: 9523–9528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Epstein JA, Glaser T, Cai J, Jepeal L, Walton DS, Maas RL. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes Dev. 1994; 8: 2022–2034 [DOI] [PubMed] [Google Scholar]
- 23. Imbert Y, Darling DS, Jumblatt MM, et al. MUC1 splice variants in human ocular surface tissues: possible differences between dry eye patients and normal controls. Exp Eye Res. 2006; 83: 493–501 [DOI] [PubMed] [Google Scholar]
- 24. Talarico EF Jr, Mangini NJ. Alternative splice variants of plasma membrane calcium-ATPases in human corneal epithelium. Exp Eye Res. 2007; 85: 869–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shaham O, Menuchin Y, Farhy C, Ashery-Padan R. Pax6: a multi-level regulator of ocular development. Prog Retin Eye Res. 2012; 31: 351–376 [DOI] [PubMed] [Google Scholar]
- 26. Azuma N, Yamaguchi Y, Handa H, Hayakawa M, Kanai A, Yamada M. Missense mutation in the alternative splice region of the PAX6 gene in eye anomalies. Am J Hum Genet. 1999; 65: 656–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Biamonti G, Bonomi S, Gallo S, Ghigna C. Making alternative splicing decisions during epithelial-to-mesenchymal transition (EMT). Cell Mol Life Sci. 2012; 69: 2515–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet. 2011; 12: 715–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell. 2009; 33: 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Warzecha CC, Shen S, Xing Y, Carstens RP. The epithelial splicing factors ESRP1 and ESRP2 positively and negatively regulate diverse types of alternative splicing events. RNA Biol. 2009; 6: 546–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dittmar KA, Jiang P, Park JW, et al. Genome-wide determination of a broad ESRP-regulated posttranscriptional network by high-throughput sequencing. Mol Cell Biol. 2012; 32: 1468–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Horiguchi K, Sakamoto K, Koinuma D, et al. TGF-beta drives epithelial-mesenchymal transition through deltaEF1-mediated downregulation of ESRP. Oncogene. 2012; 31: 3190–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Warzecha CC, Jiang P, Amirikian K, et al. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J. 2010; 29: 3286–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Alpatov R, Munguba GC, Caton P, et al. Nuclear speckle-associated protein PNN/DRS binds to the transcriptional corepressor CtBP and relieves CtBP-mediated repression of the E-cadherin gene. Mol Cell Biol. 2004; 24: 10223–10235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Costa E, Canudas S, Garcia-Bassets I, et al. Drosophila dSAP18 is a nuclear protein that associates with chromosomes and the nuclear matrix, and interacts with pinin, a protein factor involved in RNA splicing. Chromosome Res. 2006; 14: 515–526 [DOI] [PubMed] [Google Scholar]
- 36. Araki-Sasaki K, Ohashi Y, Sasabe T, et al. An SV40-immortalized human corneal epithelial cell line and its characterization. Invest Ophthalmol Vis Sci. 1995; 36: 614–621 [PubMed] [Google Scholar]
- 37. Xu W, Seok J, Mindrinos MN, et al. Human transcriptome array for high-throughput clinical studies. Proc Natl Acad Sci U S A. 2011; 108: 3707–3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004; 5: R80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004; 3: Article3 [DOI] [PubMed] [Google Scholar]
- 40. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995; 57: 289–300 [Google Scholar]
- 41. Seok J, Xu W, Gao H, Davis RW, Xiao W. JETTA: junction and exon toolkits for transcriptome analysis. Bioinformatics. 2012; 28: 1274–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shapiro IM, Cheng AW, Flytzanis NC, et al. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 2011; 7: e1002218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Landgren H, Carlsson P. FoxJ3, a novel mammalian forkhead gene expressed in neuroectoderm, neural crest, and myotome. Dev Dyn. 2004; 231: 396–401 [DOI] [PubMed] [Google Scholar]
- 44. Mattson MP. Neurobiology: ballads of a protein quartet. Nature. 2003; 422: 385, 387 [DOI] [PubMed] [Google Scholar]
- 45. Joo JH, Lee YJ, Munguba GC, et al. Role of Pinin in neural crest, dorsal dermis, and axial skeleton development and its involvement in the regulation of Tcf/Lef activity in mice. Dev Dyn. 2007; 236: 2147–2158 [DOI] [PubMed] [Google Scholar]
- 46. Azuma N, Tadokoro K, Asaka A, et al. The Pax6 isoform bearing an alternative spliced exon promotes the development of the neural retinal structure. Hum Mol Genet. 2005; 14: 735–745 [DOI] [PubMed] [Google Scholar]
- 47. Davis N, Yoffe C, Raviv S, et al. Pax6 dosage requirements in iris and ciliary body differentiation. Dev Biol. 2009; 333: 132–142 [DOI] [PubMed] [Google Scholar]
- 48. Duncan MK, Kozmik Z, Cveklova K, Piatigorsky J, Cvekl A. Overexpression of PAX6(5a) in lens fiber cells results in cataract and upregulation of (alpha)5(beta)1 integrin expression. J Cell Sci. 2000; 113 (Pt 18); 3173–3185 [DOI] [PubMed] [Google Scholar]
- 49. Chauhan BK, Yang Y, Cveklova K, Cvekl A. Functional interactions between alternatively spliced forms of Pax6 in crystallin gene regulation and in haploinsufficiency. Nucleic Acids Res. 2004; 32: 1696–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang W, Cveklova K, Oppermann B, Kantorow M, Cvekl A. Quantitation of PAX6 and PAX6(5a) transcript levels in adult human lens, cornea, and monkey retina. Mol Vis. 2001; 7: 1–5 [PMC free article] [PubMed] [Google Scholar]
- 51. Jaworski C, Sperbeck S, Graham C, Wistow G. Alternative splicing of Pax6 in bovine eye and evolutionary conservation of intron sequences. Biochem Biophys Res Commun. 1997; 240: 196–202 [DOI] [PubMed] [Google Scholar]
- 52. Leu S, Ouyang P. Spatial and temporal expression profile of pinin during mouse development. Gene Expr Patterns. 2006; 6: 620–631 [DOI] [PubMed] [Google Scholar]
- 53. Lapuk A, Marr H, Jakkula L, et al. Exon-level microarray analyses identify alternative splicing programs in breast cancer. Mol Cancer Res. 2010; 8: 961–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. de Almeida SF, Grosso AR, Koch F, et al. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat Struct Mol Biol. 2011; 18: 977–983 [DOI] [PubMed] [Google Scholar]
- 55. Kim S, Kim H, Fong N, Erickson B, Bentley DL. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc Natl Acad Sci U S A. 2011; 108: 13564–13569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010; 327: 996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.