Abstract
The tanshinone natural products possess a variety of pharmacological properties including anti-bacterial, anti-inflammatory, anti-oxidant, and anti-neoplastic activity. The molecular basis of these effects, however, remains largely unknown. In the present study, we explored the direct effect of tanshinones on the enzyme telomerase. Telomerase is up-regulated in the majority of cancer cells and is essential for their survival, making it a potential anti-cancer drug target. We found that the ortho-quinone tanshinone II-A inhibits telomerase in a time- and DTT-dependent fashion, and the hydrogen peroxide scavenger catalase protected telomerase from inactivation. These findings demonstrate that ortho-quinone containing tanshinones can inhibit telomerase owing to their ability to generate reactive oxygen species. The results also provide evidence that telomerase is directly and negatively regulated by reactive oxygen species.
Keywords: telomerase, ortho-quinones, hydrogen peroxide, reactive oxygen species, tanshinones
Tanshinones, diterpenes isolated from the root of Salvia miltiorrhiza, have been widely used in Asia for the treatment of numerous disorders (cardiovascular and inflammatory diseases, hepatitis, and cancer) and exhibit diverse pharmacological properties (anti-oxidant, anti-inflammatory, and anti-cancer activity) (Figure 1).1 Although the mechanisms of these effects have been explored, accurate details remain elusive. Proposed mechanisms for the biological activity of tanhinones include DNA binding2 and induced intracellular production of reactive oxygen species.3 Despite these recent advances, the molecular basis of tanshinone activity remains poorly understood. Because telomerase is up-regulated in the majority of cancer cells and can directly affect apoptosis, we hypothesized that telomerase may be a key molecular target of tanshinones. Indeed, it was recently reported that tanshinone treatment can lead to decreased cellular telomerase activity commensurate with reduced levels of hTERT mRNA, the catalytic telomerase subunit.4 However, the pleiotropic effects of tanshinones preclude potential direct effects on telomerase to be determined from current reports.
Figure 1.
Tanshinones tested in this study.
Telomerase is a ribonucleoprotein complex that uses its RNA subunit to synthesize the repetitive G-rich DNA, 5′-TTAGGG/3′-AATCCC in humans, found at each 3′ chromosome end.5 Telomerase activity is present in the overwhelming majority of cancer cells and normal proliferative cells such as stem cells, but tends to be absent from human somatic cells.6 In the absence of telomerase, the termini of chromosomes, called telomeres, are not efficiently replicated.7 As a result of this end-replication problem, cells require active telomerase for persistent replication.8 Telomerase promotes tumorigenicity in part by helping maintain telomeric DNA length to overcome replicative senescence. Additionally, other functions of telomerase beyond the chromosome ends have been suggested to contribute to tumorigenesis.9 Notably, telomerase appears capable of inhibiting apoptosis by mechanisms independent of telomeric DNA synthesis10, 11 and can promote proliferation through the Wnt pathway.12 Pharmacological agents that inhibit telomerase with defined mechanisms would be useful agents to study telomerase biology and may serve as leads for therapeutic development. Here, we describe our characterization of the direct effect of several tanshinones on telomerase enzymatic activity.
We used a telomerase assembly assay13 to test the effect of tanshinones on telomerase. The assembly assay allows identification of small molecules that affect telomerase through several mechanisms including direct inhibition of its enzymatic activity, blocking specific required protein-RNA interactions,13 and inhibiting the chaperone hsp90, which is required to reconstitute telomerase activity in viro.14, 15 In telomerase assembly assays, Tan II-A (75% inhibition at 50 µM), Tan I (65% inhibition), and cryptotanshinone (75% inhibition), which contain ortho-quinone C rings, inhibited telomerase, whereas neo-tanshinlactone (25% inhibition), which has a lactone C ring, was a significantly less effective inhibitor. This suggests that the ortho-quinone moiety found in some tanshinones is a critical requirement for telomerase inhibition. The results also showed that an aromatic A ring (compare tanshinone I to Tan II-A) and a heteroaromatic D ring (compare Tan II-A to cryptotanshinone) are not required for telomerase inhibition.
Since Tan II-A directly inhibited telomerase, we determined if Tan II-A cytotoxicity correlated with telomerase. As expected, Tan II-A was a potent cytotoxin of a telomerase positive breast cancer cell line, MCF-7 (EC50 = 0.90 µM), and a prostate cancer cell line, PC-3 (EC50 = 1.2 µM), consistent with previous reports.1, 16–18 Furthermore, telomerase activity was reduced 50% in MCF-7 cells 24 hr after exposure to 3 µM Tan II-A (data not shown), confirming results with the structurally related Tan I-A reported by Liu et al.4 Telomerase negative GM847 and Saos-2 cell lines were less sensitive to Tan II-A cytotoxicity (EC50 of >20 µM).
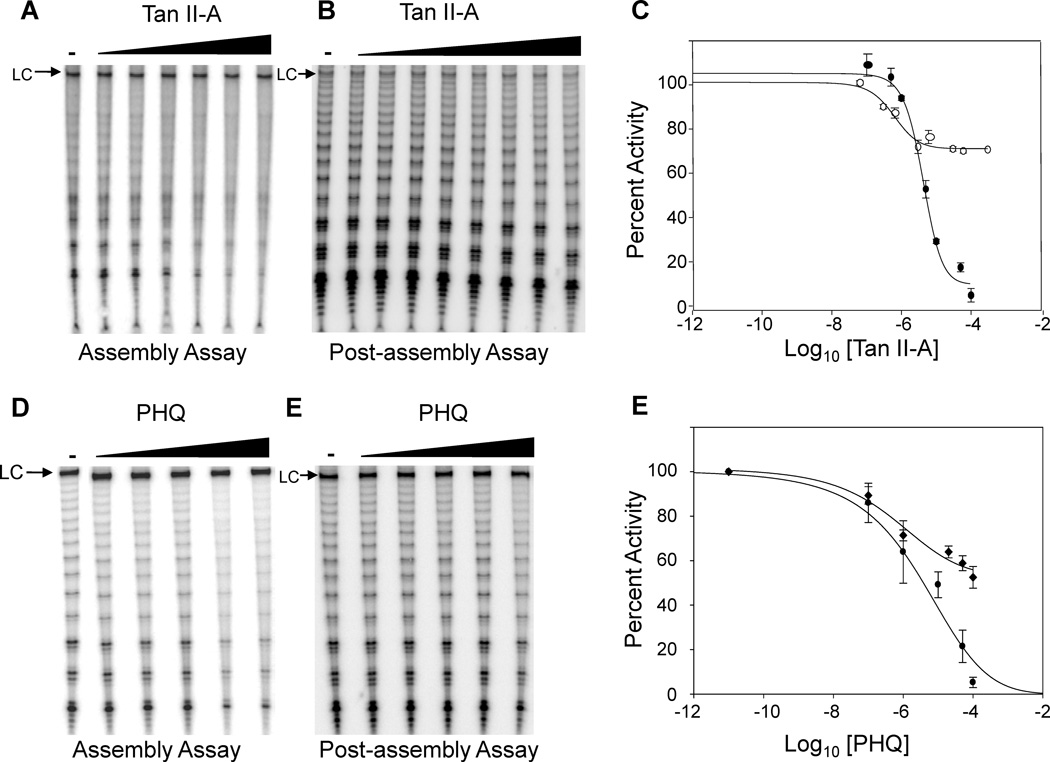
To examine the importance of the ortho-quinone moiety, we examined telomerase inhibition by Tan II-A and the simple ortho-quinone 9,10-phenanthrenequinone (PHQ). Tan II-A and PHQ were equipotent inhibitors (IC50 = 5.0 µM in the assembly assay) of telomerase (Figure 2). Both Tan II-A and PHQ were also inhibitors of pre-assembled telomerase (IC50 = 0.5 µM) (Figure 2). However, the compounds only partially inhibited telomerase activity when added to preassembled telomerase with the maximum amount of inhibition ~30% for Tan II-A ~50% for PHQ, compared to >95% when added before assembly.
Figure 2.
Tan II-A and PHQ inhibit telomerase. (A) Telomerase inhibition by Tan II-A in an assembly assay. hTERT and hTER were incubated with Tan II-A (0.1, 0.5, 1, 5, 10, 50 and 100 µM) in an assembly assay. hTERT was translated in rabbit reticulocyte lysates and incubated with in vitro transcribed hTER 1.5 h in the presence of inhibitor. Telomerase activity was then determined by direct primer extension assay.13 Telomerase products were resolved by gel electrophoresis, imaged by phosphorimaging, and quantified using ImageQuant. Percent telomerase activity was determined by comparison with a positive control containing DMSO as a carrier and products were normalized to a loading control (LC), which is a 100 nt 32P-labeled DNA oligonucleotide used to control for recovery and loading irregularities. (B) Inhibition of assembled telomerase by Tan II-A. Telomerase was reconstituted in rabbit reticulocyte lysate then assayed (post-assembly) by the direct primer-extension protocol in the presence of Tan II-A (0.06, 0.3, 0.6, 3, 6, 30, 60 and 300 µM). (C) Quantification of telomerase assays. The curve with ● symbols is data from the assembly assay, while the curve with ○ symbols is data from the post-assembly assay. Raw activity levels were obtained by gel analysis using ImageQuant and normalized to loading controls. (D) Telomerase inhibition by PHQ in an assembly assay. hTERT and hTER were incubated with PHQ (0.001, 0.1, 1, 10, 50 and 100 µM) for 1.5 h before telomerase activity was detected by direct primer extension assay. (E) Inhibition of pre-assembled telomerase by PHQ. Telomerase was reconstituted in rabbit reticulocyte lysate then assayed by direct primer-extension in the presence of PHQ (0.001, 0.1, 1, 10, 50 and 100 µM). (F) Quantification of telomerase assays. The curve with ● symbols is data from the assembly assay, while the curve with ♦ symbols is data from the post-assembly assay. Data in C and E are from at least three independent experiments and standard deviation is shown.
It is well documented that reactive oxygen species (ROS) are formed in the presence of a reducing agent (such as DTT), oxygen, and catalytic amounts of ortho-quinones capable of undergoing redox cycling.19, 20 This lead to the hypothesis that redox cycling is responsible for Tan II-A inhibition of telomerase. If redox cycling is responsible for inhibition, we would expect inhibition to be time and DTT dependent. To test this, we incubated purified pre-assembled telomerase with PHQ and varying concentrations of DTT for increasing time periods followed by a direct telomerase assay. Telomerase activity was inhibited by ~35% when pre-incubated with DTT (5 mM) for 5 min; however, it was >95% inhibited when telomerase was incubated in the presence of DTT for 60 min (Figure 3). We also found that DTT concentration directly correlated with extent of inhibition (Figure 3B). These findings are in agreement with results from Bova et al. showing that inactivation of protein tyrosine phosphate α by ortho-quinone compounds is greatly enhanced by DTT and that inhibition correlated with hydrogen peroxide generation.21 The time and DTT dependence of PHQ inhibition of telomerase are consistent with the hypothesis that telomerase inhibition by ortho-quinones is mediated by ROS. The concentration of free thiols, in particular glutathione, in cells is 1–5 mM, so the concentration of DTT used here is relevant to cellular conditions.
Figure 3.
Inhibition of telomerase by PHQ is time and DTT dependent. (A) Reconstituted telomerase was pre-incubated with or without PHQ (50 µM) and DTT (5 mM) for either 5 min or 60 min before telomerase activity was determined. Lane 1, DMSO only; lane 2, 5 min incubation with DTT and PHQ; lane 3, 60 min incubation with DTT and PHQ. (B) Reconstituted telomerase was incubated with or without PHQ (50 µM) and increasing concentrations of DTT for 60 min prior to telomerase assay. Lane 1, DMSO and PHQ; lane 2, PHQ and 500 nM DTT; lane 3, PHQ and 50 µM DTT; lane 4, PHQ and 5 mM DTT. Telomerase activity was measured by direct primer extension assay, percent telomerase activity was determined by comparison with the DMSO-only control, and products were normalized to a loading control, LC as described in the legend of Figure 2. Percent activities are from three independent experiments (average SD < 10%).
Because catalase is a hydrogen peroxide scavenger, we investigated its ability to protect telomerase from ortho-quinone generated ROS. We pre-incubated purified telomerase with PHQ, DTT, and catalase for 1 hour before performing a telomerase assay. As predicted, catalase completely protected telomerase (Figure 4). In order to test if the inactivation of telomerase by PHQ is reversible, purified telomerase was pre-incubated with PHQ and DTT for 1 hour prior to the addition of catalase. We found that the addition of catalase after 1 hour did not rescue telomerase activity, suggesting that inactivation is irreversible (Figure 4, lane 4).
Figure 4.
Catalase protects telomerase from inhibition by PHQ in the presence of DTT. Reconstituted telomerase was incubated with PHQ in the presence or absence of catalase, and telomerase activity was determined by direct primer extension assay. Lane 1, DMSO only; lane 2, telomerase was incubated with PHQ (50 µM), DTT (5mM), and catalase (3.5U) for 1hr; lane 3, telomerase was incubated with PHQ (50 µM) and DTT (5mM) for 1hr; lane 4, telomerase was incubated with PHQ (50 µM) and DTT (5mM) for 1hr prior to addition of catalase (3.5U). Percent telomerase activity was determined by comparison with the DMSO-only control, and products were normalized to a loading control, LC as described in the legend of Figure 2. Percent activities are from three independent experiments (average SD < 10%).
The observation that catalase can protect telomerase from ortho-quinone inhibition raised the possibility that ortho-quinone tanshinones produce reactive oxygen species such as superoxide and hydrogen peroxide that could subsequently inactivate telomerase. To examine this possibility, we incubated Tan II-A and PHQ separately with DTT in PBS and monitored the production of hydrogen peroxide. Tan II-A generated hydrogen peroxide in a time-dependent fashion that plateaued at 60 min resulting in the formation of ~2.5 mM hydrogen peroxide (Figure 5). In contrast, when Tan II-A was incubated in PBS alone without DTT, hydrogen peroxide formation was not observed. Additionally, incubation with catalase completely blocked hydrogen peroxide production as expected.
Figure 5.
Tan II-A produces hydrogen peroxide. (A) Tan II-A (20 µM) was incubated in PBS with 5 mM DTT (blue bar), with 5 mM DTT and 30 U catalase (red bar), or in the absence of DTT (white bar) for the indicated times. Production of hydrogen peroxide was measured using a hydrogen peroxide kit from Assay Designs. Data are from two independent experiments, columns represent the mean and the error bars represent the standard deviation. Increased hydrogen peroxide at 60 min compared to 30 min was confirmed by one way ANOVA.
The results of this study reveal that ortho-quinone containing tanshinones are telomerase inhibitors owing to their ability to engage in redox cycling leading to hydrogen peroxide production. These findings have important implications for the clinical use of tanshinones and reveal for the first time that telomerase itself is a biologically significant and direct target of ROS. The results also add to the growing evidence that induction of ROS is a primary mechanism of Tan II-A biological activity.3, 22 Given that the ROS induction we observed is directly related to the concentration of reducing equivalent DTT, we predict that the ability to generate ROS in cells may be similarly controlled by cellular redox environment, which may help to explain the pleiotropic effects of Tan II-A and other tanshinones. The cell-based assays reported here suggest that Tan II-A is cytotoxic to both telomerase-positive and telomerase-negative cells, but that telomerase-positive cell lines were more sensitive to Tan II-A than telomerase-negative cells. Initially we considered that this was due to oncogenic addiction of telomerase by the telomerase-dependent cancer cells. However, the realization that Tan II-A is redox active leads to the hypothesis that the cytotoxicity profile observed is dependent on the levels of reducing agents and thus the redox environment of the cells. It is known that the redox environment of cancer cells is imbalanced so it is likely that redox active compounds like Tan II-A will have varying effects on cells dependent on redox status.23 Interestingly, tanshinones act as anti-oxidants in some cells, perhaps by affecting transcription of key anti-oxidant genes.24
It is clear that Tan II-A inhibits in vitro reconstituted recombinant telomerase through direct ROS damage, but the effects of Tan II-A on cellular telomerase may be an indirect or a direct effect of ROS. Tan II-A has been shown to affect several biological targets and has diverse biological activity.1 For example, Tan II-A inhibits expression of genes required for osteoclast differentiation and activity while not inducing apoptosis.25 Another study showed that Tan II-A-induced apoptosis of human hepatocellular carcinoma cells coincides with up-regulation of pro-apoptotic proteins fas, bax, and p53 concomitant with down-regulation of bcl2, and c-myc.16 Decreased c-myc activity could account for the decreased telomerase activity in Tan II-A treated cells since c-myc regulates hTERT transcription.26 These and other cell-type dependent Tan II-A effects1 presumably arise from its ability to affect several targets perhaps through several mechanisms: ROS (such as hydrogen peroxide generation), DNA binding, and possibly direct protein binding. As a result, tanshinones and other ortho-quinone compounds capable of redox-cycling are poor leads for a target-based drug discovery approach since ROS undoubtedly affects many cellular targets.
We conclude that the mechanism of telomerase inhibition by Tan II-A is ROS dependent, providing evidence that telomerase is directly regulated by ROS. It is known that increased ROS can accelerate telomere loss in diverse cell types.27 Although telomere loss occurs naturally with each round of DNA replication, telomere shortening can be accelerated by the presence of ROS, which has been attributed to oxidation of telomeric DNA. In addition, telomerase activity was reported to be sensitive to oxidative stress in endothelial28–30 and vascular smooth muscle cells,31 commensurate with accelerated telomere loss. Several mechanisms for ROS-dependent telomerase inhibition can be considered and there is evidence for decreased transcription of telomerase RNA32 and hTERT.33 In addition, the potential effects of cellular ROS on telomerase are complicated by the observation that the subcellular location of telomerase is ROS sensitive.34, 35 For example, hTERT appears excluded from or transported out of the nucleus and can localize to the mitochondria in mildly stressed cells. Evidence that telomerase can, in different cells, protect the mitochondria from ROS36 or enhances ROS damage to mitochondrial DNA37 have been reported. Telomeric DNA has also been shown to be sensitive to oxidative stress,38, 39 but there is no reported evidence suggesting that ROS can directly inhibit telomerase.
The results here contribute to the mechanistic understanding of ROS induced telomere dysfunction by showing that ROS directly inhibits telomerase. While ROS can affect many cellular targets, it is likely that mild ROS induced telomere dysfunction is an important contributor to negative ROS effects. ROS-induced telomere dysfunction appears to occur by multiple mechanisms: inhibition of telomerase, changes in the subcellular telomerase location, and damage to telomeric DNA. Current studies are underway to identify the redox-sensitive residues in the telomerase complex and to examine the biological significance of the telomerase sensitivity to ROS. The findings reported here suggest a critical role of oxidative damage to telomerase in cellular aging and in cancer cell biology and portend a role of ROS and perhaps reactive nitrogen species in regulating telomerase.
Supplementary Material
Acknowledgments
This work was supported in part by NSF grant MCB-0751372 (MBJ) and a Graduate Student Research Grant from AFAR (JS). The authors wish to thank Drs. Lea Harrington, Sheila Stewart, and Robert Weinberg for sharing regents and cell lines.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data
Experimental procedures associated with this article can be found in the online version.
References and notes
- 1.Wang X, Morris-Natschke SL, Lee KH. Med. Res. Rev. 2007;27:133. doi: 10.1002/med.20077. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Z, Zhang J, Jin L, Song T, Wu G, Gao J. Biol. Pharm. Bull. 2008;31:2342. doi: 10.1248/bpb.31.2342. [DOI] [PubMed] [Google Scholar]
- 3.Lee WY, Liu KW, Yeung JH. Cancer Lett. 2009;285:46. doi: 10.1016/j.canlet.2009.04.040. [DOI] [PubMed] [Google Scholar]
- 4.Liu XD, Fan RF, Zhang Y, Yang HZ, Fang ZG, Guan WB, Lin DJ, Xiao RZ, Huang RW, Huang HQ, Liu PQ, Liu JJ. Int. J. Mol. Sci. 2010;11:2267. doi: 10.3390/ijms11062267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Autexier C, Lue NF. Annu. Rev. Biochem. 2006;75:493. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- 6.Shay JW, Bacchetti S. Eur. J. Cancer. 1997;33:787. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 7.Moon IK, Jarstfer MB. Front. Biosci. 2007;12:4595. doi: 10.2741/2412. [DOI] [PubMed] [Google Scholar]
- 8.Aubert G, Lansdorp PM. Physiol. Rev. 2008;88:557. doi: 10.1152/physrev.00026.2007. [DOI] [PubMed] [Google Scholar]
- 9.Cong Y, Shay JW. Cell Res. 2008;18:725. doi: 10.1038/cr.2008.74. [DOI] [PubMed] [Google Scholar]
- 10.Cao Y, Li H, Deb S, Liu JP. Oncogene. 2002;21:3130. doi: 10.1038/sj.onc.1205419. [DOI] [PubMed] [Google Scholar]
- 11.Rahman R, Latonen L, Wiman KG. Oncogene. 2005;24:1320. doi: 10.1038/sj.onc.1208232. [DOI] [PubMed] [Google Scholar]
- 12.Park JI, Venteicher AS, Hong JY, Choi J, Jun S, Shkreli M, Chang W, Meng Z, Cheung P, Ji H, McLaughlin M, Veenstra TD, Nusse R, McCrea PD, Artandi SE. Nature (London) 2009;460:66. doi: 10.1038/nature08137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keppler BR, Jarstfer MB. Biochemistry. 2004;43:334. doi: 10.1021/bi035583e. [DOI] [PubMed] [Google Scholar]
- 14.Keppler BR, Grady AT, Jarstfer MB. J. Biol. Chem. 2006;281:19840. doi: 10.1074/jbc.M511067200. [DOI] [PubMed] [Google Scholar]
- 15.Holt SE, Aisner DL, Baur J, Tesmer VM, Dy M, Ouellette M, Trager JB, Morin GB, Toft DO, Shay JW, Wright WE, White MA. Genes Dev. 1999;13:817. doi: 10.1101/gad.13.7.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan SL, Wei YQ, Wang XJ, Xiao F, Li SF, Zhang J. World J. Gastroenterol. 2004;10:2024. doi: 10.3748/wjg.v10.i14.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Bastow KF, Sun CM, Lin YL, Yu HJ, Don MJ, Wu TS, Nakamura S, Lee KH. J. Med. Chem. 2004;47:5816. doi: 10.1021/jm040112r. [DOI] [PubMed] [Google Scholar]
- 18.Ryu SY, Lee CO, Choi SU. Planta Med. 1997;63:339. doi: 10.1055/s-2006-957696. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q, Dube D, Friesen RW, LeRiche TG, Bateman KP, Trimble L, Sanghara J, Pollex R, Ramachandran C, Gresser MJ, Huang Z. Biochemistry. 2004;43:4294. doi: 10.1021/bi035986e. [DOI] [PubMed] [Google Scholar]
- 20.Wardman P. Free Radic. Res. Commun. 1990;8:219. doi: 10.3109/10715769009053355. [DOI] [PubMed] [Google Scholar]
- 21.Bova MP, Mattson MN, Vasile S, Tam D, Holsinger L, Bremer M, Hui T, McMahon G, Rice A, Fukuto JM. Arch. Biochem. Biophys. 2004;429:30. doi: 10.1016/j.abb.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 22.Chiu TL, Su CC. Int. J. Mol. Med. 2010;25:231. [PubMed] [Google Scholar]
- 23.Acharya A, Das I, Chandhok D, Saha T. Oxid. Med. Cell. Longev. 2010;3:23. doi: 10.4161/oxim.3.1.10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang HS, Wang SQ. Biochem. Pharmacol. 2007;73:1358. doi: 10.1016/j.bcp.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Kim HH, Kim JH, Kwak HB, Huang H, Han SH, Ha H, Lee SW, Woo ER, Lee ZH. Biochem. Pharmacol. 2004;67:1647. doi: 10.1016/j.bcp.2003.12.031. [DOI] [PubMed] [Google Scholar]
- 26.Horikawa I, Cable PL, Afshari C, Barrett JC. Cancer Res. 1999;59:826. [PubMed] [Google Scholar]
- 27.Passos JF, Saretzki G, von Zglinicki T. Nucleic Acids Res. 2007;35:7505. doi: 10.1093/nar/gkm893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furumoto K, Inoue E, Nagao N, Hiyama E, Miwa N. Life Sci. 1998;63:935. doi: 10.1016/s0024-3205(98)00351-8. [DOI] [PubMed] [Google Scholar]
- 29.Haendeler J, Hoffmann J, Diehl JF, Vasa M, Spyridopoulos I, Zeiher AM, Dimmeler S. Circ. Res. 2004;94:768. doi: 10.1161/01.RES.0000121104.05977.F3. [DOI] [PubMed] [Google Scholar]
- 30.Kurz DJ, Decary S, Hong Y, Trivier E, Akhmedov A, Erusalimsky JD. J. Cell Sci. 2004;117:2417. doi: 10.1242/jcs.01097. [DOI] [PubMed] [Google Scholar]
- 31.Matthews C, Gorenne I, Scott S, Figg N, Kirkpatrick P, Ritchie A, Goddard M, Bennett M. Circ. Res. 2006;99:156. doi: 10.1161/01.RES.0000233315.38086.bc. [DOI] [PubMed] [Google Scholar]
- 32.Napier CE, Veas LA, Kan CY, Taylor LM, Yuan J, Wen VW, James A, O'Brien TA, Lock RB, MacKenzie KL. Biochim. Biophys. Acta. 2010;1803:1142. doi: 10.1016/j.bbamcr.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 33.Pizzimenti S, Briatore F, Laurora S, Toaldo C, Maggio M, De Grandi M, Meaglia L, Menegatti E, Giglioni B, Dianzani MU, Barrera G. Free Radic. Biol. Med. 2006;40:1578. doi: 10.1016/j.freeradbiomed.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 34.Haendeler J, Hoffmann J, Brandes RP, Zeiher AM, Dimmeler S. Mol. Cell. Biol. 2003;23:4598. doi: 10.1128/MCB.23.13.4598-4610.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katakura Y, Yoshizaki K, Yasuda T, Tsunematsu T, Uehara N, Miura T, Fujiki T, Shirahata S. Biochem. Biophys. Res. Commun. 2005;334:450. doi: 10.1016/j.bbrc.2005.06.109. [DOI] [PubMed] [Google Scholar]
- 36.Ahmed S, Passos JF, Birket MJ, Beckmann T, Brings S, Peters H, Birch-Machin MA, von Zglinicki T, Saretzki G. J. Cell Sci. 2008;121:1046. doi: 10.1242/jcs.019372. [DOI] [PubMed] [Google Scholar]
- 37.Santos JH, Meyer JN, Van Houten B. Hum. Mol. Genet. 2006;15:1757. doi: 10.1093/hmg/ddl098. [DOI] [PubMed] [Google Scholar]
- 38.Henle ES, Han Z, Tang N, Rai P, Luo Y, Linn S. J. Biol. Chem. 1999;274:962. doi: 10.1074/jbc.274.2.962. [DOI] [PubMed] [Google Scholar]
- 39.Oikawa S, Tada-Oikawa S, Kawanishi S. Biochemistry. 2001;40:4763. doi: 10.1021/bi002721g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.