Abstract
A rose may be a rose by any other name, but when you call a dog a poodle it becomes a very different animal than if you call it a bulldog. Both the poodle and the bulldog are examples of dog breeds of which there are >400 recognized world-wide. Breed creation has played a significant role in shaping the modern dog from the length of his leg to the cadence of his bark. The selection and line-breeding required to maintain a breed has also reshaped the genome of the dog resulting in a unique genetic pattern for each breed. The breed-based population structure combined with extensive morphologic variation and shared human environments have made the dog a popular model for mapping both simple and complex traits and diseases. In order to obtain the most benefit from the dog as a genetic system, it is necessary to understand the effect structured breeding has had on the genome of the species. That is best achieved by looking at genomic analyses of the breeds, their histories, and their relationships to each other.
Introduction
The domestic dog, as a genomic subject, fills a very interesting gap between humans and a laboratory model such as the mouse. As a species, humans are numerous and wide-spread. We all share a common ancestor dating back ~150,000 yrs., but have spread across the globe inhabiting all continents and terrains (Cann et al. 1987; Conrad et al. 2006; Templeton 2002). We are exposed to many different environments, both natural and man-made, and possibly as a consequence, are susceptible to a tremendous number of diseases, for which we would like to identify causes and find cures. On the other hand, laboratory mice, as well as other model species, are derived from natural populations, but exist in any of a hundred or more different homozygous strains created by back-crossing and inbreeding to obtain a consistent genetic background on which to introduce mutation (Aylor et al. 2011; Brown 1996). They are raised in controlled environments for the purpose of reducing experimental variables. Although many diseases and disorders can be induced within these lineages, they do not develop naturally and are able to mimic the complex human conditions only in bits and pieces through individual pathways and mutations.
The domestic dog sits on the continuum between these two extremes. They, like humans, stem from a common ancestor or ancestral group some 15–30,000 years ago and have since spread into all parts of the world (Lindblad-Toh et al. 2005; Savolainen et al. 2002; Vila et al. 1997; Vonholdt et al. 2010). They have adapted to human environments and utilize the same shelters and food sources. In addition, they are susceptible to many of the same diseases, and receive diagnoses and medical treatment from veterinary health specialists. However, purebred domestic dogs exist in small somewhat homogeneous strains called breeds. These breeds were created through backcrossing and inbreeding, to fix desired traits, resulting in a much-reduced level of heterozygosity within any one breed compared to wild canids or non-breed dogs (Boyko et al. 2009; Boyko et al. 2010; Lindblad-Toh et al. 2005). While the breeds are not entirely homogeneous, there are regions of the genome, often those that contribute to specific traits, that are fixed or nearing fixation within the breed (Akey et al. 2010; Lindblad-Toh et al. 2005; Parker et al. 2009; Quilez et al. 2011; Sutter et al. 2007; Vaysse et al. 2011). This intermittent homozygosity can reduce the number of genetic mutations segregating within a breed to create a complex phenotype. Alternately, they may enhance the effect of a single mutation by increasing the chance of homozygosity, either at the allele or within the pathway, exposing genetic background effects. Thus, the dog supplies the best of both organisms: the complexity of the human species with all of its disorders and environmental effects, and the reduced complexity of the laboratory mouse with discreet strains and partially fixed backgrounds. The dual nature of the domestic dog species provides a perfect background on which to base a genomic analysis of complex traits or diseases. But in order to design such an experiment properly, it is important to understand the population structure of the dog.
History of breed relationships
Dogs can be found in ancient artwork dating back to the earliest civilizations. From Asia, Africa, southern Europe and South America, ancient people depicted dogs with very different appearances including, relative size, shape, color, ear carriage, etc. (Alderton 2002; Fogle 1993). According to drawings and sculptures, different types of dogs have been in existence for millennia. However, it was not until the Victorian era that the breeds as we recognize them today came into being. In the late 1800’s the common approach to dog breeding took a turn that remains in effect to this day. During that period clubs were formed to propagate specific types of dogs and competitions were developed to reward the creation of perfect specimens of a breed based on physical criteria (Alderton 2002; American 1998). Prior to this time dog competitions focused on the working ability of the dog such as hunting and chasing rather than on their outward appearance. In order to carry out the new competitions new rules were introduced to control breeding so that registering a dog with a breed club required that both of the dog’s parents be registered members of the breed club as well, effectively isolating each breed and reducing the available gene pool (American 1998). In addition, standards were set to describe the perfect breed representative which further encouraged line breeding to propagate the physical traits that are easier to spot early in the dog’s development. The popularity of breed clubs and the stringency of the rules and competitions created a venue for the development of many new breeds which led to the breed explosion of the 1900’s and the current ~400 breeds recognized today (Wilcox and Walkowicz 1995).
The majority dog breeds are isolated by rules and by whim but not by a physical barriers as in a laboratory strain. They are kept relatively pure but outcrossing has been known to occur, preventing complete homozygosity. In addition, the same breed in different countries may be subjected to slightly different selective pressures maintaining some level of heterozygosity across the breed as a whole. For example, the requirements for height in the Labrador retriever differ by ~2 inches when comparing US and UK standards (Wiles-Fone and Barnes 1997). In a study comparing individuals from four breeds in the US and France, it was found that some breeds show significant genomic differences between the continents even though the dogs are considered to be from the same breed and mixing between them is not prohibited (Quignon et al. 2007). In this regard the dog is unlike either mouse strains or human races. Mouse strains are physically isolated and breeding of crosses is done only under strict supervision for the purpose of scientific discovery. And while, there have been periods in our history in which groups of people have been isolated from others creating small distinct populations or races, there is no true barrier between people and the re-mixing of genomes is common place world-wide.
Microsatellite analyses of the breeds
To understand the effect of breed structure on the genomic make-up of the dog, molecular markers can be put to use both in small and large datasets. Early genetic studies concentrated on sequences from the mitochondria as these were short, abundant, and provide a relatively large amount of variation to analyze from a single assay. Mitochondrial sequences showed modest but imperfect breed clustering and were much more successful at distinguishing between species rather than breeds (Gao et al. 2004; Okumura et al. 1996; Savolainen et al. 2004; Vila et al. 1999; Vila et al. 1997; Vila et al. 2003). Mitochondrial DNA analyses have played a large role in understanding the ancient history of dogs and their descent from the grey wolf (Pang et al. 2009; Savolainen et al. 2002; Vila et al. 1997)
The first studies of genomic patterns involved small numbers of breeds and microsatellite based marker sets. From these studies it was noted that different breeds displayed different allele frequencies (Fredholm and Wintero 1995; Irion et al. 2003; Koskinen and Bredbacka 2000; Zajc et al. 1997). In addition most markers are not in Hardy Weinberg equilibrium within the breeds and they show particularly high inbreeding coefficients (Irion et al. 2003; Koskinen 2003). Because of the small numbers of breeds, little information could be gleaned regarding their relationships but they showed that each individual breed displayed its own unique pattern of marker alleles and supported the hypothesis that there was much less variation within a breed than across the species.
In 2004 a study was released examining the relationships between 85 different breeds using a dataset of 96 microsatellite markers (Parker et al. 2004). The microsatellite markers were chosen from all autosomes of the dog. Di-nucleotide repeats were chosen specifically as, in the dog, these have been shown to mutate at a rate 100 times lower than the commonly used tetra-nucleotide mapping microsatellites (Francisco et al. 1996; Ostrander et al. 1993). This is important as the alleles from these markers are more likely to be identical by decent among the breeds rather than identical by state. Calculating Wright’s F statistics based on the alleles the authors showed that more than one quarter of the genomic variation in the dog is found in differences between breeds rather than between individuals. Compare this number to the 5–10% of variation found between geographically distinct human populations and you find that dog breed definitions are more isolating than oceans. Using both a leave-one-out analysis involving a training set, and untutored clustering the dogs could be assigned to their breed of origin with up to 99% accuracy once again highlighting the extreme population stratification within the species.
The separation of the breeds being well established, the next task was to assess the similarities between the breeds. Breed clubs have developed systems of classifying dogs for registration and competition based on expected occupation or geographic origin. For instance the American Kennel Club groups the breeds into seven categories primarily based on behavior such as the Sporting group which comprises the hunting dogs such as retrievers and spaniels and the Terrier group which includes the ratters and earth dogs (American 1998). The Federation Cynologique Internationale (FCI) has 10 categories of breeds based on behavior that are further divided into 2–10 groups each based on appearance, origin, or very specific usage (Federation Cynologique 2010). For instance the FCI Sighthound category is divided into long, rough and smooth coated sighthounds while the FCI Scenthound category is divided into the scenthounds, leashed scent hounds, and scenthound related breeds. Using the 96 microsatellite dataset described above, and a combination of phylogenetic and clustering methodologies, the breeds were examined from a purely molecular viewpoint. These methods were then used to determine if the club breed groupings were based in genetic similarities stemming from common founders (Parker et al. 2004; Parker et al. 2007).
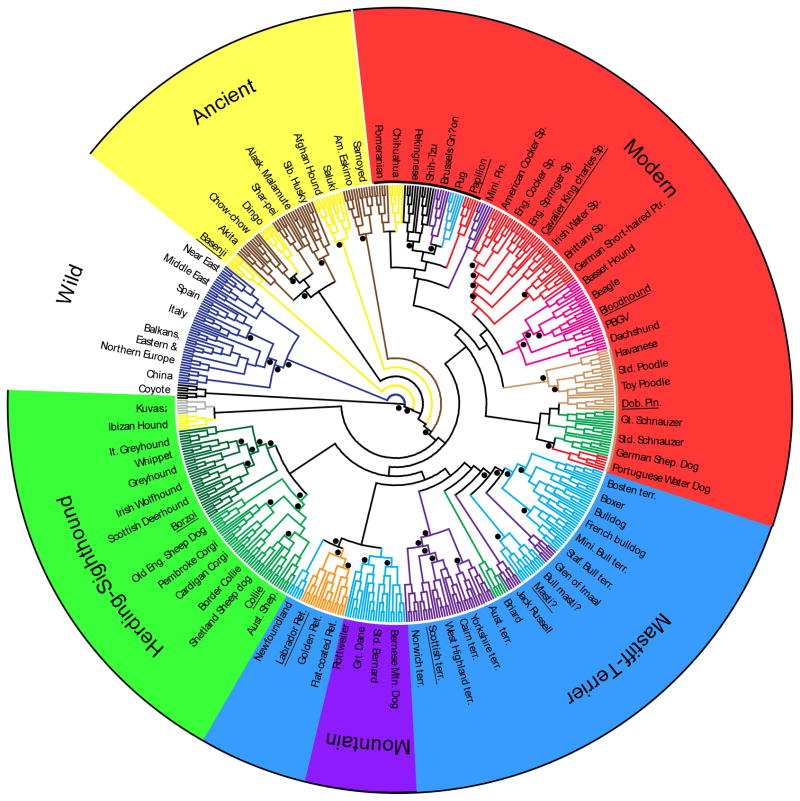
The use of phylogenetic trees to describe breed relationships is not optimal as the development of a breed and the evolution of a species are not equivalent events. While a small number of breeds may develop as an offshoot of a single founding population, most are created through hybridization of two or more original breeds or breed types (American 1998; Wilcox and Walkowicz 1995). This type of reticulated phylogeny is not well visualized using a distance matrix or through classic tree shaped dendrograms. Nevertheless, the use of such analyses can display sub-species like groupings of more closely related breeds, with the assumption that these individuals would be more similar to each other than to individuals from another population (Cornuet et al. 1999). When applied to the 85 breeds in the above study, the trees did identify small sets of closely related breeds such as the Shetland sheepdog and the collie. More interesting was the identification of a set of seemingly unrelated dogs that branched apart from the majority of breeds (Figure 1). These breeds, later referred to as the ancient breeds, comprised the Asian breeds such as Akita and Chow Chow, the northern breeds such as Siberian husky and Alaskan Malamute, the Middle Eastern saluki and Afghan hounds, and the African basenji, with the basenji being furthest removed from the modern dogs (Parker et al. 2004). While pockets of this group share obvious common origins, as a whole they are highly diverse and may offer some insight into either early or alternate domestication events taking place within the dog species. The remaining breeds, primarily of European origin, all formed a single node on the tree with little significant branching indicating a single origin for these modern breeds and/or extensive hybridization between the European breeds.
Figure 1.
Neighbor joining tree from 85 dog breeds showing the Ancient breed cluster. Ninety-six microsatellite markers were used to calculate the genetic distance between 85 breeds of dog. Using the gray wolf to root the tree, the nine breeds nearest the wolf formed statistically significant branches. The other 76 breeds form a single node indicating shared ancestry and hybridization between the branches. Figure originally published in Science (Parker et al. 2004).
To better refine the relationships between the breeds, especially within the modern “hedge”, a clustering analysis was applied to the same dataset. Clustering using the Structure program does not require calculation of distance matrices but groups all individuals based on similarities in the patterns of alleles carried (Pritchard and Rosenberg 1999; Pritchard et al. 2000). Using this method the 85 breeds were assigned to four clusters of breeds (Parker et al. 2004). The breeds listed above as the ancient breeds formed the first cluster. This group also included the wolf when a small number were added to the dataset. The second cluster comprised the mollosser type dogs such as the mastiff and bulldog. The third cluster comprised a combination of two breed types, the sighthounds and the herding dogs. Finally, the fourth and largest group was made up of hunting dogs, toy dogs and hounds. This was the first classification system for breeds that was developed through genetic data alone. These analyses clearly displayed a two-tiered population structure in the dog. Not only do breeds play a role in stratifying the individuals, but the breeds themselves are grouped by common genetic patterns.
Following the original clustering paper an additional 223 dogs were genotyped to create a dataset of 130 breeds, covering the majority of the AKC recognized breeds and including breed types that were underrepresented in the 85 breed analysis (Parker et al. 2007). The new analysis revealed a fifth breed cluster, the mountain dogs. Increasing the number of terrier breeds by 150% highlighted the common hybridization of mastiff-types with terriers to create the “bull”-terriers. In the mid 1800s dog fighting reached a peak in popularity and breeds were created specifically for the sport. The most successful cross created for this purpose combined the tenacity and energy of the terrier with the power and devotion of the molossers (Frome 1999 (rev. 2004)). These dogs, the bull-terriers, rose to popularity and remain so to this day though the sport has long since fallen from grace. Thus these two diverse breed-types are linked into a single breed cluster through their more recently created progeny rather than common ancestry.
There are also a small number of breeds that do not form unique clusters through microsatellite analysis but instead tend to group with another breed. These pairs may indicate breeds that are only recently separated such as the Norwich and Norfolk terriers which were one breed until 1964 (American 1998). Other breeds that grouped as one were the greyhound and whippet, collie and Shetland sheepdog, and bull terrier and miniature bull terrier. Based on appearance, all of these pairs could be size variations of a common type. The clustering analysis indicates that they likely have the same ancestors and may have undergone recent selection based on size. Some of the newest breeds, especially those created through hybridization such as the Australian shepherd (Wilcox and Walkowicz 1995), did not form clear single clusters but instead show the involvement of more than one founding population (Parker et al. 2004; Parker et al. 2007).
SNPs and Breeds
The first SNP analyses of dog breeds came from small datasets acquired from resequencing short chromosomal segments (Brouillette and Venta 2002; Parker et al. 2004). Calculations from SNPs genotyped in 60 breeds showed a >4 fold range in heterozygosity across the breeds and and a positive correlation with breed population size. For example, the Labrador retriever, the most popular breed in the US with annual registrations nearing 100,000 for the past 10 yrs. was among the most heterozygous breeds while the Scottish deerhound, with only 100 dogs registered each year, were among the lowest. However, deviations from Hardy-weinberg expectations within the dog breeds, such as bottlenecks, selection, and popular sire effects, create imperfections in the correlation between heterozygosity and population size.(Nielen et al. 2001) Popular breeds such as the boxer and collie can be found at the lowest end of the spectrum while uncommon breeds like the Spinone Italiano rank among the most heterozygous (Parker et al. 2004; Parker et al. 2006b).
Since the release of the whole genome sequence of the dog (Kirkness et al. 2003; Lindblad-Toh et al. 2005), many studies have turned to SNPs over microsatellites because of the ease in genotyping a bi-allelic marker and the ability to multiplex thousands of markers in one reaction. More than 2 million SNPs were identified across the dog genome from comparison of the 7.5x boxer sequence to the 3x poodle sequence and 100,000 reads from 10 diverse breeds of dog. SNP genotyping chips were developed and made publically available (Karlsson et al. 2007; Lequarre et al. 2011). These chips proved highly useful in association studies mapping morphologic traits (Cadieu et al. 2009; Drogemuller et al. 2008; Karlsson et al. 2007; Olsson et al. 2011; Parker et al. 2009; Salmon Hillbertz et al. 2007; Vaysse et al. 2011), diseases (Awano et al. 2009; Bannasch et al. 2010; Downs et al. 2011; Farias et al. 2010; Farias et al. 2011; Meurs et al. 2010; Wang et al. 2011; Wilbe et al. 2010; Zeng et al. 2011) and behaviors (Dodman et al. 2010), in individual dog breeds and in some cases across multiple breeds.
To investigate the effectiveness of a large SNP marker set in determining breed structure and relatedness, a set of 912 dogs from 85 breeds were genotyped on the Affymetrix version 2 canine SNP chip which provided alleles calls for ~48,000 SNPs across all canine autosomes (Boyko et al. 2010). These SNPs were analyzed in multiple ways including distance trees, clustering, single SNP analyses and haplotype analyses with groups of 5–15 SNPs. Relatedness was calculated across breeds and within breeds by assessing the degree of allele and haplotype sharing between individuals (Vonholdt et al. 2010). Phylogenetic analysis of 80 breeds that were represented by >6 individuals each identified the original five breed clusters and went a step further by dividing the breeds within these clusters for a final set of ten breed groups (Figure 2). For instance, the herding breeds and sighthounds formed two distinct branches on the phylogenetic tree of breeds. These branches stem from a single node showing clearly that there is ancient common ancestry between the two breed groups, but more recent common ancestry and/or hybridization between the breeds within each individual group. Similarly the terriers and mastiff-types are divided into separate branches stemming from a common node. Within the section of the tree that would make up the Modern cluster the branch support is not strong but there appears to be a distinction between the toy, spaniel, hound, working, and non-sporting breeds that were previously lumped under one banner. Once again the ancient breeds break off first, after wolf, and form four different branches according to region of origin with no single shared node. As mentioned before, wolves cluster with the ancient breeds in the microsatellite analysis. With the SNP data set, 225 wolves from four continents were genotyped and included in the population analyses. Clustering indicated that only the ancient breeds show any significant hybridization with wolves (Vonholdt et al. 2010). This may explain the increased distance between these breeds and the modern breeds as indicated by both SNP and microsatellite analyses.
Figure 2.
Clustering breeds based on SNP markers increases breed relationship definition. A neighbor joining tree build from comparisons of 10-SNP haplotypes groups 80 dog breeds into approximately 10 breed clusters. Comparable clusters from a microsatellite analysis of 130 breeds (Parker et al. 2007) are indicated by the colored bars outside of the tree. The haplotype tree of breeds was originally published in Nature (Vonholdt et al. 2010).
The primary difference between the phylogenetic analysis based on SNPs and the cluster analysis based on microsatellites is that clustering shows hybridization between groups whereas the phylogenetic tree shows a single, and apparently absolute, placement of each breed in one group. For instance, the Pekingese and shih tzu breeds group with the toy dogs on the phylogenetic tree as part of the modern breeds, but in the cluster analysis they are ~30% ancient as would be expected based on their Asian ancestry (Parker et al. 2007). Similarly, using a cluster analysis, the bloodhound displays an unexpected mixture of both mountain dog and modern breed types while the tree places it solely with the other hound breeds in the modern group. Therefore in order to obtain the most accurate view of both breed development and current relationships, it is important to consider both views.
Calculation of Wright’s F statistics based on the SNP data corroborated earlier evidence showing large amounts of variation between breeds as opposed to variation between individuals. In addition, the calculations show that nearly 4% of overall variation was found between the breed clusters (Vonholdt et al. 2010). This additional level of complexity in dog population structure can be used to design mapping studies for common disorders in the dog (Parker et al. 2006a; Parker et al. 2010). For instance, comparing disease frequency across multiple breeds of dog, T-cell lymphomas are significantly increased in the ancient breeds whereas B-cell lymphomas are common among all breed types (Modiano et al. 2005). This indicates that there may be a common mutation shared among the ancient breeds that leads to T-cell lymphomas while B-cell lymphomas are likely to be much more heterogeneous. A mapping strategy for T-cell lymphoma might include haplotype comparisons across all affected breeds within the ancient group in an effort to find an ancestral mutation. Alternately, the strategy for B-cell lymphomas would be to restrict the analysis to one breed at a time to reduce the number of loci and increase association values. Combinations of these strategies can be found in many of the genome wide association studies released in the last few years.
Mixed Breeds
Although the discussion of canine genetics often refers to the breed structure and alterations in the genome that relate to inbreeding and selection, the vast majority of dogs in the United States are of mixed heritage, also known as mutts (American 2002). What can understanding breed history tell us about these non-pedigreed companions? Boyko and colleagues examined SNP and microsatellite data from village dogs and strays collected in Africa and the Americas. These dogs, though not affiliated with any breed group, display association with the same loci controlling size and body shape as the purebred dogs (Boyko et al. 2010). In addition, the non-pedigreed dogs from the Americas show genetic signatures of a mixture of European breeds likely sharing the same ancestral backgrounds but in a currently jumbled format (Boyko et al. 2009). This implies that the same genetic mutations will be found in mixed-breed dogs as are found in pure-bred dogs.
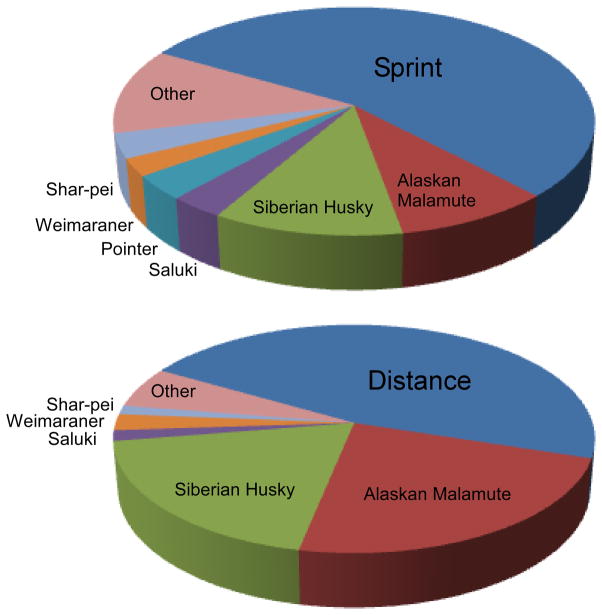
Examining mixed-breed dogs in the light of breed structure can provide an alternate method for identifying genes and mutations related to common traits. For example, in preparation for finding genes that contribute to performance attributes in working Alaskan sled dogs, researchers used genetic markers to identify the purebred contributors to the modern dogs (Huson et al. 2010). Because the Alaskan sled dog is bred entirely for working ability and not appearance, new breeds have been introduced into the lines to improve racing performance. Only those crosses that resulted in improved racing were repeated in later generations creating a selection for specific regions from purebred lines that enhance the dog’s ability to compete (Huson et al. 2011). Microsatellite markers were genotyped in 199 Alaskan sled dogs from active working kennels involved in either sprint (up to 30 miles at 18–25 mph) or long-distance (up to 1000 miles at 8–12 mph) racing. The patterns of alleles from these dogs were compared to patterns from 140 pure-breeds to ascertain the most probable outcrossings. An increase in Alaskan Malamute and Siberian husky allele patterns was found in the distance dogs compared to the sprint dogs. Alternately, a strong contribution from Pointer was identified in the sprint dogs that was absent from the distance dogs (Figure 3). The next logical step will be to identify the precise segments of the chromosomes that were retained from these crosses in order to find the genetic variants that lead to improvements in performance, be that speed, endurance, work ethic or sheer joy of running. Similar analyses can be applied to mixed breed dogs displaying diseases common to specific purebreds in order to localize the regions containing causative mutations.
Figure 3.
Breed composition of Alaskan Sled dogs. The sled dogs are divided into sprint and distance groups based on their performance level. The dogs that perform as sprinters show a considerable contribution from the Pointer breed that is missing in distance runners. The distance dogs appear to be largely mixtures of classic sled dog breeds. The other group is comprised of 9 breeds with >1% but <2% contribution to at least one group. Data is compiled from Huson et al. (Huson et al. 2010).
With their intricate breed structure and anthropomorphic lifestyle, modern domestic dogs fill a much needed role in genetic study design. They do not take the place of a laboratory strain or a human cohort, but instead add depth and understanding to both by providing real-world effects on a semi-controlled genetic background. Comparing study results across multiple models with varied histories will provide the greatest amount of information regarding the traits as well as the pathways and gene products involved in creating them. With a clear understanding of the breed structure and development to enhance the utility of the system, the dog will find a prominent place in genetic and genomic studies for years to come.
References
- Akey JM, Ruhe AL, Akey DT, Wong AK, Connelly CF, Madeoy J, Nicholas TJ, Neff MW. Tracking footprints of artificial selection in the dog genome. Proc Natl Acad Sci U S A. 2010;107:1160–1165. doi: 10.1073/pnas.0909918107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alderton D. Dogs. New York: Dorling Kindersley, Ltd; 2002. [Google Scholar]
- American Kennel Club. The Complete Dog Book. 19. New York, NY: Howell Book House; 1998. Revised ed. [Google Scholar]
- American Veterinary Medical Association. US Pet Ownership and Demographics Sourcebook. 1. Schaumburg, ILL: American Veterinary Medical Association; 2002. [Google Scholar]
- Awano T, Johnson GS, Wade CM, Katz ML, Johnson GC, Taylor JF, Perloski M, Biagi T, Baranowska I, Long S, March PA, Olby NJ, Shelton GD, Khan S, O’Brien DP, Lindblad-Toh K, Coates JR. Genome-wide association analysis reveals a SOD1 mutation in canine degenerative myelopathy that resembles amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2009;106:2794–2799. doi: 10.1073/pnas.0812297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylor DL, Valdar W, Foulds-Mathes W, Buus RJ, Verdugo RA, Baric RS, Ferris MT, Frelinger JA, Heise M, Frieman MB, Gralinski LE, Bell TA, Didion JD, Hua K, Nehrenberg DL, Powell CL, Steigerwalt J, Xie Y, Kelada SN, Collins FS, Yang IV, Schwartz DA, Branstetter LA, Chesler EJ, Miller DR, Spence J, Liu EY, McMillan L, Sarkar A, Wang J, Wang W, Zhang Q, Broman KW, Korstanje R, Durrant C, Mott R, Iraqi FA, Pomp D, Threadgill D, Pardo-Manuel de Villena F, Churchill GA. Genetic analysis of complex traits in the emerging Collaborative Cross. Genome Res. 2011;21:1213–1222. doi: 10.1101/gr.111310.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannasch D, Young A, Myers J, Truvé K, Dickinson P, Gregg J, Davis R, Bongcam-Rudloff E, Webster MT, Lindblad-Toh K, Pedersen N. Localization of Canine Brachycephaly Using an Across Breed Mapping Approach. PLoS One. 2010;5:e9632. doi: 10.1371/journal.pone.0009632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko AR, Boyko RH, Boyko CM, Parker HG, Castelhano M, Corey L, Degenhardt JD, Auton A, Hedimbi M, Kityo R, Ostrander EA, Schoenebeck J, Todhunter RJ, Jones P, Bustamante CD. Complex population structure in African village dogs and its implications for inferring dog domestication history. Proc Natl Acad Sci U S A. 2009;106:13903–13908. doi: 10.1073/pnas.0902129106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko AR, Quignon P, Li L, Schoenebeck JJ, Degenhardt JD, Lohmueller KE, Zhao K, Brisbin A, Parker HG, vonHoldt BM, Cargill M, Auton A, Reynolds A, Elkahloun AG, Castelhano M, Mosher DS, Sutter NB, Johnson GS, Novembre J, Hubisz MJ, Siepel A, Wayne RK, Bustamante CD, Ostrander EA. A simple genetic architecture underlies morphological variation in dogs. PLoS Biol. 2010;8:e1000451. doi: 10.1371/journal.pbio.1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouillette JA, Venta PJ. Within-breed heterozygosity of canine single nucleotide polymorphisms identified by across-breed comparison. Anim Genet. 2002;33:464–467. doi: 10.1046/j.1365-2052.2002.00918.x. [DOI] [PubMed] [Google Scholar]
- Brown SDM. Genetic Variants and Strains of the Laboratory Mouse. 3. New York: Oxford University Press, USA; 1996. [Google Scholar]
- Cadieu E, Neff M, Quignon P, Walsh K, Chase K, Parker HG, Vonholdt BM, Rhue A, Boyko A, Byers A, Wong A, Mosher DS, Elkahloun AG, Spady TC, Andre C, Lark KG, Cargill M, Bustamante CD, Wayne RK, Ostrander EA. Coat Variation in the Domestic Dog Is Governed by Variants in Three Genes. Science. 2009;326:150–153. doi: 10.1126/science.1177808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann RL, Stoneking M, Wilson AC. Mitochondrial DNA and human evolution. Nature. 1987;325:31–36. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- Conrad DF, Jakobsson M, Coop G, Wen X, Wall JD, Rosenberg NA, Pritchard JK. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat Genet. 2006;38:1251–1260. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- Cornuet JM, Piry S, Luikart G, Estoup A, Solignac M. New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics. 1999;153:1989–2000. doi: 10.1093/genetics/153.4.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodman NH, Karlsson EK, Moon-Fanelli A, Galdzicka M, Perloski M, Shuster L, Lindblad-Toh K, Ginns EI. A canine chromosome 7 locus confers compulsive disorder susceptibility. Mol Psychiatry. 2010;15:8–10. doi: 10.1038/mp.2009.111. [DOI] [PubMed] [Google Scholar]
- Downs LM, Wallin-Håkansson B, Boursnell M, Marklund S, Hedhammar Å, Truvé K, Hübinette L, Lindblad-Toh K, Bergström T, Mellersh CS. A Frameshift Mutation in Golden Retriever Dogs with Progressive Retinal Atrophy Endorses SLC4A3 as a Candidate Gene for Human Retinal Degenerations. PLoS One. 2011;6:e21452. doi: 10.1371/journal.pone.0021452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drogemuller C, Karlsson EK, Hytonen MK, Perloski M, Dolf G, Sainio K, Lohi H, Lindblad-Toh K, Leeb T. A Mutation in Hairless Dogs Implicates FOXI3 in Ectodermal Development. Science. 2008;321:1462. doi: 10.1126/science.1162525. [DOI] [PubMed] [Google Scholar]
- Farias FHG, Johnson GS, Taylor JF, Giuliano E, Katz ML, Sanders DN, Schnabel RD, McKay SD, Khan S, Gharahkhani P, O’Leary CA, Pettitt L, Forman OP, Boursnell M, McLaughlin B, Ahonen S, Lohi H, Hernandez-Merino E, Gould DJ, Sargan DR, Mellersh C. An ADAMTS17 Splice Donor Site Mutation in Dogs with Primary Lens Luxation. Investigative Ophthalmology & Visual Science. 2010;51:4716–4721. doi: 10.1167/iovs.09-5142. [DOI] [PubMed] [Google Scholar]
- Farias FHG, Zeng R, Johnson GS, Wininger FA, Taylor JF, Schnabel RD, McKay SD, Sanders DN, Lohi H, Seppälä EH, Wade CM, Lindblad-Toh K, O’Brien DP, Katz ML. A truncating mutation in ATP13A2 is responsible for adult-onset neuronal ceroid lipofuscinosis in Tibetan terriers. Neurobiology of Disease. 2011;42:468–474. doi: 10.1016/j.nbd.2011.02.009. [DOI] [PubMed] [Google Scholar]
- Federation Cynologique I. Standards and Nomenclature. Federation Cynologique Internationale; 2010. [Google Scholar]
- Fogle B. ASPCA Complete Dog Care Manual. New York: DK Publishing, Inc; 1993. [Google Scholar]
- Francisco LV, Langston AA, Mellersh CS, Neal CL, Ostrander EA. A class of highly polymorphic tetranucleotide repeats for canine genetic mapping. Mamm Genome. 1996;7:359–362. doi: 10.1007/s003359900104. [DOI] [PubMed] [Google Scholar]
- Fredholm M, Wintero AK. Variation of short tandem repeats within and between species belonging to the Canidae family. Mamm Genome. 1995;6:11–18. doi: 10.1007/BF00350887. [DOI] [PubMed] [Google Scholar]
- Frome JH. (rev. 2004)) Staffordshire Bull Terrier. Allenhurst, NJ: Kennel Club Books; 1999. [Google Scholar]
- Gao HW, Liang CZ, Zhang YB, Zhu LH. Polymerase chain reaction method to detect canis materials by amplification of species-specific DNA fragment. J AOAC Int. 2004;87:1195–1199. [PubMed] [Google Scholar]
- Huson HJ, Parker HG, Runstadler J, Ostrander EA. A Genetic Dissection of Breed Composition and Performance Enhancement in the Alaskan Sled Dog. BMC Genetics. 2010;11:71. doi: 10.1186/1471-2156-11-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson HJ, Vonholdt BM, Rimbault M, Byers AM, Runstadler JA, Parker HG, Ostrander EA. Breed-specific ancestry studies and genome-wide association analysis highlight an association between the MYH9 gene and heat tolerance in Alaskan sprint racing sled dogs. Mamm Genome. 2011 doi: 10.1007/s00335-011-9374-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion DN, Schaffer AL, Famula TR, Eggleston ML, Hughes SS, Pedersen NC. Analysis of genetic variation in 28 dog breed populations with 100 microsatellite markers. J Hered. 2003;94:81–87. doi: 10.1093/jhered/esg004. [DOI] [PubMed] [Google Scholar]
- Karlsson EK, Baranowska I, Wade CM, Salmon Hillbertz NH, Zody MC, Anderson N, Biagi TM, Patterson N, Pielberg GR, Kulbokas EJ, 3rd, Comstock KE, Keller ET, Mesirov JP, von Euler H, Kampe O, Hedhammar A, Lander ES, Andersson G, Andersson L, Lindblad-Toh K. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat Genet. 2007;39:1321–1328. doi: 10.1038/ng.2007.10. [DOI] [PubMed] [Google Scholar]
- Kirkness EF, Bafna V, Halpern AL, Levy S, Remington K, Rusch DB, Delcher AL, Pop M, Wang W, Fraser CM, Venter JC. The dog genome: survey sequencing and comparative analysis. Science. 2003;301:1898–1903. doi: 10.1126/science.1086432. [DOI] [PubMed] [Google Scholar]
- Koskinen MT. Individual assignment using microsatellite DNA reveals unambiguous breed identification in the domestic dog. Anim Genet. 2003;34:297–301. doi: 10.1046/j.1365-2052.2003.01005.x. [DOI] [PubMed] [Google Scholar]
- Koskinen MT, Bredbacka P. Assessment of the population structure of five Finnish dog breeds with microsatellites. Anim Genet. 2000;31:310–317. doi: 10.1046/j.1365-2052.2000.00669.x. [DOI] [PubMed] [Google Scholar]
- Lequarre AS, Andersson L, Andre C, Fredholm M, Hitte C, Leeb T, Lohi H, Lindblad-Toh K, Georges M. LUPA: a European initiative taking advantage of the canine genome architecture for unravelling complex disorders in both human and dogs. Vet J. 2011;189:155–159. doi: 10.1016/j.tvjl.2011.06.013. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ, 3rd, Zody MC, Mauceli E, Xie X, Breen M, Wayne RK, Ostrander EA, Ponting CP, Galibert F, Smith DR, DeJong PJ, Kirkness E, Alvarez P, Biagi T, Brockman W, Butler J, Chin CW, Cook A, Cuff J, Daly MJ, DeCaprio D, Gnerre S, Grabherr M, Kellis M, Kleber M, Bardeleben C, Goodstadt L, Heger A, Hitte C, Kim L, Koepfli KP, Parker HG, Pollinger JP, Searle SM, Sutter NB, Thomas R, Webber C, Baldwin J, Abebe A, Abouelleil A, Aftuck L, Ait-Zahra M, Aldredge T, Allen N, An P, Anderson S, Antoine C, Arachchi H, Aslam A, Ayotte L, Bachantsang P, Barry A, Bayul T, Benamara M, Berlin A, Bessette D, Blitshteyn B, Bloom T, Blye J, Boguslavskiy L, Bonnet C, Boukhgalter B, Brown A, Cahill P, Calixte N, Camarata J, Cheshatsang Y, Chu J, Citroen M, Collymore A, Cooke P, Dawoe T, Daza R, Decktor K, DeGray S, Dhargay N, Dooley K, Dooley K, Dorje P, Dorjee K, Dorris L, Duffey N, Dupes A, Egbiremolen O, Elong R, Falk J, Farina A, Faro S, Ferguson D, Ferreira P, Fisher S, FitzGerald M, Foley K, Foley C, Franke A, Friedrich D, Gage D, Garber M, Gearin G, Giannoukos G, Goode T, Goyette A, Graham J, Grandbois E, Gyaltsen K, Hafez N, Hagopian D, Hagos B, Hall J, Healy C, Hegarty R, Honan T, Horn A, Houde N, Hughes L, Hunnicutt L, Husby M, Jester B, Jones C, Kamat A, Kanga B, Kells C, Khazanovich D, Kieu AC, Kisner P, Kumar M, Lance K, Landers T, Lara M, Lee W, Leger JP, Lennon N, Leuper L, LeVine S, Liu J, Liu X, Lokyitsang Y, Lokyitsang T, Lui A, Macdonald J, Major J, Marabella R, Maru K, Matthews C, McDonough S, Mehta T, Meldrim J, Melnikov A, Meneus L, Mihalev A, Mihova T, Miller K, Mittelman R, Mlenga V, Mulrain L, Munson G, Navidi A, Naylor J, Nguyen T, Nguyen N, Nguyen C, Nguyen T, Nicol R, Norbu N, Norbu C, Novod N, Nyima T, Olandt P, O’Neill B, O’Neill K, Osman S, Oyono L, Patti C, Perrin D, Phunkhang P, Pierre F, Priest M, Rachupka A, Raghuraman S, Rameau R, Ray V, Raymond C, Rege F, Rise C, Rogers J, Rogov P, Sahalie J, Settipalli S, Sharpe T, Shea T, Sheehan M, Sherpa N, Shi J, Shih D, Sloan J, Smith C, Sparrow T, Stalker J, Stange-Thomann N, Stavropoulos S, Stone C, Stone S, Sykes S, Tchuinga P, Tenzing P, Tesfaye S, Thoulutsang D, Thoulutsang Y, Topham K, Topping I, Tsamla T, Vassiliev H, Venkataraman V, Vo A, Wangchuk T, Wangdi T, Weiand M, Wilkinson J, Wilson A, Yadav S, Yang S, Yang X, Young G, Yu Q, Zainoun J, Zembek L, Zimmer A, Lander ES. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Meurs K, Mauceli E, Lahmers S, Acland G, White S, Lindblad-Toh K. Genome-wide association identifies a deletion in the 3′ untranslated region of Striatin in a canine model of arrhythmogenic right ventricular cardiomyopathy. Human genetics. 2010;128:315–324. doi: 10.1007/s00439-010-0855-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modiano JF, Breen M, Burnett RC, Parker HG, Inusah S, Thomas R, Avery PR, Lindblad-Toh K, Ostrander EA, Cutter GC, Avery AC. Distinct B-cell and T-cell lymphoproliferative disease prevalence among dog breeds indicates heritable risk. Cancer Res. 2005;65:5654–5661. doi: 10.1158/0008-5472.CAN-04-4613. [DOI] [PubMed] [Google Scholar]
- Nielen AL, van der Beek S, Ubbink GJ, Knol BW. Population parameters to compare dog breeds: differences between five Dutch purebred populations. Vet Q. 2001;23:43–49. doi: 10.1080/01652176.2001.9695075. [DOI] [PubMed] [Google Scholar]
- Okumura N, Ishiguro N, Nakano M, Matsui A, Sahara M. Intra- and interbreed genetic variations of mitochondrial DNA major non-coding regions in Japanese native dog breeds (Canis familiaris) Anim Genet. 1996;27:397–405. doi: 10.1111/j.1365-2052.1996.tb00506.x. [DOI] [PubMed] [Google Scholar]
- Olsson M, Meadows JRS, Truvé K, Rosengren Pielberg G, Puppo F, Mauceli E, Quilez J, Tonomura N, Zanna G, Docampo MJ, Bassols A, Avery AC, Karlsson EK, Thomas A, Kastner DL, Bongcam-Rudloff E, Webster MT, Sanchez A, Hedhammar Å, Remmers EF, Andersson L, Ferrer L, Tintle L, Lindblad-Toh K. A Novel Unstable Duplication Upstream of HAS2 Predisposes to a Breed-Defining Skin Phenotype and a Periodic Fever Syndrome in Chinese Shar-Pei Dogs. PLoS Genet. 2011;7:e1001332. doi: 10.1371/journal.pgen.1001332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander EA, Sprague GF, Jr, Rine J. Identification and characterization of dinucleotide repeat (CA)n markers for genetic mapping in dog. Genomics. 1993;16:207–213. doi: 10.1006/geno.1993.1160. [DOI] [PubMed] [Google Scholar]
- Pang JF, Kluetsch C, Zou XJ, Zhang AB, Luo LY, Angleby H, Ardalan A, Ekstrom C, Skollermo A, Lundeberg J, Matsumura S, Leitner T, Zhang YP, Savolainen P. mtDNA data indicate a single origin for dogs south of Yangtze River, less than 16,300 years ago, from numerous wolves. Mol Biol Evol. 2009;26:2849–2864. doi: 10.1093/molbev/msp195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HG, Kim LV, Sutter NB, Carlson S, Lorentzen TD, Malek TB, Johnson GS, DeFrance HB, Ostrander EA, Kruglyak L. Genetic structure of the purebred domestic dog. Science. 2004;304:1160–1164. doi: 10.1126/science.1097406. [DOI] [PubMed] [Google Scholar]
- Parker HG, Kukekova AV, Akey DT, Goldstein O, Kirkness EF, Baysac KC, Mosher DS, Aguirre GD, Acland GM, Ostrander EA. Breed relationships facilitate fine-mapping studies: a 7.8-kb deletion cosegregates with Collie eye anomaly across multiple dog breeds. Genome Res. 2007;17:1562–1571. doi: 10.1101/gr.6772807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HG, Meurs KM, Ostrander EA. Finding cardiovascular disease genes in the dog. J Vet Cardiol. 2006a;8:115–127. doi: 10.1016/j.jvc.2006.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HG, Shearin AL, Ostrander EA. Man’s best friend becomes biology’s best in show: genome analyses in the domestic dog. Annu Rev Genet. 2010;44:309–336. doi: 10.1146/annurev-genet-102808-115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HG, Sutter NB, Ostrander EA. Understanding Genetic Relationships among Purebred Dogs: The PhyDo Project. In: Ostrander EA, Giger U, Lindblad-Toh K, editors. The Dog and its Genome. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006b. pp. 141–157. [Google Scholar]
- Parker HG, VonHoldt BM, Quignon P, Margulies EH, Shao S, Mosher DS, Spady TC, Elkahloun A, Cargill M, Jones PG, Maslen CL, Acland GM, Sutter NB, Kuroki K, Bustamante CD, Wayne RK, Ostrander EA. An expressed fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science. 2009;325:995–998. doi: 10.1126/science.1173275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Rosenberg NA. Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quignon P, Herbin L, Cadieu E, Kirkness EF, Hedan B, Mosher DS, Galibert F, Andre C, Ostrander EA, Hitte C. Canine population structure: assessment and impact of intra-breed stratification on SNP-based association studies. PLoS ONE. 2007;2:e1324. doi: 10.1371/journal.pone.0001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilez J, Short AD, Martinez V, Kennedy LJ, Ollier W, Sanchez A, Altet L, Francino O. A selective sweep of >8 Mb on chromosome 26 in the Boxer genome. BMC Genomics. 2011;12:339. doi: 10.1186/1471-2164-12-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon Hillbertz NHC, Isaksson M, Karlsson EK, Hellmen E, Pielberg GR, Savolainen P, Wade CM, von Euler H, Gustafson U, Hedhammar A, Nilsson M, Lindblad-Toh K, Andersson L, Andersson G. Duplication of FGF3, FGF4, FGF19 and ORAOV1 causes hair ridge and predisposition to dermoid sinus in Ridgeback dogs. Nat Genet. 2007;39:1318–1320. doi: 10.1038/ng.2007.4. [DOI] [PubMed] [Google Scholar]
- Savolainen P, Leitner T, Wilton AN, Matisoo-Smith E, Lundeberg J. A detailed picture of the origin of the Australian dingo, obtained from the study of mitochondrial DNA. Proc Natl Acad Sci U S A. 2004;101:12387–12390. doi: 10.1073/pnas.0401814101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen P, Zhang YP, Luo J, Lundeberg J, Leitner T. Genetic evidence for an East Asian origin of domestic dogs. Science. 2002;298:1610–1613. doi: 10.1126/science.1073906. [DOI] [PubMed] [Google Scholar]
- Sutter NB, Bustamante CD, Chase K, Gray MM, Zhao K, Zhu L, Padhukasahasram B, Karlins E, Davis S, Jones PG, Quignon P, Johnson GS, Parker HG, Fretwell N, Mosher DS, Lawler DF, Satyaraj E, Nordborg M, Lark KG, Wayne RK, Ostrander EA. A single IGF1 allele is a major determinant of small size in dogs. Science. 2007;316:112–115. doi: 10.1126/science.1137045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton A. Out of Africa again and again. Nature. 2002;416:45–51. doi: 10.1038/416045a. [DOI] [PubMed] [Google Scholar]
- Vaysse A, Ratnakumar A, Derrien T, Axelsson E, Rosengren Pielberg G, Sigurdsson S, Fall T, Seppala EH, Hansen MS, Lawley CT, Karlsson EK, Bannasch D, Vila C, Lohi H, Galibert F, Fredholm M, Haggstrom J, Hedhammar A, Andre C, Lindblad-Toh K, Hitte C, Webster MT. Identification of genomic regions associated with phenotypic variation between dog breeds using selection mapping. PLoS Genet. 2011;7:e1002316. doi: 10.1371/journal.pgen.1002316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila C, Maldonado JE, Wayne RK. Phylogenetic relationships, evolution, and genetic diversity of the domestic dog. J Hered. 1999;90:71–77. doi: 10.1093/jhered/90.1.71. [DOI] [PubMed] [Google Scholar]
- Vila C, Savolainen P, Maldonado JE, Amorim IR, Rice JE, Honeycutt RL, Crandall KA, Lundeberg J, Wayne RK. Multiple and ancient origins of the domestic dog. Science. 1997;276:1687–1689. doi: 10.1126/science.276.5319.1687. [DOI] [PubMed] [Google Scholar]
- Vila C, Walker C, Sundqvist AK, Flagstad O, Andersone Z, Casulli A, Kojola I, Valdmann H, Halverson J, Ellegren H. Combined use of maternal, paternal and bi-parental genetic markers for the identification of wolf-dog hybrids. Heredity. 2003;90:17–24. doi: 10.1038/sj.hdy.6800175. [DOI] [PubMed] [Google Scholar]
- Vonholdt BM, Pollinger JP, Lohmueller KE, Han E, Parker HG, Quignon P, Degenhardt JD, Boyko AR, Earl DA, Auton A, Reynolds A, Bryc K, Brisbin A, Knowles JC, Mosher DS, Spady TC, Elkahloun A, Geffen E, Pilot M, Jedrzejewski W, Greco C, Randi E, Bannasch D, Wilton A, Shearman J, Musiani M, Cargill M, Jones PG, Qian Z, Huang W, Ding ZL, Zhang YP, Bustamante CD, Ostrander EA, Novembre J, Wayne RK. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature. 2010;464:898–902. doi: 10.1038/nature08837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Zangerl B, Werner P, Mauldin E, Casal M. Familial cutaneous lupus erythematosus (CLE) in the German shorthaired pointer maps to CFA18, a canine orthologue to human CLE. Immunogenetics. 2011;63:197–207. doi: 10.1007/s00251-010-0499-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbe M, Jokinen P, Truve K, Seppala EH, Karlsson EK, Biagi T, Hughes A, Bannasch D, Andersson G, Hansson-Hamlin H, Lohi H, Lindblad-Toh K. Genome-wide association mapping identifies multiple loci for a canine SLE-related disease complex. Nat Genet. 2010;42:250–254. doi: 10.1038/ng.525. [DOI] [PubMed] [Google Scholar]
- Wilcox B, Walkowicz C. Atlas of Dog Breeds of the World. 5. Neptune City, NJ: T.F.H. Publications; 1995. [Google Scholar]
- Wiles-Fone H, Barnes J. The Ultimate Labrador Retriever. Foster City, CA: Howell Book House; 1997. [Google Scholar]
- Zajc I, Mellersh CS, Sampson J. Variability of canine microsatellites within and between different dog breeds. Mammalian Genome. 1997;8:182–185. doi: 10.1007/s003359900386. [DOI] [PubMed] [Google Scholar]
- Zeng R, Farias FHG, Johnson GS, McKay SD, Schnabel RD, Decker JE, Taylor JF, Mann CS, Katz ML, Johnson GC, Coates JR, O’Brien DP. A Truncated Retrotransposon Disrupts the GRM1 Coding Sequence in Coton de Tulear Dogs with Bandera’s Neonatal Ataxia. Journal of Veterinary Internal Medicine. 2011;25:267–272. doi: 10.1111/j.1939-1676.2010.0666.x. [DOI] [PubMed] [Google Scholar]