Abstract
Several CC-motif chemokine ligands (CCLs) can block HIV-1 binding sites on CC-motif chemokine receptor 5 (CCR5) and inhibit viral entry. We studied single nucleotide polymorphisms (SNPs) in genes encoding three CCR5 ligands [CCL3 (MIP-1α), CCL4 (MIP-1β), and CCL5 (RANTES)] along with an adjacent gene encoding a CCR2 ligand [CCL2 (MCP-1)] to identify candidate markers for HIV-1 infection and pathogenesis. Analyses of 567 HIV-1 serodiscordant Zambian couples revealed that rs5029410C (in CCL3 intron 2) was associated with lower viral load (VL) in seroconverters, adjusted for gender and age (regression β=−0.57 log10, P=4×10−6). In addition, rs34171309A in CCL3 exon 3 was associated with increased risk of HIV-1 acquisition in exposed seronegatives (hazard ratio=1.52, P=0.006 when adjusted for donor VL and genital ulcer/inflammation). The CCL3 exon 3 SNP, encoding a conservative Glu-to-Asp substitution, and five neighboring SNPs in tight linkage disequilibrium all showed similar associations with HIV-1 acquisition. How these multiple CCL3 SNPs may alter the occurrence or course of HIV-1 infection remains to be determined.
Keywords: HIV-1 transmission, CCL2, CCL3, CCL4, CCL5, SNP
INTRODUCTION
The pandemic of acquired immunodeficiency syndrome (AIDS) resulting from human immunodeficiency virus (HIV-1) infection is particularly devastating in southern Africa 1. HIV-1 enters target cells through a two-step fusion process in which the CC-motif chemokine receptor 5 (CCR5) serve as a major coreceptor on human CD4+ T cells. Multiple studies have demonstrated that individuals who are homozygous for the CCR5 deletion mutation (Δ32) lack functional CCR5 and are thus highly resistant to HIV-1 infection 2–5. CC-motif chemokine ligands CCL3 (MIP-1α), CCL4 (MIP-1β), and CCL5 (RANTES) are natural ligands for CCR5; as such they may competitively inhibit binding of the receptor by HIV-1 6–10. Variants in CCR2, the gene adjacent to CCR5, have also been reported to be independently associated with HIV-1 infection, progression and transmission 11. CCL2, the natural ligand for CCR2, may also be an important factor for the HIV-1 transmission 12–14. Numerous papers studied the influence of CCR5 variants on the HIV-1 transmission, infection and disease progression. However, observations of the impact of the ligands on HIV-1 acquisition and disease control in populations of African ancestry are sparse. To complement our work on the role of the receptor polymorphisms 11, we examined the associations of single nucleotide polymorphisms (SNPs) in the CCL genes with HIV-1 heterosexual transmission and disease control in Zambia cohort.
Between 1995 and 2006 in Lusaka, Zambia, more than 10,000 couples were screened for their HIV-1 status in the Zambia-Emory HIV-1 Research Project (ZEHRP). The procedures for screening, recruitment, counseling, and follow-up visits have been described elsewhere 15, 16. The study presented here included 567 HIV-1 serodiscordant couples who were followed for at least 9 months between 1996 and 2006. Among these, 240 exposed seronegative (HESN) participants acquired phylogenetically linked HIV-1 11 from their index partners after varying lengths of follow-up (interquartile range: 6–36 months) and 327 HESN participants remained seronegative during the study intervals. Non-genetic factors, including age, gender, donor viral load, genital ulcer or inflammation in any partner, and male circumcision were retained as covariates, as established in earlier analyses of HIV-1 transmission within the study population 11, 17, 18.
In the absence of selection for pre-exposure time before enrollment, the observed genetic associations with rate of HIV-1 transmission should not have been systematically affected by duration of pre-exposure. On the other hand, because non-transmitting couples had higher frequencies of non-genetic risk factors, the observed transmission rates were higher than the overall rates in the entire cohort of discordant couples. Overall, the annual HIV-1 seroincidence (7–9 events per 100 person-years) among Zambian couples was reduced by one-half to two-thirds as a result of voluntary testing and counseling 1.
We assessed recognized SNPs reported in dbSNP to map between 1 kb upstream and 500 bp downstream of the four candidate CCL genes (CCL2, CCL3, CCL4 and CCL5). SNPs included in the analysis met at least one of the following criteria: 1) encodes a change in the amino acid sequence of the ligand; 2) occurs at a transcription binding site, an intron/exon boundary site, an alternative splicing site, promoter region, or 3’ untranslated region; or 3) has a minor allele frequency (MAF) ≥0.02 in Africans/African Americans. We also included SNPs with unknown MAF to provide additional coverage of the gene. All SNPs had to meet suitability criteria for the iPLEX SNP typing assay at the Broad Institute of MIT and Harvard 19.
RESULTS AND DISCUSSION
Overall, 63 SNPs in 4 CCL genes passed the assay design process; 52 had a call rate of over 90%; and 35 had an MAF ≥0.01 (Table S1 in Supplemental Materials). SNPs within each gene tended to have strong LD as judged by D’ and r2 values (Figure S1 in Supplemental Materials). Haplotype blocks were defined by the Gabriel algorithm in Haploview 4.2 20. Haplotype blocks 1 and 2 are in CCL2, and haplotype blocks 3, 4 and 5 are in CCL5, CCL3 and CCL4, respectively. Of the 35 SNPs, two deviated from Hardy-Weinberg equilibrium (HWE) in their distribution (Table S1). SNPs rs1719134 and rs13900 were out of HWE in the overall cohort but conform to HWE in the HESNs and index subgroups (q values at 0.014 and 0.175 respectively). Genotype frequencies of these 2 SNPs in the Zambian cohort were quite similar to those reported for other Africans documented in the dbSNP database (National Center for Biotechnology Information). Therefore, those few SNPs whose frequencies were out of HWE were most likely due to chance.
Of the few reports on CCL gene variants and HIV-1 disease progression13, 21–24, none of them addressed the association between CCL variants and HIV-1 VL. By linear regression we tested the effect of the minor SNP allele on earliest available (chronic-phase) VL in index partners and the set-point VL taken at 6 months after the imputed infection date in seroconverters 11. Carriage of one copy of the C allele of rs5029410 in CCL3 intron 2 was strongly associated with lower VL in the seroconverters (regression β=−0.57 log10, P=4×10−6) with adjustment for age and gender. However, this SNP showed no association with VL in index partners (β=0.05 log10, P=0.46). Index partners did not received antiretroviral therapy, but their average duration of infection was much longer than that of seroconverters. Thus, the SNP variant might exert its effect only early in HIV-1 infection rather than later.
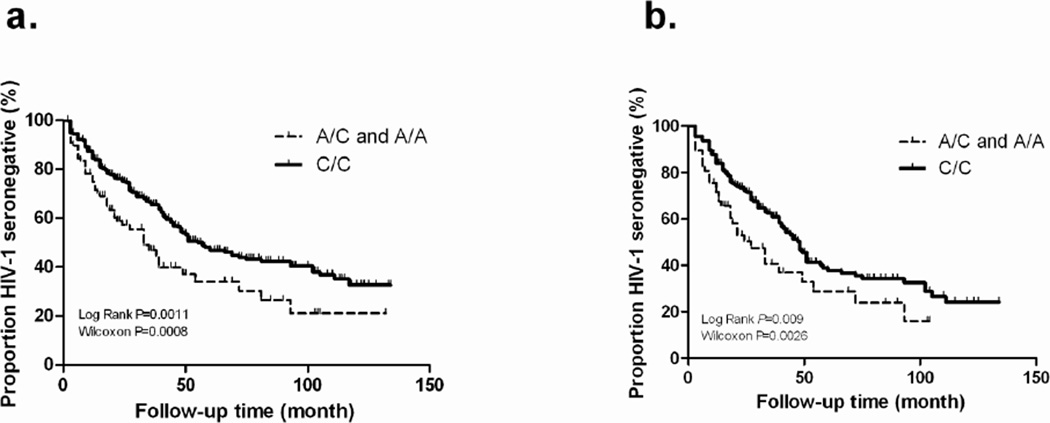
The A allele of rs34171309 in CCL3 exon 3 was associated with more rapid acquisition of HIV-1 by HESNs, as shown in an allelic proportional hazards model in the presence of non-genetic factors (HR=1.52, 95% CI 1.13–2.04, P=0.006) (Table 1). This association was also apparent in logistic regression model: the frequency of rs34171309A was higher in the SCs than HESNs (odds ratio=1.51, P=0.05). In Kaplan Meier plots, differences were seen in the overall cohort and in female HESNs (Figure 1).
Table 1.
Association of rs34171309A allele (in CCL3 exon 3) with acquisition of HIV-1 infection among exposed seronegative (HESN) Zambians before and after stratification by gender.
| All HESNs (N=567) | Female HESNs (N=295) | Male HESNs (N=272) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Factors in model | HRa | 95% CI | P | HRa | 95% CI | P | HRa | 95% CI | P |
| rs34171309Ab | 1.52 | 1.13–2.04 | 0.006 | 1.70 | 1.17–2.47 | 0.006 | 1.29 | 0.78–2.15 | 0.33 |
| GUI in both partnersc | 8.72 | 5.72–13.28 | <0.0001 | 7.85 | 4.62–13.33 | <0.0001 | 10.39 | 5.15–20.97 | <0.0001 |
| GUI in either partnerd | 3.04 | 2.12–4.35 | <0.0001 | 2.89 | 1.79–4.66 | <0.0001 | 3.50 | 1.99–6.18 | <0.0001 |
| Donor VL (per log10) | 1.37 | 1.12–1.69 | 0.003 | 1.15 | 0.86–1.53 | 0.35 | 1.66 | 1.19–2.31 | 0.003 |
Hazard ratio (HR) from a Cox proportional hazards model, adjusted for all factors in the model. CI, confidence interval; GUI, genital ulcer/inflammation; VL, plasma HIV-1 viral load (RNA copies/ml).
Also tested in logistic regression models (see text).
Genital ulcer/inflammation seen in both partners in each couple.
Genital ulcer/inflammation seen in either partner in each couple.
Figure 1.
Kaplan-Meier plots showing the relationship of rs34171309 to HIV-1 acquisition among initially HIV-1 exposed seronegative (HESN) partners of Zambia couples. (a) 567 HESNs; (b) 295 female HESNs. Genotypes AA + AC are compared with the CC genotype in order to highlight the dominant effect of the minor allele A. Rates of seroconversion depicted here for selected discordant couples are higher than rates in the entire cohort of discordant couples in the Zambia-Emory HIV-1 Research Project (ZEHRP).
The rs34171309 encodes a non-synonymous Glu-to-Asp amino acid change at position 78 in the CCL3 protein. Although this change is a conservative one, because it occurs very close to the site of CCL3 binding to the CCR5 protein, it may influence CCL3-CCR5 interaction 25. The A allele of rs34171309 and the C allele of rs5029410 were in weak LD (D’=0.44), but rs34171309A was in strong LD with five other SNPs (rs1719130, rs1719134, rs35511254, rs1634497 and rs1634499) in CCL3 (D’>0.8), and all these SNPs had similar associations with HIV-1 acquisition (data not shown). Three of them (rs1719130, rs35511254 and rs1719134) had minor alleles associated with fast HIV-1 disease progression in earlier studies 21, 26, consistent with our findings here. In an analysis of the initially HESNs stratified by gender we again found a stronger effect of rs34171309A in the HESN females than in the smaller number of males, but the effect for both groups is in the same direction (female: HR=1.70, 95% CI 1.17–2.47, P=0.006; male: HR=1.29, 95% CI 0.78–2.15, P=0.33). This difference in significance may be due to an interaction between this SNP and gender or simply to chance variation.
For the two SNPs highlighted in this study, genotypes for one (rs5029410) were validated by a TaqMan genotyping assay (Applied Biosystems, Inc.). Selective tests revealed over 98% concordance rate between TaqMan and iPLEX results. For rs34171309, however, variants could not be readily validated by alternative techniques. Based on alignments of homologous sequences from CCL3 and CCL3L1, CCL3L3 genes (Figure S2 in Supplemental Materials), only CCL3 was polymorphic at the nucleotide position corresponding to rs34171309. As a result, we infer that rs34171309A is most likely present in CCL3 and not in its homologues.
At the CCL5 locus, one SNP variant known as In1.1C (rs2280789) has been associated with higher risk of HIV-1 subtype B infection in Europeans and African Americans and with more rapid HIV-1 disease progression in African Americans6. In our study population, no such association could be established for In1.1C or its haplotypes. In particular, In1.1C had no association with HIV-1 acquisition (HR=1.05, 95% CI: 0.84–1.31, P=0.70).
We did not adjust P values for the number of tests implied by the number of SNPs eligible for analysis because a number of SNPs within each gene are in high LD with each other and the number of independent tests would actually be considerably lower. Only the strong association of rs5029410C (CCL3 intron 2 variant) would have withstood even conservative Bonferroni correction. The false discovery probability (q value) for rs5029410C and rs34171309A was 0.0001 and 0.03, respectively.
Compared with data from commercially available genome wide association chip arrays or open access databases, our study provided denser coverage of the variation in all four CCL genes studied. The 7 SNPs in CCL3 span a 5-kb genomic region. In contrast, both the Human1M-Duo DNA Analysis BeadChip (Illumina, Inc.) and the GeneChip Human SNP 6.0 Assay (Affymetrix, Inc.) each targets a single SNP in CCL3. Patterns of LD, as determined for 35 CCL SNPs in our study population may help guide future research on African cohorts, because even the latest HapMap Phase II+III dataset (Release #28, August 2010) only reports 19 SNPs in the four CCL genes studied here.
In summary, we systematically screened SNPs in the genes encoding three principal natural ligands of CCR5 (HIV-1 coreceptor) and in the adjacent gene CCR2. Data from a large prospective cohort of HIV-1 serodiscordant couples enabled us to test SNP associations with both HIV-1 acquisition and early or chronic-phase VL in a high-risk study population. Overall, two variants in CCL3 and none in other genes relevant to CCR5 or CCR2 function appeared to influence early events in the natural history of HIV-1 infection, either by altering the rate of HIV-1 infection in HESN partners or by modulating HIV-1 VL soon after seroconversion. Future functional studies of these SNPs may fully resolve their potential effects on HIV-1 acquisition and control.
Supplementary Material
Acknowledgements
We thank study participants, staff, interns, and Project Management Group members of the Zambia-Emory HIV Research Project in Lusaka, Zambia; technical staff and students at the virology laboratory at the University Teaching Hospital, Lusaka, the immunogenetics laboratory and the data analysis group in the Program in Epidemiology of Infection and Immunity at UAB School of Public Health. This study was supported primarily by the U.S. National Institute of Allergy and Infectious Diseases, through grants AI041951 and AI071906 to R.A.K., AI040951 to S.A., AI064060 to E.H., and AI076123 to J.T.
Footnotes
Supplementary Material
Refer to Web version on PubMed Central for supplementary material.
REFERENCES
- 1.UNAIDS. UNAIDS/WHO "AIDS Epidemic Update: December 2006". 2006
- 2.Liu R, Paxton WA, Choe S, Ceradini D, Martin SR, Horuk R, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86(3):367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 3.Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382(6593):722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 4.Dean M, Carrington M, Winkler C, Huttley GA, Smith MW, Allikmets R, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273(5283):1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 5.Zimmerman PA, Buckler-White A, Alkhatib G, Spalding T, Kubofcik J, Combadiere C, et al. Inherited resistance to HIV-1 conferred by an inactivating mutation in CC chemokine receptor 5: studies in populations with contrasting clinical phenotypes, defined racial background, and quantified risk. Mol Med. 1997;3(1):23–36. [PMC free article] [PubMed] [Google Scholar]
- 6.An P, Nelson GW, Wang L, Donfield S, Goedert JJ, Phair J, et al. Modulating influence on HIV/AIDS by interacting RANTES gene variants. Proc Natl Acad Sci U S A. 2002;99(15):10002–10007. doi: 10.1073/pnas.142313799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rathore A, Chatterjee A, Sivarama P, Yamamoto N, Singhal PK, Dhole TN. Association of RANTES-403 G/A-28 C/G and In1.1 T/C polymorphism with HIV-1 transmission and progression among North Indians. J Med Virol. 2008;80(7):1133–1141. doi: 10.1002/jmv.21201. [DOI] [PubMed] [Google Scholar]
- 8.Menten P, Wuyts A, Van Damme J. Macrophage inflammatory protein-1. Cytokine Growth Factor Rev. 2002;13(6):455–481. doi: 10.1016/s1359-6101(02)00045-x. [DOI] [PubMed] [Google Scholar]
- 9.Guan E, Wang J, Roderiquez G, Norcross MA. Natural truncation of the chemokine MIP-1 beta /CCL4 affects receptor specificity but not anti-HIV-1 activity. J Biol Chem. 2002;277(35):32348–32352. doi: 10.1074/jbc.M203077200. [DOI] [PubMed] [Google Scholar]
- 10.Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270(5243):1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 11.Malhotra R, Hu L, Song W, Brill I, Mulenga J, Allen S, et al. Association of chemokine receptor gene (CCR2-CCR5) haplotypes with acquisition and control of HIV-1 infection in Zambians. Retrovirology. 2011;8(1):22. doi: 10.1186/1742-4690-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eugenin EA, Osiecki K, Lopez L, Goldstein H, Calderon TM, Berman JW. CCL2/monocyte chemoattractant protein-1 mediates enhanced transmigration of human immunodeficiency virus (HIV)-infected leukocytes across the blood-brain barrier: a potential mechanism of HIV-CNS invasion and NeuroAIDS. J Neurosci. 2006;26(4):1098–1106. doi: 10.1523/JNEUROSCI.3863-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vilades C, Broch M, Plana M, Domingo P, Alonso-Villaverde C, Pedrol E, et al. Effect of genetic variants of CCR2 and CCL2 on the natural history of HIV-1 infection: CCL2-2518GG is overrepresented in a cohort of Spanish HIV-1-infected subjects. J Acquir Immune Defic Syndr. 2007;44(2):132–138. doi: 10.1097/QAI.0b013e31802b3147. [DOI] [PubMed] [Google Scholar]
- 14.Ansari AW, Bhatnagar N, Dittrich-Breiholz O, Kracht M, Schmidt RE, Heiken H. Host chemokine (C-C motif) ligand-2 (CCL2) is differentially regulated in HIV type 1 (HIV-1)-infected individuals. Int Immunol. 2006;18(10):1443–1451. doi: 10.1093/intimm/dxl078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allen S, Meinzen-Derr J, Kautzman M, Zulu I, Trask S, Fideli U, et al. Sexual behavior of HIV discordant couples after HIV counseling and testing. AIDS. 2003;17(5):733–740. doi: 10.1097/00002030-200303280-00012. [DOI] [PubMed] [Google Scholar]
- 16.Fideli US, Allen SA, Musonda R, Trask S, Hahn BH, Weiss H, et al. Virologic and immunologic determinants of heterosexual transmission of human immunodeficiency virus type 1 in Africa. AIDS Res Hum Retroviruses. 2001;17(10):901–910. doi: 10.1089/088922201750290023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKenna SL, Muyinda GK, Roth D, Mwali M, Ng'andu N, Myrick A, et al. Rapid HIV testing and counseling for voluntary testing centers in Africa. AIDS. 1997;11(Suppl 1):S103–S110. [PubMed] [Google Scholar]
- 18.Kempf MC, Allen S, Zulu I, Kancheya N, Stephenson R, Brill I, et al. Enrollment and retention of HIV discordant couples in Lusaka, Zambia. J Acquir Immune Defic Syndr. 2008;47(1):116–125. doi: 10.1097/QAI.0b013e31815d2f3f. [DOI] [PubMed] [Google Scholar]
- 19.Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet. 2009;Chapter 2 doi: 10.1002/0471142905.hg0212s60. Unit 2 12. [DOI] [PubMed] [Google Scholar]
- 20.Barrett JC. Haploview: Visualization and analysis of SNP genotype data. Cold Spring Harb Protoc. 2009;2009(10) doi: 10.1101/pdb.ip71. pdb ip71. [DOI] [PubMed] [Google Scholar]
- 21.Modi WS, Lautenberger J, An P, Scott K, Goedert JJ, Kirk GD, et al. Genetic variation in the CCL18-CCL3-CCL4 chemokine gene cluster influences HIV Type 1 transmission and AIDS disease progression. Am J Hum Genet. 2006;79(1):120–128. doi: 10.1086/505331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Modi WS, Goedert JJ, Strathdee S, Buchbinder S, Detels R, Donfield S, et al. MCP-1-MCP-3-Eotaxin gene cluster influences HIV-1 transmission. AIDS. 2003;17(16):2357–2365. doi: 10.1097/00002030-200311070-00011. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez E, Rovin BH, Sen L, Cooke G, Dhanda R, Mummidi S, et al. HIV-1 infection and AIDS dementia are influenced by a mutant MCP-1 allele linked to increased monocyte infiltration of tissues and MCP-1 levels. Proc Natl Acad Sci U S A. 2002;99(21):13795–13800. doi: 10.1073/pnas.202357499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDermott DH, Beecroft MJ, Kleeberger CA, Al-Sharif FM, Ollier WE, Zimmerman PA, et al. Chemokine RANTES promoter polymorphism affects risk of both HIV infection and disease progression in the Multicenter AIDS Cohort Study. AIDS. 2000;14(17):2671–2678. doi: 10.1097/00002030-200012010-00006. [DOI] [PubMed] [Google Scholar]
- 25.Yoshiura C, Kofuku Y, Ueda T, Mase Y, Yokogawa M, Osawa M, et al. NMR analyses of the interaction between CCR5 and its ligand using functional reconstitution of CCR5 in lipid bilayers. J Am Chem Soc. 2010;132(19):6768–6777. doi: 10.1021/ja100830f. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez E, Dhanda R, Bamshad M, Mummidi S, Geevarghese R, Catano G, et al. Global survey of genetic variation in CCR5, RANTES, and MIP-1alpha: impact on the epidemiology of the HIV-1 pandemic. Proc Natl Acad Sci U S A. 2001;98(9):5199–5204. doi: 10.1073/pnas.091056898. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.