Molecular epidemiologic analysis shows that travelers returning from Asia are the greatest source of risk.
Keywords: dengue, dengue virus, phylogenetics, importation, outbreak, Queensland, Australia, viruses
Abstract
To assess risk for importation of dengue virus (DENV) into Queensland, Australia, and sources of imported viruses, we sequenced the envelope region of DENV isolates from symptomatic patients with a history of travel during 2002–2010. The number of imported dengue cases greatly increased over the surveillance period, some of which were associated with domestic outbreaks. Patients reported traveling to (in order) Asia, Papua New Guinea, Pacific Island countries, and non–Asia-Pacific countries. By using phylogenetic methods, we assigned DENV isolates from returning residents and overseas visitors with viremia to a specific genotypic group. Genotypes circulating in Asia were extremely diverse. Genotyping and molecular clock analysis supported Asian origination of a strain that caused an outbreak of DENV-4 in Pacific Island countries during 2007–2009, and subsequently, in Innisfail, Australia, in 2009. Our findings indicate that Asia is a major source of DENVs that are imported into Australia, causing a risk for epidemics.
Queensland, a state located in the tropical and subtropical northeastern area of Australia, has a long history of dengue virus (DENV) activity. Dengue was present in the late 19th century (1) and, following a lull for most of the 20th century, dengue importation and epidemic transmission have been increasingly reported in the past 20 years (2,3). Epidemics of the disease have occurred historically in other states of Australia, but only in Queensland have epidemics been reported in recent times. These epidemics were caused by the distribution of the vector, Aedes aegypti mosquitoes. The species was once found in other Australian states, but its area of distribution has now contracted so that it lies almost exclusively within Queensland’s borders (4,5).
Despite repeated transmission events, dengue is not endemic to Queensland, and transmission requires a viremic traveler to import the virus to initiate epidemic spread (6). Rapid identification of cases and disease tracking, incorporating targeted vector surveillance, and control measures adopted rigorously to limit epidemic potential have been major factors in preventing local transmission and in reducing the cost of managing mosquito-borne disease (7).
With the apparent increasing frequency of dengue epidemics and imported cases, the disease has become a major public health issue. Exposure to multiple serotypes of DENV, of which there are 4 in total, may result in a higher probability of potentially life-threatening conditions such as dengue hemorrhagic fever and dengue shock syndrome, a potentially life-threatening condition (8). Perhaps not coincidentally, 2 fatal cases of dengue hemorrhagic fever were reported in 2004 in Queensland, and the serologic profile of the case-patients indicated secondary infection consistent with dengue shock syndrome (9). Of additional concern is the possibility that the virus may become endemic if case numbers were to rise to a point at which vector control measures became ineffectual at controlling virus spread.
Recent DENV infection is diagnosed by serologic testing, through virus isolation or by nucleic acid amplification by reverse transcription PCR (RT-PCR). The advantage of the latter is that sequencing of reaction products enables a definitive diagnosis of acute infection, identification of the virus serotype, and genotyping. As an adjunct to isolation techniques, sequencing and genotyping can provide valuable evidence of importation or can confirm local transmission and enable differentiation between multiple circulating strains and serotypes. Analysis of DENV sequence data facilitates rapid disease tracking and vector control. However, not all specimens are suitable for RT-PCR because infected persons usually exhibit a relatively short-lived viremia early in the febrile period (10). In addition, clinicians may find it difficult to obtain acute-phase samples, particularly if patients delay their initial consultation or are still in transit during their viremic phase.
As part of control measures by Queensland Public Health, we sequenced the envelope region of DENV isolates from symptomatic patients with a history of travel during 2002–2010. The proportion of the 4 DENV serotypes that were imported was determined, as well as the geographic origin of each serotype. Phylogenetic trees containing imported DENV viruses and others strains circulating throughout the world (from GenBank) were constructed by using a maximum likelihood model. From this analysis, we ascertained the likely geographic origin of imported viruses. This enabled us to assess the risk for importation of DENV from various sources by travelers entering Australia.
Materials and Methods
Virus Samples
Serum samples from patients with suspected DENV infection were referred to the Public Health Virology Laboratory, Queensland Health Forensic and Scientific Services, following the directive of Queensland Public Health medical officers, or were obtained through the public or private laboratory network. Acute-phase specimens underwent RT-PCR and serologic testing, and those that successfully yielded an RT-PCR product (after specific DENV serotype amplication) were sequenced and genotyped by phylogenetic analyses to assist public health investigations. This work was approved by the Ethics Committee of Queensland Health Forensic and Scientific Services.
Viral RNA Extraction and Nucleotide Sequencing
RNA was extracted from 200 µL of serum, either manually (QIAamp viral RNA extraction kit; QIAGEN, Hildren, Germany) or by using the EZ1 Virus Mini Kit and (QIAGEN) according to the manufacturer’s instructions. Amplification was performed for each DENV serotype by using the Superscript III/Platinum Taq High Fidelity One-Step RT-PCR System (Invitrogen, Carlsbad, CA, USA) with specific RT-PCR primers (Table 1). Nucleotide sequencing of the complete envelope gene region (DENV-1, DENV-2, and DENV-4: 1,485 bp; DENV-3: 1,479 bp) was performed by using the Big Dye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA, USA). Sequence data obtained were deposited in GenBank (Table 2).
Table 1. Amplification oligonucleotide primers for DENV genotyping RT-PCRs, Queensland, Australia, 2001–2010*†.
| DENV assay | Forward primer | Reverse primer |
|---|---|---|
| DENV-1‡ | 5′760-AACGTGGATGTCCTCTGAAGG-7803′ | 5′1600-CGAGGTCCAAGGCAGTG-15843′ |
| 5′1418-GCAACCATAACACCTCAA-14353′ | 5′2600-TGGCTGATCGAATTCCACAC-25813′ | |
| DENV-2§ | 5′-789GAAACATGCCCAGAGAATTGAAACT-8133′ | 5′-1920CCCTTCATATTGTACTCTGATAACTATTGTTCC-18883′ |
| 5′-1547AAGCTTGGCTGGTGCACAGGCAATGGTT-15743′ | 5′-2537GGGGATTCTGGTTGGAACTTGTATTGTTCTGTCC-25043′ | |
| DENV-3¶ | 5′-291TGGCTAGATGGGGTACCTTC-3103′ or5′-722GCTCCCCATGTCGGCATGGGACTGG-7463′ | 5′-1819CATCCCTTTGAGTTTCAATTTGTCCAT-17933′ |
| 5′-1685CTAGGATCTCAAGAAGGAGCAATGCA-17103′ | 5′-2550ATGGCTGTTGCCACTCTTTTGGGGGA-25253′ | |
| DENV-4# | 5′-742TGGGATTGGAAACAAGAGCTGAGACATGGATGTC-7753′ | 5′-1838CGTGTATGACATTCCCTTGATTCTCAATTTCTCCA-18043′ |
| 5′-1569CAATGGTTTTTGGACCTACCTCTACCATGG-15983′ | 5′-2539GGGGACTCTGGTTGAAATTTGTACTGTTCTGTCCA-25053′ |
*DENV, dengue virus; RT-PCR, reverse transcription PCR. †Numbering is based on DENV-1 strain DENV-1BR/90 (AF226685), DENV-2 strain New Guinea C (AF038403), DENV-3 strain Ba51 (AY858037), DENV-4 strain Dominica 1981 (AF326573). For each DENV serotype, forward primers were paired with the shown respective reverse primers in two separate genotyping RT-PCR assays (A. Pyke, unpub. method). ‡A. Pyke and D. Beasley, unpub. method. §S. Mei Lok, unpub. data. ¶I. Serafin and A. Pyke, unpub. method. #C. Howard and A. Pyke, unpub. method.
Table 2. Description of DENV virus strains analyzed, Queensland, Australia, 2002–2010*.
| Serotype/sequence name | Geographic origin | Year isolated | GenBank accession no. |
|---|---|---|---|
| DENV-1 | |||
| Bali 2003 | Bali | 2003 | JN415488 |
| Bali 2010a | Bali | 2010 | JN415489 |
| Bali 2010b | Bali | 2010 | JN415490 |
| Bali 2010c | Bali | 2010 | JN415491 |
| Bali 2010d | Bali | 2010 | JN415492 |
| Bali 2010e | Bali | 2010 | JN415493 |
| Bali 2010f | Bali | 2010 | JN415494 |
| Cairns 2003 | Cairns, Australia | 2003 | JN415495 |
| Cambodia 2007 | Cambodia | 2007 | JN415496 |
| Cook Islands 2002 | Cook Islands | 2002 | JN415497 |
| Cook Islands 2006 | Cook Islands | 2006 | JN415498 |
| East Timor 2000 | Timor-Leste | 2000 | JN415499 |
| East Timor 2008 | Timor-Leste | 2008 | JN415500 |
| East Timor 2009 | Timor-Leste | 2009 | JN415501 |
| East Timor 2010 | Timor-Leste | 2010 | JN415502 |
| Fiji 2002 | Fiji | 2002 | JN415503 |
| Fiji 2006a | Fiji | 2006 | JN415504 |
| Fiji 2006b | Fiji | 2006 | JN415505 |
| Guyana 2008 | Guyana | 2008 | JN415506 |
| India 2008 | India | 2008 | JN415507 |
| India 2010 | India | 2010 | JN415486 |
| Indonesia 2010a | Indonesia | 2010 | JN415508 |
| Indonesia 2010b | Indonesia | 2010 | JN415510 |
| Jakarta 2004 | Jakarta | 2004 | AY858983† |
| Laos 2007 | Laos | 2007 | JN415509 |
| Malaysia 1972 | Malaysia | 1972 | AF425622† |
| Malaysia 2005 | Malaysia | 2005 | JN415511 |
| Malaysia 2008 | Malaysia | 2008 | JN415512 |
| Malaysia 2010 | Malaysia | 2010 | JN415513 |
| Mareeba 2003 | Mareeba, Australia | 2003 | JN415514 |
| Palau 2000 | Palau | 2000 | JN415515 |
| Philippines 2005 | The Philippines | 2005 | JN415516 |
| Philippines 2010 | The Philippines | 2010 | JN415517 |
| PNG 2003 | Papua New Guinea | 2003 | JN415518 |
| PNG 2009 | Papua New Guinea | 2009 | JN415519 |
| Samoa 2001 | Samoa | 2001 | JN415520 |
| Singapore 2003 | Singapore | 2003 | FJ469907† |
| Singapore 2005 | Singapore | 2005 | EU081246† |
| Singapore 2005 | Singapore | 2005 | EU081247† |
| Singapore 2008 | Singapore | 2008 | JN415521 |
| Solomon Islands 2002 | Solomon Islands | 2002 | JN415522 |
| Southeast Asia 2007 | Southeast Asia | 2007 | JN415523 |
| Southeast Asia 2005 | Southeast Asia | 2005 | JN415529 |
| Sri Lanka 2004 | Sri Lanka | 2004 | JN415524 |
| Sumatra 1998 | Sumatra | 1998 | AB189121† |
| Thailand 1954 | Thailand | 1954 | D10513† |
| Thailand 1980 | Thailand | 1980 | AY732474† |
| Thailand 2001 | Thailand | 2001 | JN415525 |
| Thailand 2008a | Thailand | 2008 | JN415526 |
| Thailand 2008b | Thailand | 2008 | JN415527 |
| Thailand 2010 | Thailand | 2010 | JN415528 |
| Tonga 2008 | Tonga | 2008 | JN415530 |
| Townsville 2008 | Townsville, Australia | 2008 | JN415531 |
| Townsville 2009 | Townsville, Australia | 2009 | JN415532 |
| Venezuela 2007 | Venezuela | 2007 | EU482609† |
| Vietnam 2006 | Vietnam | 2006 | JN415533 |
| Vietnam 2006 | Vietnam | 2006 | EU482818† |
| Vietnam 2008a | Vietnam | 2008 | JN415534 |
| Vietnam 2008b | Vietnam | 2008 | JN415535 |
| Vietnam South 2008 | Vietnam | 2008 | GU131812† |
| Vietnam 2010 | Vietnam | 2010 | JN415487 |
| Yap Island 2004 | Yap Island | 2004 | AB204803† |
| DENV-2 | |||
| Bali 2009 | Bali | 2009 | JN568242 |
| Bali 2010 | Bali | 2010 | JN568243 |
| Borneo 2009 | Borneo | 2009 | JN568247 |
| Brunei 2005 | Brunei | 2005 | EU179858† |
| Cairns 2003a | Cairns, Australia | 2003 | JN568248 |
| Cairns 2003b | Cairns, Australia | 2003 | JN568249 |
| Cairns 2004 | Cairns, Australia | 2004 | JN568250 |
| Cairns 2006 | Cairns, Australia | 2006 | JN568251 |
| Cairns 2008 | Cairns, Australia | 2008 | JN568252 |
| Cairns 2010 | Cairns, Australia | 2010 | JN568253 |
| Cambodia 2003 | Cambodia | 2003 | GQ868621† |
| Cambodia 2008 | Cambodia | 2008 | GU131924† |
| China 2001 | China | 2001 | EF051521† |
| East Timor 2000 | Timor-Leste | 2000 | JN568254 |
| East Timor 2002 | Timor-Leste | 2002 | JN568255 |
| East Timor 2004 | Timor-Leste | 2004 | JN568256 |
| East Timor 2010 | Timor-Leste | 2010 | JN568257 |
| India 2001 | India | 2001 | DQ448237† |
| India 2009 | India | 2009 | JN568258 |
| India 2003 | India | 2003 | JN568260 |
| India 2010 | India | 2010 | JN568259 |
| Indonesia 2004 | Indonesia | 2004 | AY858035† |
| Kuranda 2002 | Kuranda, Australia | 2002 | JN568261 |
| Laos 2010 | Laos | 2010 | JN568244 |
| Mt Isa 2010 | Mt Isa, Australia | 2010 | JN568262 |
| New Guinea C 1944 | New Guinea | 1944 | AF038403† |
| Peru 1996 | Peru | 1996 | IQT1797† |
| Philippines 2003 | The Philippines | 2003 | JN568263 |
| Philippines 2010a | The Philippines | 2010 | JN568264 |
| Philippines 2010b | The Philippines | 2010 | JN568265 |
| PNG 2003 | PNG | 2003 | JN568266 |
| PNG 2009 | PNG | 2009 | JN568241 |
| PNG 2010a | PNG | 2010 | JN568267 |
| PNG 2010b | PNG | 2010 | JN568268 |
| PNG 2010c | PNG | 2010 | JN568269 |
| PNG 2010d | PNG | 2010 | JN568270 |
| Singapore 2008 | Singapore | 2008 | GU370051† |
| Southeast Asia 2010b | Southeast Asia | 2010 | JN568276 |
| Southeast Asia 2010a | Southeast Asia | 2010 | JN568277 |
| Sri Lanka 1996 | Sri Lanka | 1996 | FJ882602† |
| Sumatra 2009 | Sumatra | 2009 | JN568271 |
| Sumatra 2010 | Sumatra | 2010 | JN568272 |
| Taiwan 2001 | Taiwan | 2001 | DQ645541† |
| Thailand 1996 | Thailand | 1996 | AF100459† |
| Thailand 2001 | Thailand | 2001 | DQ181797† |
| Thailand 2007 | Thailand | 2007 | JN568273 |
| Thailand 2010a | Thailand | 2010 | JN568274 |
| Thailand 2010b | Thailand | 2010 | JN568275 |
| Thailand 2010c | Thailand | 2010 | JN568245 |
| Torres Strait 2003 | Torres Strait, Australia | 2003 | JN568278 |
| Townsville 1993 | Townsville, Australia | 1993 | AY037116† |
| Townsville 2010a | Townsville, Australia | 2010 | JN568279 |
| Townsville 2010b | Townsville, Australia | 2010 | JN568246 |
| Tully 2010 | Tully, Australia | 2010 | JN568280 |
| Venezuela 1990 | Venezuela | 1990 | GQ868540† |
| Vietnam 2005 | Vietnam | 2005 | FM210207† |
| Vietnam 2006 | Vietnam | 2006 | EU569721† |
| Vietnam 2010a | Vietnam | 2010 | JN568281 |
| Vietnam 2010b | Vietnam | 2010 | JN568282 |
| DENV-3 | |||
| Bali 2009 | Bali | 2009 | JN568284 |
| Bali 2010a | Bali | 2010 | JN568283 |
| Bali 2010b | Bali | 2010 | JN575560 |
| Bali 2010c | Bali | 2010 | JN575561 |
| Cairns 1998 | Cairns, Australia | 1998 | JN575562 |
| Cairns 2008a | Cairns, Australia | 2008 | JN575563 |
| Cairns 2008 | Cairns, Australia | 2008 | JN575564 |
| Cambodia 2006 | Cambodia | 2006 | JN575565 |
| East Timor 2000 | Timor-Leste | 2000 | JN575566 |
| Fiji 1992 | Fiji | 1992 | L11422† |
| India 1984 | India | 1984 | L11424† |
| Indonesia 1985 | Indonesia | 1985 | L11428† |
| Indonesia 1998 | Indonesia | 1998 | AY265857† |
| Indonesia 2004a | Indonesia | 2004 | AY858037† |
| Indonesia 2004b | Indonesia | 2004 | AY858047† |
| Indonesia 2008a | Indonesia | 2008 | JN575567 |
| Indonesia 2008b | Indonesia | 2008 | JN575568 |
| Philippines 1983 | The Philippines | 1983 | L11432† |
| Philippines 1997 | The Philippines | 1997 | AY496879† |
| Philippines 2010 | The Philippines | 2010 | JN575570 |
| PNG 2008 | Papua New Guinea | 2008 | JN575571 |
| PNG 2010a | Papua New Guinea | 2010 | JN575572 |
| PNG 2010b | Papua New Guinea | 2010 | JN575573 |
| Puerto Rico 1977 | Puerto Rico | 1977 | L11434† |
| Samoa 1986 | Samoa | 1986 | L11435† |
| Singapore 2005 | Singapore | 2005 | EU081221† |
| Southeast Asia 2008 | Southeast Asia | 2008 | JN575569 |
| Sri Lanka 1991 | Sri Lanka | 1991 | L11438† |
| Tahiti 1989 | Tahiti | 1989 | L11619† |
| Taiwan 1998 | Taiwan | 1998 | DQ675532† |
| Taiwan 1999 | Taiwan | 1999 | DQ675533† |
| Thailand 1973 | Thailand | 1973 | L11620† |
| Thailand 1987 | Thailand | 1987 | L11442† |
| Thailand 1997a | Thailand | 1997 | JN575574 |
| Thailand 1997b | Thailand | 1997 | JN575575 |
| Thailand 2010 | Thailand | 2010 | JN575576 |
| Townsville 2006 | Townsville, Australia | 2006 | JN575577 |
| Townsville 2007 | Townsville, Australia | 2007 | JN575578 |
| Townsville 2009 | Townsville, Australia | 2009 | JN575579 |
| Vietnam 2007 | Vietnam | 2007 | EU482461† |
| Vietnam 2008 | Vietnam | 2008 | JN575580 |
| DENV-4 | |||
| Bali 2010 | Bali | 2010 | JN575583 |
| Brazil 1982 | Brazil | 1982 | U18425† |
| Cairns 2002 | Cairns, Australia | 2002 | JN575584 |
| China 2001 | China | 2001 | AF289029† |
| Cook Islands 2009 | Cook Islands | 2009 | JN575582 |
| Dominica 1981 | Dominica | 1981 | AF326573† |
| East Timor 2000 | Timor-Leste | 2000 | JN575585 |
| East Timor 2007 | Timor-Leste | 2007 | JN575586 |
| El Salvador 1983 | El Salvador | 1983 | U18426† |
| Fiji 2008 | Fiji | 2008 | JN575587 |
| Indonesia 1973 | Indonesia | 1973 | U18428† |
| Indonesia 1977 | Indonesia | 1977 | U18430† |
| Indonesia 2010a | Indonesia | 2010 | JN575588 |
| Indonesia 2010b | Indonesia | 2010 | JN575589 |
| Innisfail 2009 | Innisfail, Australia | 2009 | JN575581 |
| Jakarta 2004 | Jakarta | 2004 | AY858049† |
| Malaysia 2009 | Malaysia | 2009 | JN575590 |
| New Caledonia 1984 | New Caledonia | 1984 | U18432† |
| Philippines 1984 | The Philippines | 1984 | U18435† |
| Philippines 2004 | The Philippines | 2004 | JN575591 |
| Puerto Rico 1986 | Puerto Rico | 1986 | U18436† |
| Samoa 2008 | Samoa | 2008 | JN575592 |
| Solomon Islands 2008 | Solomon Islands | 2008 | JN575593 |
| Sri Lanka 1978 | Sri Lanka | 1978 | U18437† |
| Tahiti 1985 | Tahiti | 1985 | U18439† |
| Thailand 1978 | Thailand | 1978 | U18441† |
| Thailand 1984 | Thailand | 1984 | U18442† |
| Thailand 1997 | Thailand | 1997 | AY618988† |
| Thailand 2001 | Thailand | 2001 | AY618992† |
| Thailand 2010 | Thailand | 2010 | JN575594 |
| Torres Strait 2005 | Torres Strait, Australia | 2005 | JN575595 |
| Townsville 2005 | Townsville, Australia | 2005 | JN575596 |
*DENV, dengue virus; PNG, Papua New Guinea. †Sequences obtained from GenBank.
Phylogenetic Analysis of Envelope Protein Sequence
Full-envelope protein sequences for each serotype were aligned by using the multiple alignment tool of MEGA5 (www.megasoftware.net). Unrooted trees were then constructed by using a maximum likelihood estimation with a Jukes-Cantor model and γ-distributed rates, and by constructing 1,000 replicates to generate bootstrap support values. Divergence time from a common ancestor was estimated by using the molecular clock calculator.
Results
Increasing Incidence of Dengue Outbreaks and Serotype Diversity
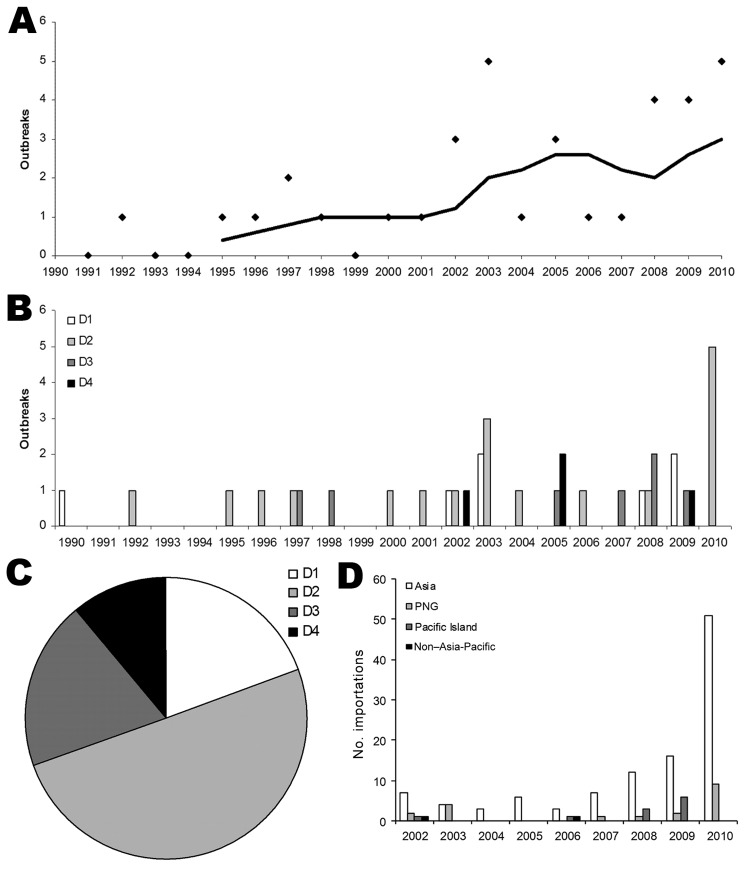
Previous reports (3,11) and anecdotal evidence indicated that there has been an increase in the number of dengue outbreaks occurring in Queensland. To investigate this apparent trend, we combined recent and historical outbreak data (3) over a 20-year period. A 5-year moving average does indeed show trends of increasing dengue outbreak incidence and increasing diversity of DENV serotypes that cause such outbreaks (Figure 1, panels A, B). A line of best fit revealed a significant increase with time (r2 = 0.48; p<0.05 by Student t test, 2-tailed). All 4 DENV serotypes caused outbreaks; DENV-2 was the most common cause (50.0%), followed by DENV1 and DENV-3 (19.4% each) and DENV-4 (11.1%) (Figure 1, panel C). The increase in outbreak incidence reflects changes in international travel over this period, which has increased 3.5-fold since the early 1990s (12). This increase is consistent with increased importations of virus carried by viremic travelers and the recognized increase in DENV infections throughout the world (13). Also of note is the dramatic increase in infections caused by imported viruses in 2010 (Figure 1, panel D).
Figure 1.
Number and diversity of dengue outbreaks in northern Queensland, Australia. A) Outbreaks of dengue causing epidemic spread in Queensland 1990–2010 showing 5-year moving average. B) Outbreaks shown as individual serotypes. C) Proportion of dengue virus serotypes responsible for the outbreaks shown in A and B. D) Geographic origins of dengue viruses imported into Queensland by viremic travelers. D1–D4, DENV-1–DENV-4; PNG, Papua New Guinea.
Geographic Origins and Diversity of Imported DENVs, 2002–2010
We ascertained the number and diversity of imported DENV serotypes from infected travelers during 2002–2010 (Table 3). This period was chosen because the most comprehensive patient sequence data were available. Information was analyzed from viremic travelers, for whom an RT-PCR amplification product and serotype designation could be obtained. The possible strain origins, which were determined after phylogenetic analyses, were compared with available travel histories to ascertain likely geographic origins of the infecting virus. The data were categorized into 4 separate regions: Asia, Papua New Guinea (PNG), the Pacific Islands, and countries outside of the Asia-Pacific region (non–Asia-Pacific).
Table 3. Number and diversity of imported dengue serotypes, Queensland, Australia, 2002–2010*.
| Region and country | No. (%) cases | Dengue serotypes (genotypes) |
|---|---|---|
| Asia | ||
| Indonesia | 37 (26.4) | 1 (I, IV), 2 (Cosmopolitan), 3 (I), 4 (II) |
| Thailand | 15 (10.7) | 1 (I), 2 (Asian genotype I), 3 (II), 4 (I) |
| Philippines | 10 (7.1) | 1 (IV), 2 (Cosmopolitan), 3 (I), 4 (I) |
| India | 9 (6.4) | 1 (V), 2 (Cosmopolitan) |
| Timor-Leste | 9 (6.4) | 1 (IV), 2 (Cosmopolitan), 4 (II) |
| Vietnam | 7 (5.0) | 1 (I), 2 (Asian genotype I), 3 (II) |
| Malaysia | 5 (3.6) | 1 (I, IV), 4 (II) |
| Laos | 2 (1.4) | 1 (I), 2 (Asian genotype I) |
| Cambodia | 2 (1.4) | 1 (I), 3 (II) |
| Singapore | 1 (0.7) | 1 (I) |
| Sri Lanka | 1 (0.7) | 1 (V) |
| Asia, not specified | 11 (7.9) | - |
| Papua New Guinea | 19 (13.6) | 1 (I, IV), 2 (Cosmopolitan), 3 (I) |
| Pacific Islands | ||
| Fiji | 4 (2.9) | 1 (IV), 4 (II) |
| Samoa | 2 (1.4) | 4 (II) |
| Solomon Islands | 1 (0.7) | 1 (ND) |
| Tonga | 3 (2.1) | 4 (ND) |
| Vanuatu | 1 (0.7) | 4 (ND) |
| Non–Asia-Pacific | ||
| Brazil | 1 (0.7) | 3 (ND) |
| Guyana | 1 (0.7) | 1 (V) |
| Total | 140 |
*ND, not determined
Most infected travelers (77.9%) reported spending time abroad in Asia. In particular, 26.4% of all virus importations could be traced to Indonesia alone. All 4 DENV serotypes were detected in the specimens sequenced from persons with a travel history to that country. Notably, patients reported traveling to other Asian countries, including Thailand and the Philippines, where all 4 DENV serotypes have been found. Most other countries to which travel was reported had at least 3 DENV serotypes (Timor-Leste, PNG, and Vietnam), 2 serotypes (Cambodia, India, Fiji, Malaysia, and Laos), or 1 serotype (Brazil, Guyana, Samoa, Singapore, Solomon Islands, Sri Lanka, Tonga, and Vanuatu). A greater degree of diversity cannot be excluded in many of these countries because sampling numbers for individual countries were often low.
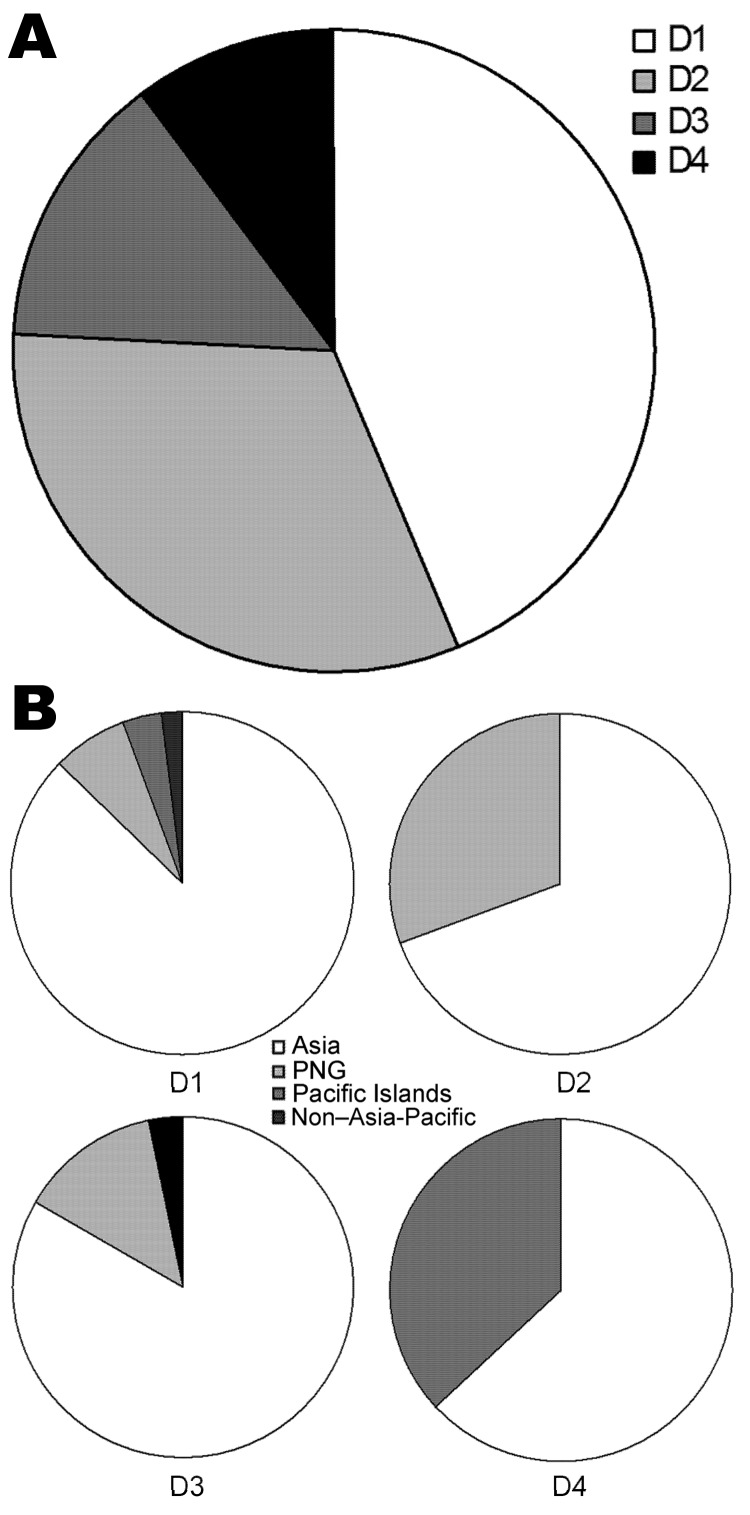
We calculated the proportion of the 4 DENV serotypes for infected travelers (Figure 2, panel A), which was slightly different from the proportion associated with outbreaks within Queensland (Figure 1, panel C). The serotype most commonly imported by travelers was DENV-1 (39.3%), followed by DENV-2 (25.7%), DENV-3 (21.4%), and DENV-4 (13.6%). Strains of all 4 DENV serotypes originated mainly in Asia (Figure 2, panel B). DENV-1 had the most diverse origins, with patients reporting travel mainly to Asia, but also to PNG, the Pacific Islands, and non–Asia-Pacific regions. In addition to Asia, DENV-2 was found to originate in PNG; DENV-3 originated in PNG and a non–Asia-Pacific area (Brazil); and DENV-4 originated in the Pacific Islands.
Figure 2.
Importation of dengue viruses (DENVs) into Queensland, Australia, 2002–2010. A) Proportion of imported DENV serotypes. B) Geographic origins of the 4 imported DENV serotypes. D1–D4, DENV-1–DENV-4; PNG, Papua New Guinea.
Origins of DENVs Imported by Returning Residents
Infected travelers were either returning Queensland residents or international visitors. Returning residents were the largest proportion of patients (96.4%) who sought treatment from the health system with dengue viremia. Further analysis was conducted on the subset of returning residents (135 of 140 patients with imported cases). Similarly to the analysis of all travelers above, infected returning residents (Table 4) reported that they had most frequently returned from Asia (77.0%), followed by PNG (13.3%), then the Pacific Islands (8.1%), and least often from non–Asia-Pacific areas (1.5%). The overall ratio was significantly different from that expected on the bases of the proportion of all Queensland residents who reported returning from dengue-endemic countries of those 4 regions, as calculated by using data from the Australian Bureau of Statistics for 2002–2010 (χ2 analysis, p = 0.0004). The proportion of patients reporting travel to Asia, and to PNG in particular, was higher than expected. The overrepresentation of cases from PNG is best explained by a recent increase in DENV activity in that country, which is consistent with a large number of importations from PNG (47.4%) during 2010 (Figure 1, panel D) and, in addition, other recent reports (14). In comparison, the proportion of patients who reported travel to the Pacific Islands was lower than expected. Because only 2 cases originated in the non–Asia-Pacific region, it was subsequently difficult to draw conclusions about this region for statistical purposes.
Table 4. Observed and expected numbers of imported DENVs by region, Queensland, Australia, 2002–2010*.
| Imported DENVs | No. (%) from Asia† | No. (%) from PNG | No. (%) from Pacific Islands | No. (%) from non–Asia-Pacific region |
|---|---|---|---|---|
| Observed | 104 (77.0) | 18 (13.3) | 11 (8.1) | 2 (1.5) |
| Expected‡ | 92 (68.2) | 11 (7.9) | 28 (20.8) | 4 (3.2) |
*χ2 is 18 (p<0.0004; 2-tailed test); DENVs, dengue viruses; PNG, Papua New Guinea. †Asia includes travelers to Indonesia, Timor-Leste, Thailand, India, Malaysia, Philippines, Vietnam, Singapore, Cambodia, Sri Lanka and Laos; Papua New Guinea; Pacific Islands includes travelers to Fiji, Samoa, Solomon Islands, Tonga, and Vanuatu; non–Asia-Pacific region was defined as all cases outside the Asia-Pacific region which included Brazil and Guyana. ‡Based on travel data from the Australian Bureau of Statistics (www.abs.gov.au/) for departing Queensland residents who named the country where they planned to spend the most time, selected for those countries designated as having an ongoing dengue transmission risk, according to the Centers for Disease Control and Prevention Dengue Map (www.healthmap.org/dengue).
Genotype Assignment of Imported DENVs
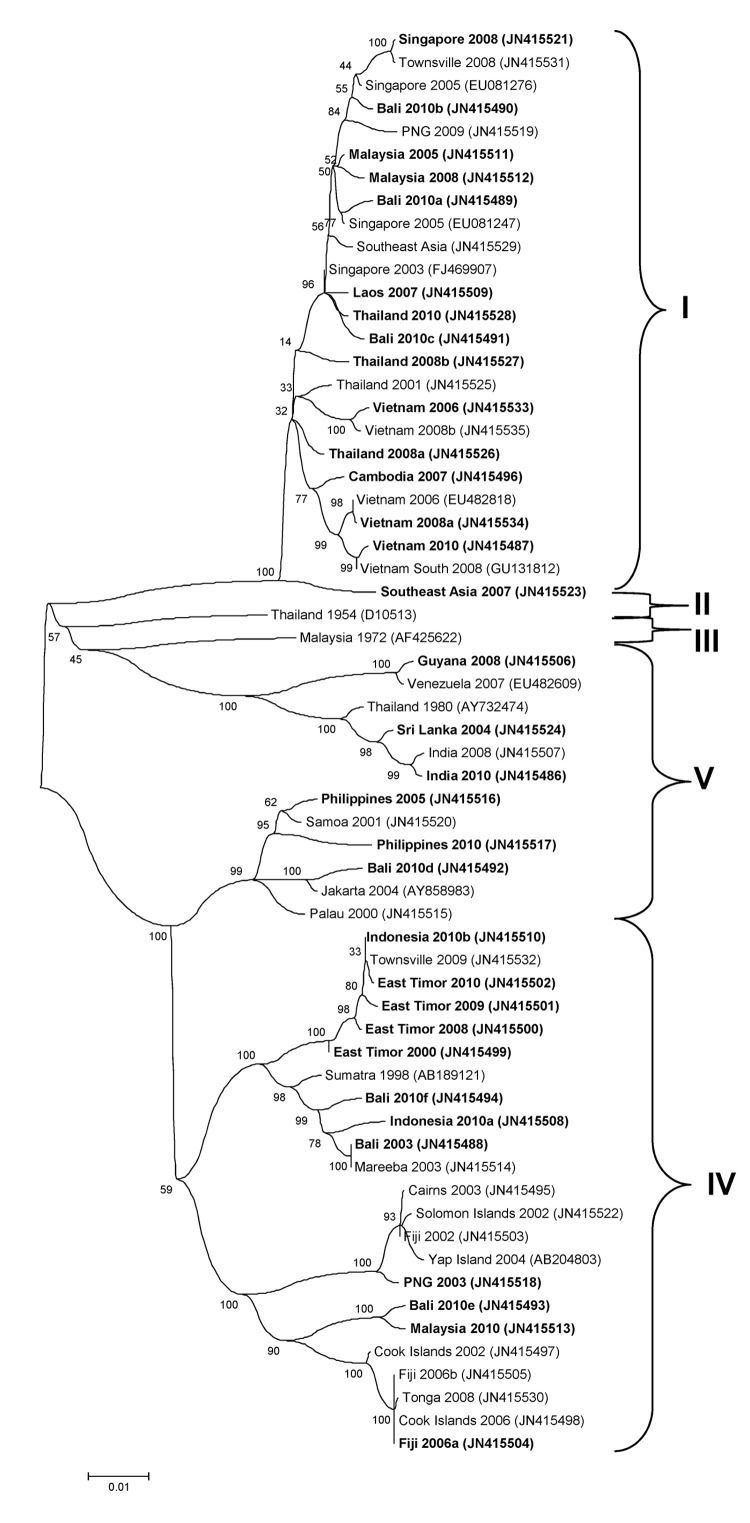
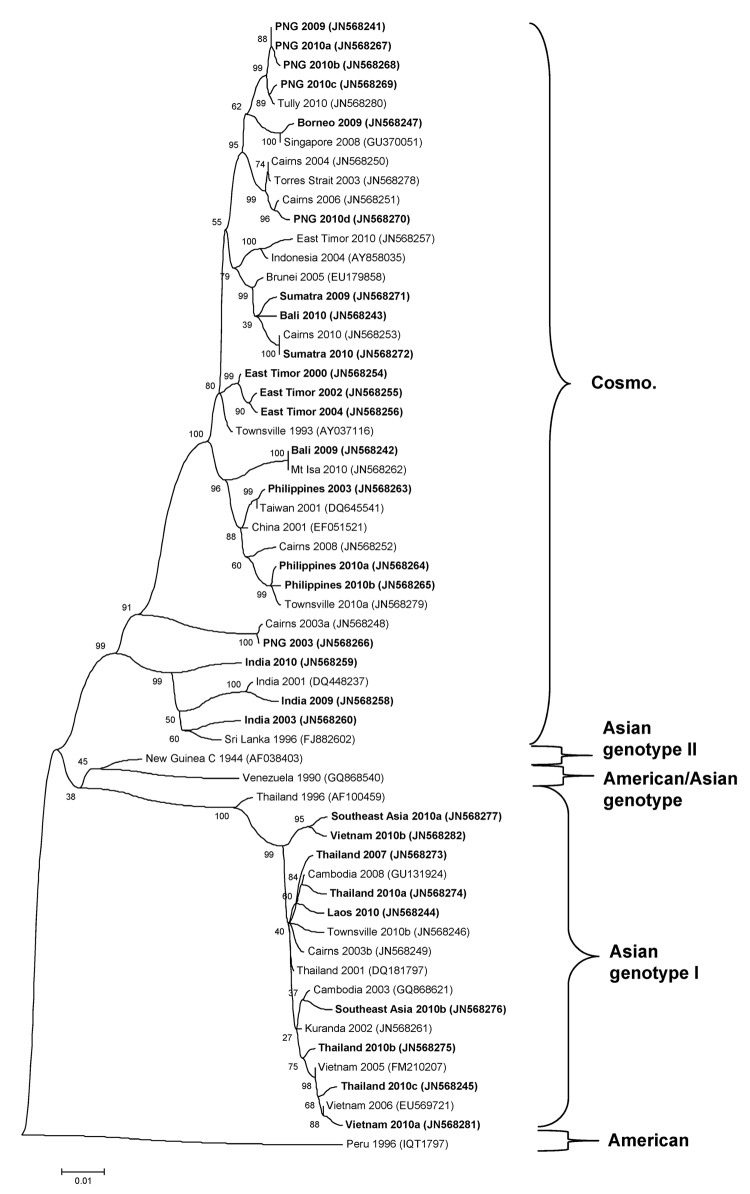
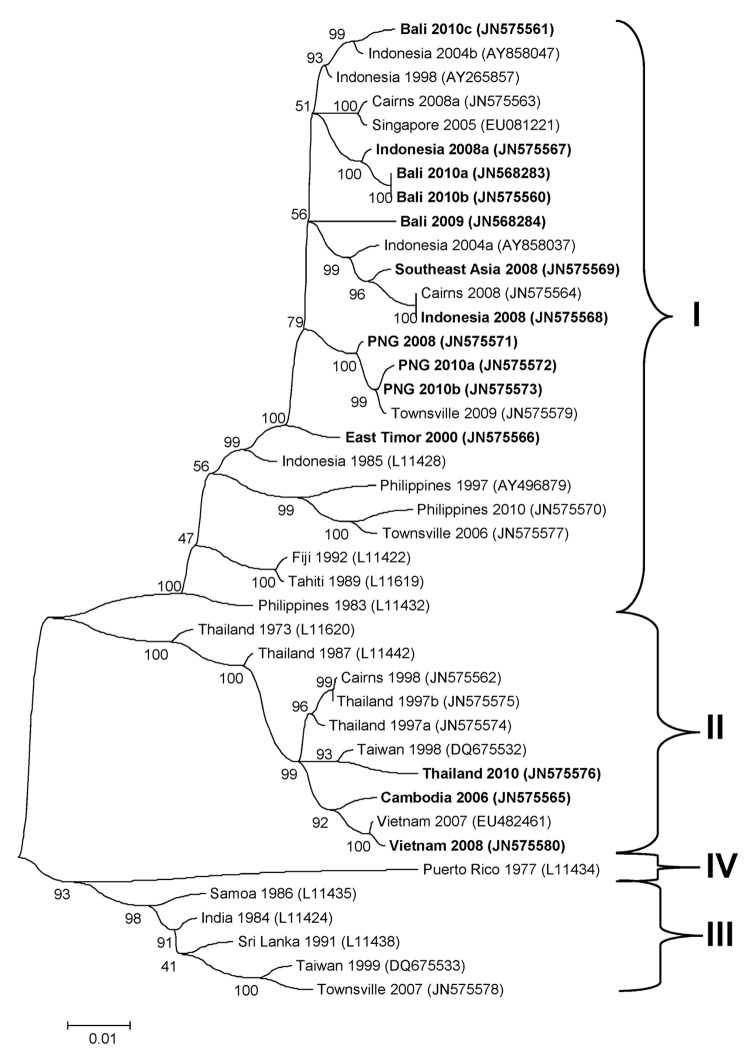
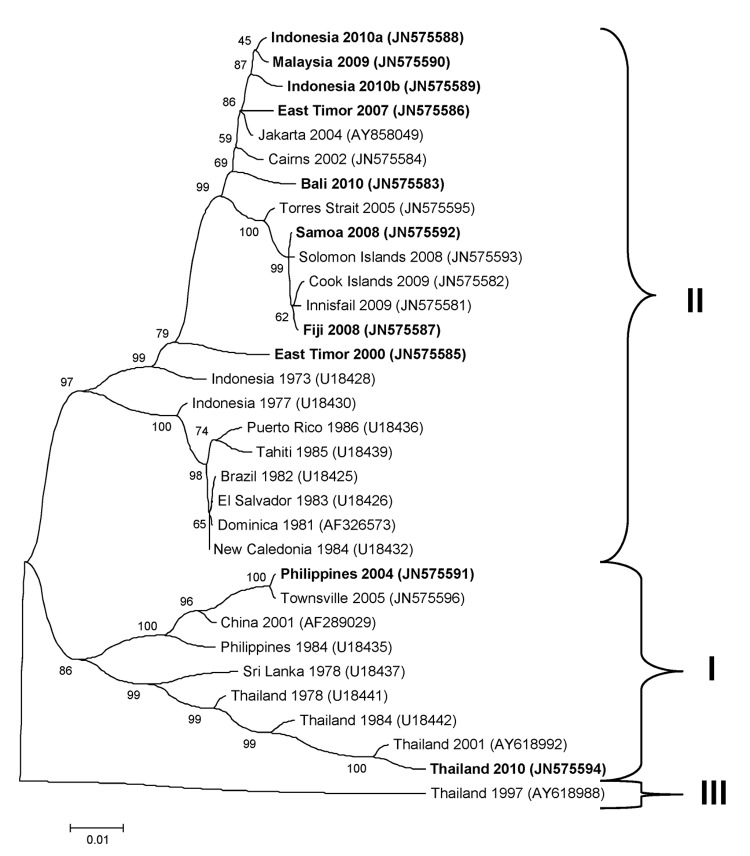
The genotypic mix for the various regions from which dengue was imported is shown in Table 3 and Figure 3, Figure 4, Figure 5, Figure 6. Across all regions, viruses could be classified into 1 of 2 genotypic groups within each serotype; the exception was DENV-1, which had 3 groups. In countries which were a source of imported viruses, generally 1 genotypic group for each serotype predominated.
Figure 3.
Phylogenetic tree showing the relationship of dengue viruses, serotype 1, imported into Queensland, Australia, 2001–2010, based on sequencing of the envelope gene. Viruses are designated according to reported origin and GenBank accession number, and imported cases are shown in boldface. Genotypes are indicated on the right. Scale bar indicates nucleotide substitutions per site.
Figure 4.
Phylogenetic tree showing the relationship of dengue viruses, serotype 2, that were imported into Queensland, Australia, 2002–2010, based on sequencing of the envelope gene. Viruses are designated according to reported origin and GenBank accession number, and imported cases are shown in boldface. Genotypes are indicated on the right. Cosmo, Cosmopolitan. Scale bar indicates nucleotide substitutions per site.
Figure 5.
Phylogenetic tree showing the relationship of dengue viruses, serotype 3, imported into Queensland, Australia, 2002–2010, based on sequencing of the envelope gene. Viruses are designated according to reported origin and GenBank accession number, and imported cases are shown in boldface. Genotypes are indicated on the right. Scale bar indicates nucleotide substitutions per site.
Figure 6.
Phylogenetic tree showing the relationship of dengue viruses, serotype 4, imported into Queensland, Australia, 2002–2010, based on sequencing of the envelope gene. Viruses are designated according to reported origin and GenBank accession number, and imported cases are shown in boldface. Genotypes are indicated on the right. Scale bar indicates nucleotide substitutions per site.
DENV genotypic groups generally circulate in particular regions (15). The viruses imported into Queensland were consistent with DENV genotypes which had previously been reported to circulate in those countries to which patients had reported travel (16–19). For example, DENV-4 genotypic group II has been reported in Indonesia, Tahiti, the Caribbean Islands, and Central and South America (17). In 2007–9, DENV-4 was introduced into the Pacific Islands, displacing DENV-1 in the process (20,21). Our genotypic analysis confirms classification of the Pacific Island DENV-4 in genoptypic group II, as recently reported (21). This was the first time this genotypic group had been reported in the Pacific region, and suggested that the origin of this strain of DENV-4 may have been Southeast Asia.
In support of this suggestion, a closely related DENV-4 strain from the Torres Strait (Figure 6, JN575595) with 99.1% envelope nucleotide identity to a DENV-4 strain from Samoa (Figure 6, JN575592), was detected before the Pacific outbreak in 2005. A maximum likelihood test of the phylogenetic tree determined that a molecular clock was applicable (Ho not rejected; p = 0.06). Using a previously published substitution rate for dengue 4 of 1 × 10−3 substitutions/site/year (22), we calculated that divergence from a common ancestor occurred in ≈2002 with an error of ± 2 years. Thus, the Pacific Island outbreak strain (Figure 6, Pacific Island clade) is geographically and temporally closely related to the Torres Strait 2005 virus. Both of these virus strains are mostly closely related to DENV-4 strains which originated in Indonesia (Figure 6, JN575583). These data support suggestions the Pacific Island outbreak strain originated in Indonesia and made its way to the Torres Strait (Australia) in 2005, probably through PNG, and into the Pacific in 2007 where it is currently circulating. A virus most closely related to the Pacific Island strain was then imported into Innisfail in northern Queensland in 2009, where it caused an outbreak (Figure 6). This incident highlights the epidemic potential of DENV strains that are imported into Queensland (3,23,24).
Discussion
In this study we have analyzed the importation of DENVs into northern Queensland, the only area within Australia where domestic epidemic spread is a risk. Two issues are apparent from these analyses. First, DENV infections, in terms of the number of importations and outbreaks, have increased in recent years. This issue is most apparent when it is considered that 42.9% of all instances of virus importation identified in this study occurred in 2010. The greatest risk was from residents returning from travel overseas, rather than overseas visitors. However, cases in the latter may be somewhat underreported because they may be more reluctant to seek medical assistance in a foreign country.
The second issue is the large degree of risk that Asia represents as a primary source of DENVs that can cause epidemics in Australia. Not only does Asia represent the biggest source of imported viruses in terms of number and serotype diversity, but it is also a source of viruses that can be imported into the Pacific region and, subsequently, a secondary source of importation into Australia as can be seen from the outbreak of the Pacific Island DENV-4 genotypic group II in Innisfail in 2009. If suggestions that the Pacific Island states are unable to sustain long-term DENV circulation are correct (20), then Asia may also be an important source of new outbreaks in the Pacific by incursions perhaps from either PNG or the islands of the Torres Strait.
To determine whether travelers returning from the 4 regions were either underrepresented or overrepresented in the dataset, we compared travelers departing Australia (using information obtained from outgoing passenger cards). A subset of Queensland residents was used as this information was available from the Australian Bureau of Statistics only for outgoing residents. The overrepresentation of infections imported from Asia and PNG relative to the Pacific Island countries may be due to higher levels of DENV activity in those countries. In the case of PNG, this result was mostly due to a large increase in imported DENVs from that country in 2010 (47.4% of all imported DENVs from PNG). Data from 2011 continue this trend to higher levels of DENVs imported from that country (data not shown). A previous study noted a decline in imported DENVS from PNG from 51% over the period 1999–2003 to 12% from 2004 to 2008 (25). The findings from this study may indicate a return to the historically higher proportion of imported DENVs from that region with the likelihood that recent dengue activity in PNG has intensified.
Little is known about overt disease in adults who acquire DENV-2 and DENV-4 infections. Disease may only be seen in those persons with previous antibody responses to another dengue serotype (26). This circumstance has implications for vaccine development because those persons with DENV antibodies may experience disease when exposed to vaccine formulations that contain apparently attenuated DENV-2 and DENV-4 (26). Susceptible adults who contract dengue while traveling represent an opportunity to study the factors associated with overt disease. To explore the pathogenicity of DENV-2 and DENV-4, serologic responses should be correlated, in the context of patient age, with molecular diagnostics in future studies of dengue surveillance.
This work clearly shows the increasing risk that viremic travelers pose to Australia, and to Queensland in particular, as a means for importing DENVs that could have substantial outbreak potential. Molecular epidemiologic studies have identified Asia as the greatest source of DENV infections that have been imported into Queensland recently. The increase in imported DENV strains and the number of outbreaks is of major public health importance and has been largely exacerbated by the heightened frequency and affordability of modern air travel. As this trend continues, the chance of the virus becoming endemic and the likelihood of the recurrence of disease also increase. Although additional studies are required to investigate the clinical implications of the imported viruses and specific patient anomalies, the sequence information presented here could assist future understanding of viral markers in relation to symptomatic disease and their association with pathogenesis.
Acknowledgments
We are grateful for the substantial contribution made by Queensland Public Health medical officers and thank private and public health practitioners for coordinating specimen collection and providing relevant clinical data and patient travel histories. We also thank the staff of Queensland Health Forensic and Scientific Services who assisted in the processing of specimens and routine diagnostics.
Biography
Dr Warrilow is currently the research and development coordinator at Public Health Virology, Queensland Health Forensic and Scientific Services. He has investigated viruses of human health importance, including arboviruses, Australian bat lyssavirus, and HIV, and his current research interests focus on RNA virus replication, diagnostics, and virus discovery.
Footnotes
Suggested citation for this article: Warrilow D, Northill JA, Pyke AT. Sources of dengue viruses imported into Queensland, Australia, 2002–2010, Emerg Infect Dis [Internet]. 2012 Nov [date cited]. http://dx.doi.org/10.3201/eid1811.120014
References
- 1.McBride JH. Dengue fever. An Australian perspective. Aust Fam Physician. 1999;28:319–23. [PubMed] [Google Scholar]
- 2.Hanna JN, Richards AR, Esmonde JV, Donohue S, Humphreys JL, Pyke AT, et al. Viraemic importations of dengue into north Queensland, 2009. Commun Dis Intell. 2010;34:57–8. [DOI] [PubMed] [Google Scholar]
- 3.Hanna JN, Ritchie SA. Outbreaks of dengue in north Queensland, 1990–2008. Commun Dis Intell. 2009;33:32–3. [DOI] [PubMed] [Google Scholar]
- 4.Beebe NW, Cooper RD, Mottram P, Sweeney AW. Australia’s dengue risk driven by human adaptation to climate change. PLoS Negl Trop Dis. 2009;3:e429. 10.1371/journal.pntd.0000429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russell RC, Currie BJ, Lindsay MD, Mackenzie JS, Ritchie SA, Whelan PI. Dengue and climate change in Australia: predictions for the future should incorporate knowledge from the past. Med J Aust. 2009;190:265–8. [DOI] [PubMed] [Google Scholar]
- 6.McBride WJH. Dengue fever: is it endemic in Australia? Intern Med J. 2010;40:247–9. 10.1111/j.1445-5994.2010.02196.x [DOI] [PubMed] [Google Scholar]
- 7.Vazquez-Prokopec GM, Chaves LF, Ritchie SA, Davis J, Kitron U. Unforeseen costs of cutting mosquito surveillance budgets. PLoS Negl Trop Dis. 2010;4:e858. 10.1371/journal.pntd.0000858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11:480–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McBride WJ. Deaths associated with dengue haemorrhagic fever: the first in Australia in over a century. Med J Aust. 2005;183:35–7. [DOI] [PubMed] [Google Scholar]
- 10.Gubler DJ, Suharyono W, Tan R, Abidin M, Sie A. Viraemia in patients with naturally acquired dengue infection. Bull World Health Organ. 1981;59:623–30. [PMC free article] [PubMed] [Google Scholar]
- 11.Fitzsimmons GJ, Wright P, Johansen CA, Whelan PI. Arboviral diseases and malaria in Australia, 2008–09: annual report of the National Arbovirus and Malaria Advisory Committee. Commun Dis Intell. 2010;34:225–40. [DOI] [PubMed] [Google Scholar]
- 12.Australian Bureau of Statistics. 3401.0–Overseas arrivals and departures, Australia, Jan 2012. [cited 2011 February]. http://www.abs.gov.au/AUSSTATS/abs@.nsf/DetailsPage/3401.0Jan%202012?OpenDocument
- 13.Kyle JL, Harris E. Global spread and persistence of dengue. Annu Rev Microbiol. 2008;62:71–92. 10.1146/annurev.micro.62.081307.163005 [DOI] [PubMed] [Google Scholar]
- 14.Senn N, Luang-Suarkia D, Manong D, Siba PM, McBride WJ. Contribution of dengue fever to the burden of acute febrile illnesses in Papua New Guinea: an age-specific prospective study. Am J Trop Med Hyg. 2011;85:132–7. 10.4269/ajtmh.2011.10-0482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weaver SC, Reisen WK. Present and future arboviral threats. Antiviral Res. 2010;85:328–45. 10.1016/j.antiviral.2009.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goncalvez AP, Escalante AA, Pujol FH, Ludert JE, Tovar D, Salas RA, et al. Diversity and evolution of the envelope gene of dengue virus type 1. Virology. 2002;303:110–9. 10.1006/viro.2002.1686 [DOI] [PubMed] [Google Scholar]
- 17.Lanciotti RS, Gubler DJ, Trent DW. Molecular evolution and phylogeny of dengue-4 viruses. J Gen Virol. 1997;78:2279–84. [DOI] [PubMed] [Google Scholar]
- 18.Twiddy SS, Farrar JJ, Vinh Chau N, Wills B, Gould EA, Gritsun T, et al. Phylogenetic relationships and differential selection pressures among genotypes of dengue-2 virus. Virology. 2002;298:63–72. 10.1006/viro.2002.1447 [DOI] [PubMed] [Google Scholar]
- 19.Wittke V, Robb TE, Thu HM, Nisalak A, Nimmannitya S, Kalayanrooj S, et al. Extinction and rapid emergence of strains of dengue 3 virus during an interepidemic period. Virology. 2002;301:148–56. 10.1006/viro.2002.1549 [DOI] [PubMed] [Google Scholar]
- 20.Li DS, Liu W, Guigon A, Mostyn C, Grant R, Aaskov J. Rapid displacement of dengue virus type 1 by type 4, Pacific region, 2007–2009. Emerg Infect Dis. 2010;16:123–5. 10.3201/eid1601.091275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao-Lormeau VM, Roche C, Aubry M, Teissier A, Lastere S, Daudens E, et al. Recent emergence of dengue virus serotype 4 in French Polynesia results from multiple introductions from other South Pacific Islands. PLoS ONE. 2011;6:e29555. 10.1371/journal.pone.0029555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vasilakis N, Holmes EC, Fokam EB, Faye O, Diallo M, Sall AA, et al. Evolutionary processes among sylvatic dengue type 2 viruses. J Virol. 2007;81:9591–5. 10.1128/JVI.02776-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanna JN, Ritchie SA, Hills SL, Pyke AT, Montgomery BL, Richards AR, et al. Dengue in north Queensland, 2002. Commun Dis Intell. 2003;27:384–9. [DOI] [PubMed] [Google Scholar]
- 24.Hanna JN, Ritchie SA, Richards AR, Humphreys JL, Montgomery BL, Ehlers GJ, et al. Dengue in north Queensland, 2005–2008. Commun Dis Intell. 2009;33:198–203. [DOI] [PubMed] [Google Scholar]
- 25.Hanna JN, Ritchie SA. An apparent recent decline in importations of dengue from Papua New Guinea into north Queensland. Commun Dis Intell. 2009;33:34–5. [DOI] [PubMed] [Google Scholar]
- 26.Vaughn DW. Invited commentary: dengue lessons from Cuba. Am J Epidemiol. 2000;152:800–3. 10.1093/aje/152.9.800 [DOI] [PubMed] [Google Scholar]