Abstract
The standard culture system for in vitro cartilage research is based on cells in a three-dimensional micromass culture and a defined medium containing the chondrogenic key growth factor, transforming growth factor (TGF)-β1. The aim of this study was to optimize the medium for chondrocyte micromass culture. Human chondrocytes were cultured in different media formulations, designed with a factorial design of experiments (DoE) approach and based on the standard medium for redifferentiation. The significant factors for the redifferentiation of the chondrocytes were determined and optimized in a two-step process through the use of response surface methodology. TGF-β1, dexamethasone, and glucose were significant factors for differentiating the chondrocytes. Compared to the standard medium, TGF-β1 was increased 30%, dexamethasone reduced 50%, and glucose increased 22%. The potency of the optimized medium was validated in a comparative study against the standard medium. The optimized medium resulted in micromass cultures with increased expression of genes important for the articular chondrocyte phenotype and in cultures with increased glycosaminoglycan/DNA content. Optimizing the standard medium with the efficient DoE method, a new medium that gave better redifferentiation for articular chondrocytes was determined.
Key words: articular cartilage, autonomous chondrocyte implantation, design of experiments, differentiation, media development, TGF-β1, tissue engineering
Introduction
In 1994, Brittberg et al.1 introduced autologous chondrocyte implantation (ACI) in clinical practice, a method in which autologous chondrocytes are used to regenerate articular cartilage. In the ACI protocol autologous chondrocytes are expanded in vitro, during which genes specific for the articular cartilage are down-regulated.2,3 After a limited number of cell divisions, the cells are induced to redifferentiate and produce new extracellular matrix (ECM) in a three-dimensional environment, through the use of a defined medium2 containing transforming growth factor β1 (TGF-β1) and dexamethasone. This medium has become the standard in basic research on chondrocyte redifferentiation. It was determined 15 years ago for the differentiation of mesenchymal stem cells into chondrocytes and was applied to adult human chondrocytes without any optimization.2,4–7
Design of experiment (DoE) methods have been applied in many fields of biotechnology over the last decades.8–13 Through the use of factorial design and response surface methodology (RSM), DoE allows for a systematic approach to optimization processes that reduces experimental bias and noise with a limited number of experiments.12,14
In DoE setups, a first factorial screen is usually performed to define the experimental domain and factors that significantly affect the outcome. This screening process often is followed by optimization steps based on more elaborate designs, which are used to determine true optimal settings for the significant factors through defining, narrowing, and redefining the experimental domain.13,14 Despite its wide use in many other parts of the biotechnology field, DoE is only starting to be recognized in the field of regenerative medicine and tissue engineering.15–19
DoE is often used to increase yield and purity in biochemical processes in which nutrients are transformed to biomass or molecular products.13,20 In cartilage tissue engineering, process yield translates to the amount of ECM produced by the cells from nutrients in the culture media. The major components of articular cartilage ECM are proteoglycans and collagens.21 Purity of the process translates to what type of ECM is produced. For the proteoglycans, aggrecan (ACAN) is up-regulated and versican (VCAN) is down-regulated as the cells transit from the dedifferentiated expansion state to the redifferentiated state and the cultures mature. In the same way, collagen II (COLII) is a marker for maturity, and collagen I (COLI) is a marker of immaturity.2 Investigating the expression ratios of ACAN:VCAN and COLII:COLI can thus be used to see how far the redifferentiation process has proceeded. The sex determining region Y-box 9 protein (SOX9) is a key marker for chondrocyte differentiation and affects the expression of several chondrocyte specific markers.22,23 A high expression of this gene is thus also important for the maturation of the articular cartilage.
The aim of this study was to optimize the standard medium in cartilage differentiation through DoE and RSM, thereby increasing the ECM yield of the chondrocytes. The purity of the ECM was ensured through investigating the ACAN:VCAN and COLII:COLI gene expression ratios, along with SOX9 expression. Through a first factorial screen of the components, significant factors affecting these expressions were determined. These factors were then optimized in two steps using more elaborate designs and RSM. To verify that an optimal medium had been determined, chondrocytes were cultured in the new formulation and the standard medium, and the degree of redifferentiation compared. The objective was further to show the usefulness of the DoE methodology in the area of cartilage tissue engineering and regenerative medicine.
Materials and Methods
Cell expansion
Surplus human chondrocytes from three patients (average age 21 years [19–23 years]) undergoing ACI were cultured in chondrocyte expansion medium consisting of Dulbecco's modified Eagle's medium/F12 (DMEM/F12, Invitrogen, Paisley, United Kingdom) supplemented with 0.1 mg/mL L-ascorbic acid (Apotekets produktionsenhet, Umeå, Sweden), 1× penicillin-streptomycin (PAA Laboratories, Pasching, Austria), 2 mM L-Glutamine (Invitrogen), and 10% human serum, at 37°C in 7% CO2 and 90% relative humidity. Medium was changed two to three times per week. Subculture was made with trypsin-EDTA solution (0.125% trypsin [Invitrogen] diluted in 0.1 M phosphate-buffered saline with 0.2 g/L EDTA) when the cells reached 80% confluence.
High-density micromass culture
Chondrocytes (passage 3) were resuspended in chondrocyte expansion medium and 2×105 cells in 50 μL were transferred to uncoated flat bottom 96-well plates (Costar, Corning, NY, USA). After adding 150 μL of the different media formulations (Table 1) to each designated well, the plate was centrifuged at 350 g for 5 min and incubated for 24 h at 37°C in 7% CO2 and 90% relative humidity to allow for micromass formation. After 24 h the culture medium was changed to the defined media formulations (Table 1). Medium was then changed three times per week for 2 weeks, where after the micromass cultures were snap-frozen in liquid nitrogen and stored at −80°C.
Table 1.
Factor Concentrations in Screen Design
| No. | TGF-β1a | ASCb | ITSc | DEXd | LINe |
|---|---|---|---|---|---|
| 1 | − | − | − | = | + |
| 2 | + | − | − | = | − |
| 3 | − | + | − | = | − |
| 4 | + | + | − | = | + |
| 5 | − | − | + | = | − |
| 6 | + | − | + | = | + |
| 7 | − | + | + | = | + |
| 8 | + | + | + | = | − |
| 9 | − | − | − | + | − |
| 10 | + | − | − | + | + |
| 11 | − | + | − | + | + |
| 12 | + | + | − | + | − |
| 13 | − | − | + | + | + |
| 14 | + | − | + | + | − |
| 15 | − | + | + | + | − |
| 16 | + | + | + | + | + |
| 17 | ◯ | ◯ | ◯ | ◯ | ◯ |
| 18 | ◯ | ◯ | ◯ | ◯ | ◯ |
| 19 | ◯ | ◯ | ◯ | ◯ | ◯ |
TGF-β1: (−), 4 ng/mL; (◯), 10 ng/mL; (+) 16 ng/mL.
Ascorbic acid (ASC): (−), 2.8 μg/mL; (◯), 14 μg/mL; (+) 25.2 μg/mL.
Insulin–transferrin–selenium solution (ITS): (−), 0.2×; (◯), 1×; (+), 1.8×.
Dexamethasone (DEX): (=), 10 nM; (◯), 100 nM; (+), 1000 nM.
HSA:LIN: (−), 0.2 mg/mL:0.4 μg/mL; (◯), 1.0 mg/mL:5.0 μg/mL; (+), 1.8 mg/mL:9 μg/mL.
No., number; TGF, transforming growth factor; HSA:LIN, human serum albumin supplemented with 5.0 μg/mL linoleic acid.
Isolation of total RNA
Micromass cultures were homogenized in 1.5-mL polypropylene tubes using tungsten beads and a TissueLyser (Quiagen, Hilden, Germany). QIAZol (Qiagen) was added to the tubes and mixed in the TissueLyser. Chloroform was added (0.2 mL/mL QIAZol) and mixed. Tubes were centrifuged at 16,000 g for 15 min at 4°C. Total RNA was isolated using the RNeasy mini kit (Qiagen) according to the manufacturers protocol. DNase I was used to remove contaminating genomic DNA from the isolated RNA (Qiagen).
Quantitative real-time PCR
All software and reagents for the analyses were purchased from Applied Biosystems (Carlsbad, CA, USA). cDNA was prepared from 200 ng of total RNA by using the High Capacity cDNA Reverse Transcription Kit. Quantitative PCR (qPCR) was performed in duplicates with cDNA corresponding to 2.5 ng of RNA and the TaqMan Universal master mixture with 1× assay-on-demand mixes of primers for the genes (assay numbers in parentheses): COL1A1 (Hs00164004_ml), COL2A1 (Hs00156568_m1), COL10A1 (Hs00166657_m1), ACAN (Hs00153936_ml), VCAN (Hs00171642_ml), SOX9 (Hs00165814_m1), and COMP (Hs00164359_m1), with CyclophillinA (Hs99999904_m1) as the reference gene. PCR was performed using the 7900HT real time PCR system (Life Technologies). Relative quantification of the target gene expression was performed according to the standard curve method calculated by the ddCq method. The experimental data were fitted to a linearization model in Modde 8.0 (Umetrics AB, Umeå, Sweden), and the different patients were added as replicates.
Experimental design of culture media formulation
Medium formulation screen
The screen included dexamethasone (Sigma, St. Louis, MO, USA), TGF-β1 (R&D systems, Minneapolis, MN, USA), human serum albumin (HSA, Equitech-Bio, Kerrville, TX, USA) supplemented with 5.0 μg/mL linoleic acid (Sigma), insulin–transferrin–selenium solution (ITS; Invitrogen), and 14 μg/mL L-ascorbic acid 2-phosphate (Sigma). To design the factorial experiments, the computer software package Modde 8.0 was used, and the five factors were set in a fraction factorial 25-1 screening design, with three center points. Concentrations of the factors in the 17 formulations factors were set according to Table 1. The factors were added to DMEM high glucose (PAA Laboratories) supplemented with 1× penicillin-streptomycin (PAA Laboratories). The different media were used to differentiate human chondrocyte micromass cultures as described above, and repeated with three patients.
Medium formulation optimization, step 1
The nonsignificant factors were kept at medium levels (see Table 1, no. 17–19). For the significant factors dexamethasone, glucose, and TGF-β1 a full factorial 23 design was made in Modde 8.0. Media formulations with low glucose levels were made in DMEM low glucose (PAA laboratories), medium levels in a 1:1 mix of DMEM high glucose and DMEM low glucose, and high levels in DMEM high glucose (Table 2). The different media were used to differentiate human chondrocyte micromass cultures as described above, and repeated with three patients.
Table 2.
Factor Concentrations in Optimization
| |
Step 1 |
Step 2 |
|||
|---|---|---|---|---|---|
| No. | TGF-β1a | DEXb | GLUc | DEXb | GLUc |
| 1 | ◯ | ≡ | − | = | ◯ |
| 2 | + | ≡ | − | ◯ | ◯ |
| 3 | ◯ | ◯ | − | = | + |
| 4 | + | ◯ | − | ◯ | + |
| 5 | ◯ | ≡ | ⊕ | = | ⊕ |
| 6 | + | ≡ | ⊕ | ◯ | ⊕ |
| 7 | ◯ | ◯ | ⊕ | − | ◯ |
| 8 | + | ◯ | ⊕ | − | + |
| 9 | ◯ | = | ◯ | − | ⊕ |
| 10 | + | = | ◯ | − | ⊕ |
| 11 | ⊕ | ≡ | ◯ | − | ⊕ |
| 12 | ⊕ | ◯ | ◯ | ||
| 13 | ⊕ | = | − | ||
| 14 | ⊕ | = | ⊕ | ||
| 15 | ⊕ | = | ◯ | ||
| 16 | ⊕ | = | ◯ | ||
| 17 | ⊕ | = | ◯ | ||
TGF-β1: (◯), 10 ng/mL; (⊕), 13 ng/mL; (+), 16 ng/mL.
DEX: (≡), 1 nM; (=), 10 nM; (−), 50 nM; (◯), 100 nM.
Glucose (GLU): (−), 1 g/L; (◯), 2.75 g/L; (⊕), 4.5 g/L; (+), 6.25 g/L.
Medium formulation optimization, step 2
Dexamethasone and glucose were added to a full 22 design in Modde 8.0 (Table 2). A 200 g/L glucose solution (Invitrogen) was used to increase glucose levels above 4.5 g/L in the DMEM high glucose medium. TGF-β1 was added reaching a concentration of 13 ng/mL in all formulations. The different media were used to differentiate human chondrocyte micromass cultures as described above, and repeated with three patients.
Medium comparison
The optimal medium contained DMEM high glucose supplemented with 0.9 g/L glucose solution, 50 nM dexamethasone, 13 ng/mL TGF-β1, 1× ITS, 1.0 mg/mL HSA supplemented with 5.0 μg/mL linoleic acid, 14 μg/mL ascorbic acid, and 1× penicillin-streptomycin. Human chondrocytes from five patients (mean age 27.5 years, range 19–42 years) were cultured for 2 weeks in micromass culture in the optimized formulation or in a control formulation consisting of DMEM high glucose (4.5 g/L glucose) supplemented with 100 nM dexamethasone, 10 ng/mL TGF-β1, 1× ITS, 1.0 mg/mL HSA supplemented with 5.0 μg/mL linoleic acid, 14 μg/mL ascorbic acid, and 1× penicillin-streptomycin. After 14 days in culture the micromasses were harvested and analyzed with qPCR for gene expression levels as described above, with biochemical methods for DNA and glycosaminoglycan content, and stained for histology comparisons.
Biochemical analysis of micromass cultures
On day 14 the cultures were digested with papain (Sigma) solution (0.3 mg papain/mL, 20 mM sodium phosphate buffer with 1 mM EDTA and 2 mM dithiothreitol) for 2 h at 60°C. The digested cultures were then mechanically dissolved by vortex and further analyzed for DNA and glycosaminoglycan (GAG) content. The amount of DNA was measured with a Hoechst 33258 (Sigma) assay, according to the manufacturer's instructions. GAG was quantified with a dimethylmethylene blue assay as previously described.24
Histology of micromass cultures
On day 14 the cultures were fixed in Histofix™ (Histolab products AB, Gothenburg, Sweden), dehydrated, and embedded in paraffin. Five-micrometer sections were cut and placed onto Super frost plus microscope slides (Menzel-Gläser, Braunschweig, Germany), deparaffinized, and stained with Alcian blue/van Gieson. Slides were examined under the microscope (Nikon Eclipse 90i, Nikon Corp., Belmont, CA, USA) and digital pictures were taken with a Nikon DXM1200F digital camera (Nikon) and the NIS Elements D software (Nikon).
Statistics
Results from the DoE were analyzed in Modde 8.0 with linear regression and ANOVA. Linear regression models were considered significant at p<0.05 and without a significant lack of fit at p>0.05. Results from the medium comparison were analyzed with a Wilcoxon paired t-test, with significance levels p<0.05 and p<0.01, as described in picted in Figures 1 and 2.
FIG. 1.
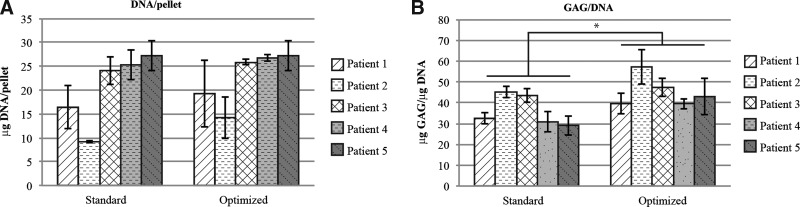
Comparison between human chondrocyte micromasses cultured in the optimized formulation and in the standard formulation, depicted as mean±standard deviation. (A) There was no significant difference in the amount of DNA in the micromass cultures from the two media formulations. (B) There was a significantly higher production of glycosaminoglycan (GAG) per cell for micromasses cultured in the optimized medium (*p<0.05).
FIG. 2.
Comparison between human chondrocyte micromasses cultured in the optimized formulation and in the standard formulation, depicted as mean±standard deviation. There were significant increases in the gene expressions of (A) the ACAN:VCAN ratio, (B) SOX9, and (C) COMP (**p<0.01). There were no differences in gene expression for (D) the COLII:COLI ratio or (E) COLX.
Results
Medium formulation screen
TGF-β1 and dexamethasone were significant factors for increasing the gene expression of SOX9 and the COLII:COLI and ACAN:VCAN ratios. TGF-β1 should be set in a range between 10 and 16 ng/mL. Dexamethasone should be set to the lower ranges, around 10 nM. Ascorbic acid, ITS, and HSA/linoleic acid did not affect these ratios significantly and were not in need of optimization. They were thus excluded from further optimization. Coefficient plots for the screen can are shown in Supplementary Figure 1.
Medium optimization, step 1
Glucose was added as a variable and included in the optimization design together with the significant factors TGF-β1 and dexamethasone. To minimize confounding between the variables, glucose was not included in the screening process to reduce the number of factors investigated.
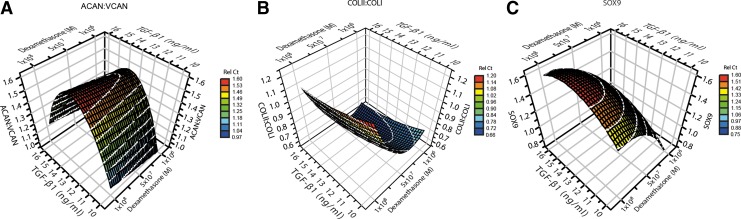
Varying TGF-β1 between 10 and 16 ng/mL showed that the highest ACAN:VCAN ratio was reached at 13 ng/mL (Fig. 3A). An optimum for dexamethasone was indicated at 10 nM. Glucose concentration had a strong impact on the ratio, with higher ratios following higher concentrations (pA:V=0.003).
FIG. 3.
Response surfaces for optimization step 1. (A) ACAN:VCAN expression ratio as a response to the factors TGF-β1 and glucose. (B) COLII:COLI expression ratio as a response to the factors TGF-β1 and glucose. (C) SOX9 gene expression as a response to the factors TGF-β1 and dexamethasone. ACAN, aggrecan; VCAN, versican; COL, collagen; SOX9, sex determining region Y-box 9 protein.
For COLII:COLI, TGF-β1 had less effect at these levels, showing an increase in concentration would result in a slightly higher ratio (Fig. 3B). The ratio increased when the concentration of dexamethasone was decreased. Low glucose concentrations gave a high ratio (pCII:CI=0.003).
SOX9 showed the same response to all three factors as the ACAN:VCAN ratio (Fig. 3C). The TGF-β1 concentration showed a peak at 13 ng/mL (pSOX9=0.027).
Due to the peak found for TGF-β1 for ACAN:VCAN and SOX9 gene expression, this variable was set at the optimal concentration 13 ng/mL and removed from further analysis.
Medium optimization, step 2
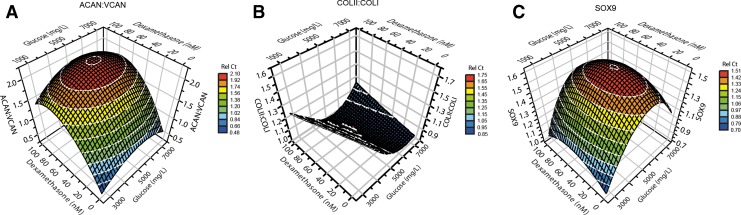
For the ACAN:VCAN ratio an optimal point for the dexamethasone concentration was determined at 50 nM. The glucose concentration gave an expression peak for both ratio and SOX9 expression at 5.4 g/L (Fig. 4A; pA:V=0.009).
FIG. 4.
Response surface for optimization step 2. (A) ACAN:VCAN expression ratio as a response to the factors dexamethasone and glucose. (B) COLII:COLI expression ratio as a response to the factors dexamethasone and glucose. (C) SOX9 gene expression as a response to the factors dexamethasone and glucose.
The COLII:COLI ratio increased with a decrease in dexamethasone. No optimal concentration was reached, however. Increasing glucose concentrations gave a decrease in the ratio (Fig. 4B; pCII:CI=0.045).
The SOX9 expression was affected similar to the ACAN:VCAN ratio, reaching peaks at the same concentrations for both factors (Fig. 4C; pSOX9=0.042).
Biochemical analysis
Culturing human chondrocytes from five patients in the two respective media showed that the optimized medium resulted in a 9% nonsignificant (p=0.102) increase (average) in DNA content in these micromass cultures (Fig. 1A). The optimized medium resulted in a significant (p=0.036) 25% increase (average) in the amount of GAG produced per cell (Fig. 1B).
Gene expression
The optimized formulation resulted in a significant increase in the ACAN:VCAN ratio (mean of all patients 62.1%, p=0.009), the SOX9 (mean of all patients 41.6%, p=0.012) expression, and the COMP expression (mean of all patients 62.7%, p=0.007), compared to the classic medium formulation (Fig. 2A,B,C, respectively). There was no significant difference between the two formulations for the COLII:COLI (p=0.321), however, with an indication of increased ratios for 3/5 of the patients (Fig. 2D), and an increase of the mean of all patients of 90%. Collagen X (COLX) followed the COLII:COLI expression (Fig. 2E; p=0.800).
Histology
Cross-sectioning of the micromass cultures showed that the optimized formulation resulted in more intensely and evenly Alcian blue van Gieson stained ECM, in patients 1 and 3, as compared to the classic formulation (Fig. 5).
FIG. 5.
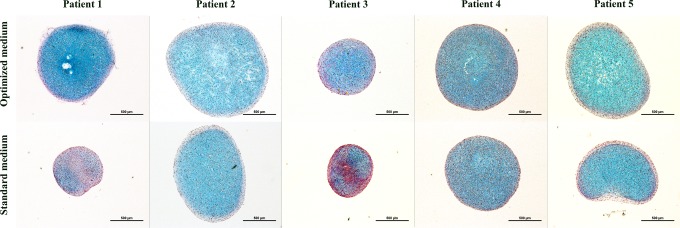
Representative histological cross sections of micromasses cultured in optimized medium above and standard medium below, stained with Alcian blue van Gieson staining. Scale bars=500 μm.
Discussion
In this study, we optimized the standard chondrogenic medium through the use of DoE and RSM. Changing concentrations of TGF-β1 (+30%), dexamethasone (−50%), and glucose (+20%) led to increased expression by the chondrocytes of genetic markers typical of articular cartilage and an increase in production of articular cartilage ECM compared to cells cultured in the standard formulation.
In the screening, three of the factors had little effect on the investigated gene expressions. L-ascorbic acid is important for the energy metabolism in growth plate chondrocytes25 and for COLII and COLX expression;25,26 however, not in a dose–response manner.27 ITS is associated with cell survival and the sustaining of metabolic activities in chondrocytes,28 while HSA/linoleic acid is a source of lipids for the cells and is needed for proliferation and chondrocyte clonal growth and as a precursor for vitamins.28,29 Taken together, the reason why these supplements did not significantly affect the system is that they are essential at basal levels for cell survival and not for the quantity of ECM production.
The significant factors TGF-β1, dexamethasone, and glucose have known effects on chondrocyte redifferentiation and anabolic processes, but our results indicated that they were not used at optimal concentrations in the standard chondrogenic formulation. The optimal concentration of TGF-β1 for chondrocyte redifferentiation was 30% above that used in the standard formulation, and above this concentration a decrease in the expression of all investigated genes was seen. This decrease is likely to be a result of feedback signaling known to be present in TGF-β1 receptor and downstream signaling pathways.30
Our results propose a reduction in dexamethasone concentrations of 50%, for an optimal concentration for ACAN:VCAN ratio and SOX9. Previously, Miyazaki et al.31 showed inhibition of COLII expression in chondrocytes by dexamethasone, and further reduction of dexamethasone could thus have resulted in significant increase in the COLII:COLI ratio. This would, however, have made the formulation divert from the optimal concentrations for the ACAN:VCAN ratio and SOX9 expression.
Glucose is known to have an anabolic effect in cartilage tissue engineering32,33 and in most research groups a high glucose concentration is used,7,34,35 but there is no general consensus.36,37
Optimizing the main factors glucose and dexamethasone for all three investigated parameters could not be done. Increasing ACAN:VCAN and SOX9 expressions would lead to decrease in COLII:COLI expression. To increase this ratio simultaneously would require addition of other growth factors or implementation or other culture environments. This, however, lies outside the scope of this paper. Since SOX9 is an essential transcription factor for articular cartilage,23 a decision was made to optimize the medium in a second step on that parameter, along with the ACAN:VCAN ratio. In this step a peak concentration for glucose was determined. With this final step in the optimization process, the resulting new formulation favors the production of noncollagenous ECM over the collagenous ECM.
With the ability to systematically vary several factors at the same time, DoE is a powerful tool to affect a specific gene or set of genes and induce cells to elicit a specific response. Previously, Pritchett et al.38 showed in a similar way this strength of DoE through specifically targeting glycosylation of cystatin C, at the expense productivity in a yeast model. Tissue engineered constructs can in this way be tailored to fit their purpose, be it ECM content, cellularity, or mechanical integrity. In proceeding steps, new growth factors such as bone morphogenetic protein-2 or insulin-like growth factor-1 could easily be implemented in new designs, as well as using the system for other cell types, such as embryonic stem cells, mesenchymal stem cells, or even induced pluripotent stem cells.
An issue with ACI is the relatively low amount of starting material that can be retrieved from one biopsy; further, the chondrocytes should not be expanded for more than six to eight population doublings before being initiated to redifferentiate, which limits the defect size that can be treated.39,40 The optimized formulation showed higher biomass yield per cell, with a significant increase in proteoglycan production without an increase in proliferation, and thus a more efficient use of the chondrocytes.
That the medium formulation determined in this study is the optimal medium for chondrogenic differentiation in vitro is a strong statement. There are still unknown factors affecting the system and the strength of the response of the cells varied somewhat from patient to patient. Studies including mechanical integrity of the constructs and genome-wide arrays would be the next step in the optimization process, along with the addition of other growth factors for the optimization of the COLII:COLI expression. However, we find it remarkable that improvements of this formulation have not previously been published. In this article, DoE has shown to be a useful method for optimization processes. and we believe that implementing it in these types of biotechnology research projects would help improve efficiency in the field of tissue engineering and regenerative medicine.
Supplementary Material
Author Disclosure Statement
The authors declare that no competing financial interests exist.
References
- 1.Brittberg M. Lindahl A. Nilsson A, et al. Treatment of deep cartilage defects in the knee with autologous chondrocyte transplantation. N Engl J Med. 1994;331:889–895. doi: 10.1056/NEJM199410063311401. [DOI] [PubMed] [Google Scholar]
- 2.Benya PAD. Shaffer JD. Dedifferentiated chondrocytes reexpress the differentiated collagen phenotype when cultured in agarose gels. Cell. 1982;30:215–224. doi: 10.1016/0092-8674(82)90027-7. [DOI] [PubMed] [Google Scholar]
- 3.Watt FM. Effect of seeding density on stability of the differentiated phenotype of pig articular chondrocytes in culture. J Cell Sci. 1988;89(Pt 3):373–378. doi: 10.1242/jcs.89.3.373. [DOI] [PubMed] [Google Scholar]
- 4.Johnstone B. Hering T. Caplan A. In vitro chondrogenesis of bone marrow-derived mesenchymal progenitor cells. Exp Cell Res. 1998;238:265–272. doi: 10.1006/excr.1997.3858. [DOI] [PubMed] [Google Scholar]
- 5.Grogan SP. Barbero A. Diaz-Romero J, et al. Identification of markers to characterize and sort human articular chondrocytes with enhanced in vitro chondrogenic capacity. Arthritis Rheum. 2007;56:586–595. doi: 10.1002/art.22408. [DOI] [PubMed] [Google Scholar]
- 6.Dehne T. Karlsson C. Ringe J, et al. Chondrogenic differentiation potential of osteoarthritic chondrocytes and their possible use in matrix-associated autologous chondrocyte transplantation. Arthritis Res Ther. 2009;11:R133. doi: 10.1186/ar2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tallheden T. Karlsson C. Brunner A, et al. Gene expression during redifferentiation of human articular chondrocytes. Osteoarthritis Cartilage. 2004;12:525–535. doi: 10.1016/j.joca.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Haaland P. Experimental Design in Biotechnology. New York: Marcel Dekker Inc.; 1989. [Google Scholar]
- 9.Marciset O. Multifactorial experimental design for optimizing transformation: electroporation of Streptococcus thermophilus. Biotechnol Bioeng. 1994;43:490–496. doi: 10.1002/bit.260430609. [DOI] [PubMed] [Google Scholar]
- 10.Kumari P. Reddy CRK. Jha B. Comparative evaluation and selection of a method for lipid and fatty acid extraction from macroalgae. Anal Biochem. 2011;415:134–144. doi: 10.1016/j.ab.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 11.Tye H. Application of statistical “design of experiments” methods in drug discovery. Drug Discov Today. 2004;9:485–491. doi: 10.1016/S1359-6446(04)03086-7. [DOI] [PubMed] [Google Scholar]
- 12.Fisher RA. The Design of Experiments. Oxford: Oliver & Boyd; 1935. [Google Scholar]
- 13.Mandenius C-F. Brundin A. Bioprocess optimization using design-of-experiments methodology. Biotechnol Prog. 2008;24:1191–1203. doi: 10.1002/btpr.67. [DOI] [PubMed] [Google Scholar]
- 14.Montgomery D. Design and Analysis of Experiments. Hoboken, NJ: John Wiley & Sons, Inc.; 2008. [Google Scholar]
- 15.Chen Y. Bloemen V. Impens S, et al. Characterization and optimization of cell seeding in scaffolds by factorial design: quality by design approach for skeletal tissue engineering. Tissue Eng Part C Methods. 2011;17:1–11. doi: 10.1089/ten.tec.2011.0092. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y-L. Chen H-C. Lee H-P. Rational development of GAG-augmented chitosan membranes by fractional factorial design methodology. Biomaterials. 2006;27:2222–2232. doi: 10.1016/j.biomaterials.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 17.Lim M. Ye H. Drakakis E, et al. Towards information-rich bioprocessing: generation of spatio-temporal profiles through the use of design of experiments to determine optimal number and location of sensors—an example in thermal profiles. Biochem Eng J. 2008;40:1–7. [Google Scholar]
- 18.Chen WLK. Likhitpanichkul M. Ho A, et al. Integration of statistical modeling and high-content microscopy to systematically investigate cell-substrate interactions. Biomaterials. 2010;31:2489–2497. doi: 10.1016/j.biomaterials.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 19.Liu G. Kawaguchi H. Ogasawara T, et al. Optimal combination of soluble factors for tissue engineering of permanent cartilage from cultured human chondrocytes. J Biol Chem. 2007;282:20407–20415. doi: 10.1074/jbc.M608383200. [DOI] [PubMed] [Google Scholar]
- 20.Gheshlaghi R. Scharer JM. Moo-Young M, et al. Medium optimization for hen egg white lysozyme production by recombinant Aspergillus niger using statistical methods. Biotechnol Bioeng. 2005;90:754–760. doi: 10.1002/bit.20474. [DOI] [PubMed] [Google Scholar]
- 21.Kuettner KE. Biochemistry of articular cartilage in health and disease. Clin Biochem. 1992;25:155–163. doi: 10.1016/0009-9120(92)90224-g. [DOI] [PubMed] [Google Scholar]
- 22.de Crombrugghe B. Lefebvre V. Behringer R, et al. Transcriptional mechanisms of chondrocyte differentiation. Matrix Biol. 2000;19:389–394. doi: 10.1016/s0945-053x(00)00094-9. [DOI] [PubMed] [Google Scholar]
- 23.Bi W. Deng J. Zhang Z, et al. Sox9 is required for cartilage formation. Nat Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 24.Farndale RW. Buttle DJ. Barrett AJ. Improved quantitation and discrimination of sulphated glycosaminoglycans by use of dimethylmethylene blue. Biochim Biophys Acta. 1986;883:173–177. doi: 10.1016/0304-4165(86)90306-5. [DOI] [PubMed] [Google Scholar]
- 25.Shapiro IM. Leboy PS. Tokuoka T, et al. Ascorbic acid regulates multiple metabolic activities of cartilage cells. Am J Clin Nutr. 1991;54(6 Suppl):1209S–1213S. doi: 10.1093/ajcn/54.6.1209s. [DOI] [PubMed] [Google Scholar]
- 26.Clark AG. Rohrbaugh AL. Otterness I, et al. The effects of ascorbic acid on cartilage metabolism in guinea pig articular cartilage explants. Matrix Biol. 2002;21:175–184. doi: 10.1016/s0945-053x(01)00193-7. [DOI] [PubMed] [Google Scholar]
- 27.Ibold Y. Lübke C. Pelz S, et al. Effect of different ascorbate supplementations on in vitro cartilage formation in porcine high-density pellet cultures. Tissue Cell. 2009;41:249–256. doi: 10.1016/j.tice.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 28.Kisiday JD. Kurz B. DiMicco MA, et al. Evaluation of medium supplemented with insulin-transferrin-selenium for culture of primary bovine calf chondrocytes in three-dimensional hydrogel scaffolds. Tissue Eng. 2005;11:141–151. doi: 10.1089/ten.2005.11.141. [DOI] [PubMed] [Google Scholar]
- 29.Lennon D. Haynesworth S. Young R. A chemically defined medium supports in vitro proliferation and maintains the osteochondral potential of rat marrow-derived mesenchymal stem cells. Exp Cell Res. 1995;219:211–222. doi: 10.1006/excr.1995.1221. [DOI] [PubMed] [Google Scholar]
- 30.Miyazono K. Positive and negative regulation of TGF-beta signaling. J Cell Sci. 2000;113(Pt 7):1101–1109. doi: 10.1242/jcs.113.7.1101. [DOI] [PubMed] [Google Scholar]
- 31.Miyazaki Y. Dexamethasone inhibition of TGFβ-induced cell growth and type II collagen mRNA expression through ERK-integrated AP-1 activity in cultured rat articular chondrocytes. Osteoarthritis Cartilage. 2000;8:378–385. doi: 10.1053/joca.1999.0313. [DOI] [PubMed] [Google Scholar]
- 32.Phitak T. Pothacharoen P. Kongtawelert P. Comparison of glucose derivatives effects on cartilage degradation. BMC Musculoskelet Disord. 2010;11:162. doi: 10.1186/1471-2474-11-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heywood HK. Bader DL. Lee DA. Glucose concentration and medium volume influence cell viability and glycosaminoglycan synthesis in chondrocyte-seeded alginate constructs. Tissue Eng. 2006;12:3487–3496. doi: 10.1089/ten.2006.12.3487. [DOI] [PubMed] [Google Scholar]
- 34.Scotti C. Osmokrovic A. Wolf F, et al. Response of human engineered cartilage based on articular or nasal chondrocytes to IL-1β and low oxygen. Tissue Eng Part A. 2012;18:362–372. doi: 10.1089/ten.tea.2011.0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bastiaansen-Jenniskens YM. Koevoet W. de Bart ACW, et al. TGFβ Affects collagen cross-linking independent of chondrocyte phenotype but strongly depending on physical environment. Tissue Eng Part A. 2008;14:1059–1066. doi: 10.1089/ten.tea.2007.0345. [DOI] [PubMed] [Google Scholar]
- 36.van der Windt AE. Jahr H. Farrell E, et al. Calcineurin inhibitors promote chondrogenic marker expression of dedifferentiated human adult chondrocytes via stimulation of endogenous TGFbeta1 production. Tissue Eng Part A. 2010;16:1–10. doi: 10.1089/ten.TEA.2009.0082. [DOI] [PubMed] [Google Scholar]
- 37.Henderson JH. Welter JF. Mansour JM, et al. Cartilage tissue engineering for laryngotracheal reconstruction: comparison of chondrocytes from three anatomic locations in the rabbit. Tissue Eng. 2007;13:843–853. doi: 10.1089/ten.2006.0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pritchett J. Baldwin SA. The effect of nitrogen source on yield and glycosylation of a human cystatin C mutant expressed in Pichia pastoris. J Ind Microbiol Biotechnol. 2004;31:553–558. doi: 10.1007/s10295-004-0181-2. [DOI] [PubMed] [Google Scholar]
- 39.Zaucke F. Dinser R. Maurer P. Cartilage oligomeric matrix protein (COMP), collagen IX are sensitive markers for the differentiation state of articular primary chondrocytes. Biochem J. 2001;15(358(Pt 1)):17–24. doi: 10.1042/0264-6021:3580017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nelson L. Fairclough J. Archer C. Use of stem cells in the biological repair of articular cartilage. Expert Opin Biol Ther. 2010;10:43–55. doi: 10.1517/14712590903321470. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.