Abstract
Recognition of micro-RNA function and their contribution to the biology of disease has given a new insight into disease mechanisms, with these discoveries potentially improving clinical diagnostic and therapeutic options. miR-125b has been identified as an important regulator in various cancers, including prostate cancer, but the mechanism of this regulation remains incompletely understood. In these studies, the effect of castration on miR-125b serum expression was evaluated in mice, simulating androgen deprivation. Furthermore, miR-125b expression was measured by quantitative real-time polymerase chain reaction (qRT-PCR) in LNCaP prostate cancer cells treated with the antiandrogen bicalutamide. Using LNCaP cells, the effect of miR-125b modulation on apoptotic protein and NCOR2, a co-repressor of androgen receptor (AR), was examined by Western blot. A 3′-untranslated region (UTR) luciferase-binding assay was performed to confirm that miR-125b targets NCOR2. We found that surgical castration induced an initial increase in the expression of circulating miR-125b in mice, while sham surgery did not. In addition, AR blockade via bicalutamide was associated with the rapid release of miR-125b into the cell culture medium of prostate cancer cells. A previously studied target of miR-125b, a regulator in the apoptotic pathway, BAK1, could not completely account for the role of miR-125b in prostate cancer. Thus, we looked for additional targets of miR-125b and found that NCOR2, which is a repressor of AR, is a direct target of miR-125b. We found that NCOR2 protein expression was blocked by mimics of miR-125b, and a luciferase-binding assay confirmed that NCOR2 is a direct target of miR-125b. Our data provide novel evidence that miR-125b is an important regulator of the AR with specific ramification for the effectiveness of antiandrogens and other hormonal therapies in prostate cancer.
Key words: androgen receptor complex, apoptosis, castration, circulating micro-RNA, prostate cancer
Introduction
Prostate cancer is the most common non-skin cancer and the second most common cause of cancer deaths in men.1 The androgen receptor (AR) is a critical therapeutic target in prostate cancer,2 and alterations in the AR are critical to the development of this castrate-resistant state.3 Elucidating mechanisms underlying this resistance and developing new therapeutic strategies to prevent it are of great clinical interest.
Recent studies have highlighted the key regulatory role of micro-RNAs in all fundamental cellular processes, including cancer development.4 Numerous lines of investigations have shown that micro-RNAs can function as oncogenes or tumor suppressors, or sometimes as both.5 Accumulating evidence demonstrates that expression levels of micro-RNAs correlate with both stage and prognosis of human cancer,6 and the targeting of micro-RNAs represents a potential new therapeutic paradigm in oncology.7 The work of several investigators has shown that miR-21, 221, and 222 are associated with hormone-related prostate cancer growth.8–10 Additionally, Porkka et al. demonstrated a distinct pattern of micro-RNA expression comparing benign to cancerous prostate tissue and also between hormone-refractory versus hormone-sensitive prostate cancers.11 This work has focused on the identification of micro-RNA expression patterns for the purpose of classification, but not the functional role of these micro-RNAs. High expression of hsa-miR-125b (written as miR-125b herein) is observed in cancerous prostate tissue, as well as, in cell lines,6,12 and also in serum samples of patients with prostate cancer,13 although this finding has not been universally reported in prostate cancer tissues.11,14 Shi et al. have shown that overexpression of miR-125b in vitro in LNCaP cells leads to the downregulation of Bak1, an important mediator of apoptosis. However, the downregulation of the proapoptotic Bak1 alone is not sufficient to induce androgen-independent behavior, suggesting additional miR-125b targets in the development of hormonal resistance.12
In this study, we begin by assessing differences in micro-RNA expression in vivo and comparing pre- and postcastration blood levels in mice. We hypothesize that the response to androgen deprivation and eventual development of castrate-resistant prostate cancer (CRPC) are associated with alteration in miR-125b expression, consistent with these in vivo results. Our data provide novel evidence that miR-125b is an important regulator in prostate cancer by targeting the co-repressor of the AR, SMRT/NCOR2.15 We show that miR-125b directly targets the co-repressor NCOR2 and thus subsequently the AR signaling.
Materials and Methods
Cell culture, kits, and reagents
Human HEK293 and the prostate cancer cell line LNCAP (American Type Culture Collection, Manassas, VA) were grown in RPMI 1640 (Gibco, Grand Island, NY) with 10% fetal bovine serum (Gemini, Woodland, CA) plus 100 U/mL penicillin and 100 μg/mL streptomycin (Life Technologies, Grand Island, NY). All cell lines were incubated at 37°C in 5% CO2. Bicalutamide was obtained from Toronto Research Chemicals, Inc., Toronto, Canada, via Fisher Scientific, PA, and was freshly dissolved in dimethyl sulfoxide (DMSO) for use. Antibodies against Bak-1 and NCOR-2 were acquired from CalBioChem (San Diego, CA) and Thermo Scientific (Rockford, IL), respectively, and others from Cell Signaling (Beverly, MA). MiRNeasy Mini Kit, miScript Reverse Transcription Kit, miScript SYBR Green PCR Kit, QIAzol Lysis Reagent, HIPERFECT transfection reagent, and all of miScript micro-RNA primers for polymerase chain reaction (PCR) were obtained from QIAGEN (Valencia, CA). Additional primers were purchased from IDT (Coralville, IA). The information for all primer sequences is listed in Table 1. Adjusted primers for miR-331-3p and miR-21 were used for quantitative real-time polymerase chain reaction (qRT-PCR) to obtain useful melting temperatures. TRIzol® LS reagent was acquired from Invitrogen (Carlsbad, CA).
Table 1.
Primer Sequences for qRT-PCR
| Gene | Primers |
|---|---|
| Mouse miR-331-3p | 5′-GCC CCT GGG CCT ATC CTA GAA-3′ |
| Adj mouse miR-331-3p | 5′-CCC CTG GGC CTA TCC TAG AA-3′ |
| Mouse miR-21 | 5′-TAG CTT ATC AGA CTG ATG TTG A-3′ |
| Adj mouse miR-21 | 5′-GGG TAG CTT ATC AGA CTG ATG TTG A-3′ |
| Mouse miR-145 | 5′-GTC CAG TTT TCC CAG GAA TCC CT-3′ |
| Mouse miR-221 | 5′-AGC TAC ATT GTC TGC TGG GTT TC-3′ |
| Mouse miR-125b | 5′-TCC CTG AGA CCC TAA CTT GTG A-3′ |
| RNU6 for both human and mouse | Not available |
| Human miR-125b | 5′-TCC CTG AGA CCC TAA CTT GTG A-3′ |
qRT-PCR analysis of micro-RNA
Total RNA was isolated with an miRNeasy Mini Kit with QIAzol Lysis Reagent for cell pellets or TRIzol LS reagent for the cell culture medium. The isolation procedure was accomplished using a QIACube Robot, following the manufacturer's instruction. The concentration and quality of extracted RNA were assessed by NanoDrop 100 (Thermo Scientific, Waltham, MA). Equal amounts of extracted RNA (50 ng) were reverse-transcribed into cDNA in a total of 20 μL using an miScript Reverse Transcription Kit in Gene Amp PCR System 9700 (Applied Biosystems, Foster City, CA) and subjected to real-time PCR using an miScript SYBR Green PCR Kit. Detectable micro-RNA expression was normalized to small nuclear RNA (snRNA) RNU6B. Fold changes compared to controls were calculated by 2−ΔΔCt after normalizing to snRNA RNU6B, a reference gene.16
Transfection
Cells were seeded 24 h before transfection in 6-well plates or 96-well plates and transfected using the HIPERFECT transfection reagent. For micro-RNA measurement, cells were transfected for 48 h and washed with cold phosphate-buffered saline. For protein measurement, cells were transfected 3 days before collection for Western blotting. Mimics and inhibitors for miR-125b were purchased from Dharmacon (Lafayette, CO). The concentration used was 5 nM or otherwise stated.
Protein characterization
After the specified treatment, cell pellets were collected and resuspended in a lysis solution. Fifty micrograms of protein was electrophoresed and transferred to a nitrocellulose membrane (Invitrogen). The membrane was blocked and incubated overnight with the primary antibody at 4°C before being washed with Tris-buffered saline (TBS). The membrane was incubated with a secondary antibody for 1 h and again washed three times for 15 min each, before the addition of a chemiluminescent substrate (Rockford, IL). It was then immediately exposed on an X-ray film and scanned (Epson Perfection V500 Photo, Long Beach, CA) with ImageJ software (NIH online) utilized for quantification analysis of the protein expression. β-actin was used for normalization.
Luciferase untranslated region-binding experiments
The NCOR2 3′-untranslated region (UTR) containing the predicted miR-125b target sequence was amplified from genomic DNA (LNCAP cells) and cloned into the psiCheck-2 plasmid vector downstream of the Renilla luciferase stop codon (Promega, Madison, WI). The first set of primers for amplifying the NCOR2 UTR region is NCOR2 forward 5′-CCA CTG CTC TGC TCG CAG TAC GA-3′ and reverse 5′-GCT CAG TTT AGA CTT TGG TTC CAA ATG CAT-3′, and then, the PCR product was amplified again to add the needed restriction enzyme sites for cloning with the primers NCOR2 forward near stop Xho1 5′-ACT GCT CTG CTC GAG GTA CGA GAC ACT CT and NCOR2 reverse NOT1 5′-TCA GTT TAG ACT TTG CGG CCG CAT GCA T primers, which were based on this sequence of NCOR2 ref (NM 006311.2). NCOR2 3′-UTRs containing mutant target sequences were generated using primers designed to exchange a G for an A and an A for a G in the miR-125b seed region using the QuickChange-site-directed mutagenesis method (Stratagene). The specific primers for mutation are forward mut NCOR2 5′-CTG TGC AGA CCT TAC TCG GAG GAT GTT TA and reverse mut NCOR2 5′-TAA ACA TCC TCC GAG TAA GGT CTG CAC AG. Inhibition of expression of the luciferase reporter gene by miR-125b was assayed in HEK 293 cells transfected with either a vector control or a luciferase construct with the miR-125b-binding site from the NCOR2 3′-UTR. Luciferase assays were conducted using the dual luciferase reporter kit available from Promega. All methods were conducted as described by the supplier.
Surgical castration and collection of blood
Surgical castration was performed on 8-week-old male nude mice and blood was collected. Briefly, the anesthetized animal was placed in dorsal recumbence. A 2–4-mm incision was made in the scrotum, and the skin was retracted to expose the tunica. The tunica was pierced, and both testes were removed. The skin incisions were closed with stainless steel wound clips. One drop of bupivicaine (50 μL) was applied before the closure. For the sham group, the same surgical incision was made, but the testicles were not removed. Blood was collected as in the saphenous method, with no more than 200 μL of blood collected once every 2 weeks per mouse. The castrated mice were maintained for a total of 4 weeks. The serum samples from two mice were pooled at each time point to obtain an adequate blood sample size for analysis. All of these experiments were based on the approved animal protocol by Institutional Animal Care and Use Committee.
Cell viability assay
Cell viability was assessed using a tetrazolium-based assay (CellTiter 96 AQueous One Solution; Promega Corporation). Approximately 1500 cells in 100 μL of complete cell culture media per well were plated in 96-well plates in triplicate. Twenty-four hours after plating, 70 μL of serum free media with various concentrations of a mimic-plus-transfection reagent was replaced for appropriate transient transfection. Thirty microliters of serum free media with appropriate concentrations of bicalutamide in DMSO was added to achieve the desired treatment. DMSO was included in these experiments as controls. Once the treatment was complete, 20 μL of the AQueous One Solution was added to each well, yielding a final volume of 120 μL. A 96-well plate reader (Vmax Kinetic Microplate Reader; Molecular Devices, Sunnyvale, CA) was used for colorimetric analysis between 1 and 4 h (wavelength of 490 nm) after the addition of the AQueous One Solution. Cell viability assays were performed in triplicate.
Statistical analyses
The results from at least three independent experiments were expressed as the means and standard deviations. An analysis of variance, or t-test when appropriate, was used to examine potential different expressions under the designed experimental conditions. A p-value of <0.05 was considered significantly different throughout the article. Statistical software package SAS 9.2 (SAS Institute, Inc., Cary, NC) was used for all the statistical analyses.
Results and Discussions
Surgical castration induces the altered expression of micro-RNAs in mice
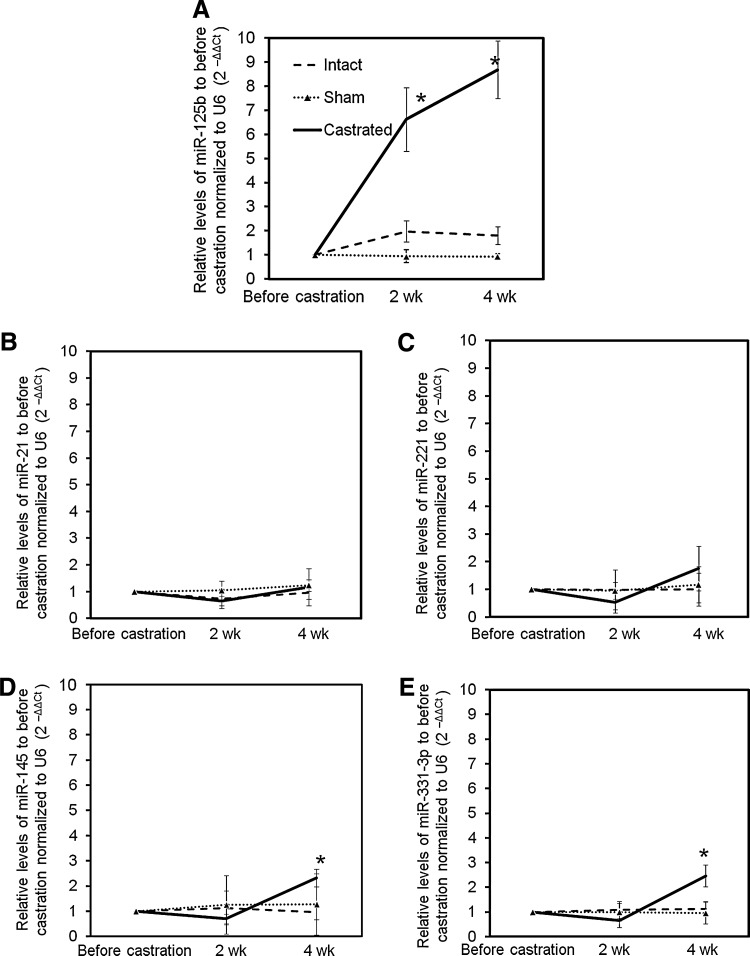
Androgen deprivation is a foundation of medical treatment for advanced prostate cancer, and studies have shown that micro-RNA regulation appears to be altered in prostate cancer. Thus, we hypothesized that the expression of specific micro-RNAs would be altered in response to androgen-deprivation therapy.17,18 To investigate this hypothesis, we began by assessing circulating levels of a subset of micro-RNAs before and after surgical castration in mice, a condition simulating the effect of androgen-deprivation therapy. Micro-RNAs mirR-21, miR-125b, miR-145, miR-221, and miR-331-3p were selected for assessment, since these micro-RNAs had previously been associated with prostate cancer.8–10,12,17–20 As shown in Figure 1, all of these micro-RNAs are detectable in the mouse sera. The circulating levels of miR-125b are increased 6.6-fold 2 weeks postcastration and continue to increase to 8.7-fold at 4 weeks, compared with precastration levels. The miR-125b levels in eugonadal (untreated) mice were not statistically altered over the course of 4 weeks (Fig. 1A). In contrast, there were no significant alterations of the circulating levels of miR-21 and miR-221 in any group of mice (Fig. 1B, C), whereas the expression of miR-145 and miR-331-3p shows mild but significant elevation (approximately two-fold increase) at 4 weeks postcastration compared with the precastration levels, and no difference was seen at the 2-week time point (Fig. 1D, E). The micro-RNAs assessed in both groups of intact and sham mice did not show any significant difference during the experimental process. The micro-RNAs that we examine in these studies are completely conserved from mouse to man as described in the Sanger database (www.mirbase.org/index.shtml). The rapid increase of miR-125b expression with castration in vivo suggests a regulatory relationship between androgen deprivation and miR-125b, providing a rationale for further examination of miR-125b's functional role related to AR signaling.
FIG. 1.
Surgical castration induces altered expression of micro-RNAs. Surgical castration was performed on 8-week-old male nude mice, and blood was collected submandibularly before and after surgery, at the time points indicated. The serum samples from two mice were collected and pooled at each time point to give an adequate blood sample for RNA extraction. Extracted total RNA was reverse-transcribed into cDNA and subjected to qRT-PCR and normalized to U6 compared with results from the corresponding mouse sera before castration, to obtain the relative fold expression using 2−ΔΔCt [miR-125b (A), miR-21 (B), miR-221 (C), miR-145b (D), and miR-331-3p (E)]. The results are expressed as mean±SD (n=3; pooled samples, total six mice per group). t-Test was used to compare the micro-RNA expression to that from the intact mouse group at each time point. *p<0.05 was considered significantly different in RNA expression. qRT-PCR, quantitative real-time polymerase chain reaction; SD, standard deviation.
Blockade of AR is associated with miR-125b secretion from cells
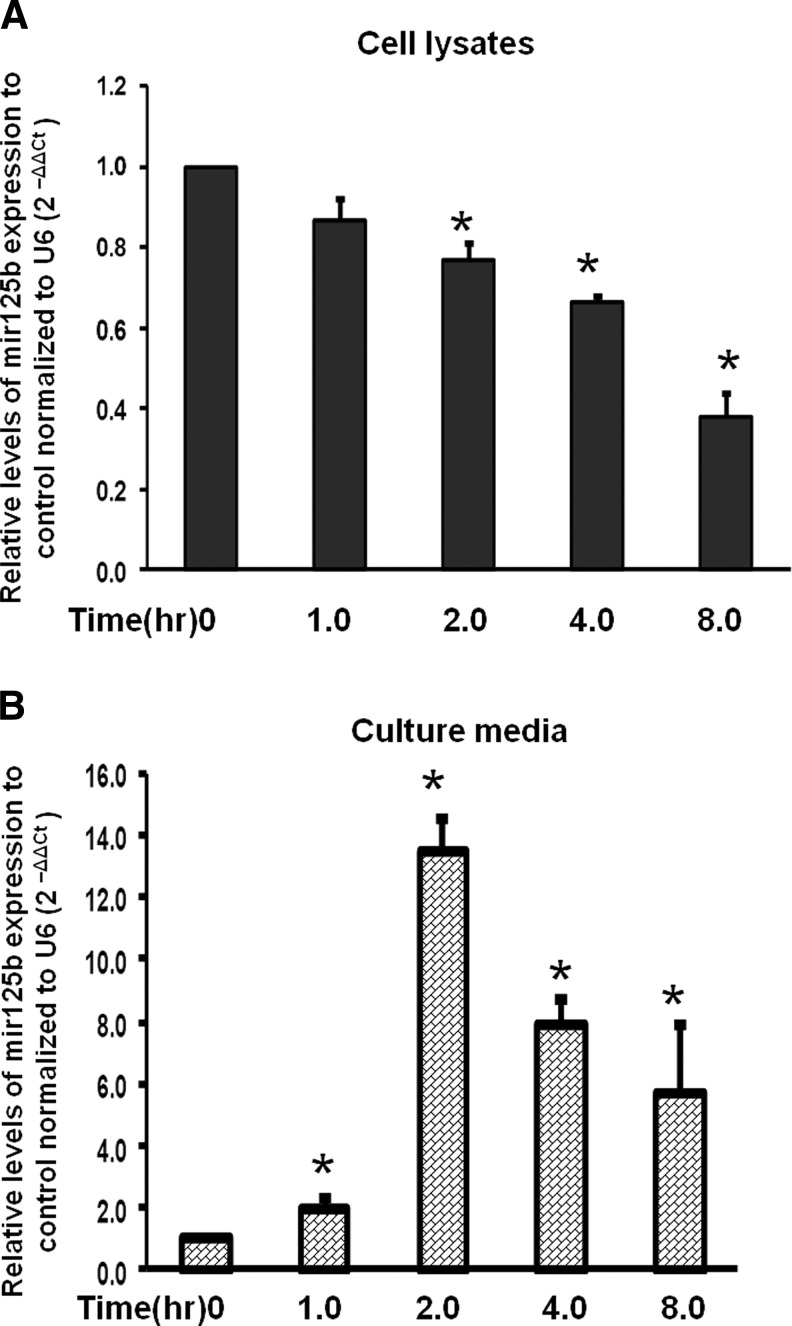
To further investigate the effect of androgen-deprivation on miR-125b expression, we treated LNCAP cells with a clinically relevant AR antagonist, bicalutamide. Figure 2 shows that bicalutamide decreased the expression of miR-125b levels in a time-dependent manner in the cell lysates (Fig. 2A). Interestingly, we found that miR-125b increased in the cell culture medium, peaking at 2 h (Fig. 2B), and these data suggest that miR-125b may be released from cells into the medium. These findings provide novel evidence that antiandrogen therapy rapidly alters the expression of cellular miR-125b and complements our finding that castration increases circulating miR-125b expression. Although the study of the molecular mechanisms of the cellular release of micro-RNAs is not the focus of the present study, this action is an active area of investigation.21–24 Circulating micro-RNAs are found in a variety of body fluids, including blood.22,23,25,26 Our results show that extracellular levels of miR-125b increase when cells are treated with antiandrogens, whereas other investigators have found that tissue levels of miR-125b decrease in the castration-resistant setting.11 miR-125b secretion out of the cells and into the media in response to bicalutamide is especially important, considering the retained functional capacity of such circulating micro-RNA.23 However, overexpression of miR-125b in cancer tissues has not been found by all studies, indicating the complexity of interpreting micro-RNA levels and potential differences between circulating and tissue levels.11
FIG. 2.
Bicalutamide differentially influences miR-125b expression in cell lysates and the culture medium in a time-dependent manner. LNCaP prostate cancer cells were treated with 32 μM bicalutamide, an antagonist of AR. miR-125b expression was normalized to the internal control U6 and then to control of no treatment to obtain the relative fold change using 2−ΔΔCt. Decreased expression was observed in the cell lysate (A) as compared to increased levels of miR-125b in the cell culture media (B) compared to baseline, respectively. The results were expressed as mean±SD (n=3). t-Test was used to compare fold changes in both samples to that of the corresponding micro-RNAs in no treatment controls. *p<0.05 was considered significantly different between the groups. AR, androgen receptor.
Overexpression of miR-125b increases LNCAP cell growth and amplifies the effect of an AR antagonist
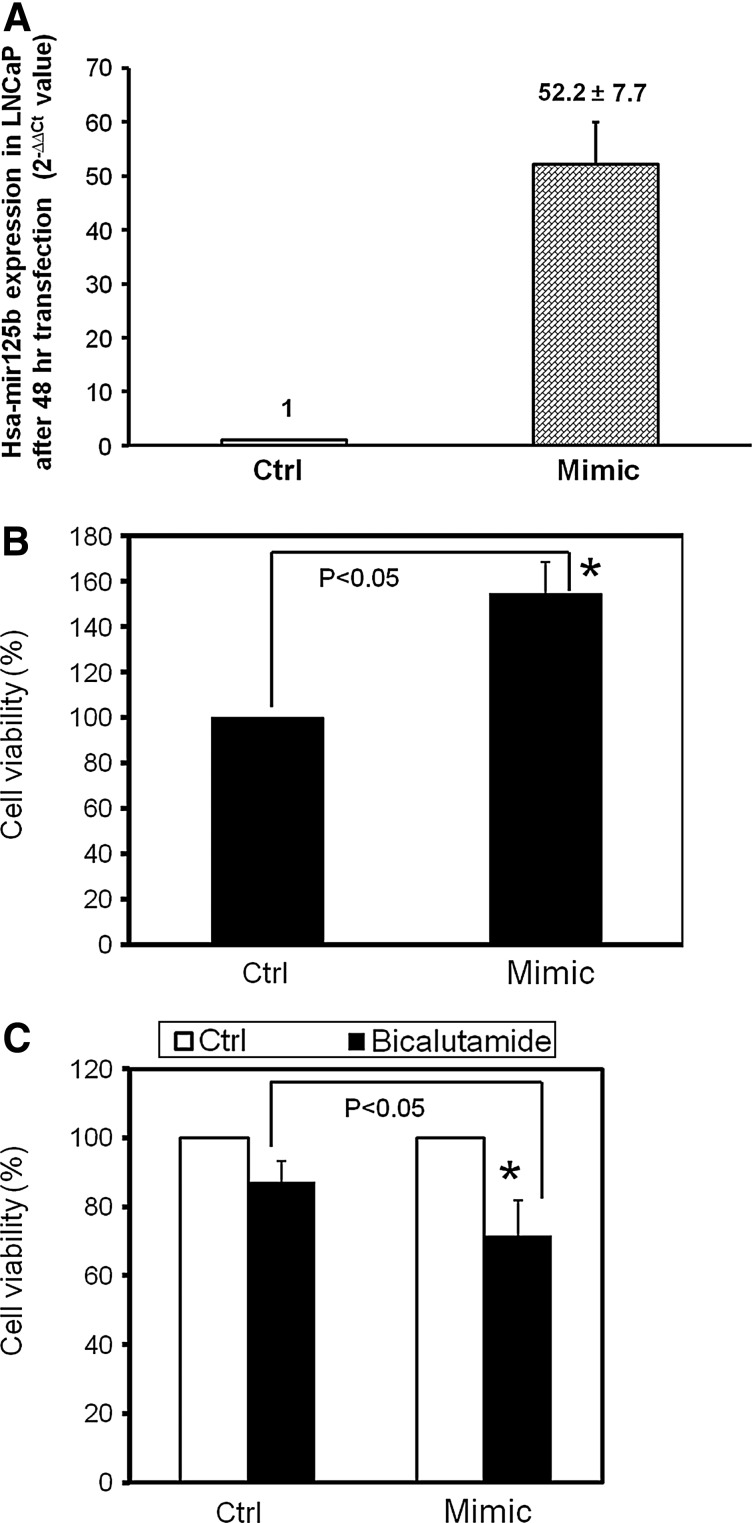
To assess the functional role of miR-125b in prostate cancer cells, we modulated miR-125b expression using a transient transfection approach and performed a cell viability assay. First, we performed the RT-PCR to validate transient transfection of the mimic of miR-125b. As shown in Figure 3A, under our conditions, this transfection increases its expression ∼50-fold. In the cell viability assay shown in Figure 3B, a mimic of miR-125b increased the cell growth 47% after a 4-day transfection, in agreement with the findings of Shi et al.12 and Zhou et al.27 Furthermore, after normalizing each group, the administration of the AR antagonist bicalutamide reduced the cell viability by 29% in the mimic setting, versus 13% in the control cells, a statistically significant difference, indicating that miR-125b mimics amplify the cell killing effect of bicalutamide (Fig. 3C). There was relatively low inhibitory activity with bicalutamide in these conditions in LNCaP cells, although these results are consistent with previous observations.28–30
FIG. 3.
Validation of transient transfection of miR-125b and the influences of miR-125b modulation on the cell growth of LNCAP cells. LNCaP cells were transfected with 25 nM miR-125b mimic. After 48 h, the cell pellets were harvested, and RNA was extracted. miR-125b levels were determined by RT-PCR and normalized by U6. RT-PCR results (A) show that this mimic increased, by 52-fold, the expression of miR-125. Cell viability assays: 1500 cells in an RPMI medium with serum were seeded in each well of 96-well plates with triplicates of each treatment in three independent repeats. After 24 h, the medium was replaced with a serum-free medium and transfected with miR-125b mimic for 72 h. Appropriate concentrations of bicalutamide in dimethyl sulfoxide were added, and the cells were incubated for another 24 h. Cell growth was assayed, comparing the treatment with non-treatment control (B) or comparing the absence and presence of bicalutamide, and normalized to corresponding treatments (C). The results were expressed as mean±SD (n=3). t-Test was used to test differences in cell growth between treatment and control. *p<0.05 was considered significantly different.
Transient transfection with miR-125b mimic influences apoptosis-related protein and AR expression
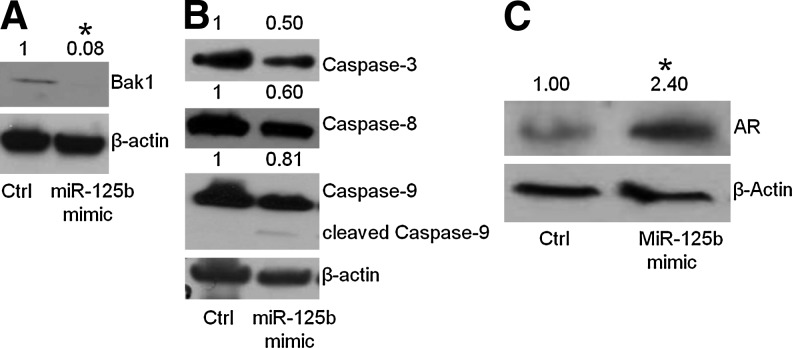
Initially, we examined the protein expression of a confirmed target of miR-125b, the proapoptotic factor, Bak1,12,27 and found that Bak1 was reduced by the mimic of miR-125b (Fig. 4A), in agreement with previous observations.12 To identify other apoptotic factors regulated by miR-125b, we further examined additional apoptosis-related proteins, including caspase-3, 8, and 9. As shown in Figure 4B, addition of miR-125b mimic does reduce these proteins, and the expected increase in the cleaved form of caspase-9 was observed (Fig. 4B and Supplementary Fig. S1), although these reductions are smaller relative to the changes observed with Bak1 (Fig. 4A). These results imply that the apoptotic pathway alone does not completely account for miR-125b's role in the development of androgen-deprivation therapy and that other proteins and additional pathways may be targets of miR-125b as well. The previous results of castrated mouse serum and bicalutamide-treated LNCaP cells imply that there is a link between miR-125b and AR. Therefore, we measured the protein level of AR under the transient transfection of miR-125b mimic. As shown in Figure 4C, the treatment of prostate cancer cells with mimics of miR-125b showed that the AR was increased in expression, indicating the alteration of AR signaling induced by the modulation of miR-125b.
FIG. 4.
miR-125b influences apoptosis-related protein expression and AR. LNCaP cells were transfected with 25 nM miR-125 mimic and inhibitor. After 72 h, the cells were collected and subjected to Western blot. The protein levels in whole-cell lysates were compared with control (no treatment) after normalization using β-actin, indicated above the gels. (A) Bak1 expression; (B) other apoptosis-related protein expression; (C) AR. t-Test was used to compare the fold change of band density for each protein, comparing the treatment and control groups. *p<0.05 was considered significantly different between treatment and control groups.
Confirmation of a repressor of AR, NCOR2, as a target of miR-125b
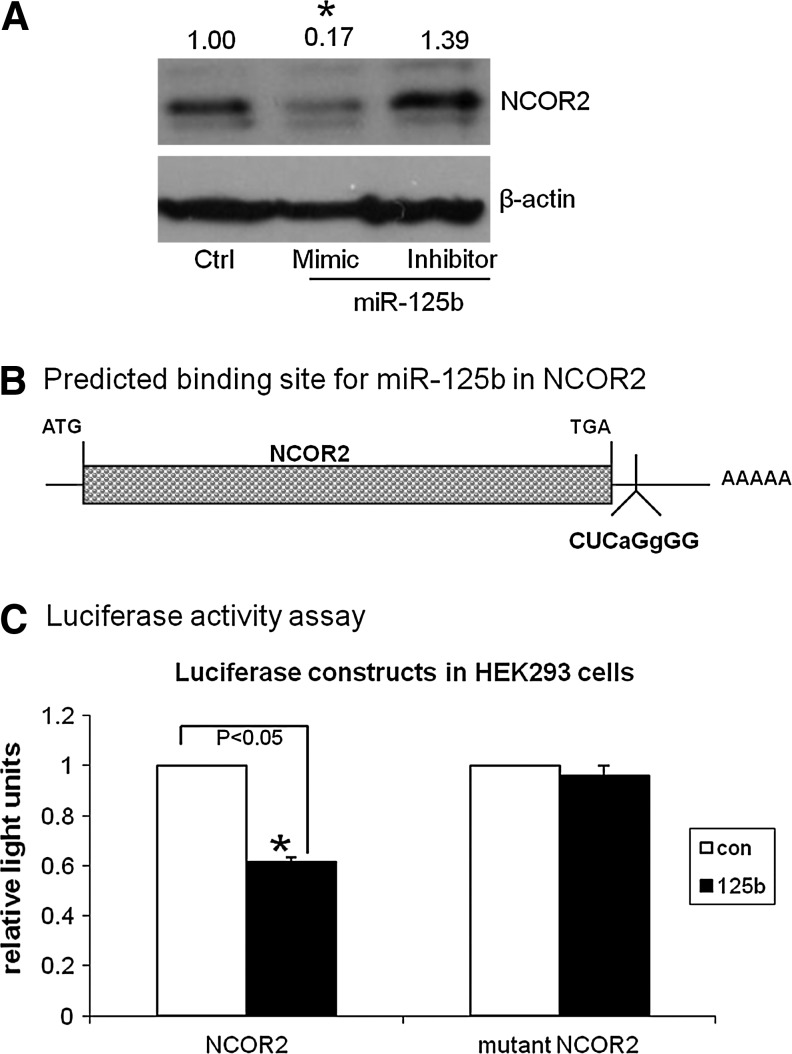
The increase in AR expression in cells treated with mimic of miR-125b suggests that a possible mechanism of AR regulation in these cells could be the loss of a repressor of AR. Thus, miR-125b could target a repressor of AR signaling; through the use of the STRING database (protein interactions) and TargetScan (www.targetscan.org/), NCOR2 was identified as a potentially relevant target of miR-125b, since NCOR2 had previously been described as a repressor of the AR,15 and TargetScan predicts that NCOR2 will be a target of miR-125b. To validate these in silico predictions, we examined the protein levels of NCOR2 after transfection of miR-125b. As shown in Figure 5A, the mimic of miR-125b decreases the expression of NCOR2. These data support a direct binding of miRNA, while the inhibitor increases the expression, supporting a regulatory role for miR-125b of NCOR2.
FIG. 5.
NCOR2 is a direct target of miR-125b. (A) LNCaP cells were transfected with miR-125b mimic and inhibitor. After 72 h, the cells were lyzed and subjected to Western blot. The protein levels of NCOR2 were compared with control (no treatment) after normalization, using β-actin, indicated by the numbers above the gels. All of the experiments were repeated three times in separate experiments. t-test was used to compare fold changes of band density of NCOR2 between the treatment and control groups. *p<0.05 was considered significantly different between the treatment and control groups. (B) NCOR2 is a predicted target of miR-125b by TargetScan 5.1 and 6.0. Shown is the seed-binding region in the 3′ UTR of NCOR2. Lower-case letters are used to designate the base pairs that were mutated to block the binding of miR-125b in the NCOR2-mutated version. (C) A luciferase reporter-binding assay confirms that NCOR2 is a direct target of miR-125b. HEK 293 cells were seeded in 12-well plates. After 24 h, the cells were transfected with the NCOR2 UTR or NCOR2 UTR-mutated plasmid and miR-125b mimic. Transfections were performed in triplicate. After 24 h, the relative luciferase activity was determined using a luminometer. Reporter activity was normalized to the firefly luciferase concentration in the cell extracts with the effect of the miR-125b mimic compared between NCOR2 and mutated NCOR2-transfected cells. The results were expressed as mean±SD (n=3). Analysis of variance was used to test for differences in the activity among the different treatments. *p<0.05 was considered significantly different.
To further interrogate the NCOR2's relationship to miR-125b, a reporter-binding assay was developed to assess the UTR of NCOR2. In addition, another construct was generated and included two base-pair changes in the seed for binding to the NCOR2 transcript (Fig. 5B). Cotransfection of miR-125b mimic and the wild-type NCOR2-UTR plasmid significantly decreases the luciferase activity in HEK 293 cells as compared with control (39% decrease) (Fig. 5C). In contrast, the effect of miR-125b mimic with the mutant NCOR2 3′-UTR demonstrated no significant change in luciferase activity. These results confirm that miR-125b directly targets NCOR2, as identified by the TargetScan database described above, and support our observations that NCOR2 protein levels are decreased in response to miR-125b mimic (Fig. 5A). Thus, these studies confirm the direct regulation of NCOR2 by miR-125b and add to our understanding of the regulation of AR after androgen deprivation.
Recently, new targets of miR-125b, p53 and core binding factor-β transcription factor, have been characterized, emphasizing the complicated, but a very well-controlled system of micro-RNA regulation.31,32 Here, we characterize another target of miR-125b, NCOR2. Modulation of the AR transcriptional complex and, specifically, of AR co-repressors is an important mechanism in the response to androgen deprivation and eventual development of CRPC.2,33–37 NCOR2 is reported to have a unique role in the mechanism of AR blockade, and bicalutamide-bound AR has been shown to recruit NCOR2, providing a direct mechanism for bicalutamide's antagonistic role against AR.15,35,38
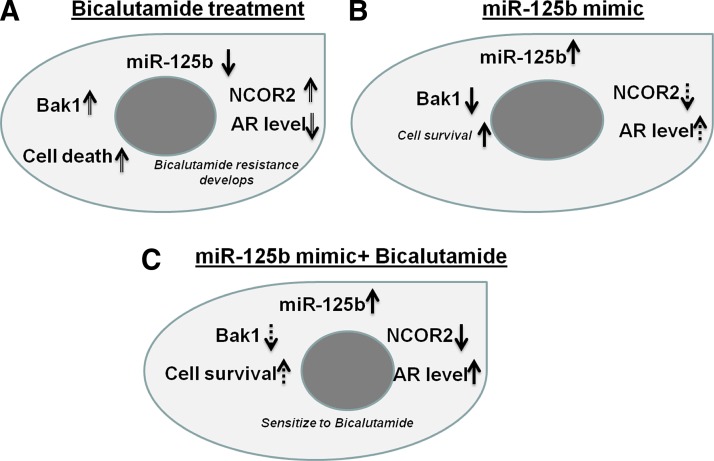
In conclusion, we began by showing increased circulating levels of miR-125b in vivo in response to castration and in vitro in response to AR blockage, supporting a regulatory relationship between AR and miR-125b. Our findings support previous reports of an association between increased miR-125b and CRPC and take this knowledge a step further by showing that miR-125b targets and effectively reduces cellular levels of NCOR2, a co-repressor of AR. In these studies, we define another mechanism for miR-125b's association with androgen deprivation and possible CRPC. In a functional assessment, we found that overexpressing miR-125b increases bicalutamide activity. This finding will require an additional investigation and study, but could potentially be explained by an increased AR protein level, then AR activity, theoretically making the cells more susceptible to bicalutamide, all due to decreased NCOR2 levels from the miR-125b mimic. These findings are summarized in the schematic diagram (Fig. 6).
FIG. 6.
Schematic description of the miR-125b's role in prostate cancer. miR-125b exerts a dual function in prostate cancer via both apoptosis and AR signaling. Three different, but consistently related observations have been found. (A) In mouse castration and bicalutamide-treated cells, both castration and deprivation causes the decrease of miR-125b within cells due to the secretion of miR-125b from tissue to circulating system or from cells to the medium. We imply that these changes would increase both Bak1 and NCOR2, which would cause both cell death and bicalutamide-related drug resistance development due to the effect of these two arms; (B), miR-125b mimic alone reduces both Bak1 and NCOR2. However, the Bak1 effect would be larger than the NCOR2 effect, in the absence of androgen-related agents. Thus, the net effect of a miR-125b mimic would be increased cell survival; (C) in the presence of bicalutamide, the addition of miR-125b decreases both Bak1 and NCOR2. However, the effect of NCOR2's decrease would be greater than the effect of Bak1's decrease in the presence of the antiandrogen agent. Thus, the addition of miR-125b decreases the AR protein levels, sensitizing the cells to the bicalutamide activity.
Supplementary Material
Acknowledgments
This work was supported by a Paul Calabresi K12 clinical scholar grant awarded to the University of Colorado Denver (K12CA086913) (T.W.F.). We also acknowledge the generous support of the Herbert Crane Endowment. The authors wish to acknowledge the technical assistance of Carol Amato.
Author Disclosure Statement
The authors declare that no competing financial interests exist.
References
- 1.Jemal A. Siegel R. Xu J, et al. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Chen CD. Welsbie DS. Tran C, et al. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10:33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 3.Damber JE. Aus G. Prostate cancer. Lancet. 2008;371:1710–1721. doi: 10.1016/S0140-6736(08)60729-1. [DOI] [PubMed] [Google Scholar]
- 4.Fusco A. MicroRNAs: a great challenge for the diagnosis and therapy of endocrine cancers. Endocr Relat Cancer. 2010;17:E3–E4. doi: 10.1677/ERC-09-0305. [DOI] [PubMed] [Google Scholar]
- 5.Negrini M. Nicoloso MS. Calin GA. MicroRNAs and cancer—new paradigms in molecular oncology. Curr Opin Cell Biol. 2009;21:470–479. doi: 10.1016/j.ceb.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 6.DeVere White RW. Vinall RL. Tepper CG, et al. MicroRNAs and their potential for translation in prostate cancer. Urol Oncol. 2009;27:307–311. doi: 10.1016/j.urolonc.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Czech MP. MicroRNAs as therapeutic targets. N Engl J Med. 2006;354:1194–1195. doi: 10.1056/NEJMcibr060065. [DOI] [PubMed] [Google Scholar]
- 8.Ribas J. Ni X. Haffner M, et al. miR-21: an androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res. 2009;69:7165–7169. doi: 10.1158/0008-5472.CAN-09-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ribas J. Lupold SE. The transcriptional regulation of miR-21, its multiple transcripts, and their implication in prostate cancer. Cell Cycle. 2010;9:923–929. doi: 10.4161/cc.9.5.10930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun T. Wang Q. Balk S, et al. The role of microRNA-221 and microRNA-222 in androgen-independent prostate cancer cell lines. Cancer Res. 2009;69:3356–3363. doi: 10.1158/0008-5472.CAN-08-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porkka KP. Pfeiffer MJ. Waltering KK, et al. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–6155. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 12.Shi XB. Xue L. Yang J, et al. An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc Natl Acad Sci U S A. 2007;104:19983–19988. doi: 10.1073/pnas.0706641104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchell PS. Parkin RK. Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ozen M. Creighton CJ. Ozdemir M, et al. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–1793. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 15.Liao G. Chen LY. Zhang A, et al. Regulation of androgen receptor activity by the nuclear receptor corepressor SMRT. J Biol Chem. 2003;278:5052–5061. doi: 10.1074/jbc.M206374200. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ. Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Coppola V. De Maria R. Bonci D. MicroRNAs and prostate cancer. Endocr Relat Cancer. 2010;17:F1–F17. doi: 10.1677/ERC-09-0172. [DOI] [PubMed] [Google Scholar]
- 18.Gandellini P. Folini M. Zaffaroni N. Towards the definition of prostate cancer-related microRNAs: where are we now? Trends Mol Med. 2009;15:381–390. doi: 10.1016/j.molmed.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 19.Epis MR. Giles KM. Barker A, et al. miR-331-3p regulates ERBB-2 expression and androgen receptor signaling in prostate cancer. J Biol Chem. 2009;284:24696–24704. doi: 10.1074/jbc.M109.030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaman MS. Chen Y. Deng G, et al. The functional significance of microRNA-145 in prostate cancer. Br J Cancer. 2010;103:256–264. doi: 10.1038/sj.bjc.6605742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibbings DJ. Ciaudo C. Erhardt M, et al. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 22.Kosaka N. Iguchi H. Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101:2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kosaka N. Iguchi H. Yoshioka Y, et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skog J. Wurdinger T. van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsujiura M. Ichikawa D. Komatsu S, et al. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer. 2010;102:1174–1179. doi: 10.1038/sj.bjc.6605608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hausler SF. Keller A. Chandran PA, et al. Whole blood-derived miRNA profiles as potential new tools for ovarian cancer screening. Br J Cancer. 2010;103:693–700. doi: 10.1038/sj.bjc.6605833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou M. Liu Z. Zhao Y, et al. MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J Biol Chem. 2010;285:21496–21507. doi: 10.1074/jbc.M109.083337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu S. Wang A. Dong Z. A novel synthetic compound that interrupts androgen receptor signaling in human prostate cancer cells. Mol Cancer Ther. 2007;6:2057–2064. doi: 10.1158/1535-7163.MCT-06-0735. [DOI] [PubMed] [Google Scholar]
- 29.Burich RA. Holland WS. Vinall RL, et al. Genistein combined polysaccharide enhances activity of docetaxel, bicalutamide and Src kinase inhibition in androgen-dependent and independent prostate cancer cell lines. BJU Int. 2008;102:1458–1466. doi: 10.1111/j.1464-410X.2008.07826.x. [DOI] [PubMed] [Google Scholar]
- 30.Colabufo NA. Pagliarulo V. Berardi F, et al. Bicalutamide failure in prostate cancer treatment: involvement of multi drug resistance proteins. Eur J Pharmacol. 2008;601:38–42. doi: 10.1016/j.ejphar.2008.10.038. [DOI] [PubMed] [Google Scholar]
- 31.Jiang L. Huang Q. Chang J, et al. MicroRNA HSA-miR-125a-5p induces apoptosis by activating p53 in lung cancer cells. Exp Lung Res. 2011;37:387–398. doi: 10.3109/01902148.2010.492068. [DOI] [PubMed] [Google Scholar]
- 32.Lin KY. Zhang XJ. Feng DD, et al. miR-125b, a target of CDX2, regulates cell differentiation through repression of the core binding factor in hematopoietic malignancies. J Biol Chem. 2011;286:38253–38263. doi: 10.1074/jbc.M111.269670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burd CJ. Morey LM. Knudsen KE. Androgen receptor corepressors and prostate cancer. Endocr Relat Cancer. 2006;13:979–994. doi: 10.1677/erc.1.01115. [DOI] [PubMed] [Google Scholar]
- 34.Berrevoets CA. Umar A. Trapman J, et al. Differential modulation of androgen receptor transcriptional activity by the nuclear receptor co-repressor (N-CoR) Biochem J. 2004;379:731–738. doi: 10.1042/BJ20031456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shang Y. Myers M. Brown M. Formation of the androgen receptor transcription complex. Mol Cell. 2002;9:601–610. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
- 36.Song LN. Coghlan M. Gelmann EP. Antiandrogen effects of mifepristone on coactivator and corepressor interactions with the androgen receptor. Mol Endocrinol. 2004;18:70–85. doi: 10.1210/me.2003-0189. [DOI] [PubMed] [Google Scholar]
- 37.Hodgson MC. Astapova I. Hollenberg AN, et al. Activity of androgen receptor antagonist bicalutamide in prostate cancer cells is independent of NCoR and SMRT corepressors. Cancer Res. 2007;67:8388–8395. doi: 10.1158/0008-5472.CAN-07-0617. [DOI] [PubMed] [Google Scholar]
- 38.Yoon HG. Wong J. The corepressors silencing mediator of retinoid and thyroid hormone receptor and nuclear receptor corepressor are involved in agonist- and antagonist-regulated transcription by androgen receptor. Mol Endocrinol. 2006;20:1048–1060. doi: 10.1210/me.2005-0324. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.