Abstract
Human embryonic stem (hES) cells are considered to be a potential source for the therapy of human diseases, drug screening, and the study of developmental biology. In the present study, we successfully derived hES cell lines from blastocysts developed from frozen and fresh embryos. Seventeen- to eighteen-year-old frozen embryos were thawed, cultured to the blastocyst stage, and induced to form hES cells using human foreskin fibroblasts. The Chula2.hES cell line and the Chula4.hES and Chula5.hES cell lines were derived from blastocysts developed from frozen and fresh embryos, respectively. The cell lines expressed pluripotent markers, including alkaline phosphatase (AP), Oct3/4, stage-specific embryonic antigen (SSEA)-4, and tumor recognition antigen (TRA)-1-60 and TRA-1-81 as detected with immunocytochemistry. The real-time polymerase chain reaction (RT-PCR) results showed that the cell lines expressed pluripotent genes, including OCT3/4, SOX2, NANOG, UTF, LIN28, REX1, NODAL, and E-Cadherin. In addition, the telomerase activities of the cell lines were higher than in the fibroblast cells. Moreover, the cell lines differentiated into all three germ layers both in vitro and in vivo. The cell lines had distinct identities, as revealed with DNA fingerprinting, and maintained their normal karyotype after a long-term culture. This study is the first to report the successful derivation of hES cell lines in Thailand and that frozen embryos maintained their pluripotency similar to fresh embryos, as shown by the success of hES cell derivation, even after years of cryopreservation. Therefore, embryos from prolonged cryopreservation could be an alternative source for embryonic stem cell research.
Key words: embryo transfer, infertility, reproductive technology, stem cells
Introduction
In human-assisted reproductive technology, the technique of cryopreservation, which involves storing an oocyte, sperm or embryo at sub-zero temperatures, is well established, effective and applied in most infertility centers around the world.1 Researchers were initially concerned with the effects of cryopreservation or cryostorage on the survival, development and molecular biology of the frozen embryo after thawing.2–4 However, recent reports have demonstrated that cryostorage does not adversely affect post-thaw survival or pregnancy outcome during in vitro fertilization (IVF) or oocyte donation in patients. Moreover, a healthy boy was born from 20-year-old cryopreserved pronuclear (PN) embryos.5,6
Embryos that display poor morphology after freeze–thawing are discarded or donated for research purposes, including human embryonic stem (hES) cell derivation. The hES cells that are derived from the pluripotent cells of a preimplantation stage embryo display unique characteristics, such as self-renewal and differentiation into all adult cell types. Thus, hES cells are not only considered to be potential sources for drug screening tests and regenerative medicine therapies, they might also serve as a model for the study of early human embryonic development.7 To date, more than 1000 hES cell lines have been derived in laboratories worldwide, and rapid advances in the knowledge and technologies associated with the culture conditions of hES cells have made it possible to derive new hES cell lines under clinical or near clinical conditions for further use.8 Although the embryos may display poor morphology, they remain a major source of starting material for the isolation of hES cells, and several methods have been applied to improve the success of hES cell derivation. Culturing poor-quality embryos in a modified culture medium increased blastocyst formation,9 and using mesenchymal stem cells as feeder cells also facilitated the derivation of hES cells from poor-quality embryos.10 However, a recent publication demonstrated that there was no correlation between the morphology of cells at the blastocyst stage and the success of hES cell derivation.11 Thus, the success of hES cell derivation can be expected even with poor-quality frozen–thawed embryos. As previously discussed, the duration of cryopreservation does not affect pregnancy outcome; thus, it would be interesting to determine whether long-term cryopreserved embryos could generate hES cell lines similar to fresh embryos.
In the present study, we aimed to derive hES cells from embryos that were previously frozen for 17–18 years under culture conditions that minimize contact with animal products. Moreover, an examination of the poor quality of fresh embryos that were not suitable for transfer was also included in this study.
Materials and Methods
Human embryos and ethical approval
The human embryos used in the present study were donated with informed consent from a couple that participated in the IVF program for infertility treatment. The isolation of hES cells was performed after obtaining the approval of the Institutional Review Board (IRB number 096/50), Faculty of Medicine, the Chulalongkorn University. The procedure was performed according to the National Guidelines for Stem Cell Research issued by the Thai Medical Council and the Ministry of Public Health.
Embryo culture
Frozen embryos at the PN stage were thawed, transferred to droplets of the global medium (LifeGlobal) supplemented with 10% serum substitute (Irvine Scientific), covered with light oil (LifeGlobal), and cultured at 37°C in 5% O2, 6% CO2, and 89% NO2. The poor-quality donated fresh embryos were cultured under the same culture conditions. After 3–4 days of culture, only the embryos that developed to the blastocyst stage were collected and transferred to the hES cell laboratory for hES cell isolation.
Preparation of the feeder layer
Commercial human foreskin-derived fibroblasts (HFF; CRL-2429, ATCC) were used. HFFs were cultured and maintained according to the manufacturer's protocol. To use HFFs as feeder cells, the confluent fibroblasts were mitotically inactivated with 10 μg/mL mitomycin C (Sigma) for 2.5–3 h, dissociated with 0.05% trypsin-ethylenediaminetetraacetic acid (Invitrogen), counted, and plated on a 0.1% gelatin-coated dish (BD Bioscience).
Isolation of the inner cell mass and the propagation of hES cells
The zona pellucida of the blastocyst was removed through a brief incubation with 0.1% acidified Tyrode's solution with close monitoring under a stereomicroscope. Zona-free blastocysts were directly plated onto the feeder cells. For propagation, the hES cells were mechanically dissociated into small pieces every 5–7 days using a 23G needle, detached from the culture dish, and plated onto new feeder cells; the culture medium was changed daily. The hES cell medium was the knockout Dulbecco's modified Eagle's medium supplemented with 20% knockout serum replacement, 1% Glutamax®, 1% nonessential amino acids, 0.1 mM 2-mercaptoethanol, 1% penicillin-streptomycin (all purchased from Invitrogen), and 8 ng/mL fibroblast growth factor (bFGF; R&D Systems).
Alkaline phosphatase staining and immunostaining
The hES cell colonies were fixed using 4% paraformaldehyde. The alkaline phosphatase (AP) activity was detected using an AP detection kit (Sigma), according to the manufacturer's protocol. The primary antibodies used were mouse antibodies against stage-specific embryonic antigen (SSEA)-4, tumor recognition antigen (TRA)-1–60 and TRA-1–81, and rabbit anti-Oct3/4 (all purchased from Abcam). For the detection of the antibodies, fluorescein isothiocyanate-conjugated goat anti-rabbit or Cy3-conjugated goat anti-mouse antibodies were used.
Real-time polymerase chain reaction
Total RNA was extracted using TRI REAGENT® (Molecular Research Center, Inc.). A total of 1 μg of total RNA was reverse transcribed using the RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas, Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. Polymerase chain reaction (PCR) was performed with PCR master mix (2×) (Fermentas, Thermo Fisher Scientific, Inc.). The PCR conditions and primers were described previously.12
Differentiation of hES cells
To determine their in vitro differentiation ability, hES cells were mechanically dissociated into small clumps and cultured in suspension in the hES cell culture medium without bFGF. Three-dimensional structures called embryoid bodies (EBs) were observed for up to 21 days. To determine their in vivo differentiation ability, a teratoma formation assay was performed. Approximately 1×106 hES cells were injected under the testicular capsule of 4- to 6-week-old nude mice. After 10–12 weeks, the mice were euthanized, and the teratomas were removed, fixed with 10% buffered formalin phosphate (Sigma), and embedded in paraffin blocks. Next, 4 μm sections were stained with hematoxylin and eosin. The care of the animals was conducted in accordance with the institutional guidelines of the Ethics Committee for Animal Laboratory Use (Approval No. 15/52).
Telomerase analysis
The telomerase activity of the hES cell lines was measured using a TRAPeze Telomerase Detection Kit (Chemicon), according to the manufacturer's instructions.
Karyotype analysis
The hES cells were incubated with 10 ng/mL of colcemid (Karyomax, Invitrogen) for 3–4 h at 37°C and 5% CO2. Subsequently, the cells were trypsinized, treated with KCl solution (Bio Industries), and fixed with fixative (3:1 methanol:acetic acid). The metaphases were spread onto microscope slides and stained using a standard G-banding technique. The chromosomes were classified according to the International System for Human Cytogenetic Nomenclature. At least 15–20 metaphases were analyzed per cell line.
DNA fingerprinting
Total genomic DNA was extracted from the undifferentiated hES cells using a DNeasy Tissue Kit (Qiagen), according to the manufacturer's instructions. Fifteen short tandem repeat (STR) loci and amelogenin were amplified using an AmpFlSTR® Identifiler® PCR Amplification Kit (Applied Biosystems), according to the manufacturer's instructions and detected using a 3100 Genetic Analyzer (Applied Biosystems).
Results
Development of embryos after long-term cryopreservation
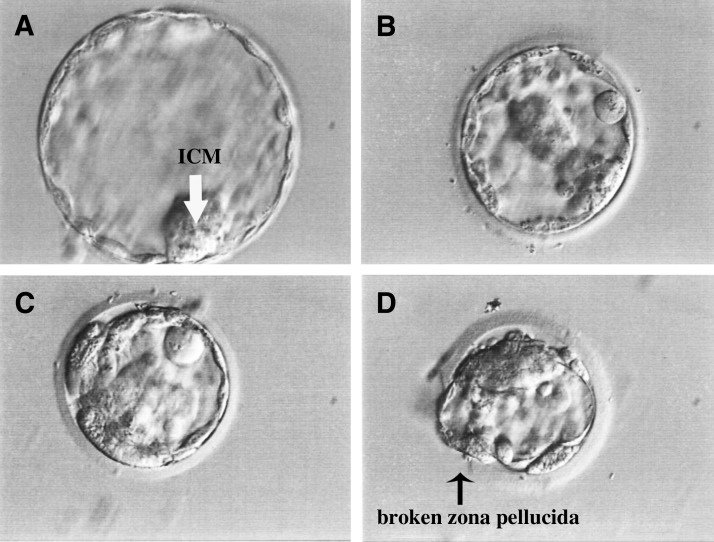
Upon approval from the ethics committee, a total of 23 embryos were thawed and cultured according to the standard protocol of the IVF Unit, King Chulalongkorn Memorial Hospital. A total of 17 of the 23 (73.9%) embryos survived after thawing: 12 (70.6%) of the surviving embryos arrested at the initial stage of development, and five (29.4%) embryos developed to the blastocyst stage after 4 days of culture. Of the five blastocysts, we only obtained one expanded blastocyst with a tightly packed inner cell mass (ICM; Fig. 1A); the other blastocysts were not fully expanded, showing thick or broken zonae pellucidae and small ICMs (Fig. 1B–D).
FIG. 1.
Morphology of the blastocysts developed from long-term cryopreserved embryos. Six days after thawing and culture, the surviving embryos developed to the blastocyst stage. Different morphologies of the blastocysts, including fully expanded blastocysts with a clear and tightly packed ICM (white arrow; A), non-fully expanded blastocysts with thick zonae pellucidae and small ICMs (B–D), and broken zona pellucida during the freezing–thawing process, were observed (black arrow; D). All of the images were obtained using a 40× objective phase-contrast microscope. ICM, inner cell mass.
Isolation of ICMs and hES cell culture
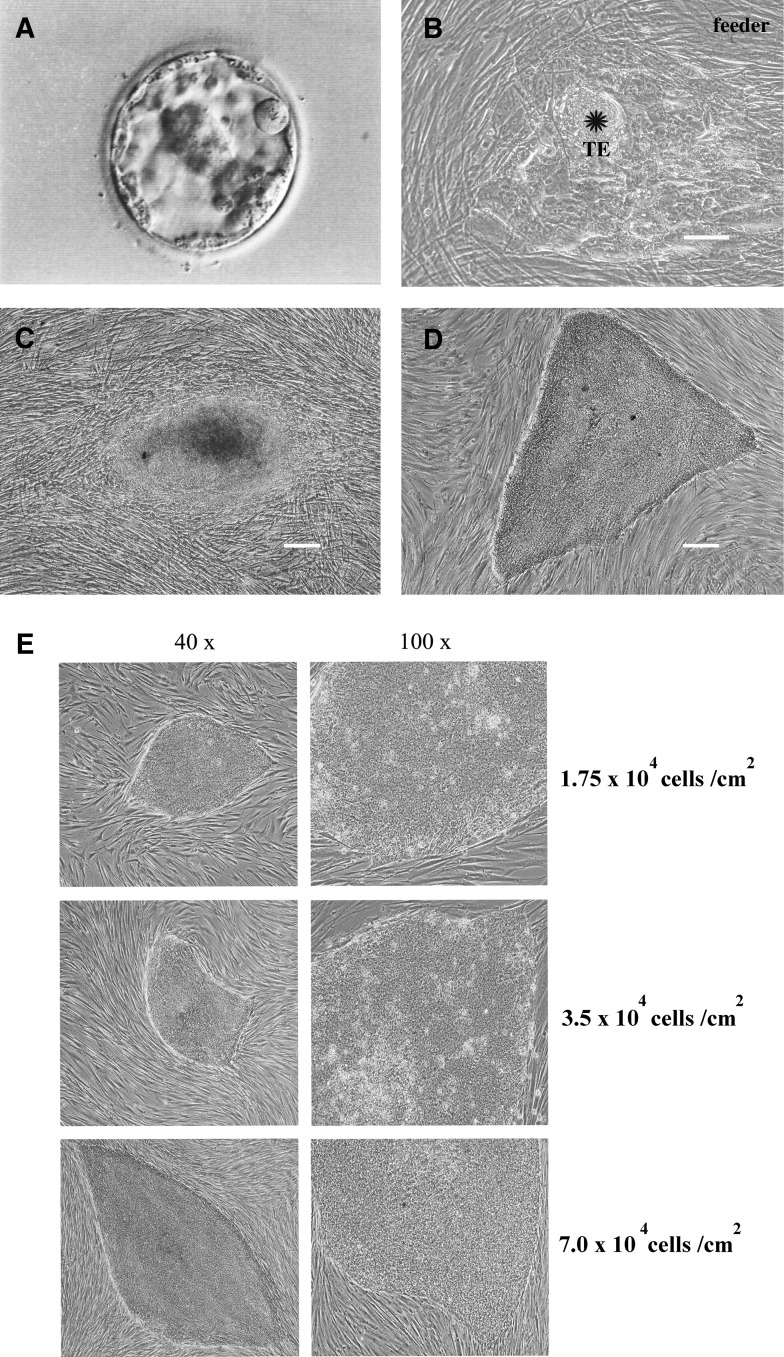
Although their quality was not high, five blastocysts from frozen–thawed embryos and eight fresh blastocysts were used for hES cell derivation. Due to the difficulty of the localization of the ICM within blastocysts (Fig. 2A), the mechanical separation of the ICM from the trophectoderm (TE) was not achieved. Thus, the zona pellucida was isolated, and the blastocysts were directly plated onto inactivated HFFs that were used as the feeders. At 24 h after the initial plating, the cultures were examined for the attachment of the blastocysts to the feeder layer. Three to four days after coculture with feeder cells, a prominent ICM surrounded by TE cells was observed (Fig. 2B). This ICM was separated from the TE using a 23G needle and plated on a layer of fresh feeder cells. Within 24 h, the ICM was attached to the new feeder cells. Seven days after the second plating of the ICM, hES-like cells began growing from the ICM clump (Fig. 2C). The newly formed hES-like cells were dissociated using a needle and plated onto fresh feeder cells. Mechanical passaging was applied to propagate the hES cells. The established hES cell line (Fig. 2D) exhibited a vigorous growth rate during a long-term culture and was routinely passaged every 5–7 days. The efficiency of the derivation of the hES cell lines from blastocysts developed from frozen–thawed and fresh embryos is summarized in Table 1.
FIG. 2.
Derivation of a hES cell line from a blastocyst developed from a frozen–thawed embryo. The images show the successful derivation of the Chula2.hES line from a poor-quality intact blastocyst developed from an 18-year- frozen embryo (A). The zona pellucida of the blastocyst was removed, and the entire blastocyst was plated on feeder cells. Three days after plating, an outgrowth of ICM (indicated by the star) surrounded by a TE was observed (B). The ICM was mechanically removed from the TE and plated onto new feeder cells. Seven days after removing the TE, an outgrowth of putative hES-like cells was observed (C), which later generated the stable hES cell line Chula2.hES (D). The morphologies of the colonies and individual Chula2.hES cells on three different feeder cell densities (E). A density of 35,000 cells/cm2 was selected for further propagation. Scale bars=100 μm, hES, human embryonic stem; TE, trophectoderm.
Table 1.
Comparison of the Efficiency of Human Embryonic Stem Cell Derivation Using Blastocysts Developed from Frozen–Thawed and Fresh Embryos
| Embryos | Blastocysts used n | Attached n (%) | ICM outgrowth n (%) | hES-like cells n (%) | Cell lines n (%) | Name |
|---|---|---|---|---|---|---|
| Frozen–thawed | 5 | 5 (100.0) | 3 (60.0) | 1 (20.0) | 1 (20.0) | Chula2.hES |
| Fresh | 8 | 7 (87.5) | 5 (62.5) | 3 (37.5) | 2 (25.0) | Chula4.hES Chula5.hES |
| Total | 13 | 12 (92.3) | 8 (61.5) | 4 (30.8) | 3 (23.1) |
ICM, inner cell mass; hES, human embryonic stem.
Morphology of hES cells on different densities of feeder cells
The cell line derived from the 18-year-old cryopreserved embryo, Chula2.hES, displayed the typical features of hES cells, including flat colonies, a high nuclear/cytoplasmic ratio with clearly distinguished nucleoli, and a defined border of colonies. At the early stage of Chula2.hES derivation (passage numbers 3–8), three different densities of feeder cells were tested. As shown in Figure 2E, after growing five subsequent cultures on three different feeder cell densities of 1.75, 3.5, and 7.0×104 cells/cm2, we found that Chula2.hES cells cultured on 1.75×104 cells/cm2 tended to differentiate more easily compared with other feeder cell densities. Moreover, the attachment of Chula2.hES cells cultured on 7.0×104 cells/cm2 decreased after subsequent passages. Thus, a feeder cell density of 3.5×104 cells/cm2 was selected for the culture of Chula2.hES cells and the derivation of the other lines, Chula4.hES and Chula5.hES.
Characterization of hES cells
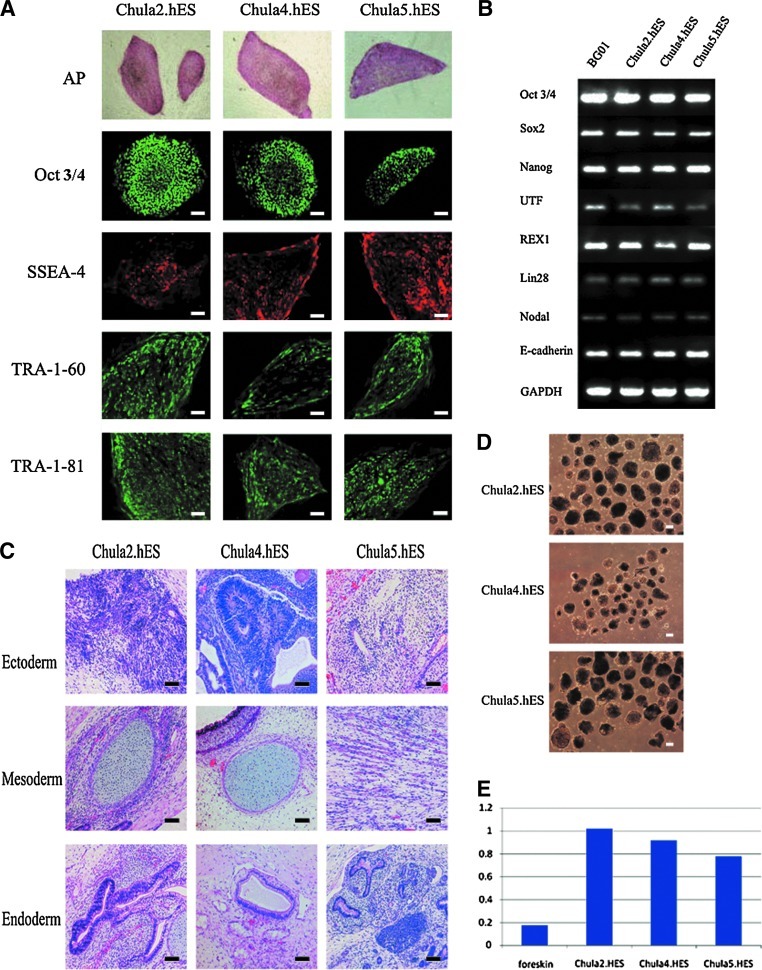
All of the hES cell lines exhibited the AP activity and expressed hES cell markers, including SSEA4, TRA-1-60, TRA-1-81, and Oct3/4 (Fig. 3A). The real-time polymerase chain reaction (RT-PCR) results confirmed that the Chula2.hES, Chula4.hES, and Chula5.hES cells are pluripotent cells, as shown by the expression of pluripotent genes, including OCT3/4, SOX2, NANOG, UTF, REX1, LIN28, NODAL, and E-Cadherin, which was similar to the NIH registered hES cell line BG01 (Fig. 3B). The ability of the hES cell lines to differentiate into embryonic germ layers was demonstrated. To evaluate their in vivo differentiation, Chula2.hES, Chula4.hES, and Chula5.hES cells were allowed to grow and differentiate for 10–12 weeks after injection into nude mice. The histopathological analysis of the teratomas revealed the presence of tissues derived from the ectoderm, endoderm, and mesoderm layers (Fig. 3C). For in vitro differentiation, the hES cell lines were mechanically dissociated into small clumps and cultured in suspension. All of the cell lines were able to form three-dimensional structures, called EBs (Fig. 3D). When the telomerase activity of the hES cell lines was analyzed, all of the cell lines displayed a high telomerase activity, which was higher than the activity of HFFs (Fig. 3E), suggesting immortal replication ability.
FIG. 3.
Characterization of the hES cell lines. The Chula2.hES, Chula4.hES, and Chula5.hES cells expressed pluripotent markers, including AP, Oct3/4, SSEA-4, TRA-1-60, and TRA-1-81, as detected with immunostaining (A), and pluripotent genes, including OCT3/4, SOX2, NANOG, UTF, REX1, LIN28, NODAL, and E-Cadherin, as detected with RT-PCR. GAPDH was used as a housekeeping gene, and the NIH registered hES cell line BG01 was used as the control (B). The in vivo differentiation of the hES cell lines was assessed using a teratoma assay. Ectodermal, mesodermal, and endodermal tissues were found in the teratomas of Chula2.hES, Chula4.hES, and Chula5.hES cells (C). The in vitro differentiation of the hES cell lines was determined using EB formation (D). The immortal replication ability of hES cell lines was measured through the high level of telomerase activity, and the levels were higher than in foreskin fibroblasts, which were used as feeder cells (E). The scale bars in (A) and (C) represent 100 μm, and the scale bar in (D) represents 10 μm. All of the images of AP in A were acquired with a 10× objective phase-contrast microscope. AP, alkaline phosphatase; SSEA, stage-specific embryonic antigen; TRA, tumor recognition antigen; RT-PCR, real-time polymerase chain reaction; GAPDH, glyceralaldehyde-3-phosphate dehydrogenase; EB, embryoid body.
Identification of hES cell lines
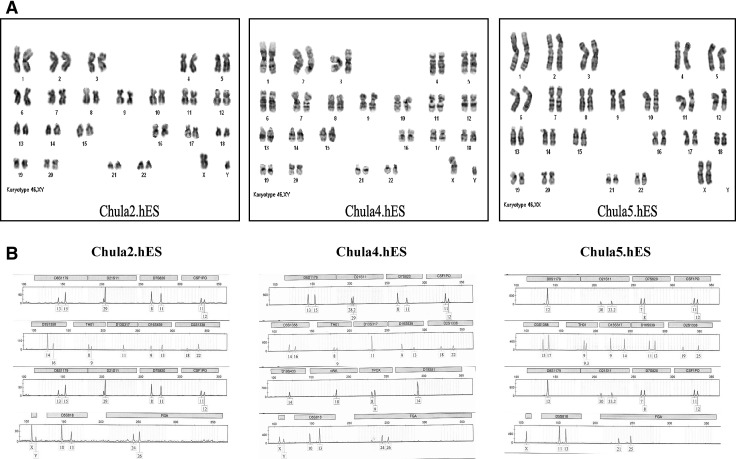
The chromosomal stability of the hES cell lines was assayed through conventional karyotyping. The cells were karyotyped using standard G-banding. At least 15–20 metaphases of each cell line were counted. Karyotyping revealed that the Chula2.hES line, which was derived from an 18-year-old frozen embryo, had a normal male chromosome content of 46 XY. The Chula4.hES and Chula5.hES cells, which were derived from blastocysts developed from fresh embryos, showed normal male and female chromosome contents of 46 XY and 46 XX, respectively (Fig. 4A). A total of 16 STR loci were analyzed for the three newly established hES cell lines, and each cell line showed distinct STR loci (Fig. 4B).
FIG. 4.
(A) The karyotype analysis of three different hES cell lines using the G-banding method and DNA fingerprinting. Chula2.hES, Chula4.hES, and Chula5.hES cells were able to maintain their normal karyotype after long-term culture, as detected with standard G-banding. The Chula2.hES, Chula4.hES, and Chula5.hES cells exhibit 46 XY, 46 XY, and 46 XX, respectively. (B) The Chula2.hES, Chula4.hES, and Chula5.hES cells displayed different DNA fingerprinting, which indicates that the three cell lines were derived from different embryos.
Discussion
In our previous study, we examined different protocols for the derivation of hES cells, but only one hES-like cell line was successfully generated from a frozen–thawed embryo using human skin fibroblasts as feeder cells.12 In the present study, a total of 12 and 11 PN stage embryos, which had been frozen in 1990 and 1991, respectively, were thawed and cultured in a commercial medium without the supplementation of growth factors or cytokines. The survival rate of the frozen–thawed embryos in our study (73.9%) was consistent with that of other studies in which the embryo culture medium was supplemented with an Rho-associated kinase (ROCK) inhibitor Y-27632 or a combination of human recombinant leukemia inhibitory factor (hrLIF) and human basic fibroblast growth factor (hbFGF).9,10 The ROCK inhibitor positively enhances the survival and proliferation of several cell types.13–15 Five embryos survived the freeze–thaw process and developed to the blastocyst stage, which demonstrated that the cryopreserved embryos in this study maintained their developmental ability, even after 17–18 years of cryostorage.
We attempted to isolate hES cells from the low-quality blastocysts derived from the frozen–thawed embryos without modifying the culture conditions. Modifying embryo culture conditions, such as culturing embryos in a modified culture medium composed of the G2.5 medium supplemented with hrLIF, hrbFGF, and human serum albumin9,16 or two additional days of culturing after reaching an early blastocyst stage,16,17 shows a beneficial effect on hES cell derivation. However, poor-quality blastocysts developed from either fresh or frozen–thawed embryos were used for hES cell derivation, and both showed the ability to generate hES cell lines. As our results showed, hES cell lines could be derived from blastocysts developed from frozen–thawed and fresh embryos with no difference in efficiency, and the cryopreservation of the embryo did not affect the efficiency of hES cell derivation.
The hES cells derived for medical purposes should not be exposed to animal components. Although the hES cell lines derived in this study were not completely xeno-free cell lines, the animal components will be removed from the culture conditions in our future studies. However, in this study, we did not add serum to the culture medium and further minimized contact with animal components in the culture conditions by using commercial HFFs as feeder cells and mechanical passage for hES cell propagation. The method for the isolation of the ICM also affects the success of hES cell derivation. To derive xeno-free hES cells, we considered mechanical methods for the isolation of the ICM or whole-blastocyst culture techniques. Both methods have previously been reported for the successful derivation of hES cell lines.18,19 However, due to their poor quality, the blastocysts contained a relatively small ICM; thus, the only method that could be used in this study was whole-blastocyst culture. Three to 4 days after the initial plating of the whole blastocyst onto feeder cells, a dome-shaped ICM was observed. This phenomenon demonstrates that our culture conditions support the growth of an ICM in poor-quality blastocysts. The complication of using this method is that the ICM is cultured in close contact with the TE. Contact with the TE might decrease the success of hES cell derivation by inducing the differentiation of the ICM.20 To avoid losing the pluripotency of the cells in the ICM, we mechanically removed the ICM outgrowth from the TE at approximately 3–4 days after the initial plating, plated the ICM on new feeder cells, and subsequently split the hES-like cells into small pieces for further propagation. Thus, three hES cell lines, Chula2.hES, Chula4.hES, and Chula5.hES, were successfully established. Other groups have also reported the early separation of the ICM from the TE.21,22 Although some reports have suggested that in whole-blastocyst culture, the ICM clump should be separated from the TE outgrowth and split into small pieces at 7–10 days after initial plating,18,20,23 no hES cell line could be derived in our hands using this method. In fact, the ICM clump should comprise a large number of pluripotent cells before the first splitting, as human pluripotent cells preferably grow as a group of cells.
Based on previous reports, the feeder cell density is an important parameter for hES cell culture. The morphology of the hES cell colonies was different when cultured on different feeder cell densities.24 To our knowledge, there are no standard criteria for the feeder cell density for the derivation and maintenance of a new hES cell line. Inzunza et al.23 suggested that when using commercially available HFFs as the feeder, a density of approximately 5.3×104 cells/cm2 could be applied for derivation, and subsequently, a density of 1.23×105 cells/cm2 could be used for hES cell maintenance. Ellerström et al.20 used a density of approximately 6.9×104 HFF cells/cm2 for the derivation of xeno-free hES cells. We tested three different feeder cell densities for the derivation of the Chula2.hES line (passage numbers 3–8). Our results demonstrated that the Chula2.hES line demonstrated the best morphology of undifferentiated cells when cultured on 3.5×104 cells/cm2 compared with their growth on 1.75 and 7.0×104 cells/cm2; thus, this density was selected for the derivation of other cell lines. In contrast to Chula2.hES cells, during the first and second passages of Chula4.hES and Chula5.hES cells, both of the cell lines tended to differentiate easily when cultured on 3.5×104 cells/cm2 of feeder cell density. The spontaneous differentiation of Chula4.hES and Chula5.hES lines can be explained by the low density of feeder cells resulting in insufficient levels of secreted factors, extracellular matrix, several molecules, and cellular contacts that are provided by the feeder cells for the maintenance of hES cells in the undifferentiated state.25 To avoid losing the pluripotent cells, we increased the density of the HFF cells from 3.5 to 5.0×104 cells/cm2, which resulted in the establishment of two stable cell lines, Chula4.hES and Chula5.hES. The 7.0×104 cells/cm2 feeder density was not selected for culturing the hES cell lines because a high feeder cell density would result in a more rapid depletion of nutrients and oxygen within the in vitro culture milieu, which could be detrimental to the growth of hES cells. In addition, a high feeder cell density could also physically hinder the attachment and growth of ES colonies during serial passages.24 Notably, our results also demonstrated that different cell lines might prefer different feeder cell densities.
The Chula2.hES line isolated from frozen–thawed embryos showed a morphology that was typical of hES cells and expressed pluripotent markers, such as AP, SSEA-4, TRA-1-60, TRA-1-81, and Oct3/4, and pluripotent genes, such as OCT3/4, SOX2, NANOG, UTF, REX1, LIN28, NODAL, and E-Cadherin. The Chula2.hES line showed high levels of telomerase activity, indicating that these cells can infinitely proliferate. The Chula2.hES cells differentiated into three embryonic germ layers through the formation of EBs, a three-dimensional structure formed in suspension culture, and teratoma tissue after injection into the testicular capsule of nude mice. The ability of differentiation strongly demonstrated the pluripotency of Chula2.hES cells. The characteristics displayed by Chula2.hES cells were similar to Chula4.hES and Chula5.hES cells, which were derived from blastocysts developed from fresh embryos. The pattern of DNA fingerprinting of all of the hES cell lines was different, confirming that the cell lines are derived from different embryos. Furthermore, all of the hES cell lines maintained their normal karyotype after prolonged culture. Taken together, our results showed that Chula2.hES, Chula4.hES, and Chula5.hES cell lines have the same pluripotency as other existing hES cell lines.7,26,27
In conclusion, this is the first report of the successful derivation of hES cell lines in Thailand. The success of hES cell derivation in the present study demonstrated that an 18-year cryopreservation did not adversely affect the pluripotency of a human embryo, and this embryo maintained its pluripotency similar to fresh embryos, as shown by the success of the derivation of Chula2.hES, Chula4.hES, and Chula5.hES lines.
Acknowledgments
The authors would like to thank Ms. Praewphan Ingrungruenglert for the RT-PCR analysis. This work was supported through funding from the Thai Government Research Budget 2007–2011 (Grant No. GRB_APS_05_54_30_01) and the National Research University Project of CHE (HR1161I).
Author Disclosure Statement
The authors have no conflicts of interest to disclose.
References
- 1.Capalbo A. Rienzi L. Buccheri M, et al. The worldwide frozen embryo reservoir: Methodologies to achieve optimal results. Ann NY Acad Sci. 2011;1221:32–39. doi: 10.1111/j.1749-6632.2010.05931.x. [DOI] [PubMed] [Google Scholar]
- 2.Balaban B. Uman B. Ata B, et al. A randomized controlled study of human day 3 embryo cryopreservation by slow freezing or vitrification: Vitrification is associated with higher survival, metabolism and blastocyst formation. Human Reprod. 2008;23:1976–1982. doi: 10.1093/humrep/den222. [DOI] [PubMed] [Google Scholar]
- 3.Boonkusol D. Gal AB. Bodo S, et al. Gene expression profiles and in vitro development following vitrification of pronuclear and 8-cell stage mouse embryos. Mol Reprod Dev. 2006;73:700–708. doi: 10.1002/mrd.20450. [DOI] [PubMed] [Google Scholar]
- 4.Park SY. Kim EY. Cui XS, et al. Increase in DNA fragmentation and apoptosis-related gene expression in frozen-thawed bovine blastocysts. Zygote. 2011;14:125–131. doi: 10.1017/S0967199406003649. [DOI] [PubMed] [Google Scholar]
- 5.Dowling-Lacey D. Mayer JF. Jones E, et al. Live birth from a frozen-thawed pronuclear stage embryo almost 20 years after its cryopreservation. Fertil Steril. 2011;95:1120e1–1120e3. doi: 10.1016/j.fertnstert.2010.08.056. [DOI] [PubMed] [Google Scholar]
- 6.Riggs R. Mayer J. Dowling-Lacey D, et al. Does storage time influence postthaw survival and pregnancy outcome? An analysis of 11,768 cryopreserved human embryos. Fertil Steril. 2010;93:109–115. doi: 10.1016/j.fertnstert.2008.09.084. [DOI] [PubMed] [Google Scholar]
- 7.Thomson JA. Itskovitz-Eldor J. Shapiro SS, et al. Embryonic stem cell line from human blastocysts. Science. 2008;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 8.Fraga AM. Souza de Araujo ES. Stabellini R, et al. A survey of parameters involved in the establishment of new lines of human embryonic stem cells. Stem Cell Rev Rep. 2011;7:775–781. doi: 10.1007/s12015-011-9250-x. [DOI] [PubMed] [Google Scholar]
- 9.Fan Y. Luo Y. Chen X, et al. A modified culture medium increase blastocyst formation and the efficiency of human embryonic stem cell derivation from poor-quality embryos. J Reprod Dev. 2010;56:533–539. doi: 10.1262/jrd.09-225m. [DOI] [PubMed] [Google Scholar]
- 10.Cortes JL. Sanchez L. Ligero G, et al. Mesenchymal stem cells facilitate the derivation of human embryonic stem cells from cryopreserved poor-quality embryos. Hum Reprod. 2009;24:1844–1851. doi: 10.1093/humrep/dep107. [DOI] [PubMed] [Google Scholar]
- 11.Strom S. Rodriguez-Wallberg K. Holm F, et al. No relationship between embryo morphology and successful derivation of human embryonic stem cell lines. PLoS One. 2010;5:e15329. doi: 10.1371/journal.pone.0015329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pruksananonda K. Rungsiwiwut R. Numchaisrika P, et al. Development of human embryonic stem cells. J Med Assoc Thai. 2009;92:443–450. [PubMed] [Google Scholar]
- 13.Watanabe K. Ueno M. Kamiya D, et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- 14.Koyanagi M. Takahashi J. Arakawa Y, et al. Inhibition of the Rho/ROCK pathway reduces apoptosis during transplantation of embryonic stem cell-derived neural precursors. J Neurosci Res. 2008;86:270–280. doi: 10.1002/jnr.21502. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L. Valdez JM. Zhang B, et al. ROCK inhibitor Y-27632 suppresses dissociation-induced apoptosis of murine prostate stem/progenitor cells and increase their cloning efficiency. PLoS ONE. 2011;6:e18271. doi: 10.1371/journal.pone.0018271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu W. Yin Y. Long X, et al. Derivation and characterization of human embryonic stem cell lines from poor quality embryos. J Genet Genomics. 2009;36:229–239. doi: 10.1016/S1673-8527(08)60110-1. [DOI] [PubMed] [Google Scholar]
- 17.Stojkovic M. Lako M. Stojkovic P, et al. Derivation of human embryonic stem cells from day 8 blastocysts recovered after three-step in vitro culture. Stem Cells. 2004;22:790–797. doi: 10.1634/stemcells.22-5-790. [DOI] [PubMed] [Google Scholar]
- 18.Kim HS. Oh SK. Park YB, et al. Methods for derivation of human embryonic stem cells. Stem Cells. 2005;23:1228–1233. doi: 10.1634/stemcells.2004-0296. [DOI] [PubMed] [Google Scholar]
- 19.Strom S. Inzunza J. Grinnemo KH, et al. Mechanical isolation of the inner cell mass is effective in derivation of new human embryonic stem cell lines. Hum Reprod. 2007;22:3015–3018. doi: 10.1093/humrep/dem335. [DOI] [PubMed] [Google Scholar]
- 20.Ellerström C. Strehl R. Moya K, et al. Derivation of a xeno-free human embryonic stem cell line. Stem Cells. 2006;24:2170–2176. doi: 10.1634/stemcells.2006-0130. [DOI] [PubMed] [Google Scholar]
- 21.Li C. Yang Y. Lu X, et al. Efficiency derivation of Chinese human embryonic stem cell lines from frozen embryos. In Vitro Cell Dev Biol Anim. 2010;46:186–191. doi: 10.1007/s11626-010-9304-4. [DOI] [PubMed] [Google Scholar]
- 22.Revazova ES. Turovets NA. Kochetkova OD, et al. HLA homozygous stem cell lines derived from human parthenogenetic blastocysts. Cloning Stem Cells. 2008;10:11–24. doi: 10.1089/clo.2007.0063. [DOI] [PubMed] [Google Scholar]
- 23.Inzunza J. Gertow K. Stromberg MA, et al. Derivation of human embryonic stem cell lines in serum replacement medium using postnatal human fibroblasts as feeder cells. Stem Cells. 2005;23:544–549. doi: 10.1634/stemcells.2004-0201. [DOI] [PubMed] [Google Scholar]
- 24.Heng BC. Liu H. Cao T. Feeder cell density—a key parameter in human embryonic stem cell culture. In Vitro Cell Dev Biol Anim. 2004;40:255–257. doi: 10.1290/0407052.1. [DOI] [PubMed] [Google Scholar]
- 25.Anisimov SV. Christophersen NS. Correia AS, et al. Identification of molecules derived from human fibroblast feeder cells that support the proliferation of human embryonic stem cells. Cell Mol Bio Lett. 2011;16:79–88. doi: 10.2478/s11658-010-0039-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inamdar MS. Venu P. Sriniyas MS, et al. Derivation and characterization of two sibling human embryonic stem cell lines from discarded grade III embryos. Stem Cell Dev. 2009;18:423–433. doi: 10.1089/scd.2008.0131. [DOI] [PubMed] [Google Scholar]
- 27.Chavez SL. Meneses JJ. Nguyen HN, et al. Characterization of six new human embryonic stem cell lines (HSF7, -8, -9, -10, -12, and -13) derived under minimal-animal component conditions. Stem Cells Dev. 2008;17:535–546. doi: 10.1089/scd.2007.0216. [DOI] [PubMed] [Google Scholar]