Abstract
Delivery inside the cells is essential for practical application of antisense technologies. The hybrid locked nucleic acid (LNA)/DNA CAAGTACTGTTCCACCA (LNA residues are underlined) was labeled by conjugation to Alexa Fluor 488 (fLNA/DNA) and tested to determine its ability to penetrate Escherichia coli cells and reach the cytoplasm. Flow cytometry analysis showed that the fLNA/DNA was associated with 14% of cells from a stationary phase culture, while association with a labeled isosequential oligodeoxynucleotide was negligible. Laser scanning confocal microscopy confirmed that the fLNA/DNA was located inside the cytoplasm.
Key words: drug discovery, drug resistance, gene therapy, nucleic acids, microbiology
Introduction
Antisense-related technologies consist of the utilization of oligonucleotides to interfere with gene expression. Although one drug, fomivirsen, has been approved by the U.S. Food and Drug Administration1 and a number of antisense drugs targeting a variety of diseases are presently in clinical trials,2–4 several challenges, such as toxicity, nonspecific effects, or inability to penetrate the target, continue to slow progress. In particular, for applications against gram-negative bacteria, uptake by the target cell has been a problem because, although a few cases have been reported,5,6 naked antisense compounds do not diffuse well inside most cells.2,7 Numerous strategies have been developed to solve this limitation. The most successful of them being the conjugation of neutral oligonucleotide analogs to cell penetrating peptides8,9 and the addition of cationic groups like N-(6-guanidinohexanoyl)piperazine to the phosphorodiamidate linker.10 Cell penetrating peptide–phosphorodiamidate morpholino oligomers were shown to penetrate bacterial cells and inhibit gene expression by blocking ribosomal function11 or acting as external guide sequences, which are short oligomers that induce RNase P-mediated cleavage of a target RNA by forming a precursor tRNA-like complex.12,13
Hybrid oligomers composed of locked nucleic acids and deoxyribonucleotides (LNA/DNA) are a new generation of chemically modified oligonucleotides that exhibit high-affinity binding to complementary DNA or RNA,14 are highly resistant to nuclease digestion,15,16 and show low toxicity.17,18 These compounds are being tested for treatment of numerous diseases and several clinical trials are under way.18,19 Although utilization of LNA/DNA as antisense antibacterials has been very limited, one such compound was recently shown to be an efficient external guide sequence eliciting RNase P-mediated degradation of aac(6′)-Ib mRNA and concomitantly inducing a significant reduction in the levels of resistance to amikacin in a hyperpermeable Escherichia coli host.15 As in most applications of antisense technologies to bacterial systems, uptake of LNA/DNA by the target cells is one of the major challenges. Attempts to conjugate LNA/DNA to cell-penetrating peptides have been largely unsuccessful, most probably due to their negatively charged nature. In our attempts to find a strategy to induce internalization of LNA/DNA co-oligomers, we found that these compounds can penetrate E. coli more efficiently than isosequential oligodeoxynucleotides.
Materials and Methods
Bacterial strains oligomers
E. coli TOP10 F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74nupG recA1araD139 Δ(ara-leu)7697 galE15 galK16 rpsL(StrR) endA1 λ− was used as host to test internalization of oligomers. Alexa Fluor 488-conjugated CAAGTACTGTTCCACCA (fLNA/DNA; LNA residues are underlined) and Alexa Fluor 488-ODN (fODN), isosequential, were used for quantification of internalization by flow cytometry and by laser scanning confocal microscopy (LSCM).
Quantification and visualization of internalized oligomers
Uptake of fLNA/DNA and fODN was quantified by flow cytometry using cells in a stationary phase that have been incubated in Lennox Luria (L) broth20 for 18 h at 37°C. Cells were washed with phosphate-buffered saline solution and incubated 15–30 min with phosphate-buffered saline and fLNA/DNA or fODN. After washing the cells, the membrane-specific stain, FM5-95, was added to a final concentration of 5 μg/mL and the samples were analyzed by flow cytometry and laser screening confocal microscopy (LSCM). Viability of cells was assessed by treating the cells as described above, but omitting the FM5-95 treatment and exposing the cells to propidium iodide. Flow cytometry data were analyzed using the CellQuest software. Visualization of incorporation of fLNA/DNA by LSCM was done in a Zeiss Pascal or Zeiss Meta confocal microscope (Zeiss) as described before.15 Statistical analysis was carried out performing an unpaired two-tailed t-test with Welch´s correction. p<0.05 was considered statistically significant.
Results and Discussion
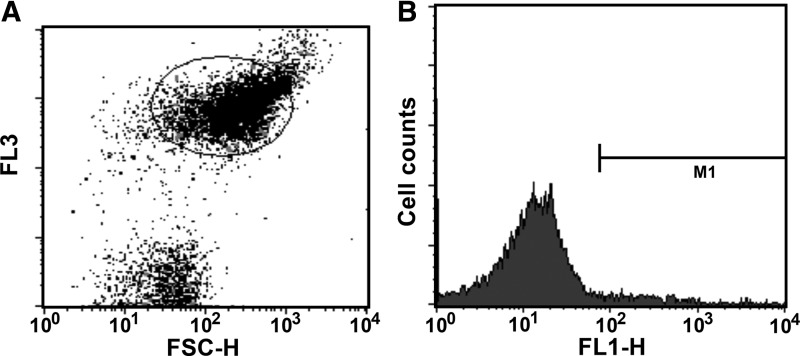
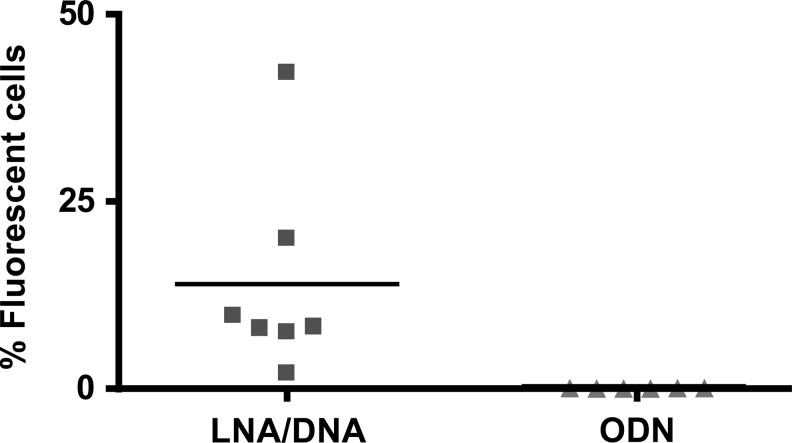
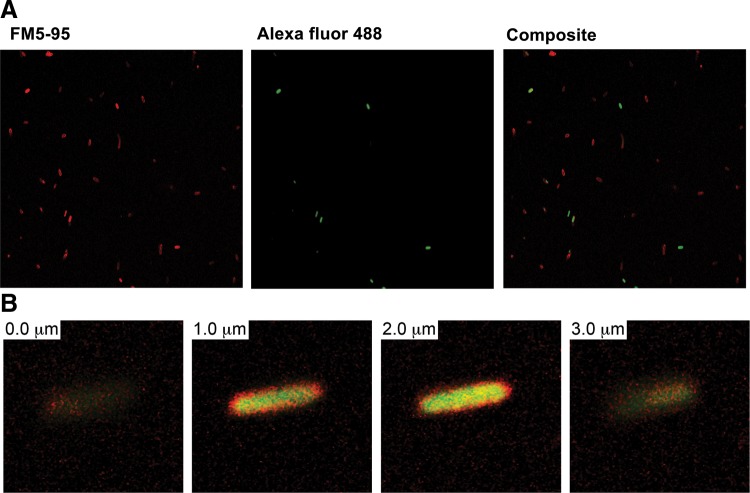
Internalization of nuclease resistant oligonucleotide analogs into bacterial cells is one of the stumbling blocks in the application of antisense technologies to prokaryotic systems. Although significant progress has been made on internalization of peptide nucleic acids and phosphorodiamidate morpholino oligomers by conjugating them to cell penetrating peptides,9,21–25 strategies to induce cell penetration by negatively charged analogs remain elusive. Since LNA/DNA oligonucleotide analogs have been recently shown to be active as external guide sequences,15 they are promising compounds to be used as antibacterials. However, before they are seriously considered as candidates for development of new antimicrobials, a strategy for delivery inside the cells must be found. Interestingly, LNA containing oligonucleotides have been recently shown to be delivered into diverse eukaryotic cells without using transfection agents, in a process called gymnosis.26,27 We tested if an LNA/DNA co-oligomer could be internalized by E. coli cells using two complementary experimental techniques, flow cytometry and LSCM. An LNA/DNA co-oligomer was labeled with the fLNA/DNA and mixed with E. coli cells in the stationary phase. The percentage of cells that took up fLNA/DNA was determined by flow cytometry. Figure 1 shows the dot plot and histogram of a typical experiment, of which seven repeats were carried out. As control, six assays were performed exposing the E. coli cells to fODN. Figure 2 shows that while it is clear that the fODN did not associate with cells, 13.96%±5.12% of them were associated with fLNA/DNA. Although these results show a significant difference of behavior of both oligomers when they get in contact with untreated E. coli cells, they do not provide enough information to know the location of the fLNA/DNA with respect to the cell. To determine if the fLNA/DNA molecules penetrated the cell wall and reach the cytosol or simply remained associated to membranes or stayed within the periplasmic space, E. coli cells were examined by microscopy after being exposed to the labeled compound. Figure 3 shows that the fLNA/DNA localizes within the cytoplasmic compartment in numerous cells. To discard the possibility that those cells that show internalization of fLNA/DNA are dead, and therefore more permeable to the entry of the oligomer, a control experiment was carried out exposing the cells to propidium iodide, a membrane impermeant dye that is excluded from viable cells, but reaches and intercalates into the DNA if cells are dead. The results showed that no more than 2% of the cells were dead in each culture.
FIG. 1.
Flow cytometry of fLNA/DNA cellular uptake. (A) Dot plot representing FSC-H (forward scattered) versus FL3. The inner circle represents the cells analyzed. (B) Histogram showing cell counters versus fluorescence from fLNA/DNA (FL1-H). M1 area represents percentage of cells containing Alexa Fluor 488.
FIG. 2.
Association of Escherichia coli cells with fLNA/DNA or fODN. The percentage of cells associated with fluorescence is compared for the fLNA/DNA (▪) and fODN (▴). The averages, shown by horizontal lines were fLNA/DNA, 13.96±5.12, fODN, 0.05±0.01. The p value is 0.0349, p<0.05 is considered significant.
FIG. 3.
Internalization of fLNA/DNA. (A) Cells in the stationary phase were incubated with fLNA/DNA as described in Materials and Methods, washed with phosphate-buffered saline, stained with FM5-95, and analyzed by laser scanning confocal microscopy. (B) Z-stack from a typical cell with internalized fLNA/DNA.
The results shown here indicate that hybrid LNA/DNA compounds are good candidates as antisense antimicrobials. However, strategies to increase the efficiency of delivery inside the cells must be developed. Furthermore, future research must address questions like the role played by the length and configuration of the LNA/DNAs in efficiency of internalization as well as the physiologic state of the recipient cells.
Acknowledgments
Financial support for this work was provided by Public Health Service grant 2R15AI047115 from the National Institutes of Health (to M.E.T.), PIP grant 11420100100152 from CONICET (to M.S.R.), and UBACyT grants X-240 and X-997 from the University of Buenos Aires. G.M.T. and C.D.S. were supported by CONICET. A.J.S.B. was supported by fellowships from the American Society for Microbiology (International Fellowship for Latin America) and CONICET. M.S.R. and A.Z. are career investigators of CONICET.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Marwick C. First “antisense” drug will treat CMV retinitis. JAMA. 1998;280:871. [PubMed] [Google Scholar]
- 2.Kole R. Krainer AR. Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nature Rev Drug Discov. 2012;11:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurreck J. Antisense technologies. Improvement through novel chemical modifications. Eur J Biochem. 2003;270:1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- 4.Rasmussen LC. Sperling-Petersen HU. Mortensen KK. Hitting bacteria at the heart of the central dogma: sequence-specific inhibition. Microb Cell Fact. 2007;6:24. doi: 10.1186/1475-2859-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chrisey LA. Walz SE. Pazirandeh M, et al. Internalization of oligodeoxyribonucleotides by Vibrio parahaemolyticus. Antisense Res Dev. 1993;3:367–381. doi: 10.1089/ard.1993.3.367. [DOI] [PubMed] [Google Scholar]
- 6.Harth G. Zamecnik PC. Tabatadze D, et al. Hairpin extensions enhance the efficacy of mycolyl transferase-specific antisense oligonucleotides targeting Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2007;104:7199–7204. doi: 10.1073/pnas.0701725104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Good L. Sandberg R. Larsson O, et al. Antisense PNA effects in Escherichia coli are limited by the outer-membrane LPS layer. Microbiology. 2000;146:2665–2670. doi: 10.1099/00221287-146-10-2665. [DOI] [PubMed] [Google Scholar]
- 8.Mellbye BL. Puckett SE. Tilley LD, et al. Variations in amino acid composition of antisense peptide-phosphorodiamidate morpholino oligomer affect potency against Escherichia coli in vitro and in vivo. Antimicrob Agents Chemother. 2009;53:525–530. doi: 10.1128/AAC.00917-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikravesh A. Dryselius R. Faridani OR, et al. Antisense PNA accumulates in Escherichia coli and mediates a long post-antibiotic effect. Mol Ther. 2007;15:1537–1542. doi: 10.1038/sj.mt.6300209. [DOI] [PubMed] [Google Scholar]
- 10.Mellbye BL. Weller DD. Hassinger JN, et al. Cationic phosphorodiamidate morpholino oligomers efficiently prevent growth of Escherichia coli in vitro and in vivo. J Antimicrob Chemother. 2010;65:98–106. doi: 10.1093/jac/dkp392. [DOI] [PubMed] [Google Scholar]
- 11.Deere J. Iversen P. Geller BL. Antisense phosphorodiamidate morpholino oligomer length and target position effects on gene-specific inhibition in Escherichia coli. Antimicrob Agents Chemother. 2005;49:249–255. doi: 10.1128/AAC.49.1.249-255.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lundblad EW. Altman S. Inhibition of gene expression by RNase P. N Biotechnol. 2010;27:212–221. doi: 10.1016/j.nbt.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Shen N. Ko JH. Xiao G, et al. Inactivation of expression of several genes in a variety of bacterial species by EGS technology. Proc Natl Acad Sci USA. 2009;106:8163–8168. doi: 10.1073/pnas.0903491106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar R. Singh SK. Koshkin AA, et al. The first analogues of LNA (locked nucleic acids): phosphorothioate-LNA and 2′-thio-LNA. Bioorg Med Chem Lett. 1998;8:2219–2222. doi: 10.1016/s0960-894x(98)00366-7. [DOI] [PubMed] [Google Scholar]
- 15.Soler Bistue AJ. Martin FA. Vozza N, et al. Inhibition of aac(6')-Ib-mediated amikacin resistance by nuclease-resistant external guide sequences in bacteria. Proc Natl Acad Sci USA. 2009;106:13230–13235. doi: 10.1073/pnas.0906529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wahlestedt C. Salmi P. Good L, et al. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc Natl Acad Sci USA. 2000;97:5633–5638. doi: 10.1073/pnas.97.10.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crinelli R. Bianchi M. Gentilini L, et al. Design and characterization of decoy oligonucleotides containing locked nucleic acids. Nucleic Acids Res. 2002;30:2435–2443. doi: 10.1093/nar/30.11.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chabot S. Orio J. Castanier R, et al. LNA-based oligonucleotide electrotransfer for miRNA inhibition. Mol Ther. 2012;20:1590–1598. doi: 10.1038/mt.2012.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun Z. Xiang W. Guo Y, et al. Inhibition of hepatitis B virus (HBV) by LNA-mediated nuclear interference with HBV DNA transcription. Biochem Biophys Res Commun. 2011;409:430–435. doi: 10.1016/j.bbrc.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J. Fritsch EF. Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- 21.Eriksson M. Nielsen PE. Good L. Cell permeabilization and uptake of antisense peptide-peptide nucleic acid (PNA) into Escherichia coli. J Biol Chem. 2002;277:7144–7147. doi: 10.1074/jbc.M106624200. [DOI] [PubMed] [Google Scholar]
- 22.Tilley LD. Hine OS. Kellogg JA, et al. Gene-specific effects of antisense phosphorodiamidate morpholino oligomer-peptide conjugates on Escherichia coli and Salmonella enterica serovar typhimurium in pure culture and in tissue culture. Antimicrob Agents Chemother. 2006;50:2789–2796. doi: 10.1128/AAC.01286-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wesolowski D. Tae HS. Gandotra N, et al. Basic peptide-morpholino oligomer conjugate that is very effective in killing bacteria by gene-specific and nonspecific modes. Proc Natl Acad Sci USA. 2011;108:16582–16587. doi: 10.1073/pnas.1112561108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geller BL. Antibacterial antisense. Curr Opin Mol Ther. 2005;7:109–113. [PubMed] [Google Scholar]
- 25.Good L. Stach JE. Synthetic RNA silencing in bacteria - antimicrobial discovery and resistance breaking. Front Microbiol. 2011;2:185. doi: 10.3389/fmicb.2011.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torres AG. Threlfall RN. Gait MJ. Potent and sustained cellular inhibition of miR-122 by lysine-derivatized peptide nucleic acids (PNA) and phosphorothioate locked nucleic acid (LNA)/2′-O-methyl (OMe) mixmer anti-miRs in the absence of transfection agents. Artif DNA PNA XNA. 2011;2:71–78. doi: 10.4161/adna.17731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stein CA. Hansen JB. Lai J, et al. Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents. Nucleic Acids Res. 2010;38:e3. doi: 10.1093/nar/gkp841. [DOI] [PMC free article] [PubMed] [Google Scholar]