Abstract
The liver is a highly resilient organ that possesses enormous regenerative capacity. This is mediated mainly through the most abundant cell type found in the liver, the hepatocyte. When the regenerative capacity of the hepatocyte is compromised, during chronic or acute liver injury, hepatic progenitor cells (HPCs) are activated to replace the damaged tissue. The HPC resides in a laminin-rich environment; as HPCs differentiate toward a hepatic or biliary fate, the extracellular matrix (ECM) composition changes, influencing cell behavior. To assess the impact that the biological ECM and the synthetic ECM have on the maintenance of hepatic stem cell gene expression, a murine hepatic stem cell line was employed. We demonstrate that hepatic stem cell gene expression could be maintained using a biological or synthetic substratum, but not on plastic alone.
Key words: biological, differentiation, liver, matrix, stem cell, synthetic
Introduction
The liver has an exceptional regenerative capacity. After acute liver injury, hepatocytes undergo mitosis, leading to regeneration of the liver mass followed by functional restoration.1 However, during chronic or severe acute liver injury, the regenerative capacity of resident hepatocytes is lost. In this situation, hepatic progenitor cells (HPCs) are activated to replace the damaged tissue. HPC populations are located at the most peripheral branches of the biliary tree, the Canals of Hering,2 in a laminin-rich environment. The laminin-rich niche regulates the stem cell behavior, maintaining the balance between quiescence, proliferation, and differentiation required for tissue homeostasis and response to injury.3
HPCs first identified in rodents are termed oval cells (OCs) due to their ovoid shape.4 Previous studies have shown that OCs are bipotent and capable of differentiating toward either hepatocytes or cholangiocytes, reminiscent of hepatoblast differentiation during fetal liver development.5–7 OCs express adult hepatocyte markers such as albumin, cytokeratin 8 (CK8), CK18, alpha-1-antitrypsin, and hepatocyte nuclear factor 4 (HNF4). They also stain positively for biliary markers, including cytokeratins CK 7, CK 14, and CK 19, the OC marker OV-6, and alpha-fetoprotein (AFP).8 Additionally, OCs express adult hematopoietic cell markers such as c-kit and Thy-1.
OCs are a plastic cell population capable of self-renewal. These attributes make them an attractive cell population for use in cell-based therapy and the development of in vitro models. The development of predictive cell-based models is important in medicine, especially as OCs have been implicated in the formation of hepatocellular carcinoma. Therefore, a better understanding of OC malignant transformation, through in vitro modeling, may serve to identify more efficacious chemotherapeutic agents.9,10
Essential to the development of cell-based therapies and predictive models is the robust delivery of stable cell populations that can be scaled up cost-effectively. The extracellular matrix (ECM) plays an essential part in this process. Therefore, the purpose of our study was to assess the suitability of a synthetic, inexpensive to manufacture, and totally defined surface for progenitor cell expansion.11 To test this, we employed a bipotent murine stem cell line (bipotent mouse oval cell line [BMOL])12 and compared the effects of different cellular substrata on stem cell gene expression.
Methods
Cell culture and seeding onto different matrices
BMOL was cultured on plastic (Corning) in the William's E medium (Gibco) supplemented with 2% fetal calf serum (Biosera) and 1% penicillin/streptomycin (Gibco). Media were changed every second day. Cells were passaged using 0.05% trypsin (Gibco). For each experiment, 1×106 cells were plated onto BD cell culture plates coated with or without laminin (BD Biosciences). For experiments with the polymer, 5×105 cells were plated onto coverslips coated with synthetic polyurethane (PU134).11 Cells were harvested by using trypsin for analysis 96 h postseeding.
RNA isolation and reverse transcription–polymerase chain reaction
About 1 μg total RNA from the different BMOL cell populations was prepared using Qiagen Kit (Qiagen) and reverse-transcribed following the manufacturer's instructions. Template cDNA, corresponding to 15 ng of RNA, was added to each polymerase chain reaction (PCR) and amplified using the QuantiFast SYBR assay (Qiagen) and QuantiTect (Qiagen). Genes used in this study are listed in Table 1. Each sample was run in triplicate for each candidate gene. Data were analyzed using LightCycler 480 Software (Roche), where expression levels of each gene of interest were normalized to peptidylprolyl isomerase A (PPIA). The two-tailed unpaired Student t-test was performed to test statistical significance, using Prism software (GraphPad Software).
Table 1.
Qiagen Primers Used for Quantitative Polymerase Chain Reaction
| Primer | Catalog number |
|---|---|
| HNF4α | QT00144739 |
| HNF1α | QT00170975 |
| HNF1β | QT00103320 |
| HNF6 | QT00297815 |
| SOX-9 | QT00163765 |
| PPIA | QT00247709 |
HNF, hepatocyte nuclear factor; PPIA, peptidylprolyl isomerase A.
Results
The effect of the cellular substrata on BMOL morphology and a stem cell marker expression
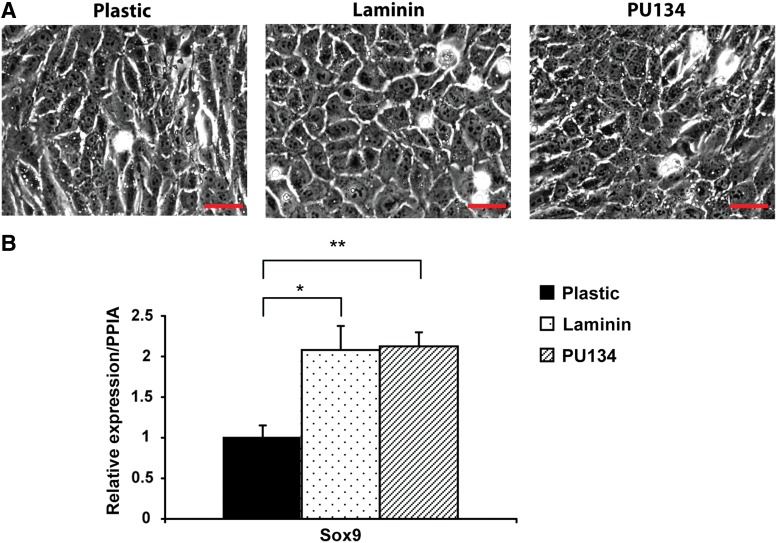
BMOLs were re-plated on different substrata: plastic, laminin, and PU134.11 By phase-contrast microscopy, we observed that BMOLs maintained on either laminin or PU134 showed the typical cobblestone-like morphology, while maintenance on plastic induced the appearance of an elongated cellular morphology (Fig. 1A). These observations indicated that maintenance of the cells on different substrata was likely affecting stem cell gene expression. As a readout of the stem cell identity, we employed a well-established stem cell marker, SOX-9.13,14 SOX-9 expression was assessed by quantitative PCR and increased significantly in cells maintained on PU134 (2.1-fold induction, p<0.01) and laminin (1.8-fold induction, p<0.05) when compared with cells maintained on plastic (Fig. 1B). These results also demonstrated that the PU134 substrate could support hepatic stem cell marker expression in a manner similar to laminin (Fig. 1B).
FIG. 1.
The effect of the cellular substrata on the bipotent mouse oval cell line (BMOL) morphology and stem cell marker. (A) Phase-contrast microscopy images representative of the BMOL morphology on different matrices at 20× magnification. The images were captured using an Axiocam MRC (Zeiss) connected to an Axiovert 200 inverted microscope (Zeiss). (B) Quantitative polymerase chain reaction of the stem cell marker SOX-9 in BMOL cultured on different surfaces, showing a significant upregulation of SOX-9 expression on cells maintained on laminin and PU134 in comparison with plastic. Relative expression refers to folds of induction of the gene compared with the endogenous gene control, peptidylprolyl isomerase A (PPIA). Data are expressed as mean±standard deviation (s.d.), *p<0.05, **p<0.01, and n=3. Scale bar represents 50 μm.
The effect of the cellular substrata on BMOL bipotential gene expression
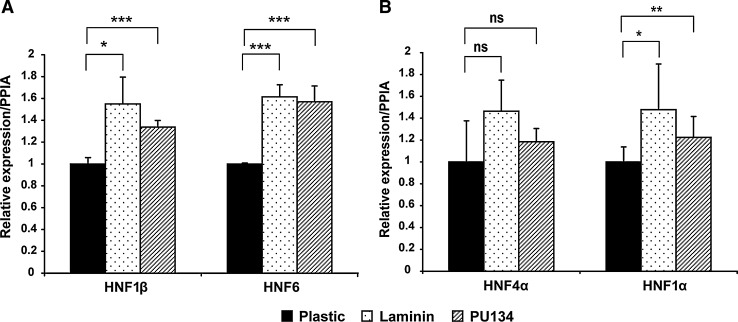
Maintaining biliary and hepatic gene expression at appropriate levels is critical within bipotential progenitor cells. To study this relationship in more detail, BMOLs were re-plated on plastic, laminin, and PU134. It has been reported that the HNF6 and HNF1β are important in biliary development,15 while HNF1α and HNF4α are important in hepatocyte specification.16,17 We assessed biliary and hepatic marker expression by quantitative PCR. We observed significant upregulation in HNF6 expression on PU134 and laminin (both 1.6-fold induction, p<0.001), and in HNF1β expression (1.4-fold, p<0.001; and 1.6-fold, p<0.05, respectively) when compared to the plastic controls (Fig. 2A). The hepatic marker HNF1α was also upregulated on laminin and PU134 (1.5-fold induction, p<0.05; and 1.2-fold induction, p<0.01, respectively). In contrast, HNF4α expression was similar between the different surfaces tested (Fig. 2B).
FIG. 2.
The effect of the cellular substrata on BMOL bipotential gene expression. Quantitative polymerase chain reaction of biliary and hepatic marker expression in BMOL cultured on plastic, laminin, and PU134. (A) Expression of the biliary markers, hepatocyte nuclear factors HNF1β and HNF6, on cells maintained on different matrices. Both HNF1β and HNF6 exhibited upregulation on laminin and PU134. (B) Expression of hepatic marker factors, HNF4α and HNF1α, on cells maintained on different matrices. HNF1α expression showed a significant upregulation on laminin and PU134. Nonsignificant (ns) change was observed for HNF4α. Relative expression refers to folds of induction of the gene compared with the endogenous gene control, PPIA. Data are expressed as mean±s.d., *p<0.05, **p<0.01, ***p<0.001, and n=3.
Discussion
This study explores the maintenance of bipotent stem cell gene expression on different ECMs. The ECM is known to play a crucial role in multiple biological processes, including liver progenitor cell self-renewal and differentiation.3 The results from our studies demonstrated that not only biological matrices have profound effects on HPCs. We employed a synthetic matrix, PU134, which is known to promote hepatocyte differentiation and long-term stable drug-inducible function.11 However, the ability of PU134 to support HPC gene expression had not been studied.
The bipotent murine oval cell line (BMOL) was employed throughout our studies. BMOLs were maintained on laminin, plastic, and PU134 surfaces. Four days post-replating, stem cell identity was determined by SOX-9 expression. SOX-9 gene expression was maintained on laminin and PU134, but not plastic (Fig. 1). In addition to stem cell marker expression, we also examined BMOL bipotential gene expression over the same time course. In contrast to plastic surfaces, BMOL cells maintained on either laminin or PU134 displayed maintenance of biliary and hepatic gene expression (Fig. 2).
The data presented demonstrates that both the laminin and PU134 supported BMOL stem cell and bipotential gene expression in vitro. These studies highlight the potential of synthetic matrices in cell biology and will likely improve cell culture definition, stability, scale-up, and reproducibility from a number of sources including: pluripotent stem cells and their derivatives; primary adult and fetal stem cells; and different somatic cell populations. This is essential if cell-based technologies are to be adopted by researchers and improve our understanding of the underlying biology in stem cell expansion, stem cell differentiation, human disease, human drug toxicity, and malignant transformation.
Acknowledgments
B.L.-V. was funded by an MRC-PhD studentship. S.P. and F.K. were supported by the EPSRC. J.P.I. was supported by an MRC Programme grant. D.C.H. was funded by an RCUK fellowship.
Author Disclosure Statement
D.C.H. is CSO of, Director of, and a shareholder in FibromEd Ltd. J.P.I. and M.B. are shareholders in FibromEd Ltd.
References
- 1.Fausto N. Campbell JS. Riehle KJ. Liver regeneration. Hepatology. 2006;43(2 Suppl 1):S45–S53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- 2.Paku S. Schnur J. Nagy P, et al. Origin and structural evolution of the earlier proliferative oval cell in rat liver. Am J Pathol. 2001;158:1313–1323. doi: 10.1016/S0002-9440(10)64082-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lorenzini S. Bird TG. Boulter L, et al. Characterisation of a stereotypical cellular and extracellular adult liver progenitor cell niche in rodents and diseased human liver. Gut. 2010;59:645–654. doi: 10.1136/gut.2009.182345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farber E. Similarities in the sequence of early histologic changes induced in the livers of rats by ethionine, 2-acetylaminofluorene and 30-methyl-4-dimethylaminoazobenzene. Cancer Res. 1956;16:142–148. [PubMed] [Google Scholar]
- 5.Vessey CJ. de la Hall PM. Hepatic stem cells: A review. Pathology. 2001;33:130–141. [PubMed] [Google Scholar]
- 6.Crosby HA. Nijjar SS. de Goyet Jde V, et al. Progenitor cells of the biliary epithelial cell lineage. Semin Cell Dev Biol. 2002;13:397–403. doi: 10.1016/s108495210200126x. [DOI] [PubMed] [Google Scholar]
- 7.Forbes S. Vig P. Poulsom R, et al. Hepatic stem cells. J Pathol. 2002;197:510–518. doi: 10.1002/path.1163. [DOI] [PubMed] [Google Scholar]
- 8.Bird TG. Lorenzini S. Forbes SJ. Activation of stem cells in hepatic diseases. Cell Tissue Res. 2008;331:283–300. doi: 10.1007/s00441-007-0542-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsia C. Evarts RP. Nakatsukasa H, et al. Occurrence of oval-type cells in hepatitis B virus-associated human hepatocarcinogenesis. Hepatology. 1992;16:1327–1333. doi: 10.1002/hep.1840160604. [DOI] [PubMed] [Google Scholar]
- 10.Libbrecht L. De Vos R. Cassiman D, et al. Hepatic progenitor cells in hepatocellular adenomas. Am J Surg Pathol. 2001;25:1388–1396. doi: 10.1097/00000478-200111000-00006. [DOI] [PubMed] [Google Scholar]
- 11.Hay DC. Pernagallo S. Diaz-Mochon JJ, et al. Unbiased screening of polymer libraries to define novel substrates for functional hepatocytes with inducible drug metabolism. Stem Cell Res. 2011;6:92–102. doi: 10.1016/j.scr.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 12.Tirnitz-Parker JE. Tonkin JN. Knight B, et al. Isolation, culture and immortalisation of hepatic oval cells from adult mice fed a choline-deficient, ethionine-supplemented diet. Int J Biochem Cell Biol. 2007;39:2226–2239. doi: 10.1016/j.biocel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Mori-Akiyama Y. Akiyama H. Rowitch DH, et al. Sox9 is required for determination of the chondrogenic cell lineage in the cranial neural crest. Proc Natl Acad Sci USA. 2003;100:9360–9365. doi: 10.1073/pnas.1631288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mori-Akiyama Y. van den Born M. van Es JH, et al. SOX9 is required for the differentiation of paneth cells in the intestinal epithelium. Gastroenterology. 2007;133:539–546. doi: 10.1053/j.gastro.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 15.Clotman F. Lannoy VJ. Reber M, et al. The one-cut transcription factor HNF6 is required for normal development of the biliary tract. Development. 2002;129:1819–1828. doi: 10.1242/dev.129.8.1819. [DOI] [PubMed] [Google Scholar]
- 16.Hakoda T. Yamamoto K. Terada R, et al. A crucial role of hepatocyte nuclear factor-4 expression in the differentiation of human ductular hepatocytes. Lab Invest. 2003;83:1395–1402. doi: 10.1097/01.lab.0000092229.93203.57. [DOI] [PubMed] [Google Scholar]
- 17.Lokmane L. Haumaitre C. Garcia-Villalba P, et al. Crucial role of vHNF1 in vertebrate hepatic specification. Development. 2008;135:2777–2786. doi: 10.1242/dev.023010. [DOI] [PubMed] [Google Scholar]