Abstract
Stage-specific embryonic antigen 3 (SSEA3) is a glycosphingolipid that has previously been used to identify cells with stem cell-like, multipotent, and pluripotent characteristics. A rare subpopulation of SSEA3-expressing cells exists in the dermis of adult human skin. These SSEA3-expressing cells undergo a significant increase in cell number in response to injury, suggesting a possible role in regeneration. These SSEA3-expressing regeneration-associated (SERA) cells were derived through primary cell culture, purified by fluorescence-activated cell sorting (FACS), and characterized. Longer in vitro culture of the primary skin cells led to lower SSEA3 expression stability after FACS-based purification, suggesting that the current culture conditions may need to be optimized to permit the large-scale expansion of SERA cells. The SERA cells demonstrated a global transcriptional state that was most similar to bone marrow- and fat-derived mesenchymal stem cells (MSCs), and the highest expressing SSEA3-expressing cells co-expressed CD105 (clone 35). However, while a rare population of MSCs was observed in primary human skin cell cultures that could differentiate into adipocytes, osteoblasts, or chondrocytes, SERA cells did not possess this differentiation capacity, suggesting that there are at least two different rare subpopulations in adult human skin primary cultures. The identification, efficient purification, and large-scale expansion of these rare subpopulations (SERA cells and MSCs) from heterogeneous adult human skin primary cell cultures may have applications for future patient-specific cellular therapies.
Key words: human dermal MSCs, regeneration, SERA cells, stage-specific embryonic antigen 3, tissue engineering
Introduction
Adult stem cell (ASC)-like subpopulations have been well characterized in the epidermis of mammalian skin.1–3 However, ASC-like subpopulations in the dermis of human skin are less well characterized. Previous research has demonstrated that the CD271 cell-surface biomarker can be used to identify and isolate a subpopulation of multipotent stem cells from primary human adherent dermal (PHAD) cells,4 although it is unclear whether these human skin multipotent stem cells play a direct role in dermal tissue regeneration. Stage-specific embryonic antigen 3 (SSEA3) is a cell-surface glycosphingolipid that plays an important role in identifying cells with either cancer-associated5–7 or stem cell/undifferentiated cell characteristics,8–14 such as human embryonal carcinoma cells,9 undifferentiated human embryo inner cell mass cells,15 human embryonic stem (ES) cells,16 and human-induced pluripotent stem cells.17 The SSEA3 epitope was discovered by Solter and colleagues in 1982 as a biomarker expressed in oocytes, embryonic cells, and cancer cells.8 In this initial report, Solter noted that SSEA3 was also expressed in erythrocytes (red blood cells), providing the first evidence that SSEA3 expression was not an exclusive cancer or stem/undifferentiated cell marker and may serve other functions in the body in a spatial, niche, and tissue-dependent manner. Reports of SSEA3-expressing cells within human tissues such as kidney,18 brain19, and skin10 have raised the possibility that the cell-surface expression of the SSEA3 epitope may facilitate the identification and isolation of ASC-like and/or undifferentiated cell subpopulations from adult human tissues. While SSEA3 has already been used to identify human skin-derived cells that are easier to reprogram to pluripotency10,13 (suggesting a less differentiated state), associated with a multi-lineage differentiation potential,11 (suggesting a multipotent or possibly pluripotent state) and correlated to the occlusion of human blood vessels20 (suggesting a possible regeneration-associated state for blood vessels), SSEA3 has not been investigated as a cell-surface biomarker facilitating the identification and isolation of human skin cells directly associated with tissue regeneration after dermal injury. The purpose of this study was to investigate whether the SSEA3 biomarker could be used to facilitate the identification and isolation of regeneration-associated cells from adult human dermis. This study is the first, to our knowledge, that presents evidence directly correlating the expression of the SSEA3 glycosphingolipid on the surface of human skin cells to significantly increased proliferation in response to a dermal tissue injury/regenerative response.
Materials and Methods
Ethics statement
Written approval for human skin biopsy procedures, human fibroblast derivation, culture, and experimental use was obtained from the Stanford University Institutional Review Board (IRB), the Stanford University Stem Cell Research Oversight (SCRO) committee, and written informed consent was obtained from each individual participant. The biopsy material used in this study was obtained and initially analyzed at Stanford University, as previously described,10 and transferred to the University of California Los Angeles (UCLA) through a material transfer agreement. Written approvals for the experiments performed in this study were obtained from the UCLA Institute Biosafety Committee (IBC) and the UCLA SCRO Committee.
In vitro tissue injury/regeneration-associated assay
Two to six biological replicates from each of three skin biopsy donors (with each biological replicate representing a dermal tissue biopsy fragment of at least 1 mm3 in volume) were analyzed for this study. The analysis of biopsy fragment cellular subpopulations was performed through cryosectioning and immunohistochemical staining, as previously described.4,10 Briefly, human skin biopsy fragments were placed into an optimal cutting temperature (OCT) compound tissue mold, frozen to −80°C, cut into sections ∼5 μm thick in a cryostat at −20°C, and analyzed via immunohistochemistry for the SSEA3 antibody. In vitro biopsy adhesion, cell migration, and extended primary cell culture (1 month) in regular cell culture media—consisting of Dulbecco's modified Eagle medium nutrient mixture F-12 (DMEM/F12) supplemented with 10% fetal bovine serum (FBS; Invitrogen), 1% MEM nonessential amino acids, 2 mM GlutaMAX™, and 100 IU/mL penicillin-streptomycin (Invitrogen)—were used as an in vitro assay for tissue injury. After 1 month of primary cell culture, biopsy fragments were manually removed from tissue culture, cryosectioned, and analyzed via immunohistochemistry.
In vitro culture of primary human skin cells
The human skin-derived (HUF1) primary cell line used in this study was obtained from a 4 mm adult skin punch biopsy, as previously described.10 All human biopsy-derived cells were cultured in DMEM/F12/FBS culture media (as previously described). The culture media was changed every 2 days. The cells were allowed to expand to 80%–90% confluency before passaging with 0.05% trypsin-EDTA (Invitrogen) and replating at a 1:3 ratio. A large bank of early-passage HUF1 cells was cryopreserved in culture media supplemented with 10% dimethyl sulphoxide (DMSO; Fisher). All research adhered to the National Academy of Sciences guidelines.
SSEA3 live cell staining and fluorescence activated cell sorting-based purification
Approximately 100 million PHAD cells per experiment were trypsinized and washed twice with ice-cold phosphate-buffered saline (PBS)+2% goat serum (PBS-G). The cells were then passed through a 40 μm filter to remove clumps. After the washes, the cells were resuspended in 0.5 mL (per 10 million cells) of ice-cold PBS-G containing 1:100 SSEA3 antibody (Millipore, mab4303) and incubated for 45 min in the dark at 4°C with gentle rocking. After primary antibody binding, the cells were washed thrice with ice-cold PBS-G, resuspended in 1 mL ice-cold PBS-G containing 1:200 Alexa 488-conjugated goat anti-rat IgM (Invitrogen, A21212), and incubated for 45 min in the dark at 4°C with gentle rocking. After secondary antibody binding, the cells were washed thrice with ice-cold PBS-G, resuspended in 2 mL of ice-cold PBS-G, passed through a 40 μm filter, and immediately analyzed and sorted on an FACSAria cell sorter (BD Biosciences). For double-staining analysis, 1:100 rat anti-human SSEA3 (Millipore, mab4303) and 1:50 mouse anti-human CD105 clone 35 (BD Biosciences, 611314) primary antibodies were used in conjunction with 1:200 DyLight 649-conjugated goat anti-rat (Jackson ImmunoResearch, 112-496-075) and 1:200 FITC-conjugated goat anti-mouse (Invitrogen, M30101) secondary antibodies, respectively. The same incubation times and parameters used for single-staining analysis were also used for double-staining analysis. 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen, D1306) was included at a 1:100 ratio for detection and removal of dead cells. In every fluorescence activated cell sorting (FACS) analysis and purification performed, a control sample of cells (exposed only to secondary antibody) was used to exclude the possibility of nonspecific binding and/or autofluorescence. Data were analyzed, DAPI-stained dead-cell exclusion and doublet-exclusion gating were performed, and viable single-cell subpopulations were sorted using BD FACSDiva Software (BD Biosciences). A bank of HUF1 cells was used to establish SSEA3NEGATIVE, SSEA3LOW, and SSEA3HIGH threshold levels for consistent categorization (Fig. 4C) throughout the experiments performed in this study. For sorting purposes, the top 10% of cells, with the highest level of SSEA3 expression, were sorted as representatives for the SSEA3HIGH subpopulation, and the bottom 10% of cells, with the lowest level of SSEA3 expression, were sorted as representatives for the SSEA3NEGATIVE subpopulation. The sorted subpopulations were allowed to adhere for 24 h before subsequent immunofluorescence, FACS analysis, transcriptional characterization, and mesenchymal stem cell (MSC)-differentiation studies were performed.
FIG. 4.
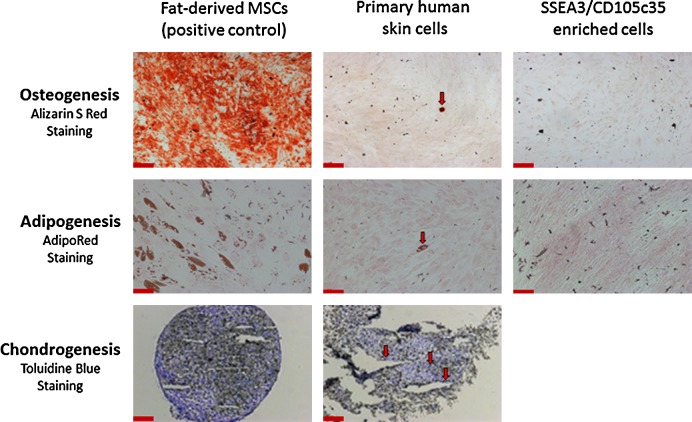
A rare population of mesenchymal stem cells (MSCs) exists in primary human adherent dermal (PHAD) cell cultures. Adipose tissue (fat)-derived MSCs, unsorted PHAD cells, and FACS purified SSEA3/CD105 (clone 35) expressing cells were investigated for osteoblast, adipocyte, and chondrocyte differentiation and analyzed using the appropriate staining. Specifically, alizarin S red for osteoblasts, AdipoRed for adipocytes, and toluidine blue O for chondrocytes. There was insufficient material to perform the three-dimensional micromass culture required for chondrogenesis after FACS-based purification. Red arrows indicate the presence of functional MSCs in PHAD cell cultures. Scale bars represent 200 μm.
Global transcriptional meta-analysis
Total RNA was purified from three biological replicates of SSEA3HIGH and SSEA3NEGATIVE FACS-purified populations (24 h postsorting) using the RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. Total RNA was used for U133 plus 2.0 microarray-based global transcriptional analysis using standard Affymetrix protocols (Affymetrix GeneChip Expression Analysis Technical Manual, rev.3. 2001). Affymetrix CEL files for the additional cells and tissues used in this study's meta-analysis were obtained from the Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo). The meta-analysis library included two replicates of adipose tissue-derived mesenchymal stem cells (AT-MSCs; GEO accession numbers GSM501001 and GSM501002 for replicates 1 and 2 respectively), bone marrow-derived mesenchymal stem cells (BM-MSCs; GSM241198 and GSM241200), CD44-expressing epithelial stem cells (GSM379269 and GSM379270), epidermal keratinocytes (GSM173532 and GSM173533), CD200-expressing hair follicle stem cells (GSM538736 and GSM538737), endothelial cells derived from skin microvasculature (GSM700326 and GSM734106) and lymphatic tissues (GSM143717 and GSM143898), skin-derived pericytes (GSM425648 and GSM425649), bone marrow-derived hematopoietic stem cells (GSM747521 and GSM747522), bone marrow-derived mono-nuclear cells (GSM794774 and GSM794775), human ES cells (GSM194307 and GSM194308), subcutaneous adipose tissue (GSM646746 and GSM646747), chondrocytes from cartilage (GSM490979 and GSM490980), fetal forebrain tissue (GSM679519 and GSM679522), metaphase II oocytes (GSM304261 and GSM304262), and liver-derived hepatocytes (GSM595063 and GSM595066). Each CEL file was uploaded to GeneSifter (VisX Labs, www.geospiza.com) using the Advanced Upload Method and normalized using the Affymetrix Microarray Analysis Suite 5.0 algorithm. Cluster analysis between groups of samples was performed through GeneSifter Project Analysis using the analysis-of-variance (ANOVA) statistical analysis (p<0.01, threshold of 100, Manhattan distance, ward linkage, and gene row centering). GeneSifter Pairwise Analysis between samples was performed using all mean normalization and t-test statistical analysis. For each Pairwise Analysis, at least two replicates from each cell line or tissue type were compared with a baseline that consisted of three replicates of the SSEA3NEGATIVE cells. Probe sets were considered significantly upregulated (compared with the SSEA3NEGATIVE cells) when the p-value (p) was <0.05, and fold change (FC) was equal or greater than 3. For the relative gene expression analysis, the significantly upregulated probe sets (compared with the SSEA3NEGATIVE cells) for each cell and tissue sample in the meta-analysis library were compared with the probe sets upregulated in the SSEA3HIGH cells. The number of upregulated probe sets shared between a sample and SSEA3HIGH cells was compared with the total number of probe sets differentially expressed in the candidate cell or tissue sample, and this value was converted to a gene expression ratio based on the average gene expression correlation (value set at 0.0) and the most similar sample analyzed (value set at 1.0). The ANOVA statistical analysis and t-test statistical analysis were performed in GeneSifter with p-values set at less than 0.05.
MSC in vitro differentiation and staining
FACS-purified SSEA3-expressing human skin cells were recovered under standard cell culture conditions, as previously described, and allowed to adhere overnight before the commencement of the MSC-differentiation protocols. Experimental controls were seeded for each MSC differentiation protocol to corroborate the differentiation of each lineage. Specifically, human AT-MSCs (courtesy of Dr. Mirko Corselli) were included as a positive MSC differentiation control, undifferentiated AT-MSCs were included as a negative control, and mixed dermal skin cells (HUF-1 p6) were included in order to assay the presence or absence of MSCs in human skin primary cell cultures before flow cytometry-based cell sorting. Osteogenesis differentiation was performed using the Stempro Osteogenesis Differentiation Kit as described by the manufacturer (Invitrogen Cat# A10486) except using 5×102 cells/cm2. After 21 days of osteogenesis differentiation, the level of in vitro mineral deposit was assayed using Alizarin Red S (Sigma Cat# A5533). Adipogenesis differentiation was performed using the Stempro Adipogenesis Differentiation Kit as described by the manufacturer (Invitrogen Cat# A10477) except using 1×103 cells/cm2. After 21 days of adipogenesis differentiation, the accumulation of intracellular triglycerides was determined with AdipoRed™ Assay Reagent (Lonza Cat# PT-7009). Chondrogenesis differentiation was performed using the Stempro Chondrogenesis Differentiation Kit as described by the manufacturer (Invitrogen Cat# A10582). Specifically, chondrogenic capacity was examined in an optimized three-dimensional micromass culture, as previously described,21 using 2.5×105 cells to form the micromass pellets, and differentiation was induced with the STEMPRO® Chondrogenesis Differentiation Kit (Invitrogen Cat# A10582). Pellets were not disturbed for the first 72 h, were harvested at day 21, fixed with 4% PFA solution for 30 min, embedded with O.C.T. (Sakura Cat# 4583), and cryosectioned. Thin cryosections (5 μm thick) were mounted and stained for glycoproteins with Toluidine blue O (Sigma Cat# 198161) 1% aqueous for 20 min at 60°C followed with three rinses with distilled water and visualization under light microscopy. Differentiation protocols were performed in 5% CO2 incubators at 37°C, culture media were changed every 2–3 days, and all light microscopy-based imaging was performed with an AxioCam HR Color Camera using AxioVision Digital Image Processing Software (Axio Observer Inverted Microscope, Carl Zeiss).
Immunofluorescence and immunohistology
Cultured cells were fixed in 4% paraformaldehyde/PBS for 15 min, washed once with PBS supplemented with 100 mM glycine for 10 min, and then washed twice with PBS for 5 min each. Blocking was performed with 4% goat serum in Casein-PBS for 1 h at room temperature. Subsequently, 1:100 SSEA3 antibody (Millipore, mab4303) was added to 4% goat serum in casein-PBS and incubated overnight at 4°C with slow nutation. The next day, the cells were washed thrice with PBS for 5 min before a fluorescent-conjugated secondary Alexa 488-conjugated goat anti-rat IgM (Invitrogen, A21212) was added at 1:200 to 4% goat serum in casein-PBS and incubated for 1 h at room temperature, protected from light. The cells were rinsed thrice with PBS, and DAPI was used to label the nuclei. A final PBS rinse for 10 min at room temperature was performed. Visualization was performed with an AxioCam MR Monocolor Camera using AxioVision Digital Image Processing Software (Axio Observer Inverted Microscope, Carl Zeiss). Human skin samples were fixed fresh either within 45–60 min of the 4 mm punch skin biopsy (preinjury samples) or after 1 month of in vitro biopsy adhesion and culture in DMEM/F12+10% FBS (postinjury samples), the latter of which was used as our in vitro assay for tissue injury. Skin biopsy fragments either pre- or postinjury were cryosectioned (as previously described) and analyzed for SSEA3 expression using the immunofluorescence protocol previously described for in vitro cultured fixed cells.
Results
SSEA3-expressing cells in human skin pre- and postinjury
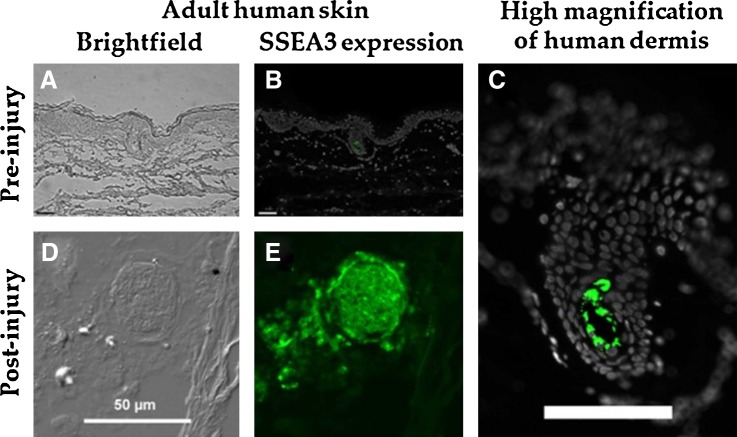
In vitro adhesion and primary cell derivation was attempted with human skin punch biopsy fragments from three donors. One of the biopsy donors did not demonstrate a successful primary cell culture and was excluded from this study. An immunohistochemical analysis of SSEA3 expression in adult human skin preinjury revealed that SSEA3-expressing cells were detected in the dermis of human skin and that these cells were extremely rare, representing less than 0.1% of the cells in the adult dermal tissue preinjury (Fig. 1A–C). However, following our in vitro model for dermal tissue injury and regeneration, a large increase in the number of SSEA3-expressing cells was observed with a greater than 20-fold local increase in the number of SSEA3-positive cells after injury (Fig. 1D, E). The SSEA3-expressing cells appeared as tightly compacted spheres of SSEA3-positive cells with more dispersed SSEA3-positive cells migrating out from this sphere (Fig. 1E). These SSEA3-expressing spheres were observed in multiple biological replicates. This result supports the hypothesis that SSEA3-expressing cells in human skin dermis are linked to the skin injury/regeneration response.
FIG. 1.
Expression of stage-specific embryonic antigen 3 (SSEA3) in adult human skin pre- and postinjury. (A) Brightfield image of human skin preinjury, (B) Immuno-histochemical analysis of SSEA3 expression in normal human skin (preinjury), (C) High-magnification view of SSEA3 expression in the dermis, (D) Brightfield image of human skin postinjury, and (E) Immuno-histochemical analysis of SSEA3 expression in human skin postinjury. SSEA3 expression in green and DAPI staining in white. Scale bars in (B, C) represent 100 μm, and scale bar in (D) represents 50 μm.
Global transcriptional meta-analysis
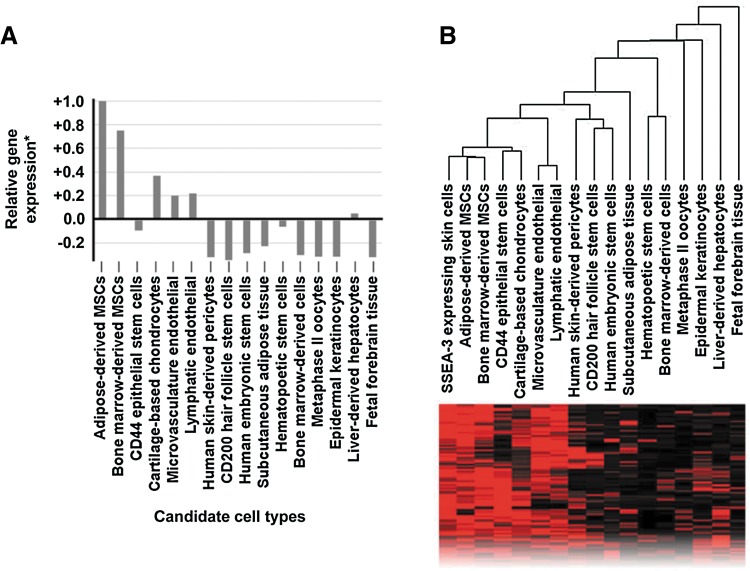
We performed a global transcriptional meta-analysis using the following cell and tissue samples in our meta-analysis library: low passage (p3) SSEA3HIGH skin-derived human somatic cells 24 h after FACS-based purification, AT-MSCs, BM-MSCs, CD44-expressing epithelial stem cells, epidermal keratinocytes, CD200-expressing hair follicle stem cells, endothelial cells derived from skin microvasculature and lymphatic tissues, skin-derived pericytes, bone marrow-derived hematopoietic stem cells, bone marrow-derived mono-nuclear cells, ES cells, subcutaneous adipose tissue, chondrocytes from cartilage, fetal forebrain tissue, metaphase II oocytes, and liver-derived hepatocytes. Pairwise analysis of the SSEA3HIGH subpopulation against the SSEA3NEGATIVE subpopulation revealed more than 200 significantly upregulated (p<0.05, FC>3) probe sets (Gene Expression Omnibus, GEO, www.ncbi.nlm.nih.gov/geo#GSE33066). A pairwise analysis of the other members of the meta-analysis library was performed (compared with the SSEA3NEGATIVE cells), and the significantly upregulated probe sets (p-value <0.05, FC>3) were compared with the previously identified SSEA3HIGH-specific probe sets. This focused gene expression analysis revealed that the SSEA3HIGH cells were more transcriptionally similar to AT-MSCs, closely followed by BM-MSCs (Fig. 2A). However, our unbiased cluster analysis also revealed that the adipose tissue and BM-MSCs were transcriptionally closer to each other than to the SSEA3-expressing cells (Fig. 2B), highlighting the possibility that the SSEA3-expressing human skin cells may be identifying a non-MSC subpopulation.
FIG. 2.
Global transcriptional meta-analysis of SSEA3-expressing human adult skin-derived cells. (A) Relative gene expression analysis in comparison to SSEA3NEGATIVE cells, (B) unbiased whole-transcriptome cluster analysis, and representative sample of transcriptome comparison heatmap. *Gene expression ratio calculated based on the average gene expression correlation (set at 0.0) compared with the most similar identified cell type (set at 1.0).
Flow cytometric analysis and purification of primary human skin cells
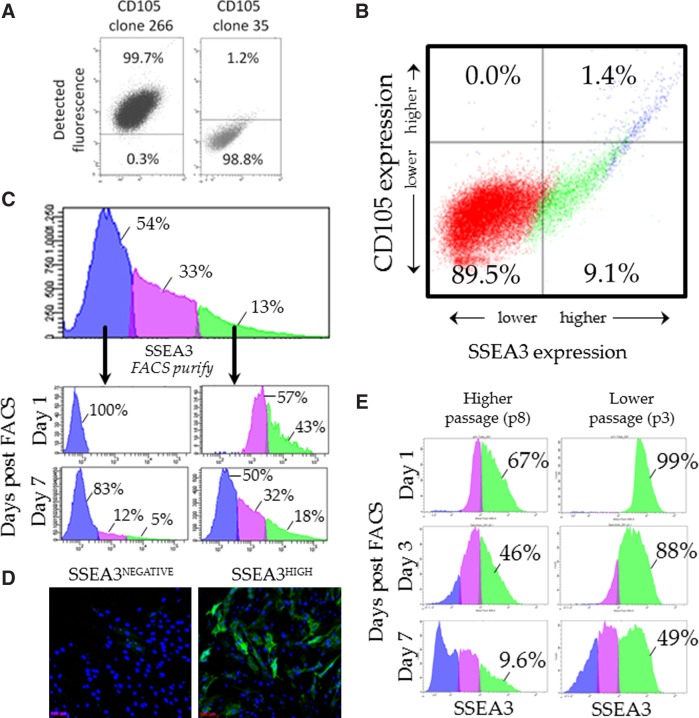
Previous research has demonstrated that SSEA3-expressing human skin cells co-express the endoglin (CD105) epitope, as detected using antibody CD105 clone 266 (BD Bioscience, 560839).11,13 These SSEA3/CD105 clone 266 identified cells are referred to as multipotent stress-enduring (MUSE) cells in reference to their multilineage differentiation capacity.11,13 However, to date, no research has examined the co-expression dynamics of SSEA3 and CD105 clone 35 (BD Biosciences, 611314). We investigated the percent of antibody bound cells using either CD105 antibody clones 266 or 35. As previously reported, CD105 clone 266 bound to almost 100% of primary cultured human skin cells11,13 (Fig. 3A, left panel). However, CD105 clone 35 (CD105c35) bound to only around 1% primary cultured human skin cells (Fig. 3A, right panel). We hypothesize that CD105c35 may be binding a rarer variant of endoglin (CD105), while the clone 266 antibody may be binding an endoglin variant ubiquitously expressed in human primary skin cells, although further evidence will be required to confirm this hypothesis. Interestingly, CD105c35 only bound to about 1% of BM-MSCs, suggesting that this antibody is not an MSC marker. Next, flow cytometric analysis was performed using both the SSEA3 antibody and the CD105c35 antibody. It was observed that there is a strong correlation between the cells that expressed the highest levels of SSEA3 and the cells which expressed CD105c35 (Fig. 3B). Approximately 1% of adult human skin-derived cells expressed both SSEA3 and CD105c35 (Fig. 3B). Next, flow cytometry was used to examine the dynamic expression of SSEA3 in primary human skin cells (using the HUF1 line previously described10). We first verified, as previously reported, that within the primary HUF1 skin culture, the cells expressed SSEA3 heterogeneously10 with 54% of the cells demonstrating no significant SSEA3 expression (SSEA3NEGATIVE), 33% demonstrating low SSEA3 expression (SSEA3LOW), and 13% of the skin cells demonstrating high SSEA3 expression (SSEA3HIGH) (Fig. 3C). FACS was then used to purify representatives of the SSEA3NEGATIVE and SSEA3HIGH subpopulations (Fig. 3C) from HUF1 cells that had been derived from the human skin biopsy fragments and maintained in culture for five passages (p5). The subpopulations were cultured under standard fibroblast culture conditions and then re-examined at intervals to assess the dynamics of SSEA3 expression. One day post-FACS, 100% of the cells in the SSEA3NEGATIVE-sorted subpopulation maintained their SSEA3NEGATIVE phenotype; notably, however, while after 1 week in culture, 83% of the cells in the SSEA3NEGATIVE-sorted subpopulation still maintained their SSEA3NEGATIVE phenotype, 12% were now SSEA3LOW and 5% were SSEA3HIGH (Fig. 3C). Even more markedly, 1 day after FACS-based purification, only 43% of the SSEA3HIGH-sorted subpopulation maintained the SSEA3HIGH phenotype, and after 1 week in culture, only 18% were still considered SSEA3HIGH, 32% were SSEA3LOW, and 50% were SSEA3NEGATIVE. This evidence suggests that expression of the SSEA3 epitope is dynamic in a cell culture. We observed no significant difference in the rate of proliferation from the SSEA3HIGH-sorted and SSEA3NEGATIVE-sorted populations, with population doubling times of 36.0±1 and 36.7±1.4 h, respectively. Immunocytochemical analysis of the SSEA3NEGATIVE-sorted and SSEA3HIGH-sorted subpopulations confirmed the heterogeneity of SSEA3 expression in both subpopulations after several days of cell culture (Fig. 3D). We hypothesized that dynamic expression of the SSEA3 molecule in vitro might reflect a culture-induced instability. Specifically, that expression of SSEA3 on the cell surface may be more stable in the skin and may be destabilized over time as the cells migrate from their original niche in the skin. To test this hypothesis, the SSEA3HIGH subpopulations from both higher passage (p8) and lower passage (p3) HUF1 cells were FACS-purified and analyzed. Higher-passage (p8) HUF1 cells had been in culture ∼1 month longer than lower-passage (p3) HUF1 cells. The higher-passage (p8) HUF1 cells quickly lost SSEA3 expression, with less than 10% of these cells still demonstrating an SSEA3HIGH phenotype after 1 week (Fig. 3E). In comparison, the lower-passage (p3) HUF1 cells demonstrated a significantly increased stability of SSEA3 expression, with nearly half of the low-passage cells maintaining an SSEA3HIGH phenotype after 1 week (Fig. 3E).
FIG. 3.
Dynamics of SSEA3 expression during in vitro culture. (A) Flow cytometry analysis of CD105 antibody clones 266 and 35 in skin biopsy-derived human cells. (B) Flow cytometry analysis of SSEA3 and CD105 clone 35 expression in skin biopsy-derived cells. (C) Fluorescence activated cell sorting (FACS)-based analysis and purification of SSEA3NEGATIVE and SSEA3HIGH subpopulations and analysis of these subpopulations 1 day and 1 week after cell sorting. (D) Immunocytochemical analysis of SSEA3HIGH and SSEA3NEGATIVE sorted cells 96 h after cell sorting. (E) FACS analysis of SSEA3HIGH cells purified from higher-passage (p8) and lower-passage (p3) cells 1, 3, and 7 days after cell sorting.
MSC analysis in adult human skin primary cultures
Human skin cells that co-expressed both SSEA3 and CD105c35 (referred to here as SSEA3-expressing regeneration-associated [SERA] cells) were purified using FACS and analyzed for MSC differentiation potential alongside human AT-MSCs (included as a positive MSC differentiation control) and mixed dermal skin cells (HUF-1 p6, included to assay the presence or absence of MSCs in human skin primary cell cultures before flow cytometry-based cell sorting). After 21 days of osteogenesis, adipogenesis, or chondrogenesis differentiation, cells were assayed for the formation of osteoblasts (observed via Alizarin Red S staining), adipocytes (observed via AdipoRed staining), or chondrocytes (observed via Toluidine blue O staining) under their respective culture conditions. The AT-MSC positive control cells demonstrated successful differentiation into osteoblasts, adipocytes, and chondrocytes (Fig. 4, left panel), demonstrating the success of the MSC differentiation protocols utilized here. While a rare (<1%) functional MSC subpopulation was observed in human skin primary cultured cells that was capable of differentiating into osteoblasts, adipocytes, and chondrocytes (Fig. 4, middle panel), the SERA cells did not demonstrate MSC differentiation capacity (Fig. 4, right panel), suggesting that the SERA cells and MSCs represent two different rare subpopulations in human dermis.
Discussion
In this study, we confirmed previous reports that identified a rare population of SSEA3-expressing cells within and derived from adult human skin.10,11,13 We examined the response of these SSEA3-expressing skin cells to tissue injury. In vitro biopsy adhesion, cell migration, and extended culture (for 1 month) were used as an in vitro assay for tissue injury. This in vitro assay for tissue injury was based on the premise that cells on the periphery of newly damaged dermal tissue (whether in vivo or in vitro) would have the same basic responses to dermal tissue damage. This premise appears to be supported by previous research that has demonstrated similarities in the in vivo and in vitro response of skin cells to tissue damage after thermally induced injury.22 Immunohistochemical analysis revealed that the SSEA3-expressing cells are extremely rare in normal human skin, representing less than 0.1% of the cells in the preinjury skin. After the in vitro injury assay, at least a 20-fold local increase in the number of SSEA3-positive cells was observed, and SSEA3-expressing cells appeared to be migrating away from the central spheres of tightly packed SSEA3-expressing cells. The function of these tightly packed SSEA3-expressing spheres is currently unknown, but they may play a role in the regeneration of human skin in response to injury. These data provide empirical support for the hypothesis that SSEA3 expression significantly correlates with tissue regeneration in human skin. Future analysis of a larger number of tissue biopsy fragments from a larger number of donors will be necessary to investigate the validity of this hypothesis. These SERA cells were derived through a primary cell culture, purified by FACS, and characterized at multiple time points over an extended in vitro culture. It was observed that the longer the primary skin cells were cultured in vitro, the lower the SSEA3 expression stability after FACS-based purification, with cells cultured an extra month in vitro demonstrating a five-fold decrease in their SSEA3 expression stability 1 week after FACS-based purification. This result supports the hypothesis that the stability of SSEA3 expression is negatively impacted by an extended in vitro culture. Whether optimization of in vitro conditions might stabilize SSEA3 expression over an extended culture is yet to be determined, but this line of research would be an important consideration for any future applications based on the expansion of pure populations of SSEA3-expressing skin cells, such as for patient-specific regenerative therapies that use SSEA3 to identify cells with putative regeneration-associated and/or reprogramming-associated10,13 cell types. Previous research by Dezawa and colleagues13 has demonstrated that skin-derived SSEA3-expressing cells represent a different population from previously identified ASC-like subpopulations in the skin, such as skin-derived precursor cells,23–25 melanocyte stem cells,26 endothelial precursors,27 pericytes,28 or neural crest-derived stem cells.29 Our results further demonstrated that the SSEA3-expressing cells shared a closer transcriptional profile with the MSCs derived from adipose tissue and bone marrow than with the CD200-expressing hair follicle stem cells,30 CD44-expressing epithelial stem cells,31 skin-derived endothelial cells,32 or skin-derived pericytes,33 supporting the hypothesis that the SSEA3-expressing cells may help in identifying an ASC-like population in the human skin. However, while a rare subpopulation of functional MSCs was observed in human skin primary cultures, the SERA cells did not demonstrate MSC differentiation capacity, suggesting that the SERA cells and MSCs represent two different subpopulations in human skin, possibly with different and/or complimentary roles in vivo. While not a canonical MSC marker, it remains possible that selection for the SSEA3 biomarker may still provide a limited degree of enhancement toward the adipogenic pathway, similar to that previously observed for SSEA4,4 although a future experimental analysis of significantly larger numbers of cells will be required to more fully investigate this hypothesis. With regard to the functional skin-derived MSCs observed in this study, it is unclear which biomarker profile will permit identification, isolation, and expansion of these rare cells from adult human skin biopsy primary cultures; however, the top MSC candidate biomarkers include CD10, CD13, CD26, CD29, CD34, CD44, CD54, CD71, CD73, CD90, CD105 (clone 266), CD106, CD146, CD166, CD271, ITGA11, STRO-1, and SSEA4.4,34,35 The potential derivation of MSCs from skin punch biopsies, which are quick, simple, and minimally invasive to perform, may be preferable for many patients compared with current MSC derivation protocols that require the surgical aspiration of adipose tissue and/or bone marrow. The identification, efficient purification, and large-scale expansion of these rare subpopulations (SERA cells and MSCs) from heterogeneous adult human skin primary cell cultures may have applications for future patient-specific cellular therapies.
Acknowledgments
The authors would like to thank Dr. Anne Chang for consenting the donors and performing the human skin biopsies, and Dr. Mirko Corselli for advice and guidance with regard to FACS analysis, the MSC antibody-based aspects of this study, and providing the AT-MSCs used in this study. This work is based on a research collaboration with Fibrocell Science, Inc. This work is supported by funding from the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research at UCLA, the National Institutes of Health (NIH RC2 HK103400 to RRP), the California Institute for Regenerative Medicine, and Fibrocell Science, Inc.
Author Disclosure Statement
J.A.B. and A.V.C. receive research funding from Fibrocell Science, Inc. J.A.B. is a scientific consultant for Fibrocell Science, Inc. J.P.A. and R.R.P. have no conflicts of interest or competing financial interests to report.
References
- 1.Fuchs E. Finding one's niche in the skin. Cell Stem Cell. 2009;4:499–502. doi: 10.1016/j.stem.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tumbar T. Guasch G. Greco V, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanpain C. Lowry WE. Geoghegan A, et al. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 4.Vaculik C. Schuster C. Bauer W, et al. Human dermis harbors distinct mesenchymal stromal cell subsets. J Invest Dermatol. 2012;132:563–574. doi: 10.1038/jid.2011.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schrump DS. Furukawa K. Yamaguchi H, et al. Recognition of galactosylgloboside by monoclonal antibodies derived from patients with primary lung cancer. Proc Natl Acad Sci USA. 1988;85:4441–4445. doi: 10.1073/pnas.85.12.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang WW. Lee CH. Lee P, et al. Expression of Globo H and SSEA3 in breast cancer stem cells and the involvement of fucosyl transferases 1 and 2 in Globo H synthesis. Proc Natl Acad Sci USA. 2008;105:11667–11672. doi: 10.1073/pnas.0804979105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohyama C. Orikasa S. Kawamura S, et al. Galactosylgloboside expression in seminoma. Inverse correlation with metastatic potential. Cancer. 1995;76:1043–1050. doi: 10.1002/1097-0142(19950915)76:6<1043::aid-cncr2820760619>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 8.Shevinsky LH. Knowles BB. Damjanov I, et al. Monoclonal antibody to murine embryos defines a stage-specific embryonic antigen expressed on mouse embryos and human teratocarcinoma cells. Cell. 1982;30:697–705. doi: 10.1016/0092-8674(82)90274-4. [DOI] [PubMed] [Google Scholar]
- 9.Kannagi R. Cochran NA. Ishigami F, et al. Stage-specific embryonic antigens (SSEA-3 and -4) are epitopes of a unique globo-series ganglioside isolated from human teratocarcinoma cells. EMBO J. 1983;2:2355–2361. doi: 10.1002/j.1460-2075.1983.tb01746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byrne JA. Nguyen HN. Reijo Pera RA. Enhanced generation of induced pluripotent stem cells from a subpopulation of human fibroblasts. PLoS One. 2009;4:e7118. doi: 10.1371/journal.pone.0007118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuroda Y. Kitada M. Wakao S, et al. Unique multipotent cells in adult human mesenchymal cell populations. Proc Natl Acad Sci USA. 2010;107:8639–8643. doi: 10.1073/pnas.0911647107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tonge PD. Shigeta M. Schroeder T, et al. Functionally defined substates within the human embryonic stem cell compartment. Stem Cell Res. 2011;7:145–153. doi: 10.1016/j.scr.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 13.Wakao S. Kitada M. Kuroda Y, et al. Multilineage-differentiating stress-enduring (Muse) cells are a primary source of induced pluripotent stem cells in human fibroblasts. Proc Natl Acad Sci USA. 2011;108:9875–9880. doi: 10.1073/pnas.1100816108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atlasi Y. Mowla SJ. Ziaee SA, et al. OCT4 spliced variants are differentially expressed in human pluripotent and nonpluripotent cells. Stem Cells. 2008;26:3068–3074. doi: 10.1634/stemcells.2008-0530. [DOI] [PubMed] [Google Scholar]
- 15.Henderson JK. Draper JS. Baillie HS, et al. Preimplantation human embryos and embryonic stem cells show comparable expression of stage-specific embryonic antigens. Stem Cells. 2002;20:329–337. doi: 10.1634/stemcells.20-4-329. [DOI] [PubMed] [Google Scholar]
- 16.Thomson JA. Itskovitz-Eldor J. Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi K. Tanabe K. Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 18.Sekine M. Yamakawa T. Suzuki A. Genetic control of the expression of two extended globoglycolipids carrying either the stage specific embryonic antigen-1 or -3 determinant in mouse kidney. J Biochem. 1987;101:563–568. doi: 10.1093/jb/101.3.563. [DOI] [PubMed] [Google Scholar]
- 19.Dasgupta S. Hogan EL. van Halbeek H. Chemical and immunological characterization of galactosyl-beta 1–3-globoside in bovine, human, and rat brain. J Neurochem. 1995;65:2344–2349. doi: 10.1046/j.1471-4159.1995.65052344.x. [DOI] [PubMed] [Google Scholar]
- 20.Zulli A. Buxton BF. Merrilees M, et al. Human diseased arteries contain cells expressing leukocytic and embryonic stem cell markers. Hum Pathol. 2008;39:657–665. doi: 10.1016/j.humpath.2007.09.022. [DOI] [PubMed] [Google Scholar]
- 21.Johnstone B. Hering TM. Caplan AI, et al. In vitro chondrogenesis of bone marrow-derived mesenchymal progenitor cells. Exp Cell Res. 1998;238:265–272. doi: 10.1006/excr.1997.3858. [DOI] [PubMed] [Google Scholar]
- 22.Coolen NA. Vlig M. van den Bogaerdt AJ, et al. Development of an in vitro burn wound model. Wound Repair Regen. 2008;16:559–567. doi: 10.1111/j.1524-475X.2008.00403.x. [DOI] [PubMed] [Google Scholar]
- 23.Fernandes KJ. McKenzie IA. Mill P, et al. A dermal niche for multipotent adult skin-derived precursor cells. Nat Cell Biol. 2004;6:1082–1093. doi: 10.1038/ncb1181. [DOI] [PubMed] [Google Scholar]
- 24.Biernaskie J. Paris M. Morozova O, et al. SKPs derive from hair follicle precursors and exhibit properties of adult dermal stem cells. Cell Stem Cell. 2009;5:610–623. doi: 10.1016/j.stem.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toma JG. Akhavan M. Fernandes KJ, et al. Isolation of multipotent adult stem cells from the dermis of mammalian skin. Nat Cell Biol. 2001;3:778–784. doi: 10.1038/ncb0901-778. [DOI] [PubMed] [Google Scholar]
- 26.Nishimura EK. Jordan SA. Oshima H, et al. Dominant role of the niche in melanocyte stem-cell fate determination. Nature. 2002;416:854–860. doi: 10.1038/416854a. [DOI] [PubMed] [Google Scholar]
- 27.Middleton J. Americh L. Gayon R, et al. A comparative study of endothelial cell markers expressed in chronically inflamed human tissues: MECA-79, Duffy antigen receptor for chemokines, von Willebrand factor, CD31, CD34, CD105 and CD146. J Pathol. 2005;206:260–268. doi: 10.1002/path.1788. [DOI] [PubMed] [Google Scholar]
- 28.Crisan M. Yap S. Casteilla L, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3:301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 29.Nagoshi N. Shibata S. Kubota Y, et al. Ontogeny and multipotency of neural crest-derived stem cells in mouse bone marrow, dorsal root ganglia, and whisker pad. Cell Stem Cell. 2008;2:392–403. doi: 10.1016/j.stem.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 30.Garza LA. Yang CC. Zhao T, et al. Bald scalp in men with androgenetic alopecia retains hair follicle stem cells but lacks CD200-rich and CD34-positive hair follicle progenitor cells. J Clin Invest. 2011;121:613–622. doi: 10.1172/JCI44478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhat-Nakshatri P. Appaiah H. Ballas C, et al. SLUG/SNAI2 and tumor necrosis factor generate breast cells with CD44+/CD24- phenotype. BMC Cancer. 2010;10:411. doi: 10.1186/1471-2407-10-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanki Y. Kohro T. Jiang S, et al. Epigenetically coordinated GATA2 binding is necessary for endothelium-specific endomucin expression. EMBO J. 2011;30:2582–2595. doi: 10.1038/emboj.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paquet-Fifield S. Schluter H. Li A, et al. A role for pericytes as microenvironmental regulators of human skin tissue regeneration. J Clin Invest. 2009;119:2795–2806. doi: 10.1172/JCI38535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mafi P. Hindocha S. Mafi R, et al. Adult mesenchymal stem cells and cell surface characterization—a systematic review of the literature. Open Orthop J. 2011;5:253–260. doi: 10.2174/1874325001105010253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halfon S. Abramov N. Grinblat B, et al. Markers distinguishing mesenchymal stem cells from fibroblasts are downregulated with passaging. Stem Cells Dev. 2011;20:53–66. doi: 10.1089/scd.2010.0040. [DOI] [PubMed] [Google Scholar]