Abstract
The Escherichia coli hlyE gene (also known as clyA or sheA) codes for a novel pore-forming toxin. Previous work has shown that the global transcription factors FNR and CRP positively regulate hlyE expression by binding at the same site. Here in vivo transcription studies reveal that FNR occupies the hlyE promoter more frequently than CRP, providing a mechanism for the moderate upregulation of hlyE expression in response to two distinct environmental signals (oxygen and glucose starvation). It has been reported that H-NS interacts with two large regions of the hlyE promoter (PhlyE), one upstream of the −35 element and one downstream of the −10 element. Here we identify two high-affinity H-NS sites, H-NS I, located at the 3′ end of the extended upstream footprint, and H-NS II, located at the 5′ end of the extended downstream footprint. It is suggested that these high-affinity sites initiate the progressive formation of higher order complexes, allowing a range of H-NS-mediated regulatory effects at PhlyE. Finally, the identification of a SlyA binding site that overlaps the H-NS I site in PhlyE suggests a mechanism to explain how SlyA overproduction enhances hlyE expression by antagonizing the negative effects of H-NS.
Recently, a novel pore-forming toxin, designated HlyE, ClyA, or SheA, was identified for Escherichia coli and Salmonella enterica serovars Typhi and Paratyphi A (2, 6, 7, 10, 16, 17, 21, 22, 35). The three-dimensional (3-D) structure of HlyE shows that it is a mostly α-helical, long (∼100 Å), rod-shaped molecule with a hydrophobic two-stranded antiparallel β-sheet at one end that is thought to allow an interaction between HlyE and target membranes (35). The HlyE protein forms pores in target membranes that appear in electron microscopy (EM) images as ring-shaped structures with internal diameters of 50 to 55 Å, when viewed from above, and as 100- to 105-Å spikes in a side view, suggesting that HlyE does not undergo large conformational changes during pore formation (35).
Two members of the CRP family of transcription factors control the expression of hlyE in E. coli K-12 by binding at the same site centered at −61.5 bp upstream of the hlyE transcriptional start (10, 36). Thus, CRP enhances hlyE expression in response to glucose starvation (36) and FNR enhances hlyE expression in response to oxygen starvation (10, 11, 23). Both CRP and FNR are ∼50-kDa homodimers that bind related inverted repeats with a TGANNNNNNTCA core motif (12). At the hlyE promoter (PhlyE), this site (TTTGATATTTATCATA) most closely resembles an FNR site (9 of 10 nucleotides match the FNR consensus, TTGATNNNNATCAA, compared to 8 of 10 matches to the CRP consensus, TGTGANNNNNNTCACA; discriminatory bases are underlined). However, it has been shown that in certain circumstances CRP can recognize FNR sites, although the affinity of CRP for an FNR site is 50-fold lower than that for an equivalent CRP site (27).
A further layer of regulation is provided by the nucleoid structuring protein H-NS (36). The H-NS protein influences the expression of many genes in E. coli K-12. It is a small ∼15-kDa protein that forms higher order complexes in a concentration-dependent manner (30, 34). The H-NS protein was shown to interact with a large region of PhlyE (from −137 to +172, relative to the transcription start site) to repress hlyE expression (36) after the observation that an hns mutant strain has a hemolytic phenotype (9).
A hemolytic phenotype was also conferred upon E. coli K-12 by the overproduction of either the E. coli or Salmonella enterica transcription factor SlyA (16, 21). The SlyA protein is a member of the MarR family of transcription factors that includes MarR and EmrR (E. coli), PecS (Erwinia chrysanthemi), HprR (Bacillus subtilis), and RovA (Yersinia enterocolitica) (20, 24). The 3-D structures of E. coli MarR and a SlyA-like protein from Enterococcus faecalis provide the structural archetypes for this family of proteins. The 3-D structures show that they are homodimers in which each subunit possesses a winged-helix DNA-binding domain (1, 37). A recent characterization of the Salmonella SlyA protein revealed that it is also a homodimer (∼32 kDa) that recognizes an inverted repeat sequence in target promoters (32). Site-directed mutagenesis of PhlyE led to the suggestion that a GC-rich sequence located between an unusual heptameric −10 element (TATGAAT) and a conventional −35 element might be the site of SlyA action (17). Here we show that the regulation of hlyE expression by H-NS is more complex than was previously thought and that the overproduction of SlyA enhances hlyE expression by antagonizing the negative effects of H-NS.
MATERIALS AND METHODS
Bacterial strains, plasmids, and microbiological methods.
Relevant characteristics of the bacterial strains and plasmids used are given in Table 1. Isogenic derivatives of M182 were constructed by P1vir-mediated transduction. Bacteria were grown in Lennox broth (L-broth; contains yeast extract, 5 g liter−1; tryptone, 10 g liter−1; and NaCl, 5 g liter−1) at 37°C supplemented with glucose (0.2% [wt/vol]), ampicillin (150 μg ml−1), tetracycline (35 μg ml−1), and chloramphenicol (20 μg ml−1), as appropriate. For β-galactosidase activity measurements (19), anaerobic cultures were grown in sealed bottles filled to the neck with medium, and aerobic cultures were grown in conical flasks (250 ml) containing medium (10 ml), with vigorous shaking (250 rpm) at 37°C. Hemolytic activities were estimated by measuring the areas of hemolysis surrounding individual colonies after 16 h of growth at 37°C on blood agar under aerobic conditions.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| M182 | Δlac | 5 |
| JRG4830 | M182 Δlac Δcrp | 5 |
| JRG4864 | M182 Δlac Δhns; Cmr | This work |
| JRG2631 | M182 Δlac Δfnr Δcrp; Tetr | Jeff Cole |
| JRG4865 | M182 Δlac Δfnr Δhns; Cmr Tetr | This work |
| JRG4732 | M182 Δlac Δcrp Δhns; Cmr | This work |
| JRG4866 | M182 Δlac Δfnr Δcrp Δhns; Cmr Tetr | This work |
| JRG4747 | M182 Δlac Δfnr; Cmr | This work |
| JRG3212 | M182 Δlac Δfnr Δcrp; Cmr | This work |
| JRG4385 | JM109 (pGS1482); Apr | 32 |
| Plasmids | ||
| pRS415 | Medium-copy-number vector (15 to 20 copies per cell; pBR322 origin of replication) for construction of hlyE::lacZ reporter plasmid pGS1629; Apr | 29 |
| pGS1629 | pRS415 containing PhlyE (−299 to +82); Apr | This work |
| pRW50 | Low-copy-number (2 to 5 copies per cell; pSC101 origin of replication), lac reporter vector; Tetr | 15 |
| pGS1065 | pRW50 containing PhlyE (−97 to +61); Tetr | 10 |
| pGS1064 | pUC118 containing PhlyE (−97 to +61, start codon is +1); Apr | 10 |
| pGS1482 | pGEX-KG (13) containing the slyA coding region; Apr | 32 |
| pGS1657 | pBluescript containing S. enterica slyA coding region and promoter; Apr | This work |
Gel shift assays.
Initial H-NS gel shifts used a PCR-amplified 696-bp fragment of PhlyE containing all of the previously reported sequences protected by H-NS from DNase I digestion (36) plus additional upstream and downstream sequences. In subsequent experiments, this fragment was further resolved into four subfragments by restriction digestion with SspI, BsaMI, and DraI. Target DNAs (200 to 500 ng) were incubated with H-NS (0 to 6 μM) in 25 mM HEPES, pH 7.6, containing 0.1 mM EDTA, 10% (vol/vol) glycerol, 5 mM dithiothreitol, 50 mM KCl, and 0.01 mg of poly(dI-dC) ml−1 for 15 min at 25°C in a total volume of 10 or 20 μl. Complexes were separated in 6% (wt/vol) nondenaturing Tris-borate-EDTA-buffered polyacrylamide gels for the long PhlyE fragment or in 12.5% (wt/vol) gels for the dissected PhlyE fragments. DNA was visualized by staining with ethidium bromide. The interaction of H-NS and SlyA with PhlyE was also investigated by using PhlyE DNA amplified by PCR with pGS1064 (10) (Table 1) as the template and with pUC/M13 forward and reverse primers. The product was digested with BamHI and radiolabeled with Klenow enzyme and [32P]α-dGTP (26). For H-NS, ∼10 ng of DNA and 0.04 to 2.0 μM H-NS were coincubated for 5 min at 25°C in a solution containing 10 mM Tris-HCl (pH 7.6), 50 mM MgCl2, 2 mM spermidine, and 15 mM potassium glutamate (total incubation volume, 10 μl). For SlyA, ∼10 ng of DNA and 0.06 to 0.63 μM SlyA protein were coincubated for 2 min at 25°C in a solution containing 10 mM Tris-HCl (pH 9.0), 50 mM KCl, and 0.1% (vol/vol) Triton X-100 (total incubation volume, 10 μl). Reactions were then loaded onto gels for autoradiographic analysis.
DNase I footprinting.
For H-NS, the reactions (total volume, 10 μl) contained radiolabeled PhlyE (∼10 ng), H-NS (1 μM), 10 mM Tris-HCl (pH 7.6), 50 mM MgCl2, 2 mM spermidine, and 15 mM potassium glutamate. For SlyA, the reactions (total volume, 10 μl) contained radiolabeled PhlyE (∼10 ng), SlyA (2.0 and 4.0 μM), 20 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 10 mM dithiothreitol, and 5% (vol/vol) glycerol. The mixtures were incubated for 2 to 5 min at 25°C, followed by digestion with DNase I (1 μl of a 1-U μl−1 solution for 5 to 60 s at 25°C). Reactions were stopped by the addition of 200 μl of 0.3 M sodium acetate (pH 5.2) containing 20 mM EDTA, followed by phenol-chloroform extraction. The DNA was ethanol precipitated and resuspended in 10 μl of loading buffer (80% [vol/vol] formamide, 0.1% [wt/vol] sodium dodecyl sulfate, 10% [vol/vol] glycerol, 8 mM EDTA, 0.1% [wt/vol] bromophenol blue, and 0.1% [wt/vol]) xylene cyanol) for electrophoretic fractionation on 6% (wt/vol) polyacrylamide-urea gels and autoradiographic analysis. Maxam and Gilbert G tracks were used to provide a calibration (18).
Other methods.
The H-NS protein was provided by C. F. Higgins (MRC Clinical Sciences Centre, Hammersmith Hospital, London, United Kingdom), and the SlyA protein was isolated from E. coli strain JRG4385 (Table 1) as previously described (32). The manipulation of DNA, PCR, and plasmid constructions was achieved by conventional methods (26). Plasmid copy numbers were estimated by the method of Taylor and Brose (33).
RESULTS
Effects of the global transcription factors FNR, CRP, and H-NS on hlyE expression in liquid culture.
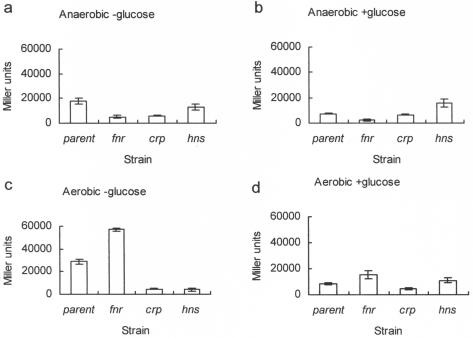
Previous studies have shown that FNR, CRP, and H-NS contribute to hlyE expression when E. coli is grown on a solid medium (10, 36). Both FNR and CRP make positive contributions, whereas H-NS acts negatively under these conditions. Because bacteria grown on solid media are exposed to different microenvironments due to the generation of concentration gradients of nutrients and oxygen within colonies, the starting point for this work was to investigate the effects of all three global regulators on hlyE expression in liquid cultures by using a plasmid-based hlyE::lacZ gene fusion (pGS1629; Table 1). The reporter plasmid contained a fragment of DNA stretching from −299 bp to +82 bp relative to the hlyE transcript start site ligated with pRS415 (Table 1). Initially, transcriptional activity from PhlyE was estimated for anaerobic cultures of parental (M182), fnr, crp, and hns strains transformed with pGS1629 (Table 1), after growth in L-broth at 37°C for 16 h. The data revealed that both FNR and CRP made significant positive contributions to hlyE expression, whereas the hns mutation resulted in slightly reduced hlyE transcription under these conditions (Fig. 1a). It was predicted that the addition of glucose (0.2% [wt/vol]) to the medium would abolish the effects of CRP. Accordingly, hlyE expression was reduced in cultures of the parental strain containing glucose compared to those without glucose (Fig. 1b). Moreover, while hlyE expression was reduced in the fnr strain compared to that in the parent strain in the presence of glucose, it was, as expected, unaffected in the crp mutant (Fig. 1b). The transcription of hlyE in the hns strain under anaerobic conditions in the presence of glucose is presumably driven by FNR alone. Under these conditions, hlyE expression was greater for the hns strain than for the corresponding parental cultures, suggesting that H-NS has a negative effect on FNR-driven hlyE expression (Fig. 1b). Thus, we concluded that CRP, FNR, and H-NS all contribute towards the regulation of hlyE expression.
FIG. 1.
Effects of FNR, CRP, and H-NS on in vivo transcription of hlyE. All cultures were grown in L-broth at 37°C for ∼16 h under the indicated conditions. The activity of the hlyE promoter was estimated by measuring the β-galactosidase activity associated with each culture carrying the hlyE::lacZ plasmid pGS1629. Strains: parental, M182; fnr, JRG4747; crp, JRG4830; hns, JRG4864 (Table 1). Error bars indicate 2 standard deviations from the means (n = 3).
Similar experiments were done with aerobic cultures. For these conditions, we predicted that the glucose-responsive effects of CRP would be retained, whereas the positive regulation by FNR would be abolished. The prediction for the CRP response was confirmed by the data (Fig. 1c and d). However, the data for the parental and fnr cultures suggested that even under aerobic growth conditions, some active FNR was present in the bacteria, and that even this small amount of active protein (relative to anaerobic conditions) was sufficient to interfere with CRP-mediated hlyE expression (Fig. 1c). Thus, these data suggest that under such conditions, FNR and CRP are in competition for binding at PhlyE, and that the simplest explanation of these observations is that FNR is more efficiently bound than CRP at PhlyE but is not as effective as CRP in activating hlyE transcription. Such an interpretation is consistent with the sequence of the FNR/CRP box in PhlyE, which lacks the discriminatory G-C base pairs that promote CRP specificity, and with the observation that, unlike CRP, FNR is a poor activator of class I promoters such as PhlyE (reviewed by Green et al. in reference 12). The effect of the hns mutation on hlyE expression in aerobic cultures was also interesting (Fig. 1c and d). In the absence of glucose, H-NS appeared to have a positive effect on hlyE expression (Fig. 1c). However, in the presence of glucose, H-NS appeared to have little effect on hlyE expression (Fig. 1d). Thus, under aerobic conditions, H-NS has a positive influence on CRP-mediated hlyE expression. This positive effect of H-NS was not observed under anaerobic conditions, presumably because FNR and not CRP occupies PhlyE under these conditions.
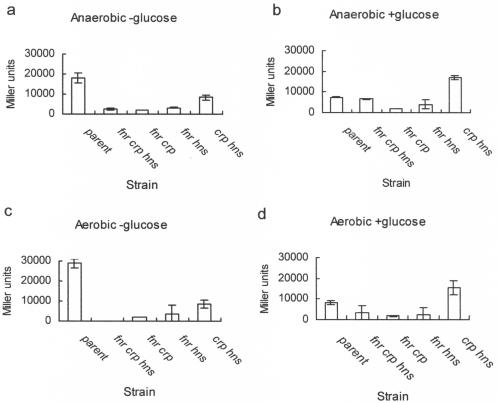
To further investigate the effects of the regulators on hlyE expression, we monitored the activity of the hlyE::lacZ fusion in pGS1629 in strains carrying multiple relevant mutations. Under anaerobic conditions in the absence of glucose, hlyE expression in an fnr crp hns triple mutant was low compared to that of the parental strain (Fig. 2a). Restoring hns did not affect hlyE expression. Similarly, restoring CRP did not enhance hlyE expression, despite the absence of glucose (Fig. 2a). However, the restoration of fnr produced a significant increase in hlyE expression under these conditions (Fig. 2a). This suggests that during anaerobic growth in liquid cultures in the absence of glucose, FNR is the major regulator of hlyE expression, and that CRP-mediated hlyE expression requires the presence of H-NS. The pattern of expression obtained in anaerobic cultures in the presence of glucose was similar to that obtained in the absence of glucose, except that hlyE expression in the fnr crp hns triple mutant and the crp hns double mutant was significantly higher than that for the corresponding cultures that lacked glucose (Fig. 2b). The hlyE expression patterns for equivalent aerobic cultures in the presence and absence of glucose were similar to those obtained under anaerobic conditions (Fig. 2c and d).
FIG. 2.
Effects of FNR, CRP, and H-NS on in vivo transcription of hlyE. All cultures were grown in L-broth at 37°C for ∼16 h under the indicated conditions. The activity of the hlyE promoter was estimated by measuring the β-galactosidase activity associated with each culture carrying the hlyE::lacZ plasmid pGS1629. Strains: parental, M182; fnr crp hns, JRG4866; fnr crp, JRG2631; fnr hns, JRG4865; crp hns, JRG4732. Error bars indicate 2 standard deviations from the means (n = 3).
In summary, the data presented here suggest that FNR and CRP are positive regulators of hlyE expression in liquid culture in response to oxygen and glucose starvation, respectively. This is in agreement with observations made with cultures grown on a solid medium (36). However, rather than the approximately eightfold enhancement in hlyE expression observed in an hns mutant on solid medium (36), in liquid culture H-NS has a positive effect on CRP-driven hlyE expression, a negative effect on FNR-driven hlyE expression, and little intrinsic regulatory activity in the absence of FNR and CRP.
H-NS binds at two regions of the hlyE promoter with high affinity.
Footprinting studies have shown that FNR and CRP activate hlyE expression from the same site centered −61.5 bp upstream of the SlyA-associated transcription start site (10, 36). A further footprinting analysis indicated that H-NS protects a large region of the hlyE promoter, extending from −137 to +172 (relative to the transcription start site) (36). To investigate how much of this extensive H-NS protection is required for the observed regulation of hlyE expression, we used two approaches as follows.
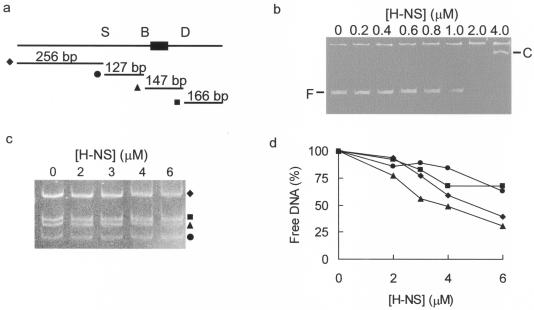
Firstly, a 696-bp region of the hlyE promoter region was amplified by PCR (Fig. 3a). Gel shift assays revealed that H-NS retarded the mobility of this fragment upon electrophoretic separation in Tris-borate-EDTA-buffered polyacrylamide gels (Fig. 3b). In the presence of 1 μM H-NS, retardation of some of the DNA target was observed as a smear in the gel. Upon the addition of further H-NS (2 μM), the free DNA was replaced entirely by a retarded smear. Only at 4 μM H-NS was a distinct retarded complex observed (Fig. 3b). The amplified fragment contains restriction sites for SspI, BsaMI, and DraI, such that a triple digest yields four fragments (Fig. 3a). Gel shift assays using a mixture of hlyE DNA fragments as the target DNA revealed that H-NS interacts with all four PhlyE fragments to some extent. However, the 147-bp DraI-BsaMI fragment was bound with the highest affinity, with 50% of this fragment retarded at H-NS concentrations of ∼3 to 4 μM (Fig. 3c and d). Thus, this region of PhlyE (−91 to +56) contains both the FNR/CRP box (−61.5) and the highest affinity H-NS site(s).
FIG. 3.
Identification of high-affinity H-NS sites within PhlyE. (a) Schematic representation of the 696-bp region encompassing all of the previously recognized regions of H-NS protection (36). The locations of relevant restriction sites (S, SspI; B, BsaMI; and D, DraI) and the sizes of the fragments released after triple digestion are indicated. The location of the FNR/CRP box is shown by the filled rectangle. (b) Interaction of H-NS with the 696-bp PhlyE fragment. The concentration of HlyE is indicated above each lane. The positions of the retarded complexes (C) and free PhlyE DNA (F) are marked. (c) The region of PhlyE containing the FNR/CRP box has the highest affinity for H-NS. The concentrations of H-NS used are indicated above each lane, and the targets were purified restriction fragments released from the 696-bp PhlyE fragment by digestion with SspI, BsaMI, and DraI. The symbols relate to the restriction fragments, as indicated in panel a. (d) Quantification of H-NS binding to different regions of PhlyE. The amount of each PhlyE fragment remaining unretarded in panel c was estimated by quantitative densitometry and plotted against the concentration of H-NS in the assay. The symbols relate to the restriction fragments, as indicated in panel a.
Secondly, for testing of whether the region of PhlyE containing the high-affinity H-NS site(s) was sufficient to account for the pattern of hlyE regulation observed in vivo, another hlyE::lacZ reporter plasmid was used (Table 1) (10). This second gene fusion (pGS1065) contains a minimal hlyE promoter beginning 18 bp downstream of the BsaMI site and ending 25 bp downstream of the DraI site and thus extends from −97 to +61 relative to the transcript start site in the low-copy-number (two to five copies per cell) vector pRW50 (Table 1). Cultures of strains M182 and M182 hns transformed with pGS1065 were grown under anaerobic conditions in the presence and absence of glucose at 37°C for ∼16 h. The transcriptional activity in vivo, as estimated by the measurement of β-galactosidase activity, indicated that this (pGS1065) low-copy-number reporter containing a minimal hlyE promoter responded in the same way as the longer medium-copy-number hlyE::lacZ fusion (pGS1629) to the presence or absence of H-NS (Table 2). Thus, in the presence of glucose, hlyE expression was enhanced in an hns mutant (365 Miller units) relative to the parent (271 Miller units) (compare with Fig. 1b). In the absence of glucose, hlyE expression was slightly lower in the hns mutant (399 Miller units) than in the parent (420 Miller units) (compare with Fig. 1a). Furthermore, there were no significant H-NS-related changes in reporter plasmid copy number (not shown), as judged by the method of Taylor and Brose (33). Thus, it was concluded that the BsaMI-DraI fragment of PhlyE contains all of the significant regulatory elements that control hlyE expression under the growth conditions used here and that the effects on hlyE expression observed with pGS1629 were not significantly affected by the copy number of the reporter plasmids.
TABLE 2.
Effect of SlyA and H-NS on transcription from a minimal hlyE promoter in vivo
| Straina | Data without glucose
|
Data with glucose
|
||
|---|---|---|---|---|
| Amt of β-galactosidase (Miller units)b | Ratioc | Amt of β-galactosidase (Miller units)b | Ratioc | |
| Parent + vector | 420 ± 11 | 1.46 | 271 ± 6 | 1.14 |
| Parent + SlyA | 614 ± 10 | 310 ± 21 | ||
| hns + vector | 399 ± 10 | 0.91 | 365 ± 15 | 0.82 |
| hns + SlyA | 365 ± 17 | 299 ± 38 | ||
| crp fnr + vector | 257 ± 6 | 0.87 | 152 ± 2 | 1.11 |
| crp fnr + SlyA | 223 ± 7 | 169 ± 2 | ||
Strains containing the low-copy-number minimal hlyE::lacZ promoter fusion plasmid (pGS1065) were grown under anaerobic conditions for 16 h at 37°C in L-broth with or without glucose (0.2% [wt/vol]) as indicated. The strains used were as follows: parent, M182(pGS1065); hns, JRG4864(pGS1065); crp fnr, JRG3212(pGS1065) (Table 1). SlyA indicates the presence of multicopy slyA (pGS1657), and vector indicates the presence of the vector (pBluescript).
Means (n = 3) and standard deviations are given.
Ratio of β-galactosidase activities obtained with multicopy slyA and vector transformants.
Identification of the high-affinity H-NS sites in PhlyE.
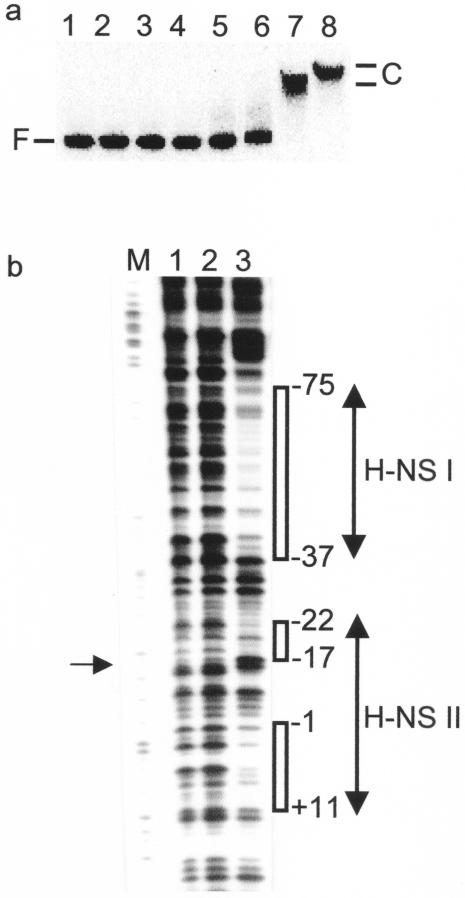
For determination of the number of H-NS sites within the minimal PhlyE sequence from pGS1065, further gel shift and footprinting assays were undertaken. The gel shifts showed that H-NS bound to PhlyE at concentrations close to those observed for other H-NS-regulated genes (∼1 μM) (3) and that two distinct retarded complexes were formed (Fig. 4a). The locations of the H-NS binding sites were determined by DNase I footprinting (Fig. 4b). Two protected regions were detected: H-NS I, consisting of an A-T-rich region (20 A-T/24 bp) stretching from −75 to −37 which overlaps the FNR/CRP box centered at −61.5; and H-NS II, between −22 and +11, overlapping the −10 element of PhlyE. The H-NS II site contains a hypersensitive base at position −17 that is separated by several unprotected bases (−16 to +1) from another protected A-T-rich region (8 A-T/10 bp) (Fig. 4b). Based upon the size and properties of H-NS and the extent of the DNase I footprint, it is likely that several H-NS molecules bind PhlyE.
FIG. 4.
Location of H-NS binding sites in PhlyE. (a) H-NS binds at PhlyE. Lane 1, no protein; lanes 2 to 8, H-NS (0.04, 0.08, 0.12, 0.25, 0.5, 1.0, and 2.0 μM, respectively). The positions of the retarded complexes (C) and free PhlyE DNA (F) are indicated. (b) DNase I footprints of H-NS at PhlyE. Lane M, Maxam and Gilbert G track; lanes 1 and 2, no protein; lane 3, H-NS (1 μM). The regions of H-NS protection (open boxes) and the hypersensitive base (arrow at the left) are indicated. All numbering is relative to the SlyA-associated transcription start site (36).
Effects of H-NS on SlyA-promoted hlyE expression.
Overproduction of the E. coli or S. enterica SlyA protein confers a hemolytic phenotype on E. coli K-12 by enhancing hlyE expression (16, 17, 21). To test the effects of H-NS on SlyA-driven hlyE expression, we measured β-galactosidase activities from cultures of isogenic parental and hns strains carrying the hlyE::lacZ plasmid pGS1065 and either a multicopy S. enterica SlyA expression plasmid (pGS1657; Table 1) or, as a control, the vector (pBluescript). As expected, irrespective of the addition of glucose, the presence of multicopy slyA increased hlyE expression in cultures of the parental strain, although the enhancement was greater for cultures lacking the glucose supplement (Table 2). In contrast, in the absence of hns, SlyA overproduction did not enhance hlyE expression (Table 2). This pattern of expression was supported by qualitative studies in which the same strains were grown on blood agar plates. These revealed that multicopy slyA enhanced hemolytic activity associated with the parental strain but not with the hns strain (not shown). These results confirm and extend the observations of Westermark et al. (36) and suggest that, either directly or indirectly, SlyA activates hlyE expression by modulating H-NS binding and ultimately by counteracting the negative effects of H-NS. This suggestion was supported by the observations that multicopy slyA did not enhance hlyE expression in a crp fnr double mutant (Table 2) and that SlyA did not promote open complex formation at PhlyE in vitro (not shown). Thus, it was concluded that SlyA, like H-NS, has no intrinsic activator role at PhlyE in the absence of FNR and CRP.
Interaction of SlyA with the E. coli hlyE promoter.
The pattern of regulation reported above suggests that SlyA, like H-NS, interacts directly with the E. coli hlyE promoter. Therefore, gel shift assays were used to investigate the interaction of purified SlyA with PhlyE. These experiments revealed that the addition of 0.16 to 0.31 μM SlyA was sufficient to retard the mobility of PhlyE (Fig. 5a).
FIG. 5.
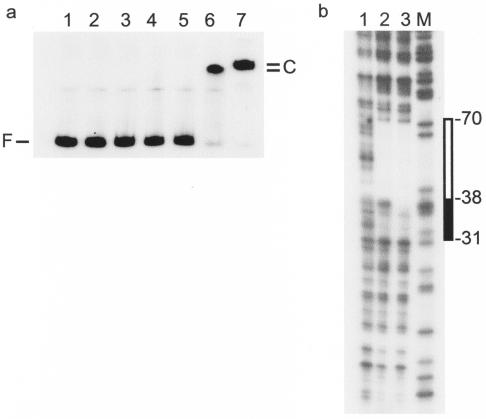
Interaction of SlyA with the hlyE promoter. (a) SlyA retards the mobility of PhlyE in band shift assays. SlyA concentrations: lane 1, 0 μM; lane 2, 0.06 μM; lane 3, 0.08 μM; lane 4, 0.1 μM; lane 5, 0.16 μM; lane 6, 0.31 μM; lane 7, 0.63 μM. The positions of the free DNA (F) and the SlyA-DNA complexes (C) are indicated. (b) DNase I footprint of SlyA at PhlyE. Lane 1, no protein; lane 2, 2.0 μM SlyA; lane 3, 4.0 μM SlyA; lane M, Maxam and Gilbert G track. The region of SlyA protection is indicated to the right (box). At the higher SlyA concentration, protection was extended, as indicated by the filled box.
A consensus SlyA DNA target was recently suggested to exist (32), and an inspection of the PhlyE sequence revealed at least two possible matches. Therefore, the location of the SlyA binding site(s) within PhlyE was investigated by DNase I footprinting (Fig. 5b). A protected region, −70 to −38 relative to the previously determined SlyA-associated transcript start site (17, 36), was observed in the presence of 2 μM SlyA. This region of protection was extended further downstream to −31 when the concentration of SlyA was increased to 4 μM (Fig. 5b). Thus, SlyA protects a region of PhlyE similar to the H-NS I region (−75 to −37; Fig. 4b). The cross sections of the E. coli MarR and E. faecalis SlyA-like protein dimers are ∼70 Å (1, 37) and therefore would be expected to protect ∼20 bp of DNA. Thus, the protection observed here (up to 40 bp) suggests that two SlyA dimers are bound at PhlyE. An inspection of the DNA sequence of the protected region revealed two related sequences with partial dyad symmetry that resemble the proposed SlyA binding consensus; they are SlyA I (−61TTATCATATTAA−50, with 8 of 12 bases (bold) matching the SlyA binding site consensus TTAGCAAGCTAA) and SlyA II (−50ATAGAAATAAAG−39, with 6 of 12 bases matching the consensus). The putative presence of two SlyA dimers at PhlyE was consistent with the two distinct complexes with retarded mobilities in the gel shift assays (Fig. 5a). The site centered at −55.5 is the better match to the SlyA binding site consensus and may be occupied in preference to the site centered at −44.5, which accordingly is less similar to the consensus.
DISCUSSION
The starting point for the work described here was the observation that lesions in hns or the overproduction of S. enterica or E. coli SlyA confers a hemolytic phenotype on E. coli K-12 by enhancing the expression of the pore-forming toxin HlyE (9, 16, 17, 21). Here we have shown that H-NS and SlyA interact directly with the hlyE promoter. Both proteins occupy a common region of PhlyE that overlaps the binding site for the global transcription factors FNR and CRP, which are known to activate hlyE expression (10, 36). In vivo and in vitro evidence suggests that SlyA activates hlyE expression by antagonizing H-NS-mediated repression.
The in vivo transcriptional evidence presented here suggests that FNR occupies the hlyE promoter more frequently than CRP even under aerobic conditions. This is evident from enhanced hlyE::lacZ expression in aerobic cultures (compare parent, fnr, and crp strains in Fig. 1c and d). These observations may be explained by a mechanism in which FNR recognizes PhlyE efficiently but acts only as a relatively poor activator of hlyE transcription (10, 11), whereas, conversely, the recognition of PhlyE by CRP is poor, but once CRP is bound, CRP-mediated activation of hlyE expression is efficient. This reciprocity of binding site recognition and transcriptional efficiency provides a mechanism for the moderate upregulation of hlyE expression in response to two distinct environmental signals (oxygen and glucose starvation) rather than, for example, the much larger degree of upregulation when FNR and CRP act synergistically by binding at different sites within the ansB promoter (28). In principle, any pair of transcription factors that recognize similar DNA sequences but have different transcriptional efficiencies could adopt this strategy.
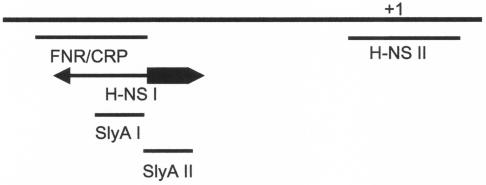
Under some of the conditions studied here, the effects of an hns lesion on hlyE::lacZ expression in liquid cultures were different from those reported by Westermark et al. for bacteria grown on a solid medium (36). This is perhaps not surprising considering the significant physiological differences between cultures grown in liquid and on solid media. In the previous report (36), H-NS acted as a strong repressor of hlyE expression, whereas here H-NS appears to have a positive effect on hlyE expression in the absence of glucose but an attenuated or negative effect on hlyE expression in the presence of glucose (Fig. 1). This suggests that, in general, H-NS inhibits FNR-driven hlyE expression (Fig. 1) but enhances CRP-driven expression in liquid cultures (compare Fig. 1b and c). Moreover, any enhancement of hlyE::lacZ expression in the hns strain observed here did not approach the eightfold increase observed on solid medium (36). Thus, it appears that H-NS modulates PhlyE activity both positively and negatively in response to the prevailing growth and/or environmental conditions, such as growth in liquid and on solid media. This may be a reflection of the formation of different H-NS-PhlyE complexes in response to environmental signals. This idea is supported by the identification of two regions within the larger previously described H-NS-PhlyE complex (36) that bind H-NS with high affinities. The first region (H-NS I) overlaps with, and extends downstream from, the FNR/CRP site (Fig. 6). The extent of protection associated with the H-NS I site (38 bp) suggests that more than one H-NS dimer is bound, and the formation of different subcomplexes could potentially act to modulate FNR/CRP-driven transcription activation either positively (occupation of the downstream portion of H-NS I; thick arrow in Fig. 6) or negatively (occupation of sequences overlapping the FNR/CRP-binding site; thin arrow in Fig. 6). While H-NS usually acts as a negative regulator of gene expression, hlyE is not the only example of a CRP-regulated gene whose expression is positively regulated by H-NS; for example, expression of both the malT and csiD genes in E. coli is stimulated by H-NS (8, 14). Thus, it would appear that a CRP family transcription factor plus H-NS is a versatile combination, providing the means to generate a range of regulatory effects.
FIG. 6.
Schematic representation of the hlyE promoter region. The locations of the FNR/CRP box and SlyA sites are shown. The high-affinity H-NS binding regions observed in DNase I footprints of PhlyE are also shown. The transcript start site (+1) is marked. The region of the H-NS I site that extends downstream of the FNR/CRP box is indicated by a thick arrow, and the part overlapping the FNR/CRP box is indicated by a thin arrow.
The second region of H-NS protection (H-NS II) overlaps the basic promoter elements, and H-NS bound at this region would be expected to repress hlyE expression by promoter occlusion. The findings that SlyA does not enhance hlyE expression in hns cultures grown in liquid medium under anaerobic conditions and that SlyA appears unable to activate hlyE expression in the absence of FNR and CRP and the determination of the location of the region of PhlyE occupied by SlyA (Fig. 6) suggest that SlyA activates hlyE expression by antagonizing the negative action of H-NS. Previous studies have indicated that the intracellular levels of H-NS change with the growth phase. However, whereas one proteomics-based study suggested a fivefold increase in H-NS in stationary-phase cultures (31), a more recent immunological analysis suggested a twofold decrease (4). In addition, we have shown that intracellular H-NS levels increase as growth temperature decreases, as evidenced by an approximately twofold increase in H-NS immunoblot signal intensity from cultures grown at 20°C compared to those grown at 37°C (N. R. Wyborn and J. Green, unpublished data). Thus, it appears that intracellular H-NS levels are influenced by the environment. Therefore, we suggest that when environmental conditions dictate a low intracellular concentration of H-NS, the H-NS I region of PhlyE is occupied to modulate the activity of the upstream activator (FNR or CRP). As the intracellular levels of H-NS increase, H-NS II is occupied and hlyE expression is downregulated by promoter occlusion. The footprinting studies described by Westermark et al. (36) indicated that further increases in H-NS levels result in the formation of higher order complexes that occupy PhlyE from the −10 element to position +172 and from just upstream of the −35 element to position −137. Thus, it appears that these higher order complexes extend from the primary sites of interaction (H-NS I and H-NS II) identified here. Such higher order complexes, which effectively silence hlyE expression, appear to have directionality in that they extend upstream from H-NS I (to as far as position −137) to occlude the FNR/CRP site and downstream from H-NS II (to as far as +172) to occlude the basic promoter elements. This leaves the region between the two primary sites of interaction unoccupied. Thus, we suggest that the formation of different H-NS-PhlyE complexes offers the opportunity for H-NS, in combination with FNR and CRP, to control hlyE expression both positively and negatively. This type of behavior, in which specific patterns of protection are replaced by general protection as the concentration of H-NS increases, has been observed for footprints of gal promoter variants (25). It was also shown previously that this general protection was dependent on the polymerization of H-NS on the DNA and that this was more likely if there was an initial nucleation event on the DNA at specific sites at low H-NS concentrations (25). It appears that the H-NS I and H-NS II regions of PhlyE are such nucleation sites. By competing with H-NS for the region of PhlyE downstream of the FNR/CRP site, SlyA may prevent the formation of the negatively acting H-NS-PhlyE complexes by blocking a primary interaction between H-NS and PhlyE, allowing FNR and CRP to continue to activate hlyE expression. Presumably, the action of the E. coli SlyA protein will prove to be similar to that of the Salmonella protein studied here, given that their primary structures are 89% identical and that the overproduction of E. coli SlyA also enhances hlyE expression (16, 21).
In summary, we have shown that the H-NS-mediated regulation of hlyE expression in E. coli K-12 is more complex than was previously suggested because H-NS can contribute positively as well as negatively to hlyE expression. This range of regulatory activity is probably associated with the progressive formation of higher order H-NS-PhlyE complexes that extend from two primary sites of interaction to ultimately bring about silencing of the hlyE gene. The H-NS and SlyA footprints suggest that H-NS regulation is overcome by the overproduction of SlyA, which by binding at a site that overlaps one of the primary H-NS sites (H-NS I), prevents the formation of repressive H-NS-PhlyE complexes. Further detailed in vitro analyses will be required to fully analyze the complex relationships between H-NS, SlyA, FNR, and CRP and their consequences for hlyE expression.
Acknowledgments
We acknowledge A. J. G. Moir for DNA sequencing. We thank Jeff Cole and Steve Busby (University of Birmingham, Birmingham, United Kingdom) and Bernt Eric Uhlin (Umea University, Umea, Sweden) for providing bacterial strains. We thank Chris Higgins (Hammersmith Hospital, London, United Kingdom) for providing the H-NS protein.
This work was supported by the Biotechnology and Biological Sciences Research Council, Swindon, United Kingdom.
REFERENCES
- 1.Alekshun, M. N., S. B. Levy, T. R. Mealy, B. A. Seaton, and J. F. Head. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 Å resolution. Nat. Struct. Biol. 8:710-714. [DOI] [PubMed] [Google Scholar]
- 2.Atkins, A., N. R. Wyborn, A. J. Wallace, T. J. Stillman, L. K. Black, A. B. Fielding, M. Hisakado, P. J. Artymiuk, and J. Green. 2000. Structure-function relationships of a novel bacterial toxin, hemolysin E. J. Biol. Chem. 275:41150-41155. [DOI] [PubMed] [Google Scholar]
- 3.Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7-17. [DOI] [PubMed] [Google Scholar]
- 4.Azam, T. A., A. Iwata, A. Nishimura, S. Ueda, and A. Ishihama. 1999. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 181:6361-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Busby, S., D. Kotlarz, and H. Buc. 1983. Deletion mutagenesis of the Escherichia coli galactose operon promoter region. J. Mol. Biol. 167:259-274. [DOI] [PubMed] [Google Scholar]
- 6.del Castillo, F. J., S. C. Leal, F. Moreno, and I. del Castillo. 1997. The Escherichia coli K-12 sheA gene encodes a 34-kDa secreted haemolysin. Mol. Microbiol. 25:107-115. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez, S. V., J. Xing, V. Kapur, S. J. Libby, R. G. Barletta, and R. A. Moxley. 1998. Regulation of the Escherichia coli sheA gene and characterization of its encoded hemolytic activity. FEMS Microbiol. Lett. 168:85-90. [DOI] [PubMed] [Google Scholar]
- 8.Germer, J., G. Becker, M. Metzner, and R. Hengge-Aronis. 2001. Role of activator site position and a distal UP-element half-site for sigma factor selectivity at a CRP/H-NS-activated σS-dependent promoter in Escherichia coli. Mol. Microbiol. 41:705-716. [DOI] [PubMed] [Google Scholar]
- 9.Gomez-Gomez, J. M., J. Blazquez, F. Baquero, and J. L. Martinez. 1996. hns mutant unveils the presence of a latent haemolytic activity in Escherichia coli K-12. Mol. Microbiol. 19:909-910. [DOI] [PubMed] [Google Scholar]
- 10.Green, J., and M. Baldwin. 1997. The molecular basis for the differential regulation of the hlyE-encoded haemolysin of Escherichia coli by FNR and HlyX lies in the improved activating region 1 contact of HlyX. Microbiology 143:3785-3793. [DOI] [PubMed] [Google Scholar]
- 11.Green, J., and M. Baldwin. 1997. HlyX, the FNR homologue of Actinobacillus pleuropneumoniae, is a [4Fe-4S]-containing oxygen-responsive transcription regulator that anaerobically activates FNR-dependent class I promoters via an enhanced AR1 contact. Mol. Microbiol. 24:593-605. [DOI] [PubMed] [Google Scholar]
- 12.Green, J., C. Scott, and J. R. Guest. 2001. Functional versatility in the CRP-FNR superfamily of transcription factors: FNR and FLP. Adv. Microb. Physiol. 44:1-34. [DOI] [PubMed] [Google Scholar]
- 13.Guan, K., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli—an improved thrombin cleavage and purification procedure of fusion proteins with glutathione-S-transferase. Anal. Biochem. 192:262-267. [DOI] [PubMed] [Google Scholar]
- 14.Johansson, J., B. Dagberg, E. Richet, and B. E. Uhlin. 1998. H-NS and StpA proteins stimulate expression of the maltose regulon in Escherichia coli. J. Bacteriol. 180:6117-6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lodge, J., R. Williams, A. Bell, B. Chan, and S. Busby. 1990. Comparison of promoter activities in Escherichia coli and Pseudomonas aeruginosa—use of a new broad host range promoter probe plasmid. FEMS Microbiol. Lett. 67:221-225. [DOI] [PubMed] [Google Scholar]
- 16.Ludwig, A., C. Tengel, S. Bauer, A. Bubert, R. Benz, H. Mollenkopf, and W. Goebel. 1995. SlyA, a regulatory protein from Salmonella typhimurium, induces a haemolytic and pore-forming protein in Escherichia coli. Mol. Gen. Genet. 249:474-486. [DOI] [PubMed] [Google Scholar]
- 17.Ludwig, A., S. Bauer, R. Benz, B. Bergmann, and W. Goebel. 1999. Analysis of the SlyA-controlled expression, subcellular localization and pore-forming activity of a 34 kDa haemolysin (ClyA) from Escherichia coli K-12. Mol. Microbiol. 31:557-567. [DOI] [PubMed] [Google Scholar]
- 18.Maxam, A. M., and W. Gilbert. 1980. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 65:499-560. [DOI] [PubMed] [Google Scholar]
- 19.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Miller, P. F., and M. C. Sulavik. 1996. Overlaps and parallels in the regulation of intrinsic multiple-antibiotic resistance in Escherichia coli. Mol. Microbiol. 21:441-448. [DOI] [PubMed] [Google Scholar]
- 21.Oscarsson, J., Y. Mizunoe, B. E. Uhlin, and D. J. Haydon. 1996. Induction of haemolytic activity in Escherichia coli by the slyA gene product. Mol. Microbiol. 20:191-199. [DOI] [PubMed] [Google Scholar]
- 22.Oscarsson, J., Y. Mizunoe, L. Li, X.-H. Lai, A. Weislander, and B. E. Uhlin. 1999. Molecular analysis of the cytolytic protein ClyA (SheA) from Escherichia coli. Mol. Microbiol. 32:1226-1238. [DOI] [PubMed] [Google Scholar]
- 23.Ralph, E. T., J. R. Guest, and J. Green. 1998. Altering the anaerobic transcription factor FNR confers a hemolytic phenotype on Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 95:10449-10452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Revell, P. A., and V. L. Miller. 2000. A chromosomally encoded regulator is required for expression of the Yersinia enterocolitica inv gene and for virulence. Mol. Microbiol. 35:677-685. [DOI] [PubMed] [Google Scholar]
- 25.Rimsky, S., F. Zuber, M. Buckle, and H. Buc. 2001. A molecular mechanism for the repression of transcription by the H-NS protein. Mol. Microbiol. 42:1311-1323. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., and D. W. Russell (ed.). 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Sawers, G., M. Kaiser, A. Sirko, and M. Freundlich. 1997. Transcriptional activation by FNR and CRP: reciprocity of binding site recognition. Mol. Microbiol. 23:835-845. [DOI] [PubMed] [Google Scholar]
- 28.Scott, S., S. Busby, and I. Beacham. 1995. Transcriptional co-activation at the ansB promoters: involvement of the activating regions of CRP and FNR when bound in tandem. Mol. Microbiol. 18:521-531. [DOI] [PubMed] [Google Scholar]
- 29.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 30.Smyth, C. P., T. Lundback, D. Renzoni, G. Siligardi, R. Beavil, M. Layton, J. M. Sidebotham, J. C. D. Hinton, P. C. Driscoll, C. F. Higgins, and J. E. Ladbury. 2000. Oligomerization of the chromatin-structuring protein H-NS. Mol. Microbiol. 36:962-972. [DOI] [PubMed] [Google Scholar]
- 31.Spassky, A., S. Rimsky, H. Garreau, and H. Buc. 1984. H1a, an E. coli DNA-binding protein which accumulates in stationary phase, strongly compacts DNA in vitro. Nucleic Acids Res. 12:5321-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stapleton, M. R., V. A. Norte, R. C. Read, and J. Green. 2002. Interaction of the Salmonella typhimurium transcription and virulence factor SlyA with target DNA and identification of members of the SlyA regulon. J. Biol. Chem. 277:17630-17637. [DOI] [PubMed] [Google Scholar]
- 33.Taylor, D. E., and E. C. Brose. 1988. Modified Birnboim-Doly method for rapid detection of plasmid copy number. Nucleic Acids Res. 16:9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ueguchi, C., T. Suzuki, T. Yoshida, K. Tanaka, and T. Mizuno. 1996. Systematic mutational analysis revealing the functional domain organization of Escherichia coli nucleoid protein H-NS. J. Mol. Biol. 263:149-162. [DOI] [PubMed] [Google Scholar]
- 35.Wallace, A. J., T. J. Stillman, A. Atkins, S. J. Jamieson, P. A. Bullough, J. Green, and P. J. Artymiuk. 2000. E. coli hemolysin E (HlyE, ClyA, SheA): X-ray crystal structure of the toxin and observation of membrane pores by electron microscopy. Cell 100:265-276. [DOI] [PubMed] [Google Scholar]
- 36.Westermark, M., J. Oscarsson, Y. Mizunoe, J. Urbonaviciene, and B. E. Uhlin. 2000. Silencing and activation of ClyA cytotoxin expression in Escherichia coli. J. Bacteriol. 182:6347-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu, R. Y., R. G. Zhang, O. Zagnitko, I. Dementieva, N. Maltzev, J. D. Watson, R. Laskowski, P. Gornicki, and A. Joachimiak. 2003. Crystal structure of Enterococcus faecalis SlyA-like transcriptional factor. J. Biol. Chem. 278:20240-20244. [DOI] [PMC free article] [PubMed] [Google Scholar]