Abstract
In Yersinia pestis, the Congo red (and hemin) binding that is characteristic of the Hms+ phenotype occurs at temperatures up to 34°C but not at higher temperatures. Manifestation of the Hms+ phenotype requires at least five proteins (HmsH, -F, -R, -S, and -T) that are organized into two separate operons: hmsHFRS and hmsT. HmsH and HmsF are outer membrane proteins, while HmsR, HmsS, and HmsT are predicted to be inner membrane proteins. We have used transcriptional reporter constructs, RNA dot blots, and Western blots to examine the expression of hms operons and proteins. Our studies indicate that transcription from the hmsHFRS and hmsT promoters is not regulated by the iron status of the cells, growth temperature, or any of the Hms proteins. In addition, the level of mRNA for both operons is not significantly affected by growth temperature. However, protein levels of HmsH, HmsR, and HmsT in cells grown at 37°C are very low compared to those in cells grown at 26°C, while the amounts of HmsF and HmsS show only a moderate reduction at the higher growth temperature. Neither the Pla protease nor a putative endopeptidase (Y2360) encoded upstream of hmsH is essential for temperature regulation of the Hms+ phenotype. However, HmsT at 37°C is sensitive to degradation by Lon and/or ClpPX. Thus, the stability of HmsH, HmsR, and HmsT proteins likely plays a role in temperature regulation of the Hms+ phenotype of Y. pestis.
Jackson and Burrows (23, 24) defined the pigmentation phenotype (Pgm+) of Yersinia pestis as the ability to form pigmented (greenish-brown) colonies on hemin agar at 26°C and to cause disease in mice without iron supplementation. Genes essential for this phenotype are encoded within a 102-kb locus (pgm locus) in Y. pestis KIM. A siderophore-dependent iron transport system (yersiniabactin), encoded by a high-pathogenicity island (HPI) that is part of the pgm locus, is required for full virulence in mice infected by peripheral routes (3, 4, 7, 14, 16, 27, 31, 61). In a separate region of the pgm locus, there is a single operon containing four genes (hmsHFRS) that are essential for the hemin storage (Hms+) phenotype. Each gene within the hmsHFRS operon (Fig. 1) is required for adsorption of hemin or Congo red (CR) to the outer membrane (OM) fraction of Y. pestis cells (30, 36, 37, 39, 40, 56). A fifth essential gene, hmsT, is located elsewhere in the Y. pestis chromosome (Fig. 1) (18, 25). HmsH and HmsF are OM proteins (36), while the cellular locations of HmsR, HmsS, and HmsT are undetermined. HmsF and HmsR possess domains found in polysaccharide biosynthetic enzymes and glycosyltransferases, respectively, and HmsT belongs to the family of GGDEF proteins (1, 25, 30). None of the Hms proteins contains recognized hemin-binding motifs.
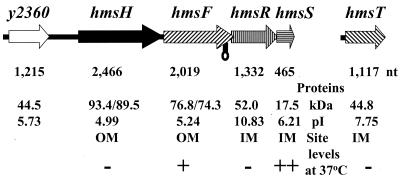
FIG. 1.
Genetic organization of the hmsHFRS and hmsT operons. Gene designations are shown above the arrows (indicating ORFs), while sizes in nucleotides are shown below the arrows. Protein characteristics (size in kilodaltons, number of amino acids, and pI) are shown as well as their location and relative levels at 37°C. The unprocessed and processed molecular masses of HmsH and HmsF are shown. The lollipop indicates a potential stem-loop structure. A minus sign indicates little or no protein at 37°C compared to 26°C, but plus signs indicate moderately reduced protein levels at 37°C.
The Hms system is responsible for the adsorption of enormous quantities of exogenous hemin to cells grown at 26°C. The “stored” hemin is not used nutritionally during periods of iron starvation, and the Hms system is not required for the virulence of bubonic plague in mammals (27, 29, 39). Additional studies have shown that the Hms+ phenotype is not needed for adhesion to or invasion of eukaryotic cells and does not enhance the survival of Y. pestis cells in neutrophils (8, 29). Instead this system is involved in blockage of the proventricular valve of infected fleas (20, 27), which likely plays a role in the transmission of plague to mammals. Hms+ cells colonize the proventriculus, a valve that separates the esophagus from the midgut, and eventually stop the flow of a blood meal into the flea midgut. In these “blocked” fleas, contaminated blood is regurgitated back into the mammalian host, where the bacteria rapidly multiply and establish an infection. Hms− mutants survive and grow in the midgut without causing blockage and flea death (20, 41).
In vitro, adsorption of hemin or CR is temperature dependent: greenish-brown or red colonies form on hemin agar or CR agar, respectively, at 26°C but not at 37°C (23, 56). On CR plates, the transition from red colonies to white colonies is gradual: at incubation temperatures of 32 to 34°C, colonies were less intensely red than those at 26 to 31°C. At 35°C, colonies were faintly pink compared to those at 37°C. This temperature-dependent effect can be overcome by increasing the copy number of the hms genes (25). An Hmsc phenotype (formation of red colonies at 37°C on CR agar) also results from a fur mutation (54). Although a putative Fur-binding site lies ∼250 bp upstream of hmsT, it is unlikely that this sequence is involved in the Fur effect on temperature regulation (25). More importantly, proventricular blockage of the flea is also temperature dependent, with incubation temperatures of 30°C preventing blockage (19). However, the mechanism of temperature regulation of the Hms+ phenotype remains to be elucidated.
In this study, we determine the cellular locations of HmsR, HmsS, and HmsT; examine expression of the hms genes; and analyze Hms protein levels at different growth temperatures. Our results indicate that expression of the Hms phenotype is not controlled at the level of transcription or by mRNA stability. Rather, the stability of select Hms proteins is affected by growth temperature.
MATERIALS AND METHODS
Bacterial strains and cultivation.
All bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli cells were grown in Luria broth (LB). Where appropriate, ampicillin (100 μg/ml), spectinomycin (100 μg/ml), tetracycline (6.25 μg/ml), streptomycin (50 μg/ml), kanamycin (50 μg/ml), or chloramphenicol (10 to 30 μg/ml) was added to cultures. Y. pestis cells were streaked onto CR plates from buffered glycerol stocks stored at −20°C and incubated at 26 to 30°C for 48 h. For most experiments, colonies were inoculated onto Tryptose blood agar base (TBA; Difco Laboratories) slants and incubated at 26 to 30°C for 24 to 48 h. Cells were washed off TBA slants with heart infusion broth (HIB; Difco Laboratories) and grown overnight with aeration in HIB at the appropriate temperature. Cultures were transferred to fresh medium the next morning. For some Western blot analyses, isolated colonies were spread onto CR plates and allowed to form a lawn (2 to 4 days). Cells were scraped off the plate and suspended in sample buffer. For studies of potential iron regulation, cells were washed off TBA slants with the deferrated, defined medium PMH2 (17), inoculated to an optical density at 620 nm (OD620) of 0.1, and incubated overnight in PMH2 at the appropriate temperature with aeration in a New Brunswick model G76 gyratory shaker water bath (200 rpm). After ∼4 generations, cells were inoculated into fresh deferrated PMH2 at an OD620 of 0.1. Growth was monitored with a Genesys5 spectrophotometer (Spectronic Instruments, Inc.), and samples were withdrawn for analysis at indicated times. For iron-replete growth, Y. pestis strains were cultivated in PMH2 supplemented with 10 μM hemin or 10 μM FeCl3. All glassware used for iron-restricted studies was soaked overnight in chromic-sulfuric acid (46.3 g of K2Cr2O7 per liter, 11.25 M sulfuric acid) or 5% Micro-90 (Cole-Parmer Instrument Co.) to remove contaminating iron and copiously rinsed in deionized water.
TABLE 1.
Bacterial strains and plasmids used in this studya
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| Y. pestis | ||
| KIM6+ | Pgm+ Hms+ Pla+ | 14 |
| KIM6 | Pgm− (Δpgm; Hms−) Pla+ | 14 |
| KIM6-2008 | Kmr Hms− (hmsH2008::mini-kan) Pla+ | 36 |
| KIM6-2011 | Kmr Hms− (hmsF2011::mini-kan) Pla+ | 36 |
| KIM6-2012 | Kmr Hms− (hmsR2012::mini-kan) Pla+ | 36 |
| KIM6-2051+ | Kmr Hms− (hmsT2051::mini-kan) Pla+ | 25 |
| KIM6-2057.1 | Hms− (ΔhmsR46, in-frame deletion) Pla+ | 29 |
| KIM6-2058.1 | Hms− (hmsH49) Pla+ | This study |
| KIM6-2093 (pKD46)+ | Apr Cmr Hms+ ΔY2360::cam Pla+ | This study |
| KIM6-2095 (pKD46)+ | Apr Kmr Hmsc Δlon clpPX::kan Pla+ | This study |
| KIM6-2096+ | Cmr Hms+inv::pTinvII hmsT::lacZ Pla+ | This study |
| KIM6-2097 | Apr Pgm− (Δpgm; Hms−) inv::pHinv hmsH::lacZ Pla+ | This study |
| KIM10+ | Hms+ Pla− (cured of pPCP1) | 40 |
| E. coli | ||
| BL21 | Strain for expression of GST fusion proteins | Pharmacia Biotech |
| DH5α | Cloning strain | 2 |
| DH5α (λpir) | Strain for maintenance of R6K ori suicide plasmids | S. C. Straley |
| M15 (pREP4) | His tag expression strain | 62; Qiagen, Inc. |
| Plasmids | ||
| pAHMS16 | 4.4 kb, AprhmsT+, 1.9-kb KpnI-FspI fragment from pAHMS14 into KpnI-HincII sites of pUC18 | 25 |
| pBluescript II KS+ | 2.9 kb, Apr, high-copy-number cloning vector | Stratagene |
| pBSlacZMCS | 7.1 kb, Apr, high-copy-number cloning vector with rrnBT1 transcriptional terminator and lacZ gene from pEU730 | This study |
| pCVD442 | 6.2 kb, AprsacB, R6K ori, suicide vector | 11 |
| pCVDHmsH49 | Apr suicide plasmid encoding hmsH49 from pNPM49 | This study |
| pEU730 | 15.2 kb, Smr Spcr, low-copy-number vector with promotorless lacZ | 15 |
| pGEX-2T | 5.0 kb, AprlacIq GST fusion protein plasmid | Pharmacia Biotech |
| pGEX2T-HmsT | 5.6 kb, Apr, GST-′HmsT expression vector, 588-bp hmsT PCR product ligated into SmaI-BamHI sites of pGEX-2T | This study |
| pGstHmsS | 5.2 kb, Apr, GST-′HmsS expression vector, 231-bp hmsS PCR product ligated into BamHI-EcoRI sites of pGEX-2T | This study |
| pHinv | 11.2 kb, Apr, 4.5-kb EagI fragment from pHmsHlacZ ligated into corresponding site in pSucinv, lacZ expression driven by hmsH promoter region | This study |
| pHMS1.2 | 13.4 kb, AprhmsHFRS+, 9.7-kb SalI-HindIII fragment from pHMS1 into pBR322 | 36 |
| pHMS1.2SL | 13.4 kb, Apr, hmsHFRS with disrupted stem-loop in hmsF | This study |
| pHMS9 | 16.4 kb, Kmr, hmsHFRS with disrupted stem-loop in hmsF, 4.3-kb PmlI-SalI fragment from pHMS1.2SL ligated into corresponding sites of pHMS1 | This study |
| pHmsHlacZ | 7.4 kb, Apr, 314-bp PCR product from pHMS1.2 ligated into PstI-Asp718 sites of pBSlacZMCS | This study |
| pHmsTlacZ | 7.5 kb, Apr, 418-bp PCR product from pAHMS16 ligated into PacI-PmeI sites of pBSlacZMCS | This study |
| pKD3 | 2.8 kb, Cmr, template plasmid | 9 |
| pKD4 | 3.3 kb, Kmr, template plasmid | 9 |
| pKD46 | 6.3 kb, Apr, Red recombinase expression plasmid | 9 |
| pKRP10 | 2.9 kb, Apr Cmr, cam cassette plasmid | 42 |
| pLG338 | 7.3 kb, Kmr Tcr, low-copy-number cloning vector | 55 |
| pNPM49 | ∼16.4-kb, KmrhmsFRS+hmsH, spontaneous mutant | 30 |
| pQE30 | 3.5 kb, Apr, His6 tag expression vector | Qiagen, Inc. |
| pQE30H.6 | 5.6 kb, Apr, His6 tag-′HmsH expression vector; 2.2-kb NcoI-XhoI fragment from pHMS1.2 ligated into HindIII-SalI sites of pQE30 | This study |
| pQE32 | 3.5 kb, Apr, His6 tag expression vector | Qiagen, Inc. |
| pQE32F.2 | 6.0 kb, Apr, His6 tag ′HmsF expression vector; 2.5-kb PmlI-PvuII fragment from pHMS1.2 ligated into SmaI site of pQE32 | This study |
| pRI203 | 9.0 kb, Apr, encodes Y. pseudotuberculosis inv | 22; V. L. Miller |
| pSUC1 | 4.7 kb, Apr, suicide vector derived from pCVD442, sacB, R6K ori | 12 |
| pSucinv | 6.7 kb, Apr, suicide vector with 2.0-kb StuI-XhoI fragment of Y. pseudotuberculosis inv gene from pRI203 ligated into SmaI-SalI sites of pSUC1 | This study |
| pSucinvII | 6.1 kb, Cmr, suicide vector with Y. pseudotuberculosis inv, pSucinv with bla replaced by 0.8-kb EcoRI-HincII fragment from pKRP10 encoding cam | This study |
| pTinvII | 10.6 kb, Cmr, 4.6-kb EagI fragment from pHmsTlacZ ligated into corresponding site in pSucinvII, lacZ expression driven by hmsT promoter region | This study |
All Y. pestis strains are avirulent due to lack of the low-calcium response plasmid pCD1. Strains with a plus sign possess an intact 102-kb pgm locus containing the genes for the hemin storage (hms) and yersiniabactin (Ybt) iron transport system. All other Y. pestis strains have either a pgm deletion or a mutation within the pgm locus. Apr, Cmr, Kmr, Smr, Spcr, and Tcr indicate resistance to ampicillin, chloramphenicol, kanamycin, streptomycin, spectinomycin, and tetracycline, respectively.
Plasmids and recombinant DNA and RNA techniques.
All plasmids used in this study are listed in Table 1. Plasmids were purified from overnight cultures by alkaline lysis (5) and further purified when necessary by polyethylene glycol precipitation (21). Standard cloning and recombinant DNA methods (46, 47) were used to construct the various plasmids listed in Table 1. A standard CaCl2 procedure was used to introduce plasmids into E. coli (47). Y. pestis cells were transformed by electroporation as previously described (13). When necessary, plasmid DNA or PCR products were sequenced by either Retrogen, Inc., or in our laboratory. Sequencing reactions in our laboratory were performed via the dideoxynucleotide chain termination method (48) by using [35S]dATP (Amersham/USB), Sequenase version 2.0 (Amersham/USB), and 7-deaza-dGTP (Boehringer-Mannheim Biochemicals). Samples were electrophoresed through a 6% polyacrylamide gel containing 8.3 M urea cast in Tris-borate-EDTA buffer (47). Dried gels were exposed at room temperature to Kodak Biomax MR film. Synthetic oligonucleotide primers were purchased from Integrated DNA Technologies.
For RNA work, glassware and plasticware were cleaned in 5% Micro-90 (Cole-Parmer Instrument Co.), soaked in 0.1% diethyl pyrocarbonate overnight, copiously rinsed in deionized water, and autoclaved. Bacterial cultures were grown at 26 and 37°C in HIB to the late exponential phase. Cells were resuspended in a mixture of cold TRIzol reagent (Gibco BRL) and zirconia-silica beads (BioSpec Products, Inc.), vortexed for 10 min, and then incubated for 5 to 10 min at room temperature. RNA isolation followed Gibco BRL specifications for the TRIzol Reagent. Briefly, after centrifugation, 0.2 ml of chloroform was added per ml of supernatant, and the samples were mixed and incubated at room temperature for 3 to 10 min. The supernatants were centrifuged, mixed with an equal volume of isopropanol, and incubated for 10 min at room temperature. The RNA was pelleted, rinsed with 75% ethanol, and resuspended in RNase-free water. RNA concentrations were determined spectrophotometrically (47) and confirmed by comparison of the intensities of 16S and 23S ribosomal bands after ethidium bromide staining of agarose gels.
For dot blot analysis, 1 and 0.2 μg of total RNA were suspended in a modified RNA denaturation cocktail (1% SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 6.6% formaldehyde, 50% formamide) and incubated at 65°C for 10 to 15 min (47). One microgram of RNA incubated at 37°C for 45 to 60 min with 1 μg of RNase A (Sigma) per μl was included as a control. After cooling on ice, the samples were blotted onto a Magnacharge nylon membrane (MSI Micron Separations, Inc.) using a Bio-Dot microfiltration system (Bio-Rad) and immobilized by UV cross-linking (Fisher Scientific). Membranes were probed with PCR products labeled with [α-32P]dCTP (ICN Radiochemicals) and a HexaLabel DNA labeling kit (MBI Fermentas) following the manufacturer's specifications. The PCR primers were (i) HmsHprobe F (5′-CGTGGAATCATTAGCACCAC-3′) and HmsHprobe R (5′-CGCTTATCATCTGCTTTCCA-3′) for hmsH, (ii) HmsSprobe F (5′-TTACGGAACAGCGTCTGCTA-3′) and HmsSprobe R (5′-TGAGACACGAGCCACTTTTG-3′) for hmsS, and (iii) HmsTprobe F (5′-TCATGTTACCGCTGATTGGT-3′) and HmsTprobe R (5′-GCCTAAAACACGCTGACGTA-3′) for hmsT. Hybridization was carried out at 42°C followed by high-stringency washing (47). Membranes were exposed overnight to a Phosphor screen, and the images were scanned with a Storm 860 and then quantified with Image Quant 5.0 (Molecular Dynamics Inc.).
Construction of reporter and mutant strains.
The chromosomally integrated hmsT::lacZ reporter strain (KIM6-2096+) was constructed by PCR amplification of a 418-bp fragment from pAHMS16 DNA using primers T-Pro (5′-CCTTAATTAAAATATCGTGCTGTCAGTAG-3′) and PCR-1 (5′-TTCCCGTTAACTATCTACCCAGCCCAGTA-3′). The product was digested with PacI and HpaI and cloned into the PacI-PmeI sites of pBSlacZMCS to yield pHmsTlacZ. An ∼4.6-kb EagI fragment containing the lacZ gene with an hmsT promoter was cloned from pHmsTlacZ into pSucinvII to yield pTinvII (Table 1). pTinvII was electroporated into KIM6+. Transformants were selected on TBA-chloramphenicol plates and confirmed by Southern blot analysis.
The hmsH::lacZ reporter strain (KIM6-2097) was constructed by PCR amplification of a 314-bp fragment from pHMS1.2 DNA by using primers H5b-Pro (5′-GGGGTACCAGAACACTGTATCGCAGCAT-3′) and H3-Pro (5′-AACTGCAGTATATAACCCTTAAGCCAGC-3′). The product was digested with PstI and Asp718 and cloned into corresponding sites in pBSlacZMCS, resulting in pHmsHlacZ. The ∼4.5-kb EagI fragment containing lacZ driven by the hmsH promoter was cloned from pHmsHlacZ into the EagI site of pSucinv. The resulting plasmid, pHinv, was electroporated into KIM6. Transformants were selected on TBA-ampicillin plates and confirmed by Southern blot analysis. Repeated attempts to integrate the hmsH::lacZ reporter into the chromosome of Y. pestis KIM6+ were unsuccessful.
The disrupted stem-loop structure within hmsF in pHMS9 (Table 1) was constructed with two sets of overlapping primers (SL-1A + SL-1B and SL-2A + SL-2B). SL-1A (5′-AAGGACTGGACCCAACCTGAGGGCAATAACGCCATCAGCGGCCCAATACTGGCGGGATGGATGCG-3′) and SL-1B (5′CGGGTAGTAACCAAAACTTTGCGCACCAGACAACTGCAGTTGACGCATCCATCCCGCCAG-3′) were annealed and amplified by PCR to produce a 108-bp product. Amplification of SL-2A (CAAAGTTTTGGTTACTACCCGGATAATTTTATCACTGGTGAACCGCCATTAAAAGATGTCCGCCC-3′) and SL-2B (5′-CTATCGATCATATAAAGGGTACCAGGCAGAAGACAGCACAGGGCGGACATCTTTTAATGGCGG-3′) yielded a 105-bp product. These two overlapping PCR products were then used as a template with SL-1A, SL-2B, and Pfu polymerase to produce a 192-bp PCR product with the disrupted stem-loop sequence. The 192-nucleotide (nt) PCR product was digested with Asp718 and Bsu36I and ligated into the corresponding sites in pHMS1.2, yielding pHMS1.2SL, which encodes the entire hmsHFRS operon with the disrupted stem-loop sequence. To move this mutation to the low-copy-number vector that exhibits temperature regulation of the hmsHFRS operon in Y. pestis, a 4.3-kb PmlI-SalI fragment from pHMS1.2SL was ligated into the corresponding sites of pHMS1, yielding pHMS9 (Table 1). DNA sequencing confirmed that pHMS9 contained the altered sequenced within hmsF (data not shown).
Hms− strain KIM6-2058.1 was constructed by recombination of the hmsH49 mutation into the chromosome of KIM6+. In plasmid pNPM49, the hmsH49 spontaneous mutation causes an Hms− phenotype and yields a truncated HmsH′ polypeptide of ∼44 kDa but a full-length HmsF protein (30). The hmsH49 allele was transferred to pCVD442, and the resulting suicide plasmid, pCVDHmsH49, was electroporated into KIM6+. Cells from a Cr− Apr colony were grown overnight without antibiotic selection and used to select sucrose-resistant isolates that had completed allelic exchange as previously described (4). Southern blot hybridization was used to confirm that the Hms− phenotype was not due to deletion of the pgm locus or the hmsHFRS operon.
We used Red-mediated recombination (9) to inactivate y2360 and the lon-clpX-clpP locus in Y. pestis. pKD46, encoding the Red recombinase, was introduced into Y. pestis KIM6+ by electroporation. Y. pestis KIM6(pKD46)+ cells were grown at 30°C in HIB to an OD620 of ∼0.6 and then incubated for 1.5 h with 0.2% arabinose to induce the Red recombinase. Electrocompetent cells were made from these cultures and transformed with 2 to 5 mg of purified PCR product. Cells were plated on TBA plates containing either kanamycin or chloramphenicol to select for recombinant cells. To generate KIM6-2093+, which has ∼1 kb of the y2360 gene replaced by a Cmr cassette, primers DEPKD3-1 (5′-ATGCGCTGGTGGTGACTGTTTGTGACAATTTCTATTATCTGTGTAGGCTGGAGCTGCTTC-3′) and DEPKD3-2 (5′-AGATCATGTCTTCTAACATAATTGAGCCAACCCCAATGCCATATGAATATCCTCCTTAGT-3′) were used to amplify the Cmr gene from pKD3. To generate KIM6-2095+, which has ∼6.7 kb of the lon clpPX locus replaced by a Kmr cassette, primers PL-1 (5′-TAGCTGGCAAGCAGGATTTCACTTACCAGCATTAGTTTTGTGTAGGCTGGAGCTGCTTC-3′) and PLKD4-2 (5′-AGACGGTAATGTCATACAGTGGCGAACGAGATCAATTTGCATATGAATATCCTCCTTAGT-3′) were usedto amplify the Kmr gene from pKD4. Gene replacements were confirmed by PCR. Attempts to cure the Red recombinase plasmid pKD46 from KIM6-2093+ by incubation at elevated temperature failed. KIM6-2095+ (lon clpP clpX mutant) grew slowly at 37°C, and we did not attempt to cure this strain of pKD46.
β-Galactosidase assays.
Lysates were prepared from cells with the chromosomal hmsH::lacZ or hmsT::lacZ reporters. The cells were grown in HIB or in PMH2 in the presence or absence of iron through two transfers for a total of ∼6 generations, as previously described (54). β-Galactosidase activities were measured spectrophotometrically with a Genesys5 spectrophotometer (Spectronic Instruments, Inc.) following cleavage of ONPG (4-nitrophenyl-β-d-galactopyranoside). Activities are expressed in Miller units (33).
Cellular fractionation of Y. pestis.
Y. pestis cells were grown in HIB at 26°C and harvested during exponential growth. Cellular fractions were separated according to a method described by Lucier et al. (32). Intact cells (suspended in 10 mM Tris-acetate [pH 7.8], 0.2 mM dithiothreitol [DTT], and 0.75 M sucrose) were treated with lysozyme (160 μg/ml) and EDTA to generate spheroplasts. After centrifugation, spheroplasts were resuspended in 0.25 M sucrose-10 mM Tris-acetate-5 mM EDTA-0.2 mM DTT and disrupted by sonication. Intact cells were removed by low-speed centrifugation, and the cytoplasmic fraction was separated from the membrane fraction by centrifugation (240,000 × g for 3 h). The membranes were resuspended in 0.25 M sucrose-5 mM EDTA-0.2 mM DTT and fractionated into inner membrane (IM), mixed membrane, and OM fractions by isopycnic sucrose density gradient centrifugation. Membrane fractions were washed in 60 mM Tris-10 mM MgCl2 [pH 6.8] and resuspended in phosphate-buffered saline containing 0.5% sodium dodecyl sulfate (SDS). All Hms proteins localized to membrane fractions; consequently, periplasmic and cytoplasmic fractions are not shown. Antisera against a synthetic peptide (CYESALKKANLKGYGR) from the carboxyl terminus of E. coli SecY (6), a proven IM protein, was used to evaluate contamination of OM fractions with IM components.
Production of polyclonal antibodies against Hms proteins.
Antibodies against HmsH and HmsF were raised against recombinant His-tagged proteins. For HmsH, pHMS1.2 was digested with NcoI, filled in with Klenow fragment, and ligated to HindIII linkers. A 2.2-kb HindIII-XhoI fragment was ligated into the corresponding sites of pQE30 and transformed into M15(pREP4). This construct (pQE30H.6) expresses a His6-tagged HmsH polypeptide lacking the first 130 amino acids of HmsH. For HmsF, a 2.5-kb PmlI-PvuII fragment of pHMS1.2 was ligated into the SmaI site of pQE30. The protein expressed from this plasmid (pQE32F.2) lacks the first 87 amino acids of HmsF (Table 1). Recombinant protein expression was induced with IPTG, and the proteins were purified with Ni-nitrilotriacetic acid resin following the manufacturer's instructions (Qiagen, Inc.).
Antibodies against HmsS and HmsT were raised from recombinant glutathione S-transferase (GST)-fusion polypeptides. For HmsS, a 231-bp fragment corresponding to its C-terminal 77 amino acids was amplified from pHMS1.2 with Pfu polymerase and primers 5hmsS (5′-CGGGATCCATCTGGGCCAAATACAATCAG-3′) and 3hmsS (5′-CGGAATTCTCCCTGGCGTAAATGGATCAC-3′). The PCR product was digested with BamHI and EcoRI and cloned into the corresponding sites of pGEX-2T to yield pGstHmsS (Table 1). For HmsT, a 588-bp fragment corresponding to the 195 C-terminal amino acids was amplified from pAHMS14 by PCR with Pfu polymerase and the following primers: hmsT.3BH (5′-CGTGGATCCCGTCGCACTGATAATTTCAC-3′) and hmsT.2SM (5′-CGTCCCGGGTCAAGGGGAAGACTGTAC-3′). The PCR product was digested with SmaI and BamHI and ligated into the corresponding sites of pGEX-2T to yield pGEX2T-HmsT (Table 1). For both recombinant plasmids, ligation mixtures were transformed into DH5α. Clones containing the correct inserts were confirmed by PCR, restriction enzyme digest analysis, and/or DNA sequencing and transformed into BL21 for expression of GST-HmsS and GST-HmsT proteins. Expression and purification conditions followed the manufacturer's recommendations (Amersham Pharmacia Biotech), except that recombinant proteins were isolated from purified inclusion bodies as described by Williams et al. (63). Antisera against HmsH, HmsF, HmsS, and HmsT were produced in rabbits by using affinity-purified proteins separated by preparative SDS-polyacrylamide gel electrophoresis (PAGE).
Using the Genetics Computer Group (GCG) program PeptideStructure, a region of HmsR corresponding to the C-terminal 20 amino acids (CKRKRARWVSPDRGIGRVKS) was selected for the production of antibodies. The oligopeptide was conjugated to keyhole limpet hemocyanin (KLH) and used to produce polyclonal antiserum from rabbits (Research Genetics, Inc.).
Western blot analysis.
For Western blot analysis, equal protein concentrations of whole-cell extracts of Y. pestis cells or cellular fractions were separated on polyacrylamide gels containing SDS and immunoblotted to polyvinylidene fluoride membranes (Immobilon P; Millipore). For detection of HmsR, samples were not boiled. A modification of the procedure of Towbin et al. (58) was used for immunodetection. Briefly, membranes were blocked with 5% nonfat dry milk in 10 mM Tris-HCl (pH 7.6)-137 mM NaCl (TBS) with 0.1% Tween 20 (TBST) and then incubated with the appropriate antisera diluted in TBST. Following incubation with horseradish peroxidase (HRP)-conjugated protein A (Amersham Pharmacia Biotech), the immunoreactive proteins were detected with the ECL enhanced chemiluminescence Western blotting detection reagent (Amersham Pharmacia Biotech) and visualized on Kodak Biomax Light film. Levels of Hms and SecY proteins were quantitated with Scion Image.
Sequence analysis.
Potential IM-spanning domains for HmsR, HmsS, and HmsT were identified with TMHMM2.0, TMpred, and HMMTOP 2.0 (26, 34, 59, 60). Mfold Secondary Structure from SeqWeb version 1.1 of the GCG was used to calculate ΔG values for potential secondary structures in hmsF.
RESULTS AND DISCUSSION
Cellular locations of HmsR, HmsS, and HmsT.
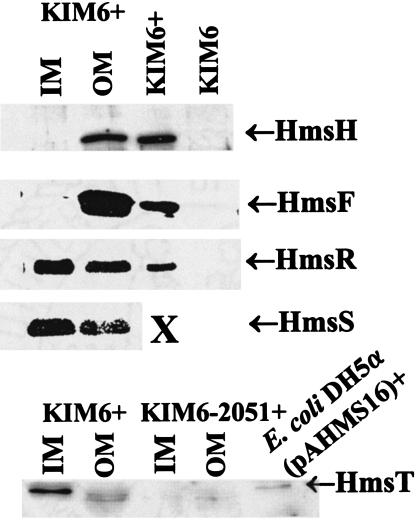
Previously, 125I labeling of whole cells identified HmsH and HmsF as OM proteins (36, 37). Sequence analysis of HmsR, HmsS, and HmsT with TMHMM2.0, TMpred, and HMMTOP 2.0 (26, 34, 59, 60) predicted that all three are IM proteins with five, five, and two transmembrane domains, respectively. Western blot analysis of cellular fractions was used to confirm the cellular locations of all five Hms proteins. Figure 2 shows that HmsH and HmsF were also found in the OM fraction by cellular fractionation. HmsR, HmsS, and HmsT were present predominately in the IM fraction (Fig. 2). Although a significant amount of HmsR and HmsS appeared in the OM fraction, this probably represents contamination of the OM with IM components. Antiserum against the carboxyl terminus of E. coli SecY, an IM protein (6), demonstrated that between 30 and 41% of IM proteins were in the OM fraction (data not shown). Thus, HmsH and HmsF are likely the only two identified Hms proteins that interact with the extracellular environment.
FIG. 2.
Cellular location of Hms proteins. All Hms proteins localized to membrane fractions; consequently, cytoplasmic and periplasmic fractions are not shown. KIM6+, E. coli DH5α(pAHMS16), and KIM6 lanes contain whole-cell extracts used as positive and negative controls. X indicates that unfractionated KIM6+ cell extracts were not tested with antibody against HmsS.
Operon structure of hmsHFRS.
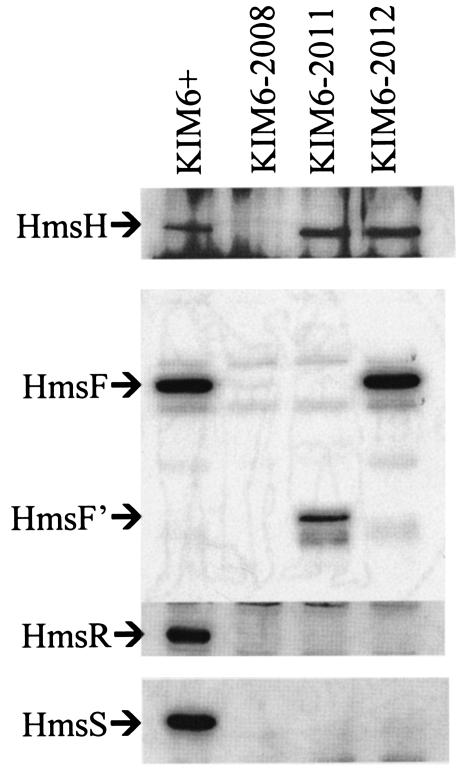
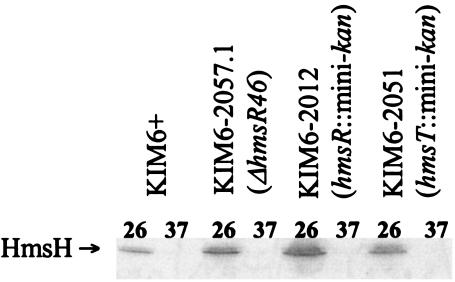
Both sequence and mutational analyses of the hmsHFRS genes indicate that they form a polycistronic operon (30, 36, 37). Western blot analysis of extracts from Y. pestis strains containing polar mini-kan insertions into different hms genes confirmed this. The mini-kan insertions in hmsH, hmsF, and hmsR prevented expression of downstream gene products (Fig. 3). Thus, hmsHFRS constitutes a polycistronic operon.
FIG. 3.
Western blots showing polarity of hmsH, hmsF, and hmsR mutations. Blots were reacted with antiserum against HmsH, HmsF, HmsR, or HmsS. The band designated HmsF′ in panel A is presumably truncated HmsF due to insertion of mini-kan into hmsF. Strains KIM6-2008, KIM6-2011, and KIM6-2012 have hmsH2008::mini-kan, hmsF2011::mini-kan, and hmsR2012::mini-kan insertions, respectively.
Transcriptional regulation of hmsHFRS and hmsT.
Both the adsorption of hemin and inorganic iron by the Hms system and the presence of a potential Fur-binding site upstream of hmsT (25, 39) suggest that the iron status of cells might affect the expression of hms genes. To test this, the promoter regions for hmsHFRS and hmsT were fused to lacZ to examine transcriptional regulation during iron deprivation compared to that under iron- or hemin-surplus growth conditions. The hmsHFRS::lacZ and hmsT::lacZ reporters were integrated into the disrupted chromosomal inv gene of Y. pestis (49) to avoid copy-number effects of reporter plasmids. Repeated attempts to integrate the hmsHFRS::lacZ reporter into a Pgm+ strain failed for unknown reasons. Thus, the hmsHFRS::lacZ reporter studies were conducted in a Δpgm strain. However, Western blot analysis indicates that lack of the hmsHFRS operon does not affect regulation (see below).
Cells containing the appropriate reporter construct were grown at 26 or 37°C to mid-exponential phase in PMH2 with no additions (iron deficient), 10 μM FeCl3, or 10 μM hemin and analyzed for β-galactosidase activity. Transcription from the hmsHFRS and the hmsT promoter regions was not regulated by iron or hemin (Table 2), indicating that the expression of the hms genes is not affected by the iron status of the cell or the availability of exogenous hemin. It also suggests that the putative Fur-binding sequence upstream of hmsT (25) is not functional or controls the expression of a divergently transcribed gene encoded upstream of hmsT.
TABLE 2.
β-Galactosidase activities of Y. pestis containing either the hmsHFRS::lacZ or hmsT::lacZ reportera
| Strain | Mean β-galactosidase activity: (Miller units) of cells grown atb:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 26°C in deferrated PMH2
|
37°C in deferrated PMH2
|
26°C in HIB
|
37°C in HIB
|
|||||||||
| NAc | 10 μM FeCl3 | 10 μM Hemin | NA | 10 μM FeCl3 | 10 μM Hemin | Exponential | Early stationary | Late stationary | Exponential | Early stationary | Late stationary | |
| KIM6-2097 (hmsHFRS::lacZ) | 11,192 ± 1,653 | 8,422 ± 1,733 | 7,614 ± 1,618 | 15,012 ± 2,861 | 13,180 ± 2,530 | 11,772 ± 3,054 | 3,411 ± 426 | 5,438 ± 769 | 8,959 ± 2,023 | 4,584 ± 589 | 6,709 ± 444 | 8,526 ± 1,127 |
| KIM6-2096+ (hmsT::lacZ) | 7,071 ± 1,281 | 6,686 ± 2,840 | 5,873 ± 1,985 | 9,198 ± 2,437 | 7,933 ± 993 | 7,410 ± 1,156 | 2,325 ± 478 | 2,724 ± 787 | 4,256 ± 763 | 6,056 ± 1,584 | 7,499 ± 3,708 | 12,844 ± 1,068 |
Cells were harvested during exponential, early-stationary, and late-stationary phases of growth (HIB cultures) or only during exponential-phase growth (PMH2 cultures).
Enzyme activities are expressed in Miller units (33). The values shown (mean±standard deviations) are the average of two or more individual reactions from three or more independent cultures.
NA, no additions
The effect of growth temperature and cell density on transcription from the hmsHFRS and hmsT promoters was also examined. Cultures were grown at 26 or 37°C, and β-galactosidase activities of mid-exponential-phase cells were compared to those of early- and late-stationary-phase cells. Growth temperature had no significant effect on the β-galactosidase activity of cells with the hmsHFRS::lacZ reporter during any phase of growth (Table 2). However, transcriptional activity from the hmsT promoter was ∼threefold higher at 37°C relative to that at 26°C at all stages of growth (Table 2). This temperature effect was observed in cells grown in HIB but not in those grown in PMH2. In addition, transcription from the hmsHFRS promoter was significantly higher in PMH2 than in HIB at both temperatures (compare exponential-phase cultures in HIB to PMH2 + 10 μM FeCl3 in Table 2). Similar results were seen with the hmsT reporter at 26°C. This suggests that unidentified components of the media can affect the expression of these two operons. In HIB, late-stationary-phase cells consistently had the highest β-galactosidase activities; however, the differences were less than threefold for the hmsHFRS promoter and less than twofold for the hmsT promoter compared to activity in exponential-phase cells. These modest increases could be due to continuous expression of the lacZ gene and accumulation of the relatively long-lived β-galactosidase protein. Overall, these results indicate that temperature regulation of the Hms+ phenotype does not occur at the level of transcription.
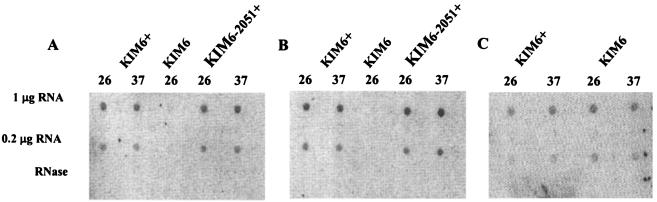
RNA dot blot analysis indicated that the mRNA levels for hmsH, hmsS, and hmsT from cultures grown in HIB were not significantly affected by growth temperature (Fig. 4). The ∼threefold increase observed with the hmsT reporter for cells grown at the higher temperature was not apparent in the RNA dot blot studies. This could be due to differences in the sensitivities of the assays. Alternatively, the reporter gene studies may give aberrant values as a result of the selection of the promoter region and insertion site of the reporter gene. However, these results conclusively demonstrate that transcription of the hmsHFRS and hmsT operons does not control the temperature-dependent expression of the Hms+ phenotype and also eliminates significant mRNA turnover as a potential control mechanism.
FIG. 4.
RNA dot blot. Total RNA from cells of KIM6+ (Hms+), KIM6 (Δpgm), and KIM6-2051+ (hmsT2051::mini-kan) cultured at 26 or 37°C were transferred to nylon membranes and hybridized against probes for hmsH (A), hmsS (B), or hmsT (C). RNase indicates hybridization against 1 μg of RNA treated with RNase A.
Posttranscriptional regulation of the Hms+ phenotype.
There are a number of posttranscriptional mechanisms that may prevent CR adsorption at 37°C. The F1 glycoprotein capsule, which is highly expressed at 37°C but not at 26°C (38), may block CR adsorption. Consequently, we examined the ability of two F1− mutants (K25 Fra− Lcr− and M23+ Fra−) to bind CR at 26 and 37°C. Both mutants showed a normal temperature-regulated Hms+ phenotype (data not shown). Thus, the F1 capsule does not physically prevent CR binding by the Hms system.
We had previously identified a stem-loop structure toward the end of hmsF and just before the start of hmsR (Fig. 1). This structure has some of the characteristics of an antiterminator and might affect message elongation, mRNA stability, or translation of hmsR and hmsS. Since this structure lies within the hmsF open reading frame (ORF), we altered nucleotides that would greatly reduce base pairing in the stem without altering the amino acid sequence of HmsF or introducing unusual codon usage for these amino acids. These nucleotide changes reduced the calculated ΔG from −14.1 to −8.6 kcal mol−1 and were incorporated into pHMS1, which contains the entire hmsHFRS operon on a low-copy plasmid. The resulting plasmid was designated pHMS9 (Table 1). Like KIM6(pHMS1) cells, KIM6(pHMS9) cells maintained a normal Hms+ phenotype: formation of red colonies at 26°C and white colonies at 37°C on CR agar (data not shown). Thus, this stem-loop structure does not seem to play a key role in temperature regulation.
Western blot analysis of Hms protein expression.
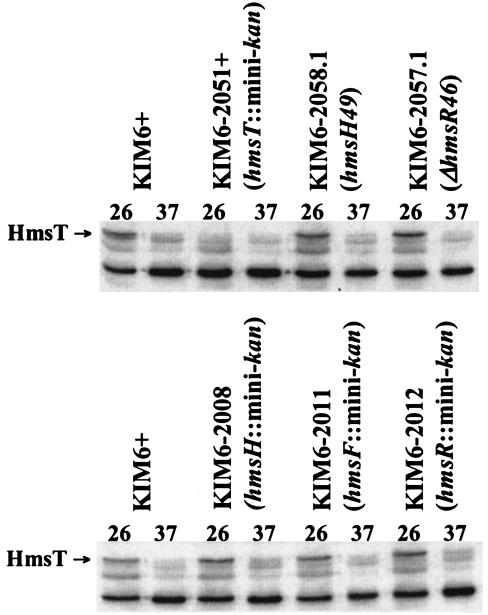
We used Western blot analysis to examine the expression of all five identified Hms proteins. Steady-state levels of HmsH, HmsF, and HmsS did not vary significantly under iron-deficient (no additions) or iron-surplus (FeCl3 or hemin) growth conditions in PMH2 medium (data not shown), confirming the lack of transcriptional regulation by the iron status of cells (Table 2). A comparison of HmsH levels in Hms+ cells, a fur mutant, and various hms mutants grown at 26 and 37°C (Fig. 5) (data not shown) supports three conclusions. First, HmsH levels at 37°C were drastically reduced and barely detectable compared to levels at 26°C. Second, the fur mutation did not increase expression of HmsH at 37°C (data not shown). Third, mutations in hmsR and hmsT did not significantly alter HmsH levels at 37°C (Fig. 5). Similarly, an hmsF::mini-kan mutation does not affect the amount of HmsH present at 37°C (data not shown). Since hmsF::mini-kan and hmsR::mini-kan mutations are polar, we can conclude that the hmsS gene product is also not involved in regulating expression of HmsH. Like HmsH, the levels of HmsT were barely detectable at 37°C (Fig. 6). We also tested whether mutations in hms genes affected levels of the HmsT protein. None of the hms mutations examined affected HmsT protein levels at 26 or 37°C.
FIG. 5.
Mutations in hmsR, hmsS, and hmsT do not affect levels of HmsH protein after growth at 26 or 37°C. Equal concentrations of whole-cell lysates were separated by SDS-PAGE; immunoblots were reacted with the antiserum against HmsH. HmsS is not produced in KIM6-2012 due to polarity of the mini-kan insertion into hmsR, which is upstream of hmsS.
FIG. 6.
Mutations in hmsH, hmsF, hmsR, and hmsS do not affect levels of HmsT after growth at 26 or 37°C. Equal concentrations of whole-cell lysates were separated by SDS-PAGE; immunoblots were reacted with the antiserum against HmsT. The antibody against HmsT cross-reacted with other proteins: one of these bands at 37°C is near the molecular mass of HmsT. Cells of KIM6-2008, KIM6-2011, and KIM6-2012 will not produce HmsS due to polarity of the mini-kan insertions into the hmsHFRS operon.
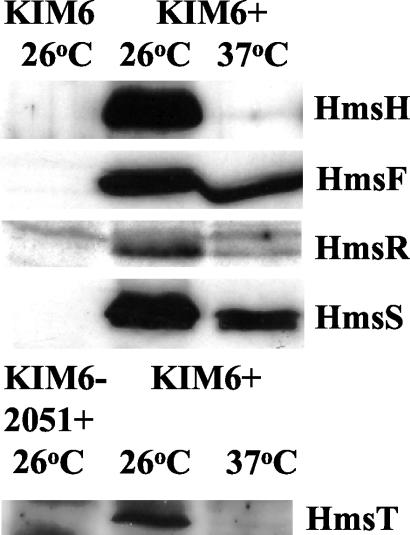
Steady-state levels of all of the Hms proteins were analyzed in HIB cultures grown to the stationary phase overnight at 37 or 26°C. Figure 7 shows that HmsH, HmsR, and HmsT were barely detectable after growth at 37°C. However, HmsF and HmsS levels were only moderately affected by growth temperature. Thus, the protein levels from the first and third genes of the hmsHFRS operon were drastically reduced at the higher growth temperature, while those from the second and fourth genes were only moderately affected (Fig. 1). This alternating pattern of expression makes it unlikely that translational control is responsible for the temperature-dependent expression of the Hms+ phenotype.
FIG. 7.
Western blot analysis of expression of HmsH, HmsF, HmsR, HmsS, and HmsT proteins from Y. pestis KIM6+ (Hms+) and KIM6 (Δpgm; i.e., Hms−) or KIM6-2051+ (hmsT2051::mini-kan) cells grown at 26 or 37°C. Equal concentrations of whole-cell lysates were separated by SDS-PAGE; immunoblots were reacted with the antiserum against individual Hms proteins. Relevant proteins are labeled. HmsT is encoded outside the pgm locus and therefore still expressed in KIM6 cells; consequently, KIM6-2051+ cell extracts were used as a negative control.
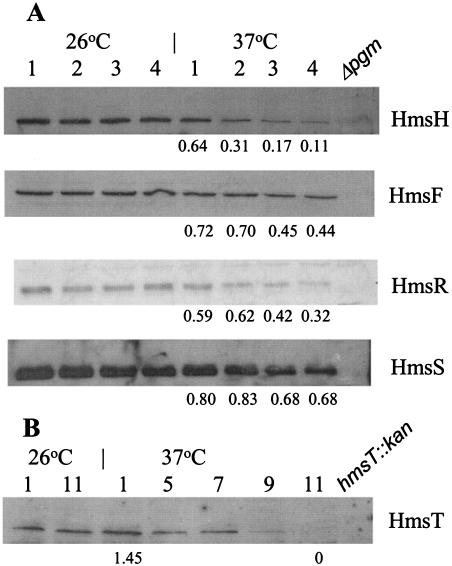
To examine turnover of the different Hms proteins at 37°C, we grew a culture of KIM6+ at 26°C to the mid-exponential phase in HIB. Half of the culture was maintained at 26°C, and the other half was shifted to 37°C. Samples for Western blot analysis were taken at hourly intervals for 4 h after the temperature shift. The levels of HmsH and HmsR were significantly reduced by 1 h of growth at 37°C and barely detectable by 3 or 4 h (11 and 32%, respectively, of the levels in 26°C cultures; Fig. 8). In contrast, HmsF and HmsS levels showed only moderate decreases by 3 to 4 h of incubation, with the levels of HmsF dropping slightly more than that of HmsS (44 and 68%, respectively, of the levels in 26°C cultures; Fig. 8). Surprisingly, HmsT was stable over the 4-h incubation period. In experiments extending the incubation times, we found that HmsT protein levels remained relatively stable up to 7 h after the temperature shift, but were essentially undetectable after 11 h of incubation at 37°C (Fig. 8).
FIG. 8.
Western blot analysis of Hms proteins in Y. pestis KIM6+ (Hms+). Cells were grown at 26°C and then shifted to 37°C or maintained at 26°C. Samples were taken at the indicated times (in hours) after the temperature shift. Equal amounts of whole-cell lysates were separated by SDS-PAGE; immunoblots were reacted with the antiserum against individual Hms proteins. Relevant proteins are labeled. Levels of Hms proteins were quantitated by using Scion Image: numbers below blots indicate the ratio of the indicated protein after growth at 37°C compared to that at 26°C.
Proteolysis of Hms proteins.
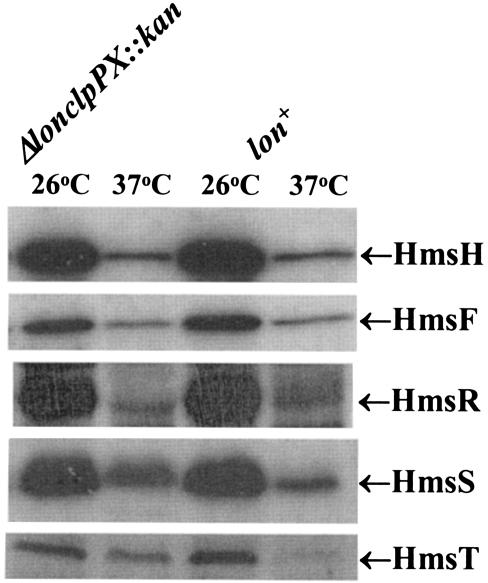
Loss of Hms proteins could be due to increased susceptibility to proteolysis or to increased expression of a protease at 37°C. One candidate is plasminogen activator (Pla) that is encoded on pPCP1. At 37°C, Pla degrades the C3 component of complement as well as some Yersinia outer proteins (Yops) and converts plasminogen to plasmin (28, 51, 52). However, Y. pestis KIM10+ cells, which lack pPCP1 (40, 50), still formed white colonies at 37°C on CR agar. A second candidate is y2360, which is located upstream of hmsH (Fig. 1) (10) and potentially encodes an endopeptidase. Deletion of this gene also failed to cause CR+ colony formation at 37°C. Thus neither Pla nor Y2360 is essential for the loss of CR binding at 37°C. However, a strain (KIM6-2095+) in which the lon and clpPX genes were replaced with a kanamycin resistance cassette (Δlon clpPX::kan) formed reddish colonies at 37°C on CR plates. From the apparent intensity, the mutant bound much more CR than its parent at 37°C but still bound less at 37°C than at 26°C. Due to the poor growth of this mutant in liquid media, lawns of cells were harvested from CR plates after 2 to 4 days of incubation at 37°C. Western blot analysis showed increased levels of HmsT at 37°C compared to those in KIM6+. Even after 4 days of growth of KIM6-2095+ at 37°C, HmsT levels in the mutant remained dramatically increased compared to levels from a 2-day culture of the parental strain (Fig. 9) (data not shown). However, the apparent levels of HmsH, HmsF, HmsR, and HmsS at 37°C were not increased compared to that in KIM6+ (Fig. 9). These results suggest that the levels of HmsT may play a key role in the temperature regulation of the Hms+ phenotype.
FIG. 9.
Effect of lon clpPX protease mutation in KIM6-2095(pKD46)+ on levels of Hms proteins after growth at 26 and 37°C. Equal concentrations of whole-cell lysates were separated by SDS-PAGE; immunoblots were reacted with antiserum against individual Hms proteins. Relevant proteins are labeled. The control lon+ derivative is KIM6(pKD46)+.
Temperature regulation of the Hms+ phenotype.
Our studies indicate a posttranscriptional mechanism is responsible for the lower concentrations of HmsH, HmsR, and HmsT in Hms+ cells grown at 37°C compared to 26°C. Growth temperature had only a moderate effect on the levels of HmsF and HmsS (Fig. 6). Thus, there is an alternating pattern of temperature control of Hms proteins in the hmsHFRS operon (see Fig. 1). Unless individual ORFs are separately affected, this would appear to eliminate translational initiation and elongation as regulatory mechanisms for temperature control of the Hms+ phenotype. However, it remains possible that RNase processing of the hmsHFRS mRNA results in expression of only HmsF and HmsS at 37°C.
Barely detectable or greatly reduced levels of HmsH, HmsR, and HmsT after overnight growth at 37°C compared to 26°C may be one key to temperature regulation. The levels of HmsH and HmsR were significantly lower after 4 h of incubation at 37°C. Curiously, HmsT was stable for up to 7 h at 37°C, followed by complete turnover thereafter. Although it is possible that upon temperature shift, newly synthesized Hms proteins are degraded but membrane-bound forms are stable, the level of HmsH after 4 h at 37°C (Fig. 8) is too low for this to be the exclusive mechanism. An altered tertiary structure at 37°C may make HmsH, HmsR, and HmsT susceptible to proteolytic degradation. On the other hand, they may be sensitive to degradation by a protease that is more highly expressed or active at 37°C than at lower temperatures. We have demonstrated that Lon, ClpXP, and/or ClpAP is involved in the degradation of HmsT at 37°C. Although HmsT is an IM protein, portions of the protein are likely exposed to the cytoplasm and thus susceptible to Lon or Clp proteases. Turnover of HmsH and HmsR clearly requires a different protease that could reside in the periplasm. Increased levels of HmsT in the Δlon clpPX::kan mutant at 37°C result in CR binding. Likewise, moderate- to high-copy-number plasmids carrying hmsHFRS genes cause an Hms+ phenotype at 37°C. This constitutive phenotype corresponds to increased levels of HmsH, -F, -R, and -S at both 26 and 37°C (data not shown).
While the stability of each of the HmsH, HmsR, and HmsT proteins likely plays a role in temperature regulation of the Hms+ phenotype, HmsT may be a key regulatory element. HmsT contains a GGDEF domain that is similar to the catalytic domain of adenylyl cyclase and is predicted to function as a diguanylate cyclase to synthesize cyclic-di-GMP (1, 35, 57). HmsT is 48% identical and 58% similar to AdrA of Salmonella enterica serovar Typhimurium. In Salmonella, AdrA regulates the synthesis of cellulose: cellulose biosynthetic genes are constitutively expressed, but cellulose is only made when adrA is expressed (43, 65). Similarly, in Acetobacter xylinus, the level of cyclic-di-GMP controls the synthesis of a biofilm containing cellulose (44, 45). HmsR possesses a glycosyltransferase domain related to enzymes involved in synthesis of polysaccharides for the formation of a biofilm. In Y. pestis, HmsT may be involved in regulating the activity of HmsR, which may synthesize a biofilm. It is likely that this biofilm, rather than Hms proteins directly, is responsible for CR binding (53, 64) and participates in the blockage of fleas. Clearly, reduced levels of HmsR and possibly HmsH may also participate in temperature regulation of the Hms+ phenotype. Further studies will be needed to conclusively demonstrate that Y. pestis forms a biofilm and the role of the Hms proteins in its development and regulation.
Acknowledgments
This study was supported by Public Health Service grant AI25098 from the National Institutes of Health.
We thank James W. Lillard, Jr., for construction of Y. pestis KIM6-2058.1 and Timothy L. Yahr for providing antiserum against E. coli SecY.
REFERENCES
- 1.Ausmees, N., R. Mayer, H. Weinhouse, G. Volman, D. Amikam, M. Benziman, and M. Lindberg. 2001. Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol. Lett. 204:163-167. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1987. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 3.Bearden, S. W., J. D. Fetherston, and R. D. Perry. 1997. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect. Immun. 65:1659-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bearden, S. W., and R. D. Perry. 1999. The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol. Microbiol. 32:403-414. [DOI] [PubMed] [Google Scholar]
- 5.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brundage, L., C. J. Fimmel, S. Mizushima, and W. Wickner. 1992. SecY, SecE, and band 1 form the membrane-embedded domain of Escherichia coli preprotein translocase. J. Biol. Chem. 267:4166-4170. [PubMed] [Google Scholar]
- 7.Buchrieser, C., M. Prentice, and E. Carniel. 1998. The 102-kilobase unstable region of Yersinia pestis comprises a high-pathogenicity island linked to a pigmentation segment which undergoes internal rearrangement. J. Bacteriol. 180:2321-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowan, C., H. A. Jones, Y. H. Kaya, R. D. Perry, and S. C. Straley. 2000. Invasion of epithelial cells by Yersinia pestis: evidence for a Y. pestis-specific invasin. Infect. Immun. 68:4523-4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng, W., V. Burland, G. Plunkett III, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, S. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fetherston, J. D., V. J. Bertolino, and R. D. Perry. 1999. YbtP and YbtQ: two ABC transporters required for iron uptake in Yersinia pestis. Mol. Microbiol. 32:289-299. [DOI] [PubMed] [Google Scholar]
- 13.Fetherston, J. D., J. W. Lillard, Jr., and R. D. Perry. 1995. Analysis of the pesticin receptor from Yersinia pestis: role in iron-deficient growth and possible regulation by its siderophore. J. Bacteriol. 177:1824-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fetherston, J. D., P. Schuetze, and R. D. Perry. 1992. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol. Microbiol. 6:2693-2704. [DOI] [PubMed] [Google Scholar]
- 15.Froehlich, B., L. Husmann, J. Caron, and J. R. Scott. 1994. Regulation of rns, a positive regulatory factor for pili of enterotoxigenic Escherichia coli. J. Bacteriol. 176:5385-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gehring, A. M., E. DeMoll, J. D. Fetherston, I. Mori, G. F. Mayhew, F. R. Blattner, C. T. Walsh, and R. D. Perry. 1998. Iron acquisition in plague: modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem. Biol. 5:573-586. [DOI] [PubMed] [Google Scholar]
- 17.Gong, S., S. W. Bearden, V. A. Geoffroy, J. D. Fetherston, and R. D. Perry. 2001. Characterization of the Yersinia pestis Yfu ABC inorganic iron transport system. Infect. Immun. 69:2829-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hare, J. M., and K. A. McDonough. 1999. High-frequency RecA-dependent and -independent mechanisms of Congo red binding mutations in Yersinia pestis. J. Bacteriol. 181:4896-4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinnebusch, B. J., E. R. Fischer, and T. G. Schwan. 1998. Evaluation of the role of the Yersinia pestis plasminogen activator and other plasmid-encoded factors in temperature-dependent blockage of the flea. J. Infect. Dis. 178:1406-1415. [DOI] [PubMed] [Google Scholar]
- 20.Hinnebusch, B. J., R. D. Perry, and T. G. Schwan. 1996. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273:367-370. [DOI] [PubMed] [Google Scholar]
- 21.Humphreys, G. O., G. A. Willshaw, and E. S. Anderson. 1975. A simple method for the preparation of large quantities of pure plasmid DNA. Biochim. Biophys. Acta 383:457-463. [DOI] [PubMed] [Google Scholar]
- 22.Isberg, R. R., D. L. Voorhis, and S. Falkow. 1987. Identification of invasin: a protein that allows enteric bacteria to penetrate cultured mammalian cells. Cell 50:769-778. [DOI] [PubMed] [Google Scholar]
- 23.Jackson, S., and T. W. Burrows. 1956. The pigmentation of Pasteurella pestis on a defined medium containing haemin. Br. J. Exp. Pathol. 37:570-576. [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson, S., and T. W. Burrows. 1956. The virulence-enhancing effect of iron on non-pigmented mutants of virulent strains of Pasteurella pestis. Br. J. Exp. Pathol. 37:577-583. [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, H. A., J. W. Lillard, Jr., and R. D. Perry. 1999. HmsT, a protein essential for expression of the haemin storage (Hms+) phenotype of Yersinia pestis. Microbiology 145:2117-2128. [DOI] [PubMed] [Google Scholar]
- 26.Krogh, A., B. Larsson, G. von Heijne, and E. L. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 27.Kutyrev, V. V., A. A. Filippov, O. S. Oparina, and O. A. Protsenko. 1992. Analysis of Yersinia pestis chromosomal determinants Pgm+ and Psts associated with virulence. Microb. Pathog. 12:177-186. [DOI] [PubMed] [Google Scholar]
- 28.Lähteenmäki, K., P. Kuusela, and T. K. Korhonen. 2001. Bacterial plasminogen activators and receptors. FEMS Microbiol. Rev. 25:531-552. [DOI] [PubMed] [Google Scholar]
- 29.Lillard, J. W., Jr., S. W. Bearden, J. D. Fetherston, and R. D. Perry. 1999. The haemin storage (Hms+) phenotype of Yersinia pestis is not essential for the pathogenesis of bubonic plague in mammals. Microbiology 145:197-209. [DOI] [PubMed] [Google Scholar]
- 30.Lillard, J. W., Jr., J. D. Fetherston, L. Pedersen, M. L. Pendrak, and R. D. Perry. 1997. Sequence and genetic analysis of the hemin storage (hms) system of Yersinia pestis. Gene 193:13-21. [DOI] [PubMed] [Google Scholar]
- 31.Lucier, T. S., and R. R. Brubaker. 1992. Determination of genome size, macrorestriction pattern polymorphism, and nonpigmentation-specific deletion in Yersinia pestis by pulsed-field gel electrophoresis. J. Bacteriol. 174:2078-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lucier, T. S., J. D. Fetherston, R. R. Brubaker, and R. D. Perry. 1996. Iron uptake and iron-repressible polypeptides in Yersinia pestis. Infect. Immun. 64:3023-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, J. H. 1992. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Möller, S., M. D. R. Croning, and R. Apweiler. 2001. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17:646-653. [DOI] [PubMed] [Google Scholar]
- 35.Park, Y. W., and H. D. Yun. 1999. Cloning of the Escherichia coli endo-1, 4-d-glucanase gene and identification of its product. Mol. Gen. Genet. 261:236-241. [DOI] [PubMed] [Google Scholar]
- 36.Pendrak, M. L., and R. D. Perry. 1991. Characterization of a hemin-storage locus of Yersinia pestis. Biol. Metals 4:41-47. [DOI] [PubMed] [Google Scholar]
- 37.Pendrak, M. L., and R. D. Perry. 1993. Proteins essential for expression of the Hms+ phenotype of Yersinia pestis. Mol. Microbiol. 8:857-864. [DOI] [PubMed] [Google Scholar]
- 38.Perry, R. D., and J. D. Fetherston. 1997. Yersinia pestis—etiologic agent of plague. Clin. Microbiol. Rev. 10:35-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perry, R. D., T. S. Lucier, D. J. Sikkema, and R. R. Brubaker. 1993. Storage reservoirs of hemin and inorganic iron in Yersinia pestis. Infect. Immun. 61:32-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perry, R. D., M. L. Pendrak, and P. Schuetze. 1990. Identification and cloning of a hemin storage locus involved in the pigmentation phenotype of Yersinia pestis. J. Bacteriol. 172:5929-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pollitzer, R. 1954. Plague. WHO Monogr. Ser. 22:1-698. [Google Scholar]
- 42.Reece, K. S., and G. J. Phillips. 1995. New plasmids carrying antibiotic-resistance cassettes. Gene 165:141-142. [DOI] [PubMed] [Google Scholar]
- 43.Römling, U. 2002. Molecular biology of cellulose production in bacteria. Res. Microbiol. 153:205-212. [DOI] [PubMed] [Google Scholar]
- 44.Ross, P., Y. Aloni, C. Weinhouse, D. Michaeli, P. Weinberger-Ohana, R. Meyer, and M. Benziman. 1985. An unusual guanyl oligonucleotide regulates cellulose synthesis in Acetobacter xylinum. FEBS Lett. 186:191-196. [DOI] [PubMed] [Google Scholar]
- 45.Ross, P., R. Mayer, H. Weinhouse, D. Amikam, Y. Huggirat, M. Benziman, E. de Vroom, A. Fidder, P. de Paus, L. A. J. M. Sliedregt, G. A. van der Marel, and J. H. van Boom. 1990. The cyclic diguanylic acid regulatory system of cellulose synthesis in Acetobacter xylinum. Chemical synthesis and biological activity of cyclic nucleotide dimer, trimer, and phosphothioate derivatives. J. Biol. Chem. 265:18933-18943. [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simonet, M., B. Riot, M. Fortineau, and P. Berche. 1996. Invasin production by Yersinia pestis is abolished by insertion of an IS200-like element within the inv gene. Infect. Immun. 64:375-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sodeinde, O. A., and J. D. Goguen. 1988. Genetic analysis of the 9.5-kilobase virulence plasmid of Yersinia pestis. Infect. Immun. 56:2743-2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sodeinde, O. A., and J. D. Goguen. 1989. Nucleotide sequence of the plasminogen activator gene of Yersinia pestis: relationship to ompT of Escherichia coli and gene E of Salmonella typhimurium. Infect. Immun. 57:1517-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sodeinde, O. A., A. K. Sample, R. R. Brubaker, and J. D. Goguen. 1988. Plasminogen activator/coagulase gene of Yersinia pestis is responsible for degradation of plasmid-encoded outer membrane proteins. Infect. Immun. 56:2749-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spiers, A. J., S. G. Kahn, J. Bohannon, M. Travisano, and P. B. Rainey. 2002. Adaptive divergence in experimental populations of Pseudomonas fluorescens. I. Genetic and phenotypic bases of wrinkly spreader fitness. Genetics 161:33-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Staggs, T. M., J. D. Fetherston, and R. D. Perry. 1994. Pleiotropic effects of a Yersinia pestis fur mutation. J. Bacteriol. 176:7614-7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stoker, N. G., N. F. Fairweather, and B. G. Spratt. 1982. Versatile low-copy-number vectors for cloning in Escherichia coli. Gene 18:335-341. [DOI] [PubMed] [Google Scholar]
- 56.Surgalla, M. J., and E. D. Beesley. 1969. Congo red-agar plating medium for detecting pigmentation in Pasteurella pestis. Appl. Microbiol. 18:834-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tal, R., H. C. Wong, R. Calhoon, D. Gelfand, A. L. Fear, G. Volman, R. Mayer, P. Ross, D. Amikam, H. Weinhouse, A. Cohen, S. Sapir, P. Ohana, and M. Benziman. 1998. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J. Bacteriol. 180:4416-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tusnády, G. E., and I. Simon. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849-850. [DOI] [PubMed] [Google Scholar]
- 60.Tusnády, G. E., and I. Simon. 1998. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 283:489-506. [DOI] [PubMed] [Google Scholar]
- 61.Une, T., and R. R. Brubaker. 1984. In vivo comparison of avirulent Vwa− and Pgm− or Pstr phenotypes of yersiniae. Infect. Immun. 43:895-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Villarejo, M. R., and I. Zabin. 1974. β-Galactosidase from termination and deletion mutant strains. J. Bacteriol. 120:466-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williams, J. A., J. A. Langeland, B. S. Thalley, J. B. Skeath, and S. B. Carroll. 1995. Expression of foreign proteins in E. coli using plasmid vectors and purification of specific polyclonal antibodies, p. 15-58. In D. M. Glover and B. D. Hames (ed.), DNA cloning 2: expression systems. IRL Press, New York, N.Y.
- 64.Wood, P. J. 1980. Specificity in the interaction of direct dyes with polysaccharides. Carbohydr. Res. 85:271-287. [Google Scholar]
- 65.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Römling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452-1463. [DOI] [PubMed] [Google Scholar]