Abstract
An in-depth analysis of the intracellular metabolite concentrations, metabolic fluxes, and gene expression (metabolome, fluxome, and transcriptome, respectively) of lysine-producing Corynebacterium glutamicum ATCC 13287 was performed at different stages of batch culture and revealed distinct phases of growth and lysine production. For this purpose, 13C flux analysis with gas chromatography-mass spectrometry-labeling measurement of free intracellular amino acids, metabolite balancing, and isotopomer modeling were combined with expression profiling via DNA microarrays and with intracellular metabolite quantification. The phase shift from growth to lysine production was accompanied by a decrease in glucose uptake flux, the redirection of flux from the tricarboxylic acid (TCA) cycle towards anaplerotic carboxylation and lysine biosynthesis, transient dynamics of intracellular metabolite pools, such as an increase of lysine up to 40 mM prior to its excretion, and complex changes in the expression of genes for central metabolism. The integrated approach was valuable for the identification of correlations between gene expression and in vivo activity for numerous enzymes. The glucose uptake flux closely corresponded to the expression of glucose phosphotransferase genes. A correlation between flux and expression was also observed for glucose-6-phosphate dehydrogenase, transaldolase, and transketolase and for most TCA cycle genes. In contrast, cytoplasmic malate dehydrogenase expression increased despite a reduction of the TCA cycle flux, probably related to its contribution to NADH regeneration under conditions of reduced growth. Most genes for lysine biosynthesis showed a constant expression level, despite a marked change of the metabolic flux, indicating that they are strongly regulated at the metabolic level. Glyoxylate cycle genes were continuously expressed, but the pathway exhibited in vivo activity only in the later stage. The most pronounced changes in gene expression during cultivation were found for enzymes at entry points into glycolysis, the pentose phosphate pathway, the TCA cycle, and lysine biosynthesis, indicating that these might be of special importance for transcriptional control in C. glutamicum.
The gram-positive bacterium Corynebacterium glutamicum is widely used for the industrial production of different amino acids (4). Hereby lysine is one of the major products, with worldwide production of about 400,000 tons per year and an annual market increase of about 10 to 15% (6). Extensive investigation and optimization of lysine-producing strains of C. glutamicum have been done for several decades. Recent DNA technologies give the opportunity for the rational strain improvement of C. glutamicum by the targeted modification of genes (30). The large potential of this approach was illustrated by a recent study in which a tremendous increase in lysine production was obtained by the mutation of only three genes in the wild-type C. glutamicum strain ATCC 13032 (29). One of the key tasks in targeted strain optimization is the identification of genetic modifications that lead to improved strain characteristics. The experience of the past clearly shows that a detailed quantitative knowledge of metabolic physiology is required for the rational design of superior production strains. Especially for amino acid production by C. glutamicum, which is characterized by a close connection between central metabolism and product biosynthetic pathways, an understanding of global metabolic regulation has turned out to be crucial for effective strain improvement.
Extensive research has been used to sequence the whole genome of C. glutamicum and to investigate its genetic repertoire (2, 9, 23, 27, 35). Metabolic reconstruction via functional gene annotation revealed fascinating insights into this organism, including functional predictions for >60% of the identified genes (14). Gene expression (transcriptome) analysis with C. glutamicum has recently been realized by the development of specific DNA microarrays (14) and was used to investigate gene expression during the growth of C. glutamicum on glucose and acetate (10, 26) and during the production of valine (18). Expression profiles of selected genes of central metabolism (19) and amino acid production (7) of C. glutamicum were determined. For proteome analysis of C. glutamicum, two-dimensional gel electrophoresis was recently used to identify different proteins (11, 12, 13, 31) and to study the influence of nitrogen starvation on the proteome (32). For the quantification of metabolic fluxes (the fluxome), comprehensive approaches combining 13C tracer experiments, metabolite balancing, and isotopomer modeling have been developed (16, 42, 43, 48) and applied to C. glutamicum, involving, e.g., comparative fluxome analysis during growth, glutamate, and lysine production (21, 34), during lysine production in batch cultures (45), of different mutants of a lysine-producing strain genealogy (47), during growth on acetate and/or glucose (40), and during lysine production on different carbon sources (17).
For a full description of the physiological state of a biological system, not one, but all, components (the genome, transcriptome, proteome, intracellular metabolite concentrations [metabolome], and fluxome) have to be analyzed. The different profiling tools have, however, mainly been applied separately to C. glutamicum. Very few studies have used the combined application of different profiling techniques (7). Therefore, our knowledge about metabolic control in C. glutamicum involving the understanding of the links between its different components, e.g., between the transcriptome (expression level of a certain gene) and the fluxome (flux catalyzed by the corresponding enzyme), is still limited.
For the present work, in-depth profiling of lysine production by C. glutamicum was performed with batch cultures by the combined analysis of the metabolome, transcriptome, and fluxome. The characterization was performed at different phases of the cultivation so that the alteration of gene expression, metabolic fluxes, and intracellular metabolite concentrations during the process could be simultaneously estimated.
MATERIALS AND METHODS
Bacterial strain.
Lysine-producing C. glutamicum ATCC 13287 was obtained from the American Type Culture Collection (Manassas, Va.). This homoserine auxotrophic strain was derived from wild-type C. glutamicum ATCC 13032 by UV mutagenesis and selection (28). It overproduces lysine in the absence of threonine due to the release of aspartokinase from concerted feedback inhibition by threonine and lysine.
Media and growth conditions.
Precultures from cells grown on LB5G agar plates (36) were incubated overnight with shaking at 150 rpm at 30°C in a 500-ml baffled shake flask containing 50 ml of LB5G medium. Subsequently, cells were harvested by centrifugation (10,000 × g, 2 min), washed twice, resuspended in 0.9% NaCl, and used as an inoculum for the main culture. The main culture was performed in a 250-ml bioreactor (Meredos, Bovenden, Germany) at 30 ± 0.1°C and shaking at 800 rpm with 125 ml of PMB medium (36) containing 80 mM glucose, 1.3 mM threonine, and 0.2 mM methionine, with the amino acids being added due to the auxotrophy of C. glutamicum ATCC 13287 for homoserine. In tracer experiments, a mixture of 41.4 mM [13C6]glucose and 40.3 mM [1-13C]glucose was used as the carbon source. The pH was controlled at 7.00 ± 0.05 by the addition of 2 M NaOH. For aeration of the bioreactor, pressurized air from a compressor supply system was used. The gas buffer tank was periodically refilled with compressed air from outside, which led to periodic carbon dioxide concentration changes in the aeration gas between about 400 and 800 ppm. As shown below, this input of nonlabeled carbon dioxide was considered in the metabolic model. The aeration rate was maintained at 125 ml min−1 by a mass flow controller (Brooks Instruments, Veenendaal, The Netherlands).
Intracellular metabolite extraction.
For the extraction of intracellular metabolites, cells with between 1 and 4 mg of cell dry mass (CDM) contained in about 0.2 to 2 ml of culture broth were separated from the medium by fast vacuum filtration, four washing steps with 4 ml of 0.9% NaCl each (cellulose nitrate, 0.45-μm-pore-size, 25-mm-diameter filter; Sartorius, Göttingen, Germany), and incubation of the filter with the attached cells for 15 min in 2 ml of boiling water as previously described (49). The extracts were used directly for amino acid quantification with high-performance liquid chromatography (HPLC) or were further processed for labeling analysis with gas chromatography-mass spectrometry (GC/MS) (47).
Analytics.
The optical density (OD) and CDM were determined as described previously (17). The correlation of OD to CDM was as follows: CDM = 0.353 × OD (grams per liter). The CDM was converted into a cell volume for calculation of intracellular concentrations via a correlation factor of 1.95 μl of cytoplasm mg of CDM−1 (8). For the quantification of extracellular concentrations, about 1 ml of culture supernatant was separated by filtration (polyvinylidene fluoride membrane, 0.45-μm pore size; Roth, Karlsruhe, Germany) and diluted 1:10 on an analytical balance. The analysis of organic acids and amino acids was done by HPLC (17). Trehalose and glucose were measured by use of an HPLC apparatus (Biotek, Neufahrn, Germany) equipped with an Aminex HPX 87 H 300- by 7.8-mm column (Bio-Rad, Hercules, Calif.) as the stationary phase and 5 mM H2SO4 as the mobile phase and by refractive index detection. Glycerol and dihydroxyacetone were quantified enzymatically (Boehringer-Mannheim, Darmstadt, Germany). Mean relative errors were 5% (for OD and CDM) and 3% (for sugars, amino acids, and organic acids).
The labeling patterns of free intracellular amino acids glutamate, valine, alanine, and lysine in cell extracts of C. glutamicum were analyzed by GC/MS after derivatization with N-methyl-N-t-butyldimethyl-silyl-trifluoroacetamide (MBDSTFA; Macherey-Nagel, Düren, Germany) (46). For the analysis, 400 μl of cell extract was lyophilized, dissolved in 20 μl of dimethylformamide (0.1% pyridine), and derivatized with 20 μl of MBDSTFA. Mass isotopomer distributions of the derivatized amino acids were quantified by selective ion monitoring at m/z 432 to 437 (glutamate), m/z 232 to 234 (alanine), m/z 288 to 293 (valine), and m/z 431 to 437 (lysine). For glutamate, valine, and lysine, the measured ion clusters resulted from the release of a t-butyl group from the derivatization residue and therefore contained the entire carbon skeleton of the amino acid, whereas the alanine ion cluster represented a fragment ion containing alanine carbons C-2 and C-3. The relative measurement errors of single mass isotopomer fractions, defined as M0 (relative amount of nonlabeled mass isotopomer fraction), M1 (relative amount of singly labeled mass isotopomer fraction), and corresponding terms for higher levels of labeling, were 0.29% (M0), 0.27% (M1), and 0.36% (M2) for alanine; 0.24% (M0), 0.10% (M1), 0.16% (M2), 0.16% (M3), 0.16% (M4), and 0.18% (M5) for valine; 0.24% (M0), 0.16% (M1), 0.13% (M2), 0.13% (M3), 0.10% (M4), and 0.19% (M5) for glutamate; and 1.45% (M0), 0.71% (M1), 0.37% (M2), 0.66% (M3), 0.44% (M4), 0.49% (M5), and 1.09% (M6) for lysine.
Calculation of yields.
As described previously, yields were calculated for each time point from sampling points before and after the examined time point (36). To minimize the effect of measurement errors for single data points, we used the Spline Toolbox of Matlab (Mathworks Inc., Natick, Mass.) to smooth concentrations.
Fluxome analysis.
The distributions of intracellular fluxes during cultivation were estimated by a tracer experiment combined with GC/MS labeling analysis, metabolite balancing, and isotopomer modeling (47). In order to estimate actual flux distributions at different time points, we considered mass isotopomer distributions of free intracellular amino acids. These pools are permanently renewed and thus reflect the actual flux state of the cell. In addition to labeling data, stoichiometric data were included in the flux estimation. These were derived by calculating actual yields for secreted products and actual stoichiometric demands for anabolic precursors at each examined time point from measured extracellular concentrations, as previously described (36). Metabolic fluxes were calculated with a metabolic network model in MATLAB 6.1 and SIMULINK 3.0 (Mathworks Inc.). The mathematical details of the model were described by Wittmann and Heinzle (44, 47). In addition to previous work, an influx of nonlabeled CO2 was included in the model to reflect the relatively high CO2 content in the aeration gas, which contributed to the actual CO2 labeling in the fermentor and, via CO2 incorporation by carboxylating reactions, also to the labeling of intracellular metabolites. The consideration of this reaction markedly improved the fit in all cases. For the calculation of anabolic precursor demands, the cellular composition of C. glutamicum according to Marx et al. (20) was used. For parameter estimation, the boundaries for flux reversibilities were set at 0 (irreversible) and 25 (highly reversible). Thus, a back flux can reach the 25-fold value compared to the net flux of the regarded reversible reaction.
Transcriptome analysis.
Samples for RNA extraction were taken at different time points during fermentation. Cells were harvested by centrifugation at the cultivation temperature (30°C, 10,000 × g, 1 min), separated from the supernatant, and rapidly frozen in dry ice-acetone (−50°C). The samples were stored at −70°C until analysis. Preparation of the DNA arrays was done as follows. Based on sequence information provided by BASF AG (Ludwigshafen, Germany), the amplification of annotated open reading frames from C. glutamicum involved the design and synthesis (Eurogentec, Seraing, Belgium) of specific primer pairs for the amplification of DNA fragments of approximately 500 bp from each protein-specifying gene (26). Genomic DNA (0.5 ng) was used as a template in standard PCRs. Thirty-five cycles of denaturation at 95°C for 30 s, annealing at 55°C for 60 s, and polymerization at 72°C for 60 s were conducted. A 2-μl aliquot of each PCR was analyzed by agarose gel electrophoresis. More than 97% of the resultant PCR products displayed bands of the correct sizes. cDNA glass arrays were subsequently produced by spotting of the PCR products with a Microgrid II robot (Biorobotics, Cambridge, United Kingdom) equipped with an SMP3 pin (ArrayIt, Sunnyvale, Calif.). The array layout included 384 PCR products from Saccharomyces cerevisiae with no homology to C. glutamicum as negative controls. Additionally, each C. glutamicum open reading frame was present at two separate positions on the array. For transcriptome analysis, 15 μg of RNA from each sample was labeled with a Cyscribe direct labeling kit (Amersham, Little Chalfont, United Kingdom) according to the manufacturer's protocol. Each sample was competitively hybridized to the cDNA array together with a z-pool consisting of a pool of all samples. For each time point of the cultivation, duplicate transcriptome analyses were performed. Scanning was performed with a microarray scanner from Agilent Technologies (Waldbronn, Germany). Each gene's measured intensity was divided by its control channel value for each sample. If the control channel value was <0.1, then 0.1 was used instead. If the control channel and signal channel values were both <0.1, then no data were reported. Values below 0 were set to 0. The values given in this publication are the geometric means of two ratios (sample to z-pool) determined in independent measurements.
Chemicals.
Tryptone and yeast extract were purchased from Difco (Detroit, Mich.). All other chemicals were of analytical grade and were purchased from Grüssing (Filsum, Germany), Acros Organics (Geel, Belgium), Merck (Darmstadt, Germany), Aldrich (Steinheim, Germany), and Fluka (Buchs, Switzerland). The tracer substrates 99% [1-13C]glucose and 99% [13C6]glucose were supplied by Campro Scientific (Veenendaal, The Netherlands).
RESULTS AND DISCUSSION
Cultivation profile of lysine-producing C. glutamicum.
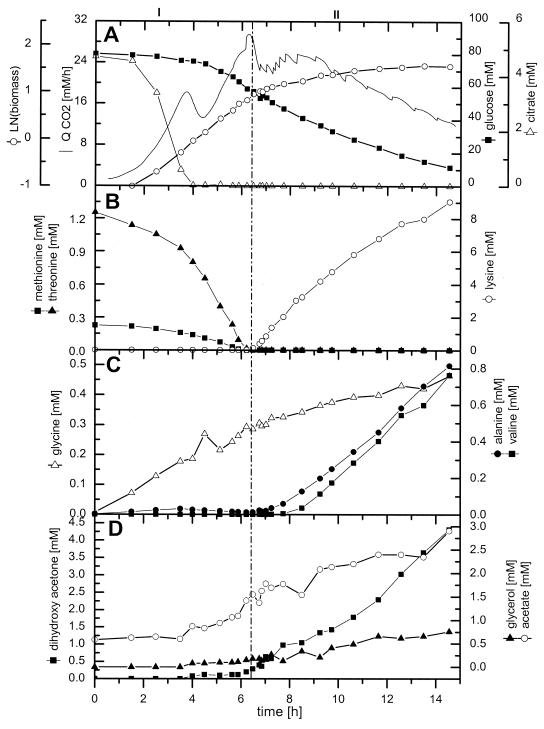
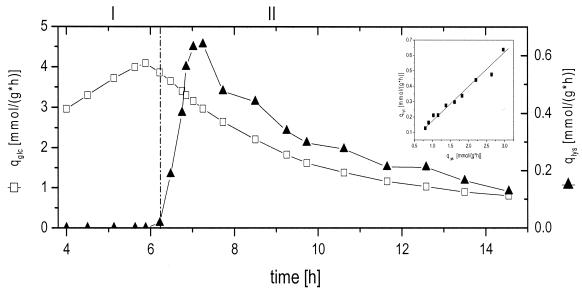
The cultivation profile of C. glutamicum ATCC 13287 is displayed in Fig. 1. During 15 h of cultivation, 9.0 mM lysine and 4.5 g of CDM liter−1 were produced from 70.2 mM glucose. The corresponding yields for lysine and biomass were 0.13 cmol cmol−1 and 0.04 cmol cmol−1, respectively. The cultivation can be divided into two major phases. For the first 6 h of cultivation (growth phase), exponential growth was observed. With the depletion of essential threonine and methionine from the medium, growth was markedly reduced and the production of lysine started (production phase). The shift between the two phases is indicated by a vertical line in Fig. 1. During the growth phase, the specific growth rate was 0.40 h−1, corresponding to a doubling time of 2.1 h. The biomass yield during this phase was 0.08 cmol cmol−1. The growth phase was further characterized by the accumulation of different by-products, with glycine (0.3 mM) and acetate (1.0 mM) showing the largest increases. After 4 h of cultivation, the depletion of citrate, present in the medium as a complexing agent, led to a transient drop in the volumetric carbon dioxide evolution rate, QCO2 (Fig. 1A). After 6.2 h, the essential amino acids threonine and methionine were completely consumed from the medium, resulting in a drastic alteration of the cultivation profile (Fig. 1B). This was due to the fact that the depletion of threonine leads to the release of aspartokinase from concerted feedback inhibition by threonine and lysine (15). The shift in metabolism was indicated by distinct changes in the QCO2 (Fig. 1A). Despite the depletion of threonine and methionine from the medium, cells still continued to grow. During the production phase, the specific growth rate, however, was only 0.07 h−1 and thus significantly lower than that of the growth phase. Interestingly, the accumulation of lysine in the medium was delayed about 30 min compared to threonine and methionine consumption. The biomass yield during this second phase was as follows: YX/S = 0.02 cmol cmol−1. It was thus significantly lower than that for the growth phase. The lysine yield during this phase was as follows: YP/S = 0.18 cmol cmol−1. Together with lysine secretion, other by-products, such as alanine, valine, dihydroxyacetone, and glycerol, accumulated in the medium (Fig. 1C and D). This might have been due to an overflow of central metabolic pathways and was probably a consequence of the disturbed growth of C. glutamicum. The dominant by-product was dihydroxyacetone, with a final concentration of 4.3 mM. Significant accumulation of this compound was previously observed during lysine production of C. glutamicum ATCC 21526 on fructose and glucose (17) and during growth of C. glutamicum ATCC 17865 on fructose (5). Enzymes catalyzing the formation of dihydroxyacetone, such as a dihydroxyacetone phosphatase or dihydroxyacetone kinase, have not yet been clearly identified. Currently, two database entries relate to dihydroxyacetone kinase, whereas dihydroxyacetone phosphatase has not yet been annotated for C. glutamicum (http://www3.ncbi.nlm.nih.gov/Taxonomy/). In addition, other organic acids and alcohols such as glycerol and acetate were observed. Trehalose and lactate accumulated to final concentrations of 0.4 and 0.5 mM, respectively, independently of the cultivation phase (data not shown). The secretion of alanine and valine was restricted to the production phase, whereas glycine was mainly accumulated during the growth phase (Fig. 1C). The calculation of specific rates of glucose consumption (qGlc) and lysine production (qLys) provided additional insights into the cultivation profile (Fig. 2). Between 5 and 6 h of cultivation, qGlc remained almost constant, at 4 mmol g−1 h−1. However, it was significantly reduced during the lysine production phase. Directly with the consumption of threonine and methionine, qGlc started to decrease and approached a value of 1 mmol g−1 h−1 towards the end of the cultivation. The specific lysine production rate, qLys, was zero during the growth phase. After 6 h, it increased to a maximum value of 0.65 mmol g−1 h−1 within 60 min, followed by a subsequent decrease. As shown by the insert in Fig. 2, qGlc and qLys were closely correlated during the production phase between 7 and 15 h. The product yields at different time points of the cultivation are given in Table 1. The metabolism of C. glutamicum concerning the stoichiometry of growth and product formation changed markedly over time. The biomass yield during the growth phase after 5.8 h was 86.1 mg of CDM mmol of glucose−1 and was thus significantly higher than the values during the phase shift and lysine production. This was mainly due to the auxotrophy of C. glutamicum ATCC 13287 leading to growth reduction with the depletion of threonine and methionine in the medium. The most remarkable difference between the growth and lysine production phases was observed for the lysine yield, which increased from 0 to about 185 mmol mol glucose−1. Interestingly, the actual lysine yield was almost constant at different time points of the lysine production phase. Despite the reduced specific glucose uptake rate, the cells maintained a constant relative flux towards the desired product. In addition to lysine, different by-products were formed. These by-products stemmed from different parts of the central metabolism, such as glucose 6-phosphate (trehalose), upper glycolysis (glycine, glycerol, and dihydroxyacetone), pyruvate (alanine, valine, and pyruvate), acetyl-coenzyme A (CoA) (acetate), and the tricarboxylic acid (TCA) cycle (oxoglutarate and succinate). The overall secretion of by-products was relatively low during the growth phase but increased during further cultivation. Linked to the biomass yield, the anabolic demand of different precursor metabolites showed a distinct decrease from the growth phase to the production phase (Table 2).
FIG. 1.
Profile of batch cultivation of lysine-producing C. glutamicum ATCC 13287. (A) Volumetric carbon dioxide production rate (QCO2), concentrations of glucose and citrate, and CDM. (B) Concentrations of methionine, threonine, and lysine. (C) Concentrations of glycine, alanine, and valine. (D) Concentrations of glycerol, dihydroxyacetone, and acetate. The beginning of the lysine production phase is marked by a vertical line.
FIG. 2.
Specific glucose uptake rate (qGlc) and lysine production rate (qLys), shown in millimoles per gram per hour, during batch cultivation of lysine-producing C. glutamicum ATCC 13287. The beginning of the lysine production phase is marked by a vertical line. The insert in the figure displays the relationship between qGlc and qLys during hours 7 to 15 of cultivation.
TABLE 1.
Yields of biomass and secreted products of lysine-producing C. glutamicum ATCC 13287 at different time points of batch cultivation
| Product | Yield (mmol mol−1) at indicated timea
|
|||
|---|---|---|---|---|
| 5.8 h | 6.9 h | 8.1 h | 9.2 h | |
| Biomass | 86.1 | 68.1 | 55.5 | 56.2 |
| Lysine | 0.0 | 191.0 | 181.9 | 184.0 |
| Valine | 0.0 | 0.0 | 8.6 | 15.2 |
| Alanine | 0.0 | 0.0 | 3.1 | 17.4 |
| Glycine | 8.3 | 0.0 | 1.7 | 3.9 |
| Glutamate | 0.0 | 0.0 | 0.0 | 0.0 |
| Glycerol | 6.5 | 75.7 | 54.1 | 62.0 |
| Trehalose | 3.9 | 7.9 | 3.1 | 7.4 |
| α-Ketoglutarate | 0.6 | 1.7 | 0.5 | 1.0 |
| Succinate | 11.4 | 0.0 | 12.3 | 0.0 |
| Acetate | 24.1 | 50.8 | 23.7 | 31.1 |
| Lactate | 0.9 | 0.0 | 0.0 | 0.0 |
| Pyruvate | 0.0 | 0.0 | 0.0 | 0.0 |
Data for biomass are given in milligrams of dry biomass per millimole.
TABLE 2.
Anabolic demand for intracellular metabolites of lysine-producing C. glutamicum ATCC 13287 at different time points of batch cultivation
| Precursor | Demand for precursor (mmol mol of glucose−1) at indicated timea
|
|||
|---|---|---|---|---|
| 5.8 h | 6.9 h | 8.1 h | 9.2 h | |
| Glucose 6-phosphate | 17.7 | 14.0 | 11.4 | 11.5 |
| Fructose 6-phosphate | 6.1 | 4.8 | 3.9 | 4.0 |
| Pentose 5-phosphate | 75.7 | 59.9 | 48.8 | 49.4 |
| Erythrose 4-phosphate | 23.1 | 18.3 | 14.9 | 15.1 |
| Glyceraldehyde 3-phosphate | 11.1 | 8.8 | 7.2 | 7.2 |
| 3-Phosphoglycerate | 111.3 | 88.1 | 71.7 | 72.7 |
| Pyruvate-phosphoenolpyruvate | 171.6 | 135.7 | 110.6 | 112 |
| α-Ketoglutarate | 147.2 | 116.5 | 94.9 | 96.1 |
| Oxaloacetate | 77.8 | 61.6 | 50.2 | 50.8 |
| Acetyl-CoA | 215.3 | 170.2 | 138.7 | 140.5 |
| Diaminopimelateb | 12.6 | 9.9 | 8.1 | 8.2 |
| Lysineb | 17.4 | 13.8 | 11.2 | 11.4 |
The estimation of precursor demands was based on the actual biomass yield at the different time points of the cultivation (Table 1) and the biomass composition previously measured for C. glutamicum (20).
Diaminopimelate and lysine are regarded as separate anabolic precursors. This is due to the fact that anabolic fluxes from pyruvate and oxaloacetate into diaminopimelate (cell wall) and lysine (protein) contribute, in addition to the flux of lysine secretion, to the overall flux through the lysine biosynthetic pathway.
Metabolome profile of lysine-producing C. glutamicum.
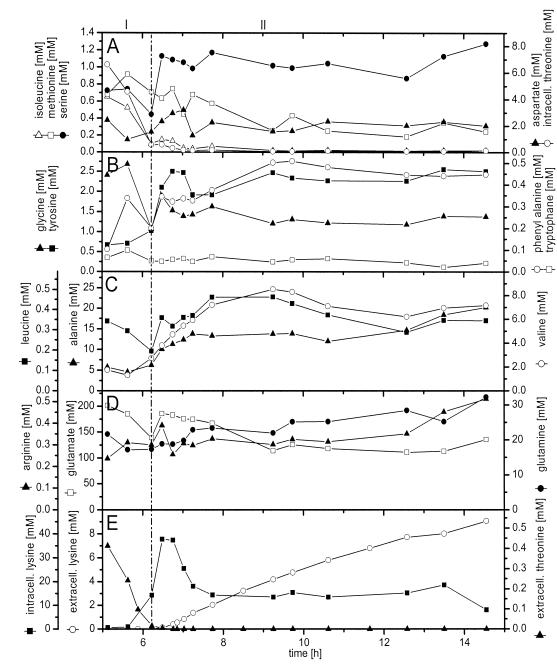
Intracellular concentrations of free amino acids in lysine-producing C. glutamicum were assessed by metabolite extraction and HPLC (Fig. 3). Below we describe the observed dynamics whereby the measured amino acids were grouped according to their biosynthetic pathways.
FIG. 3.
Intracellular and extracellular amino acid concentrations during batch cultivation of lysine-producing C. glutamicum ATCC 13287. (A) Intracellular concentrations of isoleucine, threonine, aspartate, methionine, and serine. (B) Intracellular concentrations of tyrosine, tryptophan, and phenylalanine. (C) Intracellular concentrations of alanine, leucine, and valine. (D) Intracellular concentrations of glutamine, arginine, and glutamate. (E) Intracellular concentration of lysine and extracellular concentrations of lysine and threonine. The beginning of the lysine production phase is marked by a vertical line.
(i) Aspartate family.
A fascinating insight into the regulation of lysine biosynthesis was provided by the intracellular and extracellular concentrations of threonine and lysine (Fig. 1B and 3A and E). Linked to the depletion of threonine from the medium, the intracellular threonine level also decreased, from 7 mM after 5 h to <2 mM after 6 h, and approached almost zero towards the end of the cultivation. During the initial growth phase, the intracellular lysine level remained at about 1 mM. About 60 min before lysine accumulated in the medium, marked changes in the intracellular lysine concentration were observed. Within 1 h, the intracellular lysine pool drastically increased to about 40 mM. It stayed at this elevated level for about 30 min before it again dropped to about 15 mM and remained almost constant during the lysine production phase. Hereby the intracellular decrease coincided with the secretion of lysine into the medium. The actual beginning of lysine production was therefore much earlier than was suggested by extracellular accumulation. Isoleucine, which is solely derived from supplemented threonine by the homoserine dehydrogenase-negative strain C. glutamicum ATCC 13287, had a profile that was very similar to that of threonine, characterized by a decrease during the growth phase and a constant pool during the production phase. In contrast to threonine, methionine was rather constant, at about 0.8 mM, during the growth phase and exhibited a minor decrease until the end of the cultivation. Aspartate, one of the precursors of lysine, was maintained at an almost constant level of about 2 mM, even during the phase shift with the drastically increasing demand for lysine formation. The amount of lysine which accumulated between 5.8 and 6.2 h was about 10 times larger than the actual pool size of aspartate, indicating an effective supply of aspartate in C. glutamicum.
(ii) Glutamate family.
Glutamate exhibited by far the highest intracellular concentration among all of the amino acids (Fig. 3D). During the growth phase, the glutamate pool was rather constant, at about 190 mM. Between 5.8 and 6 h, the initial period of intracellular lysine accumulation, the glutamate pool decreased about 40 mM. This might have been caused by the increase in lysine biosynthesis and the corresponding demand for glutamate as an ammonium donor. The intracellular lysine pool increased about 15 mM, which is equivalent to a glutamate demand of 15 mM via the dehydrogenase pathway and 30 mM via the succinylase pathway. The glutamate pool, however, was rapidly refilled after this initial decrease. The concentrations of glutamine and arginine remained almost constant during the cultivation.
(iii) Pyruvate family.
Intracellular alanine and valine stayed constant during the growth phase, at 5 and 2 mM, respectively (Fig. 3C). After the phase shift, both pools increased. Interestingly, the intracellular accumulation of alanine and valine correlated well with their extracellular accumulation, a behavior that was very different from that of lysine. During the lysine production phase, intracellular alanine and valine were maintained at elevated levels of about 14 and 7 mM. It seems that both compounds formed from pyruvate are overflow metabolites and may accumulate because of the down-regulation of central metabolic pathways during reduced growth.
(iv) 3-Phosphoglycerate family.
The intracellular profiles of glycine and serine, both originating from 3-phosphoglycerate, showed a strong correlation (Fig. 3A and B). The two amino acids exhibited a decrease just before the start of intracellular lysine accumulation, a fast replenishment of their intracellular pools during the following phase shift, and constant pools during the rest of the process. The level of serine during the growth phase (0.7 mM) was lower than that during the lysine production phase (1.2 mM). The opposite was found for glycine.
(v) Aromatic amino acid family.
The pool sizes of tyrosine, phenylalanine, and tryptophan were small throughout the whole cultivation (Fig. 3B). Tryptophan levels remained almost constant during the process. Tyrosine and phenylalanine exhibited a slight increase during the phase shift and stabilized at about 2.5 and 0.4 mM, respectively, towards the end of the cultivation.
In summary, the metabolic switch from growth to lysine production in C. glutamicum was accompanied by drastic changes in intracellular amino acid pools. The alterations of the different amino acids did not follow a general trend, but instead revealed a complex pattern. In contrast, the lysine production phase was characterized by rather constant intracellular pools until the end of the cultivation.
Fluxome profile of lysine-producing C. glutamicum. (i) 13C labeling analysis of free intracellular amino acids by MS.
For an estimation of intracellular flux distributions during the cultivation of lysine-producing C. glutamicum, an MS analysis of free intracellular amino acids in cell extracts, taken at different time points from the reactor, was performed (Table 3). Glutamate, alanine, and valine were detected with high signal intensities in all samples (data not shown). Lysine labeling was quantified in all samples except the first one during the growth phase, which is likely caused by the initially low intracellular lysine concentration (Fig. 3E). The easy measurability of glutamate, alanine, and valine is probably due to their high intracellular pool sizes. Signals of other amino acids with lower intracellular abundance were less intense. Such low-intensity signals were not considered for the flux calculation due to the increased probability of background interference (3).
TABLE 3.
Relative mass isotopomer fractions of intracellular alanine, valine, glutamate, and lysine of lysine-producing C. glutamicum ATCC 13287 cultivated in a 1:1 mixture of [1− 13C]- and [13C6]glucose
| Time (h) or data typea | Relative mass isotopomer fractions for amino acid
|
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alanine
|
Valine
|
Glutamate
|
Lysine
|
|||||||||||||||||||
| M0 | M1 | M2 | M0 | M1 | M2 | M3 | M4 | M5 | M0 | M1 | M2 | M3 | M4 | M5 | M0 | M1 | M2 | M3 | M4 | M5 | M6 | |
| 5.8 | ||||||||||||||||||||||
| Exp | 0.289 | 0.263 | 0.448 | 0.080 | 0.118 | 0.193 | 0.243 | 0.162 | 0.204 | 0.043 | 0.104 | 0.204 | 0.263 | 0.203 | 0.157 | *b | ||||||
| Calc | 0.291 | 0.242 | 0.467 | 0.080 | 0.119 | 0.198 | 0.245 | 0.163 | 0.196 | 0.043 | 0.101 | 0.208 | 0.259 | 0.231 | 0.159 | |||||||
| 6.9 | ||||||||||||||||||||||
| Exp | 0.300 | 0.246 | 0.455 | 0.086 | 0.118 | 0.194 | 0.242 | 0.161 | 0.199 | 0.039 | 0.100 | 0.203 | 0.263 | 0.235 | 0.160 | 0.037 | 0.082 | 0.147 | 0.205 | 0.216 | 0.186 | 0.127 |
| Calc | 0.296 | 0.238 | 0.466 | 0.084 | 0.118 | 0.197 | 0.243 | 0.160 | 0.199 | 0.039 | 0.101 | 0.205 | 0.263 | 0.233 | 0.160 | 0.033 | 0.086 | 0.147 | 0.208 | 0.211 | 0.184 | 0.132 |
| 8.1 | ||||||||||||||||||||||
| Exp | 0.290 | 0.245 | 0.465 | 0.082 | 0.118 | 0.194 | 0.243 | 0.163 | 0.200 | 0.037 | 0.097 | 0.201 | 0.264 | 0.239 | 0.162 | 0.035 | 0.081 | 0.148 | 0.207 | 0.217 | 0.186 | 0.126 |
| Calc | 0.291 | 0.240 | 0.468 | 0.080 | 0.117 | 0.196 | 0.244 | 0.162 | 0.201 | 0.037 | 0.098 | 0.203 | 0.263 | 0.237 | 0.163 | 0.030 | 0.080 | 0.146 | 0.209 | 0.217 | 0.188 | 0.130 |
| 9.2 | ||||||||||||||||||||||
| Exp | 0.288 | 0.248 | 0.465 | 0.081 | 0.117 | 0.194 | 0.243 | 0.164 | 0.201 | 0.036 | 0.096 | 0.200 | 0.264 | 0.241 | 0.163 | 0.032 | 0.079 | 0.146 | 0.205 | 0.219 | 0.190 | 0.129 |
| Calc | 0.288 | 0.242 | 0.470 | 0.079 | 0.117 | 0.196 | 0.243 | 0.163 | 0.202 | 0.036 | 0.097 | 0.202 | 0.264 | 0.239 | 0.164 | 0.030 | 0.079 | 0.146 | 0.208 | 0.219 | 0.190 | 0.131 |
Experimental GC/MS data (exp) and values predicted by the solution of the mathematical model corresponding to the optimized set of fluxes (calc) are shown.
*, at this time point the intracellular lysine concentration was too small to obtain reliable mass isotopomer fractions.
(ii) Estimation of intracellular flux distributions.
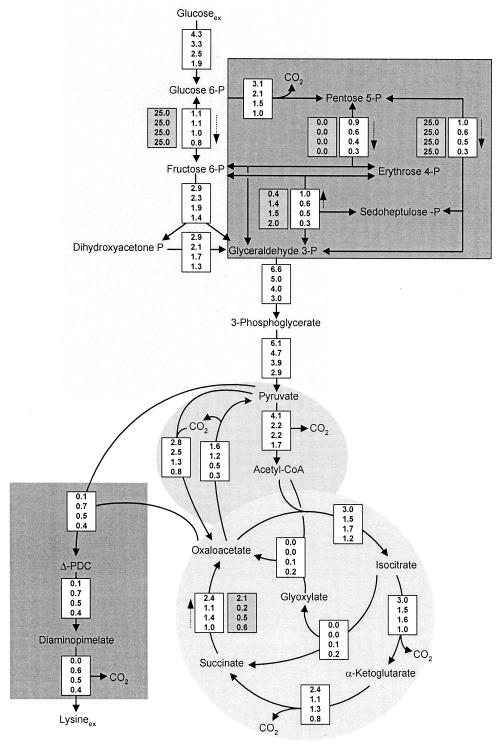
Some of the key tasks of the present work were the quantification of metabolic fluxes at different time points of the cultivation and the elucidation of dynamic changes in the metabolic activity of C. glutamicum. For these purposes, experimental data obtained after 5.8 h (growth phase), 6.9 h (phase shift), and 8.2 and 9.2 h (lysine production phase) of cultivation were used to calculate metabolic flux distributions. For the assessment of the actual fluxome at different time points, free intracellular pools of alanine, valine, lysine, and glutamate were used for labeling measurements. The flux calculations are based on the assumption of a metabolic and isotopic steady state. Clearly, the labeling state of free amino acids is more responsive and much better suited to fit dynamic phenomena than the labeling state of amino acids in cell hydrolysate, which is usually used for metabolic flux analyses. However, some factors that potentially influenced the fluxes obtained in the present study should be mentioned. Whereas a metabolic pseudo-steady-state in free amino acid pools is probably reached quickly, the isotopic steady state might take longer, especially for multiply labeled isotopomers. The time constants of intracellular pools of alanine and lysine in C. glutamicum, calculated from the pool sizes and anabolic demands (49), are in the range of about 4 min and are thus relatively low. Alanine, valine, and lysine are overproduced during the major part of the cultivation, so that efficient renewal of their intracellular pools is guaranteed. The time constant for glutamate in C. glutamicum, however, is about 1 h and is thus significantly higher (49). Although it can be expected that this number is reduced in practice due to various transamination reactions, this time constant might still be nonnegligible. Of further potential relevance is protein turnover, which could (to a certain extent) result in back feeding of historically labeled amino acids into free metabolite pools. We consider the chosen simplified approach to be justified for the phases before and after the shift from growth to lysine production, as each was characterized by relatively constant cultivation parameters, i.e., intracellular amino acid pools. This holds true for flux estimations at 5.8 h (growth phase before the shift) and at 8.2 and 9.2 h (production phase after the shift). More significant changes (e.g., in intracellular amino acid pools) were observed during the phase shift. Thus, the influence of intracellular concentration changes on flux estimation might be more pronounced for the flux estimation at 6.9 h. The flux data obtained at 6.9 h, directly after the shift, might therefore not be as accurate as the data for the other time points, which should be considered when the flux data are evaluated. To account for the potential influence of system dynamics, we used all flux data with one digit less than is usually done. We are aware of the fact that a complete flux analysis in a dynamic case would require the description of all important metabolic steps with their complete kinetics, involving measurement of the dynamics of concentrations and labeling patterns of all significant metabolites. Such complete dynamic analyses, however, entail a tremendous experimental and computational effort.
Approaches based on the labeling of free intracellular amino acids have proven useful in previous studies of metabolic fluxes of C. glutamicum in batch cultures. Nuclear magnetic resonance (NMR)-measured labeling of free intracellular glutamate was included in a 13C flux analysis of C. glutamicum grown on fructose (5), whereas free intracellular alanine and glutamate were analyzed by NMR for a flux estimation during the growth and overproduction of amino acids of C. glutamicum in batch cultures (34). Admittedly, the fluxes estimated for the present work and for previous studies may to a certain extent be influenced by process dynamics, but they are still very valuable for investigations of metabolic physiology in batch cultures.
Parameter estimation was done by minimizing the deviation between experimental and calculated mass isotopomer fractions of intracellular glutamate, alanine, valine, and (except for the growth phase) lysine. The considered mass isotopomer distributions provided 10 labeling constraints for flux estimation during the growth phase and 15 labeling constraints during the phase shift and the lysine production phase, which led to an over-determined network in all cases that could be solved by a least-squares approach. The approach we performed utilized metabolite balancing during each step of the optimization, involving stoichiometric data on product formation (Table 1) and stoichiometric data on precursor demand for biomass formation (Table 2). The set of intracellular fluxes that gave the minimum deviation between experimental and simulated labeling patterns was taken as the best estimate for the intracellular flux distribution. For all time points, identical flux distributions were obtained with multiple initialization values for the flux parameters, suggesting that global minima were identified in all examined cases. Obviously, a high level of agreement between experimentally determined and calculated mass isotopomer ratios was achieved (Table 3). The actual intracellular flux distributions during the cultivation of lysine-producing C. glutamicum are shown in Fig. 4. All values are expressed as specific fluxes, in millimoles per gram per hour. The influxes of nonlabeled CO2 from the aeration gas (see above) were determined to be 8.8 mmol g−1 h−1 (5.8 h), 2.6 mmol g−1 h−1 (6.9 h), 2.7 mmol g−1 h−1 (8.2 h), and 2.2 mmol g−1 h−1 (9.2 h).
FIG. 4.
Intracellular flux distribution of lysine-producing C. glutamicum after 5.8, 6.9, 8.1, and 9.2 h of cultivation (displayed in this order from top to bottom for each reaction). All fluxes are given in millimoles per gram per hour. For reversible reactions, dashed arrows indicate the direction of the net flux and the values in the shaded boxes are the obtained reversibilities of the corresponding enzymes.
(iii) Glucose uptake flux.
The flux for glucose uptake revealed a tremendous decrease during the process. The glucose uptake flux was 4.3 mmol g−1 h−1 after 5.8 h but significantly decreased after the depletion of threonine and methionine (Fig. 4). After 9.2 h, it was reduced almost 60%. The uptake flux for the sole carbon and energy source can be regarded as an important indicator of cellular activity. For lysine-producing C. glutamicum, the deregulation of lysine biosynthesis, which is caused by the absence of essential amino acids, was obviously coupled to a loss of metabolic activity. This had a strong impact on the productivity of the strain, because, despite an almost constant relative flux to lysine biosynthesis during the production phase (Table 1), the corresponding absolute flux markedly decreased (Fig. 4).
(iv) Flux to glycolysis and PPP.
The flux partitioning between glycolysis and the pentose phosphate pathway (PPP) is of special importance for lysine production because PPP is the major source of NADPH, which is required in large amounts for lysine biosynthesis. During the later growth phase, at 5.8 h, relatively large fluxes through glycolysis and PPP resulted (Fig. 4). The switch to lysine production was accompanied by a strong reduction in the PPP flux within 1 h. During this time, the glycolytic flux towards fructose 6-phosphate, however, was kept almost constant. The reduction in the glucose uptake flux during the rest of the cultivation mainly caused a reduction in the PPP flux, which dropped by about 70% of its initial value, whereas the glycolytic flux to fructose 6-phosphate decreased only slightly. The decrease in the absolute flux to PPP resulted in a 70% reduction of the net flux through transaldolase and transketolase to PPP (Fig. 4). During the production phase, the enhanced secretion of dihydroxyacetone and glycerol caused an additional withdrawal of carbon from glycolysis. These compounds were previously observed as by-products of C. glutamicum, especially when grown on fructose, but also to a smaller extent when grown on glucose (5, 17). Previously, a reduction in the flux capacity of glyceraldehyde dehydrogenase was regarded as the reason for the overflow of dihydroxyacetone (5). Probably as a consequence of the reduced glucose uptake, the flux through lower glycolysis decreased during cultivation. Note that these fluxes were affected immediately after the depletion of threonine and methionine in the medium.
Transaldolase and transketolase 1 fluxes were found to be reversible, whereas the transketolase 2 flux was found to be irreversible (Fig. 4). Throughout the growth phase, the phase shift, and initial lysine production, the values obtained were rather similar, whereas the reversibilities of transaldolase and transketolase 1 changed significantly after 9.2 h of cultivation. It is known that the degree of reversibility can generally be quantified only with a high level of uncertainty (41). Nevertheless, the obtained changes in flux reversibility of transaldolase and transketolase 1 after 9.2 h might have indicated changes in intracellular concentrations of PPP intermediates that might have had an influence on the reversibility of the corresponding enzyme.
(v) Flux around the pyruvate node—link between glycolysis, TCA cycle, lysine formation, and anabolism.
Pyruvate is one of the key metabolites in the central metabolism of C. glutamicum (15). The pool of pyruvate connects glycolysis, the TCA cycle, anaplerosis, lysine synthesis, and pathways with different by-products. During cultivation, drastic changes in the fluxes around the pyruvate node were observed (Fig. 4). The fraction of carbon entering the lysine biosynthetic pathway from the pyruvate node exhibited a sevenfold increase during the phase shift between 5.8 and 6.9 h. With the reduction in the absolute glucose uptake flux during further cultivation, the flux into the lysine pathway declined (Fig. 4). A relatively high anaplerotic net flux of 1.2 mmol g−1 h−1 resulted during the growth phase in order to maintain a sufficient supply of oxaloacetate for anabolism (Fig. 4). The anaplerotic net flux increased to 1.3 mmol g−1 h−1 during the phase shift, reflecting the high demand for oxaloacetate for biomass formation and lysine secretion. During further cultivation, the anaplerotic net flux declined to 0.8 mmol g−1 h−1 (at 8.2 h). Interestingly, it was significantly reduced to 0.5 mmol g−1 h−1 after 9.2 h, even though about 0.4 mmol of oxaloacetate g−1 h−1 was still required for lysine biosynthesis. This reflects the induction of the glyoxylate pathway at this stage of the cultivation, which contributed about 40% of the combined flux via anaplerotic carboxylation and the glyoxylate pathway towards oxaloacetate. The flux of pyruvate dehydrogenase decreased almost twofold during the shift from growth to lysine production. Concerning flux partitioning between pyruvate dehydrogenase and anaplerotic carboxylation, the relative flux through pyruvate dehydrogenase decreased from 78% (5.8 h) to 63% (6.9 h), which reflects a significant redirection of the carbon flux from energy metabolism to lysine production. After 9.2 h, it increased again to 77% due to the fact that a significant fraction of the flux towards oxaloacetate was channelled through pyruvate dehydrogenase and the glyoxylate pathway instead of through anaplerotic carboxylation. The reversibility of the flux between the pools of pyruvate and oxaloacetate (ratio of back flux to net flux) was at a relatively high value (1.4) during the growth phase. In contrast, the reversibility was only 1.0 (6.9 h) or 0.6 (8.2 and 9.2 h) during lysine production. This is probably linked to lysine production. Previously, a comparative flux analysis of different lysine-producing mutants of C. glutamicum revealed a clear correlation between a reduced flux reversibility between the pools of pyruvate and oxaloacetate and an increased lysine yield (47).
(vi) Flux through the TCA cycle.
During growth (5.8 h), C. glutamicum exhibited a large amount of flux through the TCA cycle, at 3.0 mmol g−1 h−1 (Fig. 4). Significantly smaller values were found during lysine production, with a sharp decrease of about 50% during the phase shift. The observed growth reduction and the corresponding decreased ATP demand were therefore directly reflected by a reduced flux through the TCA cycle. As a consequence of the reduced TCA cycle flux during lysine production, less NADPH was supplied by isocitrate dehydrogenase. The decrease in the TCA cycle net flux was accompanied by a decrease in the reversibility of the TCA cycle enzymes malate dehydrogenase (MDH) and fumarate hydratase.
(vii) Flux through the glyoxylate pathway.
The glyoxylate pathway was not active during growth and initial lysine production (Fig. 4). Small but significant fluxes, of 0.1 and 0.2 mmol g−1 h−1, were observed after 8.2 and 9.2 h, respectively. Flux partitioning between isocitrate dehydrogenase and isocitrate lyase showed that the major carbon flux (>90%) was still channelled through the TCA cycle. In previous flux studies of the lysine production phase of different C. glutamicum strains, a low level of activity of the glyoxylate pathway was observed (47).
(viii) Flux through lysine biosynthesis.
During growth, a relatively small amount of flux into the lysine biosynthetic pathway (0.1 mmol g−1 h−1) was observed. This flux was exclusively directed towards anabolism, because lysine secretion into the medium did not take place at this stage of the cultivation. During the phase shift between 5.8 and 6.9 h, the flux into lysine biosynthesis had a sevenfold increase due to the release of the strain from concerted feedback inhibition by lysine and threonine. Despite an almost constant molar lysine yield between 6.9 and 9.2 h (Table 1), the absolute flux through the lysine biosynthetic pathway decreased.
Transcriptome profile of lysine-producing C. glutamicum.
During the tracer experiment, samples were taken for transcriptome analysis after 5.6 h (growth phase), after 6.2, 6.5, 6.8, and 6.9 h (phase shift), and after 7.0, 7.3, and 7.7 h (lysine production). Expression data for selected enzymes of central carbon and energy metabolism and lysine production are shown in Tables 4 and 5. Additional transcriptome data, obtained in this study and discussed below, which could not be included in this article for the sake of brevity, are accessible online (http://www.uni-saarland.de/fak8/heinzle). All data are given as relative expression levels compared to a pool of all samples. Most expression levels showed a high level of reproducibility, as indicated by the errors given.
TABLE 4.
Expression profiles of genes of central carbon and energy metabolism during batch cultivation of lysine-producing C. glutamicum ATCC 13287a
| Enzyme | EC no. | Expression (relative level ± SD) at indicated time
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 5.63 h | 6.22 h | 6.48 h | 6.75 h | 6.85 h | 7.02 h | 7.25 h | 7.73 h | ||
| PTS uptake | |||||||||
| Glucose-specific IIABC component | 2.7.1.69 | 0.93 ± 0.03 | 1.77 ± 0.18 | 1.43 ± 0.05 | 1.04 ± 0.10 | 0.90 ± 0.04 | 0.98 ± 0.03 | 0.81 ± 0.01 | 0.62 ± 0.01 |
| β-Glycoside permease IIABC | 0.57 ± 0.02 | 1.27 ± 0.07 | 1.63 ± 0.08 | 1.25 ± 0.13 | 1.06 ± 0.04 | 1.16 ± 0.08 | 1.02 ± 0.09 | 0.83 ± 0.02 | |
| Glycolysis | |||||||||
| G6P isomerase | 5.3.1.9 | 0.72 ± 0.02 | 1.53 ± 0.07 | 1.14 ± 0.06 | 0.85 ± 0.05 | 0.69 ± 0.02 | 0.71 ± 0.08 | 0.64 ± 0.01 | 0.62 ± 0.02 |
| 6-Phosphofructokinase | 2.7.1.11 | 1.30 ± 0.00 | 1.30 ± 0.09 | 0.88 ± 0.08 | 0.92 ± 0.07 | 0.97 ± 0.01 | 1.18 ± 0.11 | 1.08 ± 0.08 | 1.26 ± 0.03 |
| FBP aldolase | 4.1.2.13 | 1.36 ± 0.06 | 1.70 ± 0.22 | 1.08 ± 0.06 | 1.01 ± 0.16 | 1.11 ± 0.15 | 0.87 ± 0.02 | 0.93 ± 0.06 | 0.93 ± 0.02 |
| TP isomerase | 5.3.1.1 | 0.99 ± 0.24 | 1.42 ± 0.11 | 1.43 ± 0.06 | 1.22 ± 0.02 | 0.99 ± 0.01 | 1.10 ± 0.08 | 0.94 ± 0.04 | 0.82 ± 0.05 |
| GA3P DH | 11.2.1.12 | 0.83 ± 0.01 | 1.37 ± 0.18 | 1.05 ± 0.08 | 1.00 ± 0.17 | 0.96 ± 0.06 | 0.99 ± 0.05 | 1.08 ± 0.06 | 1.12 ± 0.03 |
| Pyruvate DH | 1.2.2.2 | 1.62 ± 0.08 | 1.33 ± 0.08 | 1.47 ± 0.09 | 1.49 ± 0.02 | 0.96 ± 0.03 | 1.12 ± 0.02 | 0.80 ± 0.06 | 0.84 ± 0.14 |
| TCA cycle | |||||||||
| Citrate synthase | 4.1.3.7 | 2.09 ± 0.16 | 1.17 ± 0.03 | 1.04 ± 0.01 | 0.91 ± 0.15 | 0.76 ± 0.01 | 0.74 ± 0.00 | 0.78 ± 0.09 | 0.57 ± 0.04 |
| Isocitrate DH | 1.1.1.42 | 1.56 ± 0.01 | 1.33 ± 0.09 | 1.55 ± 0.15 | 1.18 ± 0.10 | 1.16 ± 0.05 | 1.17 ± 0.10 | 0.94 ± 0.03 | 0.60 ± 0.02 |
| Oxoglutarate DH E1 unit | 1.2.4.2 | 2.62 ± 0.14 | 0.99 ± 0.07 | 1.03 ± 0.11 | 0.74 ± 0.03 | 0.74 ± 0.15 | 0.78 ± 0.05 | 0.68 ± 0.02 | 0.58 ± 0.03 |
| SucCoA synthetase α-chain | 6.2.1.5 | 0.70 ± 0.02 | 1.40 ± 0.03 | 1.83 ± 0.50 | 1.34 ± 0.13 | 0.80 | 1.49 ± 0.37 | ND | 0.83 ± 0.04 |
| Malate DH (cytoplasmic) | 1.1.137 | 0.49 ± 0.02 | 1.00 ± 0.08 | 1.17 ± 0.06 | 1.01 ± 0.06 | 1.00 ± 0.17 | 0.96 ± 0.05 | 0.94 ± 0.04 | 1.14 ± 0.11 |
| Malate DH (membrane bound) | 1.1.99.16 | 1.96 ± 0.97 | 1.05 ± 0.07 | 1.13 ± 0.14 | 1.20 ± 0.25 | 2.06 ± 0.20 | 1.38 ± 0.01 | 1.24 ± 0.07 | 1.10 ± 0.10 |
| PPP | |||||||||
| G6P DH | 1.1.1.49 | 5.41 | 1.76 ± 0.12 | 0.80 ± 0.09 | 0.89 ± 0.30 | 1.09 ± 0.19 | 0.86 ± 0.03 | 1.00 ± 0.01 | 1.46 ± 0.06 |
| 6-Phosphogluconate DH | 1.1.1.44 | 1.38 ± 0.11 | 0.94 ± 0.04 | 0.72 ± 0.02 | 0.88 ± 0.04 | 0.80 ± 0.02 | 0.94 ± 0.06 | 0.92 ± 0.03 | 1.18 ± 0.06 |
| Transaldolase | 2.2.1.2 | 1.70 ± 0.13 | 1.14 ± 0.02 | 0.78 ± 0.06 | 0.75 ± 0.09 | 0.83 ± 0.05 | 0.92 ± 0.01 | 1.04 ± 0.01 | 1.21 ± 0.05 |
| Transketolase | 2.2.1.1 | 2.15 ± 0.15 | 1.50 ± 0.15 | 0.81 ± 0.03 | 0.67 ± 0.01 | 0.81 ± 0.02 | 0.76 ± 0.01 | 0.82 ± 0.05 | 0.88 ± 0.09 |
| Anaplerosis | |||||||||
| Pyruvate carboxylase | 6.4.1.1 | 1.99 ± 0.31 | 1.63 ± 0.02 | 1.23 ± 0.13 | 0.98 ± 0.15 | 1.56 | 1.27 ± 0.01 | ND | ND |
| PEP carboxylase | 4.1.1.31 | 2.03 ± 0.72 | 1.81 ± 0.02 | 1.53 ± 0.04 | 1.06 ± 0.10 | 1.15 ± 0.15 | 0.90 ± 0.03 | 0.90 ± 0.05 | 0.80 ± 0.03 |
| PEP carboxykinase | 4.1.1.32 | 4.25 ± 3.40 | 1.84 ± 0.04 | 1.12 ± 0.03 | 1.10 ± 0.37 | 1.11 ± 0.04 | 1.11 ± 0.17 | 1.21 ± 0.22 | 1.29 ± 0.06 |
| Malic enzyme | 1.1.1.39 | 0.69 ± 0.01 | 1.12 ± 0.04 | 1.16 ± 0.00 | 1.11 ± 0.12 | 0.99 ± 0.13 | 0.96 ± 0.06 | 1.11 ± 0.23 | 0.86 ± 0.01 |
| Isocitrate lyase | 4.1.3.1 | 3.83 ± 1.01 | 1.30 ± 0.04 | 0.93 ± 0.08 | 0.66 ± 0.04 | 0.63 ± 0.07 | 0.61 ± 0.01 | 1.00 ± 0.08 | 0.54 ± 0.02 |
| Malate synthase | 4.1.3.2 | 2.12 ± 0.11 | 2.41 ± 0.59 | 1.28 ± 0.01 | 0.75 ± 0.00 | 0.87 | ND | ND | ND |
| ATP formation | |||||||||
| ATP synthase α-chain | 3.6.1.34 | 4.11 ± 0.65 | 1.65 ± 0.01 | 0.58 ± 0.06 | 0.44 ± 0.04 | 0.58 ± 0.07 | 0.52 ± 0.01 | 0.54 ± 0.05 | 0.59 ± 0.05 |
| Respiratory chain | |||||||||
| Cytochrome c oxidase polypeptide 1 | 1.9.3.1 | 0.62 ± 0.06 | 0.65 ± 0.02 | 0.73 ± 0.03 | 1.03 ± 0.01 | 1.14 ± 0.05 | 1.27 ± 0.07 | 1.73 ± 0.11 | 1.43 ± 0.04 |
Gene expression is shown for different time points of the cultivation relating to growth phase (5.63 h), phase shift (6.22, 6.48, 6.75, and 6.85 h), and lysine production (7.02, 7.25, and 7.73 h) and is expressed as relative expression with the deviation between two replicate measurements, except for measurements where only one value was available. ND, not detected.
TABLE 5.
Expression profiles of genes of lysine biosynthesis during batch cultivation of lysine producing C. glutamicum ATCC 13287a
| Enzyme | EC no. | Expression (relative level ± SD) at indicated time
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 5.63 h | 6.22 h | 6.48 h | 6.75 h | 6.85 h | 7.02 h | 7.25 h | 7.73 h | ||
| Aspartate aminotransferase | 2.6.1.1 | 1.66 | 1.42 ± 0.05 | 0.96 | 0.62 ± 0.02 | 0.35 | ND | 0.73 | 0.93 ± 0.05 |
| Aspartate kinase | 2.7.2.4 | 0.80 ± 0.04 | 0.82 ± 0.02 | 1.06 ± 0.07 | 1.13 ± 0.02 | 1.08 ± 0.05 | 1.22 ± 0.01 | 1.15 ± 0.00 | 1.18 ± 0.05 |
| Aspartate semialdehyde DH | 1.2.1.11 | 0.99 ± 0.09 | 0.87 ± 0.09 | 1.03 ± 0.05 | 0.96 ± 0.09 | 1.02 ± 0.02 | 1.02 ± 0.01 | 1.14 ± 0.09 | 1.30 ± 0.06 |
| Dihydrodipicolinate synthase | 4.2.1.52 | 0.95 ± 0.02 | 1.21 ± 0.00 | 1.20 ± 0.09 | 1.05 ± 0.05 | 0.93 ± 0.12 | 1.00 ± 0.07 | 1.00 ± 0.05 | 1.34 ± 0.19 |
| Dihydrodipicolinate reductase | 1.3.1.26 | 0.98 ± 0.11 | 0.95 ± 0.13 | 1.20 ± 0.08 | 1.16 ± 0.05 | 1.27 ± 0.10 | 1.34 ± 0.29 | 0.95 ± 0.06 | 0.96 ± 0.07 |
| Succinyl transferase | 2.3.1.117 | 0.70 ± 0.06 | 0.57 ± 0.06 | 0.78 ± 0.13 | 0.65 ± 0.05 | 0.85 ± 0.20 | 0.70 ± 0.03 | 0.85 ± 0.13 | 0.84 ± 0.04 |
| Succinyl diaminopimelate DS | 3.5.1.18 | 1.06 ± 0.33 | 0.92 ± 0.02 | 1.27 ± 0.29 | 0.98 ± 0.03 | 0.81 ± 0.07 | 1.09 ± 0.06 | 1.04 ± 0.24 | 1.35 ± 0.03 |
| Diaminopimelate epimerase | 5.1.1.7 | 0.63 ± 0.01 | 0.62 ± 0.03 | 0.83 ± 0.08 | 1.02 ± 0.08 | 0.85 ± 0.09 | 0.82 ± 0.02 | 0.76 ± 0.03 | 0.91 ± 0.01 |
| meso-Diaminopimelate DH | 1.4.1.16 | 0.84 ± 0.09 | 1.20 ± 0.01 | 1.07 ± 0.07 | 0.94 ± 0.17 | 1.02 ± 0.09 | 0.72 ± 0.05 | 0.83 ± 0.07 | 0.91 ± 0.01 |
| Diaminopimelate decarboxylase | 4.1.1.20 | 1.74 ± 0.08 | 1.38 ± 0.09 | 1.09 ± 0.01 | 0.83 ± 0.03 | 0.89 ± 0.07 | 0.84 ± 0.01 | 0.76 ± 0.01 | 0.61 ± 0.03 |
| Lysine exporter regulator | 0.34 ± 0.01 | 0.88 ± 0.12 | 1.38 ± 0.22 | 1.04 ± 0.02 | 1.32 ± 0.00 | 1.28 ± 0.12 | 1.45 ± 0.26 | 1.33 ± 0.08 | |
| Lysine exporter | 0.32 ± 0.03 | 0.61 ± 0.03 | 1.14 ± 0.02 | 1.44 ± 0.11 | 1.48 ± 0.09 | 1.70 ± 0.28 | 1.14 ± 0.05 | 1.22 ± 0.03 | |
Gene expression is shown for different time points of the cultivation relating to growth phase (5.63 h), phase shift (6.22 and 6.48 h), and lysine production (6.75, 6.85, 7.02, 7.25, and 7.73 h) and is expressed as relative expression. The deviation between two replicate measurements is shown, except for measurements where only one value was available. ND, not detected.
(i) Expression of genes of glucose uptake.
In C. glutamicum, glucose is taken up by a phosphotransferase system (PTS). The glucose-specific IIABC component and the β-glycoside permease IIABC, which encode parts of the glucose PTS system, had parallel expression profiles during cultivation, which indicates a concerted regulation of gene expression for these PTS components (Table 4). The switch from growth to lysine production was characterized by a strong increase in the expression levels of these two genes. During further cultivation, their expression levels gradually decreased.
(ii) Expression of glycolytic genes.
The expression of glycolytic genes exhibited distinct dynamics during cultivation (Table 4). During the growth phase, genes encoding glucose 6-phosphate isomerase, triosephosphate isomerase, and glyceraldehyde 3-phospate dehydrogenase had distinct increases in transcript level. Other genes, such as those for phosphofructokinase and fructosebisphosphate aldolase, exhibited relatively constant expression levels during this phase. The phase shift was characterized by a general decrease in the expression of glycolytic genes. This correlated with the decreasing substrate uptake flux and the decreasing expression levels of the PTS genes. During the lysine production phase, the levels of transcription of glycolytic genes were more or less unchanged.
(iii) Expression of PPP genes.
The genes coding for glucose 6-phosphate dehydrogenase, 6-phosphogluconate dehydrogenase, transketolase, and transaldolase showed similar expression characteristics (Table 4). This is especially interesting with regard to the fact that the genes are located in different regions of the C. glutamicum genome (http://gib.genes.nig.ac.jp/). The transcript levels of all genes decreased during the switch from growth to lysine production. Glucose 6-phosphate dehydrogenase, catalyzing the entry step into the PPP, had the largest decline. Between 5.6 and 6.5 h of cultivation, the expression level of this gene decreased about sevenfold, whereas the decreases for the other PPP genes were only two- and threefold. During the production phase, the PPP gene levels slightly increased.
(iv) Expression of anaplerotic genes.
The genes for pyruvate carboxylase and phosphoenolpyruvate (PEP) carboxylase were simultaneously expressed during growth, the phase shift, and lysine production (Table 4). Both genes exhibited a gradual decrease in expression during the cultivation. Concerning the back flux from the TCA cycle to glycolysis, expression profiles of PEP carboxykinase and malic enzyme were obtained. Both enzymes were expressed in all samples. The expression level of PEP carboxykinase decreased 3.8-fold between 5.6 and 6.5 h. In contrast, the transcript level for the malic enzyme increased about 1.7-fold during this phase. Both genes thus strongly responded to the depletion of essential threonine and methionine. During further cultivation, the expression levels of PEP carboxykinase and malic enzyme were relatively stable. The expression levels of isocitrate lyase and malate synthase were highest at the beginning of the fermentation and decreased towards the end (Table 4).
(v) Expression of TCA cycle genes.
Different TCA cycle genes were highly expressed during the growth phase at 5.6 h and were down-regulated during the shift from growth to lysine production (Table 4). For example, the expression levels of the genes for citrate synthase, oxoglutarate dehydrogenase, and membrane-bound MDH decreased about twofold between 5.6 and 6.5 h. Towards the end of the cultivation, transcript levels for citrate synthase, isocitrate dehydrogenase, and oxoglutarate dehydrogenase decreased, correlating with the gradually decreasing TCA cycle flux. Exceptions to these results were found with the expression levels of cytoplasmic MDH and succinyl-CoA synthetase, which were enhanced 100% during the phase shift. The two subunits of succinate dehydrogenase had slightly different expression levels (http://www.uni-saarland.de/fak8/heinzle). For the flavoprotein, an almost constant expression level was observed. The transcript level for the iron-sulfur protein slightly decreased during the initial phase of the cultivation and remained rather stable during the phase shift and the lysine production phase. Fumarate hydratase and aconitate hydratase had relatively constant expression levels during the whole process (http://www.uni-saarland.de/fak8/heinzle).
(vi) Expression of lysine biosynthetic genes.
Genes encoding proteins for lysine biosynthesis and secretion were of special importance because they lead to the desired product (Table 5). Only a low basal expression level was observed for the lysine exporter and the lysine exporter-regulating protein during the growth phase. Within a short time after the release of aspartokinase from feedback inhibition, the expression levels of these two genes increased dramatically. Obviously, the strong intracellular accumulation of lysine, which is known to induce its own exporter system, caused the increased transcription of the two genes. Towards the end of the process, the exporter and its regulator exhibited stable expression levels, probably maintained by the constant intracellular lysine concentration. It is known that two alternative pathways are present in C. glutamicum for lysine synthesis. As shown in Table 5, enzymes of both pathways were expressed at all examined time points, confirming previous findings that both pathways contribute to lysine production (33, 45). In addition to the lysine export genes, different genes of lysine biosynthesis were subjected to changes in transcript level during the phase shift. A slight enhancement of gene expression between 5.6 and 6.5 h resulted for aspartokinase, dihydropicolinate synthase, dihydropicolinate reductase, and diaminopimelate epimerase. Rather constant expression levels during the whole process were observed for aspartate semialdehyde dehydrogenase, succinyl diaminopimelate desuccinylase, and meso-diaminopimelate dehydrogenase. Exceptions to the rather unaffected gene expression levels were aspartate aminotransferase and diaminopimelate decarboxylase, which showed clear decreases in expression during lysine production compared to the growth phase. Diaminopimelate, one of the intermediates of the lysine pathway, is an important precursor for cell wall synthesis in C. glutamicum. The reduction of growth at the beginning of lysine production, resulting in less demand for cell wall constituents, could trigger the reduction of aspartate aminotransferase expression, which is the entry point into this pathway. This might indicate that parts of the lysine pathway are controlled by the growth state of C. glutamicum.
(vii) Expression of genes related to electron transport and ATP formation.
The genes for the different ATP synthase subunits, as exemplified by the α-chain (Table 4), had very high expression levels during the growth phase. This underscores the importance of this enzyme for the metabolic functioning of growing C. glutamicum cells. The end of exponential growth due to the depletion of essential amino acids caused a dramatic down-regulation of ATP synthase transcription. The expression level dropped by about 90% between 5.6 and 6.5 h and remained at a low level to the end of cultivation. Other ATP synthase subunits had very similar expression profiles (http://www.uni-saarland.de/fak8/heinzle). In contrast, the expression level for cytochrome c oxidase was higher during the phase shift and lysine production than during the initial growth phase.
Coordination of transcriptome, fluxome, and metabolome.
For the present work, an in-depth profiling of lysine production by C. glutamicum was performed with batch cultures by a combined analysis of the metabolome, transcriptome, and fluxome. The characterization was performed at different phases of the cultivation, so that alterations of gene expression, metabolic fluxes, and intracellular metabolite concentrations during the process could be simultaneously analyzed. As already shown, the change of C. glutamicum from pure growth to lysine production is reflected in changes in expression levels of various genes of the central metabolism and in metabolic fluxes catalyzed by the corresponding proteins. The combination of transcription, metabolome, and flux profiling carried out in the present work allows for the identification of correlations between transcript levels, metabolite concentrations, and metabolic activities in the metabolism of C. glutamicum.
(i) Growth during production phase.
Despite the depletion of threonine and methionine from the medium, cells of C. glutamicum still continued to grow. This raises the question of which growth-related processes are still active during lysine production. The same phenomenon of prolonged growth after the consumption of threonine and methionine was previously observed for lysine-producing C. glutamicum ATCC 21253, a strain directly derived from C. glutamicum ATCC 13287 and thus very similar to it (36). After limitation of the essential amino acids, C. glutamicum ATCC 21253 had an increase in total soluble proteins, indicating that protein synthesis still continued. To further address this point, we additionally analyzed the transcription of genes related to translation, which involves different ribosomal proteins and translation and elongation factors (http://www.uni-saarland.de/fak8/heinzle). All of these genes showed significant expression during the whole cultivation. Some ribosomal protein genes exhibited a decrease during the shift from growth to lysine production and a relatively constant expression during subsequent cultivation. Other ribosomal protein genes maintained stable expression levels during the whole process. Most of the translation-elongation factor genes were expressed at relatively constant levels in all samples. This shows that the protein synthesis machinery was present in C. glutamicum during the lysine production phase. Previously, it was concluded that probably either scavenging of threonine reserves or accretion of cellular constituents is related to the observed increase in biomass during lysine production (36). The present study shows that intracellular levels of free threonine and methionine decrease to very low values during the lysine production phase. Therefore, intracellular peptides or recycled proteins probably contribute to the supply of the required intracellular pools of threonine and methionine. Genes involved in glycogen formation and degradation were expressed throughout the whole cultivation, indicating the simultaneous formation and cleavage of glycogen (http://www.uni-saarland.de/fak8/heinzle). During the phase shift, gene expression levels for glycogen-forming enzymes increased, whereas the expression levels of glycogen-cleaving enzymes decreased. This indicates that, at least to a certain extent, an increase in the net formation of glycogen may take place during lysine production. In this context, genetic engineering of glycogen metabolism might be of interest in order to redirect the carbon flux from the production of storage material toward the production of lysine. However, the elemental composition of C. glutamicum ATCC 21253 remained constant during the growth and lysine production phases (36), which indicates that the formation of storage carbohydrates occurs only to a limited extent.
(ii) Lysine production.
The threshold level of intracellular threonine at which aspartokinase was released from feedback inhibition was in the range of 2 to 3 mM. With the beginning of intracellular lysine accumulation due to the deregulated aspartokinase, the intracellular lysine level increased because the cells lacked a functional exporter protein. The transient accumulation of intracellular lysine prior to its secretion is clear evidence for the induction of the active lysine exporter, which was previously shown to be present in C. glutamicum (37). As shown by the delayed decrease in intracellular lysine, transcription, translation, and integration of the lysine exporter into the cell membrane lasted for about 30 min. The sharp decrease in the size of the lysine pool suggested that, once it was functional, the exporter worked effectively.
(iii) Relationship of expression level and metabolic flux.
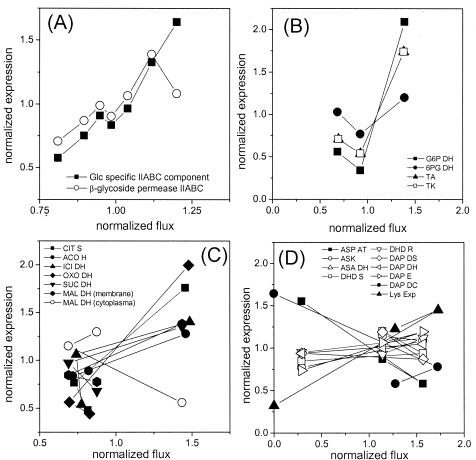
The direct relationship between metabolic flux and expression level was calculated for different time points of the cultivation. Flux values for each reaction and expression levels for each gene were normalized to the mean value of all time points to allow a direct comparison of all data. The coordination of the fluxome and transcriptome for substrate uptake by the PTS system, the PPP, the TCA cycle, and lysine biosynthesis is shown in Fig. 5. The expression of the PTS genes for glucose clearly correlated with the specific glucose uptake rate (Fig. 5A). A higher substrate uptake activity was linked to a higher gene expression level for the corresponding transporters. A similar picture was also obtained for the PPP genes glucose 6-phosphate dehydrogenase, transaldolase, and transketolase, for which an increase in gene expression corresponded to an increase in metabolic flux (Fig. 5B). The transcription of 6-phosphogluconate dehydrogenase followed the same trend, but it was much less pronounced. This is probably due to different catalytic properties of the PPP enzymes, which might be subject to additional regulation at the metabolic level. NADPH and NADP are important regulators of glucose 6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase in C. glutamicum (25). The coordination of expression and metabolic flux for the different TCA cycle genes is shown in Fig. 5C. For most enzymes, the maximum flux correlated with maximum gene expression and a decrease in flux during cultivation responded to a decrease in the corresponding expression level. Down-regulation at the transcriptional level of different TCA cycle enzymes obviously plays an important role in the reduction of the flux through the TCA cycle during the cultivation of lysine-producing C. glutamicum. An exception to this was found for cytoplasmic MDH, which showed an increase in expression level although the net flux through the TCA cycle decreased. As was previously shown, C. glutamicum possesses two different MDH enzymes, with the membrane-bound MDH (MQQ) being more important for the physiology of the strain (24). Regarding the coordination of expression level and metabolic flux, MQQ had a behavior that is typical for most of the TCA cycle enzymes. Therefore, this enzyme might play an important role in the cyclic action of the TCA cycle. The function of MDH seems to be different. With in vitro studies, MDH and MQQ were found to be capable of cyclic interconversion of malate and oxaloacetate, leading to the net oxidation of NADH (24). In this context, the increased expression of MDH may contribute to the regeneration of excess NADH, which is not needed under conditions of reduced growth and reduced energy demand. The observed increase in gene expression for succinyl-CoA synthetase is probably due to the demand of succinyl-CoA for the biosynthesis of lysine. Succinyl-CoA is required for the succinylase pathway. Deregulated lysine formation might therefore cause an increased demand for this precursor.
FIG. 5.
Coordination of gene expression and metabolic fluxes in glycolysis (A), the TCA cycle (B), and lysine biosynthesis (C) during batch cultivation of lysine-producing C. glutamicum ATCC 13287. For substrate uptake, glucose uptake flux (Fig. 2) and the expression of PTS components (Table 4) were considered. In the other cases (B to D), the intracellular flux data obtained after 5.8, 6.9, and 8.1 h (Fig. 4) were related to expression levels at 5.6, 6.8, and 7.7 h (Table 5), respectively.
Surprisingly, most of the genes in the lysine biosynthetic pathway had almost unaffected expression levels, despite the drastically increased flux with the beginning of lysine production (Fig. 5D). This involved genes for aspartokinase, aspartate semialdehyde dehydrogenase, dihydrodipicolinate synthase, dihydrodipicolinate reductase, and genes of both alternative lysine branches. Obviously, the metabolic capacities of these enzymes were not limited by the levels of their mRNAs. C. glutamicum was able to direct a sevenfold increase in flux through these enzymes without a large change in transcript levels. For aspartate aminotransferase and diaminopimelate decarboxylase, increased flux was linked to reduced expression. A threefold higher metabolic flux could be channelled through these enzymes even though gene expression levels were threefold lower. It is possible that the reduction of growth triggered the down-regulation of these enzymes. The lysine exporter was the only enzyme for which enhanced gene expression was linked to an increased metabolic flux. The lysine secretion flux was a clear response to the induction of the transporter.
The correlation between expression level and metabolic flux for pyruvate carboxylase, PEP carboxylase, PEP carboxykinase, and malic enzyme could not be directly determined for the present work because only the overall forward and backward flux between glycolysis, but not the contribution of the different enzymes, could be resolved in the flux analysis (Fig. 4). The expression levels of PEP carboxykinase and malic enzyme showed contrary changes during the phase shift. The transcript level of PEP carboxykinase decreased 3.8-fold, whereas that of malic enzyme increased 1.7-fold. This could indicate changing in vivo contributions of the two corresponding enzymes, which encode parallel reactions between the TCA cycle and glycolysis. The reaction with malic enzyme is linked to the formation of NADPH. PEP carboxykinase withdraws the lysine precursor oxaloacetate. Assuming that the gene expression profiles of these enzymes are an indication of their in vivo activities, a decrease of PEP carboxykinase and an increase of malic enzyme would both favor the formation of lysine. For glycolytic genes, the general decrease in expression matched the decreasing flux through the glycolytic chain during the phase shift. Surprisingly, genes for glucose 6-phosphate isomerase, triosephosphate isomerase, glyceraldehyde 3-phosphate dehydrogenase, and 3-phosphoglycerate kinase had increased expression levels during the growth phase, despite the decreased glycolytic flux. This may indicate that regulation at the metabolic level is involved in control of the glycolytic flux.
It was previously shown that isocitrate lyase and malate synthase are induced in C. glutamicum grown on glucose at a certain level of acetate (39). The concentration of acetate in the medium, which was in the millimolar range, was obviously high enough to trigger the gene expression of isocitrate lyase and malate synthase. However, only at elevated acetate levels during lysine production did these enzymes have in vivo activities. The secretion of acetate still continued after the glyoxylate pathway was activated. Therefore, only a certain fraction of acetate produced was recycled into the central metabolism by this pathway.
(iv) Distribution of transcriptional control.
An overall view on the distribution of transcriptional regulation within the central metabolism of C. glutamicum can be obtained by comparing the relative changes in gene expression throughout the cultivation. Taking the ratio between the highest and the lowest expression levels found for a gene during cultivation as a measure for its regulatory importance, an interesting picture is yielded for different pathways of C. glutamicum. For glycolysis, the highest relative changes in gene expression during the cultivation were found for glucose 6-phosphate isomerase and pyruvate dehydrogenase, which encode the initial and terminal steps, respectively, of the pathway (Table 4). The same picture was found for the lysine biosynthetic pathway, in which aspartate aminotransferase, diaminopimelate decarboxylase, and the lysine exporter had the highest relative expression changes by far (Table 5). Accordingly, major changes in gene expression were found for citrate synthase, the entry point into the TCA cycle (Table 4). These findings may indicate that the entry points into pathways and linking points between different pathways are of special importance for transcriptional control in C. glutamicum.
(v) NADPH metabolism.
Based on metabolic fluxes, an insight into the NADPH metabolism of C. glutamicum was gained. Hereby a demand of 15.5 mmol of NADPH g of CDM−1 and of 4 mol mol of glucose−1 was assumed for the production of biomass and lysine, respectively (20). Glucose 6-phosphate dehydrogenase, 6-phosphogluconate dehydrogenase, and isocitrate dehydrogenase were considered catalysts of NADPH-supplying reactions. After 5.8 h, 5.8 mmol of NADPH g−1 h−1 was required for anabolic demands, whereas 5.8 mmol of NADPH g−1 h−1 (PPP enzymes) and 3.0 mmol of NADPH g−1 h−1 (isocitrate dehydrogenase) was supplied. Overall, a relatively high apparent surplus of 3.0 mmol of NADPH g−1 h−1 was present during the growth of C. glutamicum ATCC 13287. A high apparent excess of NADPH was also observed for wild-type C. glutamicum ATCC 13032 (47). Obviously, this apparent NADPH excess is a typical phenomenon of C. glutamicum under conditions of pure growth. Possibly the NAPDH generated is directed towards the respiratory chain of C. glutamicum under conditions of growth (22). This appears logical considering the point that isocitrate dehydrogenase, one of the NADPH-producing enzymes, is located in the TCA cycle. After 6.9 h of cultivation, the total demand of NADPH for anabolism and lysine production was 6.0 mmol of NADPH g−1 h−1 and was thus almost equal to the NADPH supply of 6.1 mmol of NADPH g−1 h−1. A possible limitation of lysine production by NADPH at this stage of the cultivation can therefore not be excluded. Lysine production after 8.2 and 9.2 h of cultivation was again linked to apparent excesses of NADPH of 1.4 mmol of NADPH g−1 h−1 and 0.9 mmol of NADPH g−1 h−1, respectively. The role of malic enzyme could not be directly estimated from the analysis of the fluxome. This was due to the fact that only the total back flux from the TCA cycle could be determined, but the contributions of the single enzymes involved could not be resolved because they are linked by identical stoichiometries and identical labeling patterns. Malic enzyme was expressed during growth and lysine production (Table 4). Therefore, an additional supply of NADPH by this reaction seems possible.
Concluding remarks.
The data of the present work provide a detailed insight into the metabolic functioning and regulation of lysine-producing C. glutamicum. Key metabolic properties of the examined strain of C. glutamicum which were important for lysine overproduction were the release of aspartokinase from feedback inhibition by the depletion of threonine and the strong induction of the lysine exporter. Lysine overproduction demanding high amounts of NADPH was probably favored by the high apparent NADPH excess. As described above, the combination of different profiling techniques provided detailed quantitative information for a biological network analysis and therefore displayed an important prerequisite for targeted strain improvement. It appears rather straightforward to additionally include proteome analysis in future studies and to extend the metabolome measurements to intermediary metabolites of pathways, e.g., glycolysis, PPP, and the TCA cycle, in order to complete the set of information and to provide extensive and detailed data about metabolic functioning and regulation in C. glutamicum. The high potential of approaches integrating different profiling tools has recently been shown for different organisms (1, 38, 50).
REFERENCES
- 1.Al Zaid Siddiquee, K., M. J. Arauzo-Bravo, and K. Shimizu. 2004. Metabolic flux analysis of pykF gene knockout Escherichia coli based on 13C-labeling experiments together with measurements of enzyme activities and intracellular metabolite concentrations. Appl. Microbiol. Biotechnol. 63:407-417. [DOI] [PubMed] [Google Scholar]
- 2.Bathe, B., J. Kalinowski, and A. Pühler. 1996. A physical and genetic map of the Corynebacterium glutamicum ATCC 13032 chromosome. Mol. Gen. Genet. 252:255-265. [DOI] [PubMed] [Google Scholar]
- 3.Dauner, M., and U. Sauer. 2000. GC-MS analysis of amino acids rapidly provides rich information for isotopomer balancing. Biotechnol. Prog. 16:642-649. [DOI] [PubMed] [Google Scholar]
- 4.de Graaf, A. A. 2000. Metabolic flux analysis of Corynebacterium glutamicum, p. 506-555. In K. Schügerl and K. H. Bellgardt (ed.), Bioreaction engineering. Springer Verlag, Berlin, Germany.
- 5.Dominguez, H., C. Rollin, A. Guyonvarch, J.-L. Guerquin-Kern, and M. Cocaign-Bousquet. 1998. Carbon-flux distribution in the central metabolic pathways of Corynebacterium glutamicum during growth on fructose. Eur. J. Biochem. 254:96-102. [DOI] [PubMed] [Google Scholar]
- 6.Eggeling, L., and H. Sahm. 1999. l-Glutamate and l-lysine: traditional products with impetuous developments. Appl. Microbiol. Biotechnol. 52:146-153. [Google Scholar]
- 7.Glanemann, C., A. Loos, N. Gorret, L. B. Willis, X. M. O'Brien, P. A. Lessard, and A. J. Sinskey. 2003. Disparity between changes in mRNA abundance and enzyme activity in Corynebacterium glutamicum: implications for DNA microarray analysis. Appl. Microbiol. Biotechnol. 61:61-68. [DOI] [PubMed] [Google Scholar]
- 8.Gutmann, M., C. Hoischen, and R. Krämer. 1992. Carrier mediated glutamate secretion by Corynebacterium glutamicum under biotin limitation. Biochim. Biophys. Acta 1112:115-123. [DOI] [PubMed] [Google Scholar]
- 9.Haberhauer, G., H. Schröder, M. Pompejus, O. Zelder, and B. Kröger. 2001. Corynebacterium glutamicum genes encoding proteins involved in membrane synthesis and membrane transport. Patent WO 01/00805.
- 10.Hayashi, M., H. Mizogushi, N. Shiraishi, M. Obayashi, S. Nakagawa, J. Imai, S. Watanabe, T. Ota, and M. Ikeda. 2002. Transcriptome analysis of acetate metabolism in Corynebacterium glutamicum using a newly developed metabolic array. Biosci. Biotechnol. Biochem. 66:1337-1344. [DOI] [PubMed] [Google Scholar]
- 11.Hermann, T., G. Wersch, E. M. Uhlemann, R. Schmid, and A. Burkovski. 1998. Mapping and identification of Corynebacterium glutamicum proteins by two-dimensional gel electrophoresis and microsequencing. Electrophoresis 19:3217-3221. [DOI] [PubMed] [Google Scholar]
- 12.Hermann, T., M. Finkemeier, W. Pfefferle, G. Wersch, R. Krämer, and A. Burkovski. 2000. Two-dimensional electrophoretic analysis of Corynebacterium glutamicum membrane fraction and surface proteins. Electrophoresis 21:654-659. [DOI] [PubMed] [Google Scholar]
- 13.Hermann, T., W. Pfefferle, C. Baumann, E. Busker, S. Schaffer, M. Bott, H. Sahm, N. Dusch, J. Kalinowski, A. Pühler, A. K. Bendt, R. Krämer, and A. Burkovski. 2001. Proteome analysis of Corynebacterium glutamicum. Electrophoresis 22:1712-1723. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda, M., and S. Nakagawa. 2003. The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Appl. Microbiol. Biotechnol. 62:99-109. [DOI] [PubMed] [Google Scholar]
- 15.Jetten, M. S. M., M. E. Gubler, S. H. Lee, and A. J. Sinskey. 1994. Structural and functional analysis of pyruvate kinase from Corynebacterium glutamicum. Appl. Environ. Microbiol. 60:2501-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelleher, J. K. 2001. Flux estimation using isotopic tracers: common ground for metabolic physiology and metabolic engineering. Metab. Eng. 2:100-110. [DOI] [PubMed] [Google Scholar]
- 17.Kiefer, P., E. Heinzle, O. Zelder, and C. Wittmann. 2004. Comparative metabolic flux analysis of lysine-producing Corynebacterium glutamicum on glucose and fructose. Appl. Environ. Microbiol. 70:229-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lange, C., D. Rittmann, V. F. Wendisch, M. Bott, and H. Sahm. 2003. Global expression profiling and physiological characterization of Corynebacterium glutamicum grown in the presence of l-valine. Appl. Environ. Microbiol. 69:2521-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loos, A., C. Glanemann, L. B. Willis, X. M. O′Brien, P. A. Lessard, R. Gerstmeier, S. Guillouet, and A. J. Sinskey. 2001. Development and validation of Corynebacterium DNA microarrays. Appl. Environ. Microbiol. 67:2310-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marx, A., A. A. de Graaf, W. Wiechert, G. Eggeling, and H. Sahm. 1996. Determination of the fluxes in the central metabolism of Corynebacterium glutamicum by nuclear magnetic resonance spectroscopy combined with metabolite balancing. Biotechnol. Bioeng. 49:111-129. [DOI] [PubMed] [Google Scholar]
- 21.Marx, A., K. Striegel, A. A. de Graaf, H. Sahm, and L. Eggeling. 1997. Response of the central metabolism of Corynebacterium glutamicum to different flux burdens. Biotechnol. Bioeng. 56:168-180. [DOI] [PubMed] [Google Scholar]
- 22.Matsushita, K., A. Otofuji, M. Iwahashi, H. Toyama, and O. Adachi. 2001. NADH dehydrogenase of Corynebacterium glutamicum. Purification of an NADH dehydrogenase II homolog able to oxidize NADPH. FEMS Microbiol. Lett. 203:271-276. [DOI] [PubMed] [Google Scholar]
- 23.Möckel, B., A. Weissenborn, W. Pfefferle, J. Kalinowski, B. Bathe, and A. Pühler. 1999. Genome sequencing of industrial microorganisms: the Corynebacterium glutamicum ATCC 13032 genome project. Microb. Comp. Genomics 4:111. [Google Scholar]
- 24.Molenaar, D., M. E. van der Rest, A. Drysch, and R. Yucel. 2000. Functions of the membrane-associated and cytoplasmic malate dehydrogenases in the citric acid cycle of Corynebacterium glutamicum. J. Bacteriol. 182:6884-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moritz, B., K. Striegel, A. A. de Graaf, and H. Sahm. 2000. Kinetic properties of the glucose-6-phosphate and 6-phosphogluconate dehydrogenases from Corynebacterium glutamicum and their application for predicting pentose phosphate pathway flux in vivo. Eur. J. Biochem. 267:3442-3452. [DOI] [PubMed] [Google Scholar]
- 26.Muffler, A., S. Bettermann, M. Haushalter, A. Hörlein, U. Neveling, M. Schramm, and O. Sorgenfrei. 2002. Genome-wide transcription profiling of Corynebacterium glutamicum after heat shock and during growth on acetate and glucose. J. Biotechnol. 98:255-268. [DOI] [PubMed] [Google Scholar]
- 27.Nakagawa, S., H. Mizoguchi, S. Ando, M. Hayashi, K. Ochiai, H. Yokoi, N. Tateishi, A. Senoh, M. Ikeda, and A. Ozaki. 2001. Novel polynucleotides. European patent 1,108,790.
- 28.Nakayama, K., and K. Araki. January 1973. Process for producing l-lysine. U.S. patent 3,708,395.
- 29.Ohnishi, J., S. Mitsuhashi, M. Hayashi, S. Ando, H. Yokoi, K. Ochiai, and M. Ikeda. 2002. A novel methodology employing Corynebacterium glutamicum genome information to generate a new l-lysine-producing mutant. Appl. Microbiol. Biotechnol. 58:217-223. [DOI] [PubMed] [Google Scholar]
- 30.Sahm, H., L. Eggeling, and A. A. de Graaf. 2000. Pathway analysis and metabolic engineering in Corynebacterium glutamicum. Biol. Chem. 381:899-910. [DOI] [PubMed] [Google Scholar]
- 31.Schaffer, S., B. Weil, V. D. Nguyen, G. Dongmann, K. Günther, M. Nickolaus, T. Hermann, and M. Bott. 2001. A high-resolution reference map for cytoplasmic and membrane-associated proteins of Corynebacterium glutamicum. Electrophoresis 22:4404-4422. [DOI] [PubMed] [Google Scholar]
- 32.Schmid, R., E.-M. Uhlemann, L. Nolden, G. Wersch, R. Hecker, T. Hermann, A. Marx, and A. Burkovski. 2000. Response to nitrogen starvation in Corynebacterium glutamicum. FEMS Microbiol. Lett. 187:83-88. [DOI] [PubMed] [Google Scholar]
- 33.Sonntag, K., L. Eggeling, A. A. de Graaf, and H. Sahm. 1993. Flux partitioning in the split pathway of lysine synthesis in Corynebacterium glutamicum: quantification by 13C- and 1H-NMR spectroscopy. Eur. J. Biochem. 213:1325-1331. [DOI] [PubMed] [Google Scholar]
- 34.Sonntag, K., J. Schwinde, A. A. de Graaf, A. Marx, B. J. Eikmanns, W. Wiechert, and H. Sahm. 1995. 13C NMR studies of the fluxes in the central metabolism of Corynebacterium glutamicum during growth and overproduction of amino acids in batch cultures. Appl. Microbiol. Biotechnol. 44:489-495. [Google Scholar]
- 35.Tauch, A., I. Homann, S. Mormann, S. Rüberg, A. Billault, B. Bathe, S. Brand, O. Brockmann-Gretza, C. Rückert, N. Schischka, C. Wrenger, J. Hoheisel, B. Möckel, K. Huthmacher, W. Pfefferle, A. Pühler, and J. Kalinowski. 2002. Strategy to sequence the genome of Corynebacterium glutamicum ATCC 13032: use of a cosmid and a bacterial artificial chromosome library. J. Biotechnol. 95:25-38. [DOI] [PubMed] [Google Scholar]
- 36.Vallino, J. J., and G. Stephanopoulos. 1993. Metabolic flux distributions in Corynebacterium glutamicum during growth and lysine overproduction. Biotechnol. Bioeng. 41:633-646. [DOI] [PubMed] [Google Scholar]
- 37.Vrljic, M., H. Sahm, and L. Eggeling. 1996. A new type of transporter with a new type of cellular function: l-lysine export from Corynebacterium glutamicum. Mol. Microbiol. 22:815-826. [DOI] [PubMed] [Google Scholar]
- 38.Wang, W., J. Sun, M. Hartlep, W. D. Deckwer, and A. P. Zeng. 2003. Combined use of proteomic analysis and enzyme activity assays for metabolic pathway analysis of glycerol fermentation by Klebsiella pneumoniae. Biotechnol. Bioeng. 83:525-536. [DOI] [PubMed] [Google Scholar]
- 39.Wendisch, V. F., M. Spies, D. J. Reinscheid, S. Schnicke, H. Sahm, and B. J. Eikmanns. 1997. Regulation of acetate metabolism in Corynebacterium glutamicum: transcriptional control of the isocitrate lyase and malate synthase genes. Arch. Microbiol. 168:262-269. [DOI] [PubMed] [Google Scholar]
- 40.Wendisch, V. F., A. A. de Graaf, H. Sahm, and B. J. Eikmanns. 2000. Quantitative determination of metabolic fluxes during coutilization of two carbon sources: comparative analyses with Corynebacterium glutamicum during growth on acetate and/or glucose. J. Bacteriol. 182:3088-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiechert, W., C. Siefke, A. A. de Graaf, and A. Marx. 1997. Bidirectional reaction steps in metabolic networks. II. Flux estimation and statistical analysis. Biotechnol. Bioeng. 55:118-135. [DOI] [PubMed] [Google Scholar]
- 42.Wiechert, W., M. Möllney, S. Petersen, and A. A. de Graaf. 2001. A universal framework for 13C metabolic flux analysis. Metab. Eng. 3:265-283. [DOI] [PubMed] [Google Scholar]
- 43.Wiechert, W. 2002. An introduction to 13C metabolic flux analysis. Genet. Eng. (New York) 24:215-238. [DOI] [PubMed] [Google Scholar]
- 44.Wittmann, C., and E. Heinzle. 1999. Mass spectrometry for metabolic flux analysis. Biotechnol. Bioeng. 62:739-750. [DOI] [PubMed] [Google Scholar]
- 45.Wittmann, C., and E. Heinzle. 2001. Application of MALDI-TOF MS to lysine-producing Corynebacterium glutamicum. A novel approach for metabolic flux analysis. Eur. J. Biochem. 268:2441-2455. [DOI] [PubMed] [Google Scholar]
- 46.Wittmann, C., M. Hans, and E. Heinzle. 2002. In vivo analysis of intracellular amino acid labeling by GC/MS. Anal. Biochem. 307:379-382. [DOI] [PubMed] [Google Scholar]
- 47.Wittmann, C., and E. Heinzle. 2002. Genealogy profiling through strain improvement by using metabolic network analysis: metabolic flux genealogy of several generations of lysine-producing corynebacteria. Appl. Environ. Microbiol. 68:5843-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wittmann, C. 2002. Metabolic flux analysis using mass spectrometry. Adv. Biochem. Eng. Biotechnol. 74:39-64. [DOI] [PubMed] [Google Scholar]
- 49.Wittmann, C., J. O. Krömer, P. Kiefer, T. Binz, and E. Heinzle. Impact of the cold-shock phenomenon on quantification of intracellular metabolites in bacteria. Anal. Biochem., in press. [DOI] [PubMed]
- 50.Yoon, S. H., M. J. Han, S. Y. Lee, K. J. Jeong, and J. S. Yoo. 2003. Combined transcriptome and proteome analysis of Escherichia coli during high cell density culture. Biotechnol. Bioeng. 81:753-767. [DOI] [PubMed] [Google Scholar]