Abstract
Open reading frame SCO3571 of Streptomyces coelicolor encodes a protein of the cyclic AMP (cAMP) receptor protein (CRP) superfamily of regulatory proteins. A mutant revealed a dramatic defect in germination, followed by growth delay and earlier sporulation. This phenotype correlates with those of an adenylate cyclase (cya) mutant that cannot synthesize cAMP. This finding suggests that S. coelicolor may use a Cya-cAMP-CRP system to trigger complex physiological processes such as morphogenesis.
Streptomycetes are gram-positive soil bacteria that undergo morphological differentiation. Their life cycle includes germination, vegetative mycelial growth, aerial mycelial growth, secondary metabolite synthesis, and eventually spore morphogenesis (5, 6). In the initial stages of germination, spores swell, resulting in an increase in size and decreased phase brightness followed by germ tube emergence (10). The growth of Streptomyces coelicolor in liquid media is characterized by a first phase of rapid growth (RG1), after which nutritional imbalances evoke a stress response and a shutdown of protein synthesis. This period is called the transition (T) phase and is followed by a second phase of rapid growth (RG2) and the onset of secondary metabolism (14).
In S. coelicolor, cyclic AMP (cAMP) is synthesized throughout the developmental program (16). There is a peak of cAMP accumulation during germination, and then its level increases slowly during the growth of substrate mycelium. A maximal concentration is reached during the T phase and early stages of aerial mycelium formation. Later, during RG2, the cAMP level decreases rapidly. Characterization of the adenylate cyclase mutant (cya), unable to synthesize cAMP, demonstrated that cAMP facilitates developmental processes (16). The cya mutant is defective in germination and has a growth delay of 20 h. With regard to antibiotic production, cAMP determines the structure of the final product of the actinorhodin pathway (actinorhodin red or γ-actinorhodin blue) through its effects on medium pH and also increases yield. All phenotypes specific to the cya mutant are suppressed by the exogenous addition of cAMP.
In Escherichia coli, cAMP is the effecting molecule that modulates the DNA-binding ability of the pleiotropic transcription factor cAMP receptor protein (CRP). This protein activates and in some cases represses a vast number of genes that are implicated in diverse cellular functions, including carbon metabolism and starvation, resistance to stress and acid, and motility (2-4, 12, 15). In the same way, cAMP could trigger the streptomycete lifestyle by modulating proteins such as transcription factors of the CRP/fumarate and nitrate reduction regulator (FNR) superfamily (8). Hence, we raised the question whether a cAMP receptor protein exists in Streptomyces species. Such a potential crp-like gene (hereinafter called crp) within the S. coelicolor genome has been annotated as open reading frame SCO3571, located on cosmid SCH17 (1).
Phylogenetic and comparative studies (data not shown) revealed that crp encodes a protein of 224 amino acids that clusters to the CRP subfamily of the large CRP/FNR superfamily (8, 17). These proteins present a cyclic-nucleotide monophosphate-binding domain in their N-terminal region, while the C-terminal part constitutes the DNA-binding domain. For the functional analysis of crp, we used the wild-type strain S. coelicolor A3(2) M145 (an SCP1− SCP2− prototroph) (11). Streptomyces media, culture conditions, DNA preparations, protoplast transformation, preparations of spores, and pregermination were as described by Kieser et al. (11). To demonstrate that crp is an active gene, a reverse transcription (RT)-PCR experiment was performed, and this experiment showed that crp mRNA was present in mycelia grown on a complex medium (tryptone soya broth without dextrose) (Fig. 1). The crp gene was then deleted by gene replacement by use of the PCR targeting method developed by Gust et al. (9). A PCR fragment containing short flanking regions of crp (5′-CAAGATCCTCTGAGCCGGTCGACAAGGAGAGAACTCGTGattccggggatccgtcgacc-3′ and 5′-AGAGATGTCCCTGCAACCCGGGGAGACCCCAGGGGGTCAtgtaggctggagctgcttc-3′; crp start and stop codons in italics; capital letters correspond to regions flanking the crp gene, and lowercase letters match the unique priming sites of the disruption cassette), an apramycin resistance cassette, and oriT(RK2) for efficient intergeneric conjugation was integrated in cosmid SCH17 by lambda Red-mediated recombination. The cosmid was conjugated from E. coli ET12567/pUZ8002 to S. coelicolor, where it readily undergoes recombination by double crossover. Exconjugants were selected as apramycin-resistant colonies, and the gene replacement event was verified by several PCRs with combinations of oligonucleotides that hybridized within and outside the replaced area.
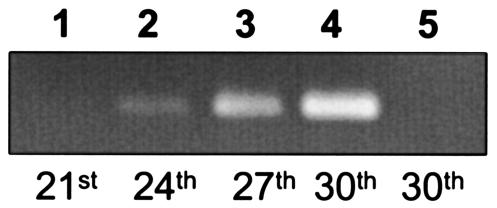
FIG. 1.
Expression of crp. A 1% agarose gel shows the crp-specific RT-PCR amplification product after the 21st, 24th, 27th, and 30th cycles of the PCR (lanes 1 to 4). No signal was obtained with RT-PCR without prior reverse transcription (control of absence of genomic DNA) (lane 5). Total RNA of S. coelicolor grown on tryptone soya brothwas prepared as described (13). A One-Step RT-PCR kit (QIAGEN) was used according to the manufacturer's instructions with the crp-specific oligonucleotides RTcrp1 (5′-AAGGCAAGGTCAAGCTCCACC-3′ [nucleotides 155 to 175]) and RTcrp2 (5′-ATGCCCTCCTCCGACTGCACG-3′ [nucleotides 486 to 506]).
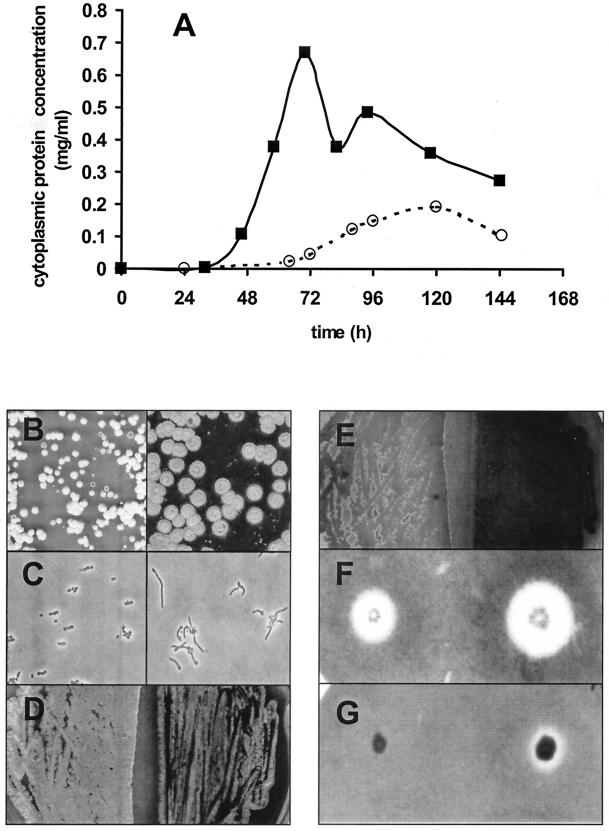
The mutant, SAF1, exhibited significantly smaller colony sizes than the wild type did (Fig. 2B). This finding correlated with a growth delay in liquid medium of about 30 h, accompanied by a twofold reduction of the final biomass (Fig. 2A). Microscopic analysis of spore germination revealed that germ tubes emerged from most wild-type spores after 8 h (Fig. 2C). Spores of the crp mutant developed into germ tubes only occasionally. Hence, the germination-negative phenotype explains the delay of growth. Interestingly, the adenylate cyclase mutant showed a similar lack of germination, which could be suppressed by the addition of 3 mM cAMP (16). In the crp mutant, since the addition of exogenous cAMP did not restore correct germination and the suppression of growth delay, one can exclude the possibility that CRP acts on cAMP biosynthesis. The life cycle was also disturbed at the sporulation step. On R2YE plates, the sporulation process of the crp mutant was achieved earlier, as plates containing this mutant were completely covered by spores about 7 days before the appearance of spores from the wild-type colonies (Fig. 2D). There was, further, a marked difference in the production of the antibiotic actinorhodin (Fig. 2E). In contrast to the wild type, which produces blue γ-actinorhodin after 2 days at 28°C, the crp strain did not synthesize this molecule before 8 days at 28°C. To further confirm that the observed phenotype was solely due to the lack of crp, SAF1 was complemented in trans. Therefore, crp was amplified by PCR with primers ScoCrp1 (5′-GCTTAATTAACTGAAAGGAGGTTAATAGTGGACGACGTTCTGCGG-3′) and ScoCrp2 (5′-ATATAAGCTTGGTCAGCGGGAGCGCTTGGC-3′) (PacI and HindIII sites are underlined, and start and stop codons are in italics) and cloned into plasmid pFT74, giving plasmid pFT169 (glkAp::crpSco). pFT74 is a pUWL-KS derivative that carries 83 bp of the glkA promoter (18). Hence, crp was cloned behind the glkA promoter. After confirmation by DNA sequencing, SAF1 protoplasts were transformed by plasmid pFT169, which resulted in the complete restoration of the observed SAF1 phenotype.
FIG.2.
The crp mutant is impaired in germination, growth, antibiotic production, and sporulation. (A) Growth curves of the S. coelicolor wild-type strain (▪) and the SAF1 mutant (○) in yeast extract-malt extract medium. (B) Difference in colony sizes between SAF1 mutant (left) and wild type (right) incubated at 28°C for 7 days. (C) Impaired germ tube emergence in the SAF1 mutant (left) relative to that of the wild type (right). Activated spores of the wild type and the SAF1 mutant were incubated in yeast extract-malt extract medium for 8 h at 28°C. Germination of spores was observed with a Zeiss light microscope (Axioplan) with phase-contrast lenses with a 100-fold oil immersion lens coupled with a charge-coupled device camera (Ikegami). (D) Enhanced sporulation in SAF1 mutant (left, view from the top). (E) After 7 days on R2YE medium incubated at 28°C, only the wild type (right) produced blue γ-actinorhodin (view from the bottom). (F and G) Effects of crp deletion on global xylanolytic activity. The xylanolytic system is still induced by xylan (F) and repressed by glucose (G) in the SAF1 mutant. The SAF1 mutant (left) and wild-type S. coelicolor strain (right) were plated on basal agar medium containing 0.5% oat spelt xylan with (G) or without (F) 1% glucose. Xylanase activity was detectable as a decolorized halo after Congo red staining applied after 5 days of incubation at 28°C. Each experiment was repeated at least three times.
As the initiation and the efficiency of the germination process are impaired and the life cycle is shortened to somehow precipitate the occurrence of the sporulation process, spores can be considered the privileged form of life of the SAF1 mutant. This “forced” predominance of the survival form of Streptomyces species may suggest that the deletion of crp somehow simulates constant stress conditions in the surrounding environment and/or inside the cell which affect all steps of the life cycle.
Since the CRP of E. coli acts as a pleiotropic gene activator on many carbohydrate operons and signals carbon catabolite repression (CCR) in response to the cAMP level (2), we asked whether this could also be the case in S. coelicolor. However, unlike an E. coli crp mutant, SAF1 was able to grow on carbon sources that are subject to CCR (starch, xylan, and cellulose). Moreover, the comparative determination of xylanase (Fig. 2F and G) cellulase, α-amylase, and mannanase (data not shown) activities on solid medium showed that these polysaccharides' hydrolytic systems were still correctly induced by their specific substrates and repressed in the presence of glucose. In liquid medium, the enzymatic activity/cytoplasmic protein concentration ratios (data not shown) were similar for the wild type and the SAF1 mutant. This finding suggests that CRP plays no role in CCR in S. coelicolor.
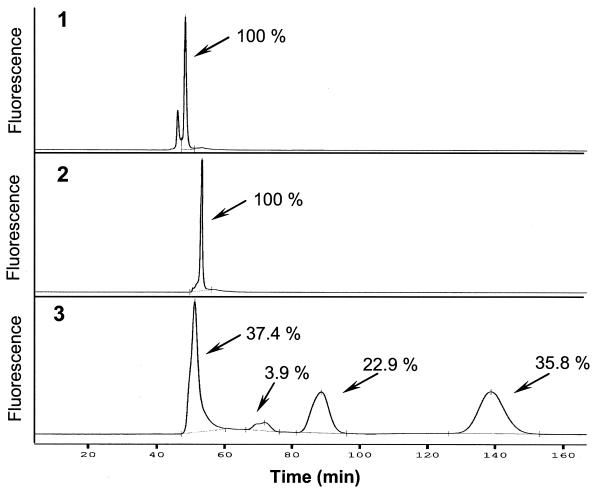
Upstream of the crp gene, we detected a partial palindromic sequence that matched the consensus sequence of a CRP-binding site (CATCCTTGTGACA*GATCACACTGTTT) (17). We found this motif (TGTGA-N4-6-TCACA) eight times in the upstream regions of S. coelicolor genes (Table 1). Hence, they could be part of a CRP regulon. Electrophoretic mobility shift assays were performed to study the possible binding of CRP to this element (7). Cytoplasmic extracts of S. coelicolor wild-type and SAF1 strains were prepared and combined with a synthetic fluorescent double-stranded oligonucleotide representing the palindrome. The binding of CRP to the probe was resolved by molecule separation on an ALFexpress sequencer. The presence of cytoplasmic wild-type extract caused three shifts of the labeled probe (Fig. 3, lane 3). The different shifts can be explained by the binding of CRP in different oligomeric forms. The application of cytoplasmic extract of the crp mutant did not produce any shift of the probe (Fig. 3, lane 2), suggesting either that CRP is the protein that binds this palindromic site or that it is required for the transcriptional activation of the trans-acting factor that interacts with the crp promoter.
TABLE 1.
Intergenic regions containing the cis consensus sequencea bound by proteins from the CRP/FNR superfamily in S. coelicolor
| Geneb | Putative CRP-FNR cis sequence | Annotation (length in amino acids) |
|---|---|---|
| SCO2545 | GTATGTGACCTAGATCACATCA | Possible transmembrane efflux protein (449) |
| SCO3571 | ATCCTTGTGACAGATCACACTGTT | crp, probable transcriptional regulator (224) |
| SCO2914 | ACTGTGACGTGTCACACA | Probable amino acid permease (475) |
| SCO3945 | GTGAATGTGAACGCGTTCACAAGCGT | cydA, cytochrome oxidase subunit I (501) |
| SCO4280 | TTCGTTGTGACTTGAGTCACAAGGGG | Probable reductase (202) |
| SCO4281(c) | Hypothetical protein (533) | |
| SCO4562 | TTGGGCTTGTGACCTGCTTCACATGTTCGC | nuoA, NADH dehydrogenase subunit (119) |
| SCO4561(c) | Possible NLP/P60 family protein (277) | |
| SCO5029 | GATGTGAGCTTTCACACG | Possible secreted protein (238) |
| SCO5864 | GAGTCGCCTGTGTGATCGATCACAGCGGAGGCTT | Unknown (98) |
TGTGA-N4-6-TCACA.
(c), open reading frame on the complementary strand. Nucleotides shown in boldface are in the searched sequence, and those underlined form the palindrome.
FIG. 3.
Electrophoretic mobility shift assays. The DNA probe (the putative crp-like cis-acting element 110 nucleotides upstream of the GTG start codon) together with an excess (1,500-fold) of nonspecific salmon sperm DNA (Sigma) was diluted in DNA-binding buffer (10 mM Tris-HCl [pH 7.5], 50 mM KCl, 1 mM Na2EDTA, 5% glycerol) and added to 55 μg of total cytoplasmic extracts. Lane 1, DNA probe alone; lane 2, DNA probe plus cell extract from SAF1 mutant; lane 3, DNA probe plus cell extract from S. coelicolor wild type. Percentages indicate the proportions of the various shifts. Each experiment was repeated at least three times.
In conclusion, the observed physiological effects of the crp mutation correlate with those observed in the adenylate cyclase mutant (16). This finding suggests that S. coelicolor may use a Cya-cAMP-CRP system to trigger complex physiological processes such as morphogenesis. Future research is required to unequivocally demonstrate the binding of cAMP to CRP and to elucidate all CRP-binding sites in the genome.
ADDENDUM IN PROOF
After this paper was accepted, Brekasis and Paget (D. Brekasis and M. S. B. Paget, EMBO J. 22:4856-4865, 2003) reported that the Rex protein, a novel sensor of the NADH/NAD+ redox poise, binds to the putative cis elements of the cyd and nuo operons, which we have identified as putative CRP-FNR cis sites. The fact that Rex alone could not regulate the nuo operon led to the conclusion that at least one other regulatory system is likely to be involved. Hence, it might be possible that CRP constitutes this regulator, which would indicate that Rex and CRP are inextricably linked in their regulation capacities.
Acknowledgments
The Belgian government supported this work as part of the Interuniversity Poles of Attraction (grant PAI P5/33), the Fund for Joint Basic Research of Belgium (contract no. 2.4530.03), and the Walloon Region (convention no. 114930). The work was also supported through grant Signal Mechanisms of C-Regulation SFB473 of the Deutsche Forschungsgemeinschaft. A.D. and G.M. are research fellows of the Fund for Research in Industry and Agriculture (FRIA), S.R. is a postdoctoral researcher of the Walloon Region, and J.D. is a research associate of the National Fund for Scientific Research (FNRS) of Belgium.
We thank B. Gust for training in the PCR targeting technique at the John Innes Center (Norwich, United Kingdom). We also thank Fabrizio Giannotta, Catherine Raskin, and Anne Famerie for valuable assistance and kind support.
REFERENCES
- 1.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 2.Brückner, R., and F. Titgemeyer. 2002. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 209:141-148. [DOI] [PubMed] [Google Scholar]
- 3.Busby, S., and R. H. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 4.Castanie-Cornet, M. P., and J. W. Foster. 2001. Escherichia coli acid resistance: cAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology 147:709-715. [DOI] [PubMed] [Google Scholar]
- 5.Chater, K. F. 2001. Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex? Curr. Opin. Microbiol. 4:667-673. [DOI] [PubMed] [Google Scholar]
- 6.Chater, K. F. 1998. Taking a genetic scalpel to the Streptomyces colony. Microbiology 144:1465-1478. [DOI] [PubMed] [Google Scholar]
- 7.Filée, P., M. Delmarcelle, I. Thamm, and B. Joris. 2001. Use of an ALFexpress DNA sequencer to analyze protein-nucleic acid interactions by band shift assay. BioTechniques 30:1044-1048, 1050-1051. [DOI] [PubMed] [Google Scholar]
- 8.Green, J., C. Scott, and J. R. Guest. 2001. Functional versatility in the CRP-FNR superfamily of transcription factors: FNR and FLP. Adv. Microb. Physiol. 44:1-34. [DOI] [PubMed] [Google Scholar]
- 9.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hardisson, C., M. B. Manzanal, J. A. Salas, and J. E. Suarez. 1978. Fine structure, physiology and biochemistry of arthrospore germination in Streptomyces antibioticus. J. Gen. Microbiol. 105:203-214. [DOI] [PubMed] [Google Scholar]
- 11.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 12.Marschall, C., V. Labrousse, M. Kreimer, D. Weichart, A. Kolb, and R. Hengge-Aronis. 1998. Molecular analysis of the regulation of csiD, a carbon starvation-inducible gene in Escherichia coli that is exclusively dependent on sigma s and requires activation by cAMP-CRP. J. Mol. Biol. 276:339-353. [DOI] [PubMed] [Google Scholar]
- 13.Nothaft, H., S. Parche, A. Kamionka, and F. Titgemeyer. 2003. In vivo analysis of HPr reveals a fructose-specific phosphotransferase system that confers high-affinity uptake in Streptomyces coelicolor. J. Bacteriol. 185:929-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puglia, A. M., J. Vohradsky, and C. J. Thompson. 1995. Developmental control of the heat-shock stress regulon in Streptomyces coelicolor. Mol. Microbiol. 17:737-746. [DOI] [PubMed] [Google Scholar]
- 15.Soutourina, O., A. Kolb, E. Krin, C. Laurent-Winter, S. Rimsky, A. Danchin, and P. Bertin. 1999. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J. Bacteriol. 181:7500-7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Süsstrunk, U., J. Pidoux, S. Taubert, A. Ullmann, and C. J. Thompson. 1998. Pleiotropic effects of cAMP on germination, antibiotic biosynthesis and morphological development in Streptomyces coelicolor. Mol. Microbiol. 30:33-46. [DOI] [PubMed] [Google Scholar]
- 17.Vollack, K. U., E. Hartig, H. Korner, and W. G. Zumft. 1999. Multiple transcription factors of the FNR family in denitrifying Pseudomonas stutzeri: characterization of four fnr-like genes, regulatory responses and cognate metabolic processes. Mol. Microbiol. 31:1681-1694. [DOI] [PubMed] [Google Scholar]
- 18.Wehmeier, U. F. 1995. New multifunctional Escherichia coli-Streptomyces shuttle vectors allowing blue-white screening on XGal plates. Gene 165:149-150. [DOI] [PubMed] [Google Scholar]