Abstract
XylS controls the expression of the meta-cleavage pathway for the metabolism of benzoates in Pseudomonas putida KT2440. The xylS gene is expressed from two promoters, Ps1 and Ps2. Transcription from Ps2 is low and constitutive, whereas transcription from Ps1 is induced in the presence of toluene. In this study, we also show that translation of mRNA generated from Ps1 is 10 times more efficient than that generated from Ps2. This pattern of transcription and translation of xylS gives rise to two modes of activation of the promoter of the meta pathway operon (Pm) according to the concentration of XylS in the cell. In cells growing with benzoate, with small amounts of XylS, the activated XylS regulator binds the effector and stimulates transcription from Pm, whereas in cells growing with toluene, the high levels of XylS suffice to stimulate transcription from Pm even in the absence of XylS effectors.
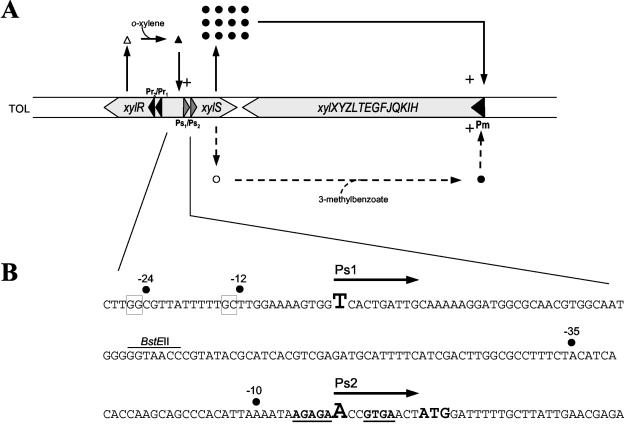
The xyl genes of the Pseudomonas putida TOL plasmid encode the genetic information required for the degradation of toluene and related aromatic compounds. The upper and meta pathway operons, expressed from the Pu and Pm promoters, respectively, comprise the xyl structural genes, whereas the xylR and xylS genes, expressed from the Pr and Ps promoters, respectively, encode the regulatory proteins of the catabolic operons (12, 16) (Fig. 1A).
FIG. 1.
(A) Schematic representation of the meta-cleavage pathway regulatory circuits. The regulatory loops are explained in the text. Dotted lines reflect the situation in the presence of a meta-cleavage pathway effector (i.e., 3-methylbenzoate), and solid lines describe the situation in the presence of upper pathway effectors (i.e., o-xylene). Triangles, XylR; circles, XylS; open symbols, regulator unable to stimulate transcription; solid symbols, regulator able to stimulate transcription. +, stimulation of transcription. (B) Nucleotide sequence of the xylS promoters and leader regions. Arrows above the sequence indicate the two transcripts, with start sites for Ps1- and Ps2-derived transcripts in oversized letters. The −10 and −35 and −12 and −24 positions for Ps2 and Ps1, respectively, are indicated by dots. The first ATG of the xylS open reading frame is in boldface, oversized letters. The two suggested RBSs for the two transcripts are in boldface and underlined. The BstEII site used for insertion of the Ω-Km interposon in pMAR3 is indicated.
The XylR protein is the master regulator in the control of TOL plasmid catabolic operons for the metabolism of toluene (5, 6, 8, 15, 17). Expression of XylR occurs from two tandem promoters, Pr1 and Pr2, the activity of which is high regardless of the growth phase and growth conditions (7, 11). The XylS protein is the regulator that stimulates expression of the meta-cleavage pathway from the Pm promoter (17). The xylS gene is expressed from a single promoter or from two tandem promoters, depending on the growth conditions (4). In the absence of aromatic hydrocarbons, the xylS gene is mainly expressed at low constitutive levels from the σ70-dependent promoter Ps2 (4). In the presence of toluene, an active XylR protein bound to target upstream-activator sequences stimulates transcription from the σ54-dependent Ps1 promoter, without actually varying the transcription level of Ps2 (1, 4). Furthermore, in an rpoN-null mutant, in which expression from Ps1 was completely abolished, Ps2 activity remains constant, and the XylS protein synthesized from this transcript is able to promote transcription from Pm in the presence of 3-methylbenzoate (4).
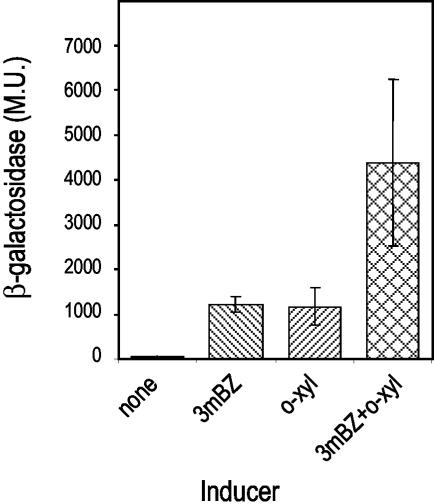
This regulatory system can be faithfully reproduced in P. putida without the TOL plasmid but bearing a plasmid with a transcriptional fusion of the Pm promoter to the lacZ gene and a compatible plasmid carrying the divergent xylS and xylR genes (17). These constructs also reproduced the pattern of expression in a heterologous host such as Escherichia coli. In accordance with previous data, in E. coli cells bearing pJLR107 (Pm:′lacZ) (18) and pKT570 (xylR xylS) (3) and growing in the absence of effector, expression from Pm showed basal levels (50 Miller units), whereas it increased more than 20-fold when it grew in the presence of the meta-cleavage pathway substrate 3-methylbenzoate (Fig. 2). In the presence of the gratuitous upper pathway effector o-xylene, transcription from Pm increased about 20 times (Fig. 2). The following addition of the XylS effector 3-methylbenzoate further increased expression from the Pm promoter to 80 times the basal level. According to the model shown in Fig. 1, these results could be explained as follows. In the presence of a substrate of the meta-cleavage pathway, XylS is synthesized at a low level from the Ps2 promoter (Fig. 1B) and becomes active to stimulate transcription of the meta-cleavage operon, leading to the 20-fold increase in activity (Fig. 1A and 2). When cells are grown in the presence of upper pathway effectors, activation of the XylR- and σ54-dependent promoter Ps1 leads to the overexpression of XylS, which, even in the absence of a specific effector, is able to activate transcription from Pm (Fig. 1).
FIG. 2.
Expression from the Pm promoter in cells bearing xylR and xylS genes growing in the presence of different aromatic compounds. E. coli ET8000 cells bearing pKT570 (xylS xylR) and pJLR107 (Pm::′lacZ) were grown overnight on Luria-Bertani medium containing the appropriate antibiotics. Cultures were diluted 100-fold in the same medium without addition or supplemented with 1 mM 3-methylbenzoate (3mBZ), 1 mM o-xylene (o-xyl), or 1 mM each effector, as indicated. After 1 h of incubation, β-galactosidase activity (Miller units [M.U.]) was determined in permeabilized whole cells as previously described (14). Data are the average of eight independent determinations. The standard deviation is depicted as error bars.
The activity of the two xylS promoters in P. putida TOL plasmid has been determined before (4, 5, 13). In the absence of an effector, or in the presence of meta-cleavage pathway effectors, transcription from both Ps2 and Ps1 is low and reaches similar levels. In the presence of upper pathway effectors, Ps2 remains constant at low levels or increases slightly, whereas Ps1 is induced 5-to 10-fold through XylR-dependent activation (13). However, the primer extension analyses described in reference 13 showed that the differences in expression observed between the two promoters and under the different conditions were not enough to account for the observed differences in XylS-dependent Pm expression between the uninduced and the o-xylene-activated conditions shown in Fig. 2. These observations lead us to consider that in addition to the transcriptional control of xylS expression, there should be a supplementary regulation step at the postranscriptional level that could be exerted through the different translation efficiencies of the two transcripts obtained from the different xylS promoters.
Analysis of the sequence upstream from the first ATG of xylS is of interest to explain the potential differences in translation efficiencies of the two xylS mRNAs. We found that the mRNA derived from Ps1 had a 140-nucleotide leader region with a putative ribosome-binding site (RBS) located 10 nucleotides upstream of the xylS start codon (Fig. 1B), whereas Ps2-derived mRNA presented a short 10-nucleotide leader region with a putative RBS only 3 nucleotides upstream of the first AUG (20) (Fig. 1B). To confirm our hypothesis, we constructed xylS transcriptional and translational fusions to the reporter gene lacZ. The transcriptional fusion was constructed by inserting a 2,440-bp fragment from the TOL plasmid containing xylR and the two promoters Ps1 and Ps2, in pIC552 in front of a galK′-′lacZ fusion bearing the well-characterized translational signal of the galT leader sequence (9), to obtain pMAR1 (Fig. 3). To construct the translational fusion, the same fragment was first cloned in pMLB1034 to obtain an in-frame fusion of the first 25 codons of xylS to the sixth codon of lacZ (19). The fusion was further subcloned in pIC552 to render pMAR2 (Fig. 3). Plasmids pMAR1 and pMAR2 were introduced into E. coli ET8000, and β-galactosidase activity was determined in the presence and in the absence of the XylR effector o-xylene. In parallel, reverse transcription analysis of the mRNAs produced from the two constructs under both conditions was carried out. Figure 3 shows that in the transcriptional fusion pMAR1, the presence of the effector produced a strong induction of both β-galactosidase activity and mRNA synthesis from Ps1, in agreement with previous results. In contrast, in the translational fusion pMAR2, β-galactosidase activity was induced almost 100 times, although mRNA synthesis from Ps1 was similar to the level observed for the transcriptional fusion. Taking into account that in the presence of effectors Ps2 transcription remained almost unchanged and Ps1 is induced almost 10-fold, the increase in β-galactosidase activity over the expected values (almost 10-fold) is probably due to a better translation of Ps1-derived transcripts.
FIG. 3.
Levels of β-galactosidase expressed from different xylS fusions in the presence and in the absence of o-xylene. E. coli ET8000 cells bearing pMAR1, pMAR2, or pMAR3, which include the sequence for the xylR regulator (data not shown) and bear the promoter fusions described in the text and illustrated in the figure, were grown overnight on Luria-Bertani medium containing the appropriate antibiotics. Cultures were diluted 100-fold in the same medium supplemented (+) or not (−) with 1 mM o-xylene. After 4 h of incubation, cells had reached a turbidity at 660 nm of about 1.0 to 1.2. Samples were taken for mRNA analysis or to determine β-galactosidase activity in permeabilized whole cells as previously described (14). β-Galactosidase data are the average of six to nine independent determinations, with standard deviations below 15% of the given values. mRNA reverse primer extension was carried out with excess primer as described previously (10). 32P-labeled cDNA products corresponding to Ps1- and Ps2-derived transcripts are shown. The arrow below the autoradiography fragments shows the electrophoretic mobility. T-leader represents the translational signal of the galT leader sequence (9).
In order to experimentally distinguish between expression coming from Ps1 and Ps2 mRNAs, we inserted a ΩKm interposon (2) in the unique BstEII site located between Ps1 and Ps2 in pMAR2 (Fig. 1B). In this construct (pMAR3), transcripts coming from Ps1 are truncated by the presence of a terminator sequence at each end of the interposon, and therefore only Ps2-derived xylS′-′lacZ transcripts are formed. pMAR3 was introduced in ET8000, and β-galactosidase activity was determined as described above. Figure 3 shows that activity generated from Ps2 and translated by using the translation signals of the short Ps2-derived transcript was lower than the level of activity in pMAR2 in the presence of o-xylene. These results indicate that the translational signals of Ps1-derived long transcripts are more efficient than those of Ps2-derived short mRNA and play an important role in XylS expression. The leader region of Ps2 mRNA (10 nucleotides) is shorter than 12 to 15 nucleotides, the minimum value for the stable binding of ribosomes (21), and is probably responsible for the poor translation levels of this mRNA. In addition, our results suggest that Ps1-derived mRNA translation is very efficient, since higher activity levels are obtained with this wild-type mRNA (pMAR2) than with the galK translation signals present in pMAR1 (Fig. 3).
Therefore, we can conclude that the overexpression of xylS in the presence of upper pathway effectors that leads to a strong induction of Pm is the consequence of both a higher transcription from Ps1 and a more efficient translation of its derived mRNA. The 140-nucleotide leader region of this mRNA, which includes an RBS located at a good distance from the first start codon, is probably responsible for this efficiency. The regulation model could then be summarized as follows: in the absence of the upper pathway effector, the xylS gene is expressed at low level from Ps2, and it is translated at a low rate to render basal amounts of XylS protein, able to induce Pm only when activated by a meta-cleavage pathway effector. In the presence of an upper pathway effector, xylS is transcribed mainly from Ps1 promoter, but the transcript produced is efficiently translated into high XylS protein levels, which by themselves are able to promote the highest transcription levels observed from Pm. This allows coordinate expression of the two consecutive segments of the pathway and would not require the upper pathway substrate to be transformed into a benzoate derivative to initiate transcription activation of the meta pathway. This would lead to a maximal efficiency in the complete degradation of the aromatic compound entering the upper part of the pathway.
Acknowledgments
This study was supported by grant QLRT2001-01923 from the European Commission, grant BMC2001-0515 from the MCYT, and grant BIO2003-00515 from the CICYT.
We thank Eduardo Santero for plasmid vectors and helpful discussion of this study and Patricia Marín for valuable technical assistance.
REFERENCES
- 1.de Lorenzo, V., M. Herrero, M. Metzke, and K. N. Timmis. 1991. An upstream XylR and IHF induced nucleoprotein complex regulates the σ54 dependent Pu promotor of TOL plasmid. EMBO J. 10:1159-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 3.Franklin, F. C. H., M. Bagdasarian, M. M. Bagdasarian, and K. N. Timmis. 1981. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc. Natl. Acad. Sci. USA 78:7458-7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallegos, M. T., S. Marqués, and J. L. Ramos. 1996. Expression of the TOL plasmid xylS gene in Pseudomonas putida occurs from a σ70-dependent promoter or from σ70- and σ54-dependent tandem promoters according to the compound used for growth. J. Bacteriol. 178:2356-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holtel, A., M. A. Abril, S. Marqués, K. N. Timmis, and J. L. Ramos. 1990. Promoter upstream activator sequences are required for expression of the xylS gene and upper pathway operon on the Pseudomonas TOL plasmid. Mol. Microbiol. 4:1551-1556. [DOI] [PubMed] [Google Scholar]
- 6.Inouye, S., A. Nakazawa, and T. Nakazawa. 1983. Molecular cloning of regulatory gene xylR and operator promoter regions of the xylABC and xylDEGF operons of the TOL plasmid. J. Bacteriol. 155:1192-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inouye, S., A. Nakazawa, and T. Nakazawa. 1985. Determination of the transcription initiation site and identification of the protein product of the regulatory gene xylR for xyl operons on the TOL plasmid. J. Bacteriol. 163:863-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inouye, S., A. Nakazawa, and T. Nakazawa. 1987. Expression of the regulatory gene xylS on the TOL plasmid is positively controlled by the xylR gene product. Proc. Natl. Acad. Sci. USA 84:5182-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macián, F., I. Perez-Roger, and M. E. Armengod. 1994. An improved vector system for constructing transcriptional lacZ fusions: analysis of regulation of the dnaA, dnaN, recF and gyrB genes of Escherichia coli. Gene 145:17-24. [DOI] [PubMed] [Google Scholar]
- 10.Manzanera, M., I. Aranda-Olmedo, J. L. Ramos, and S. Marqués. 2001. Molecular characterization of Pseudomonas putida KT2440 rpoH gene regulation. Microbiology 147:1323-1330. [DOI] [PubMed] [Google Scholar]
- 11.Marqués, S., A. Holtel, K. N. Timmis, and J. L. Ramos. 1994. Transcriptional induction kinetics from the promoters of the catabolic pathways of TOL plasmid pWW0 of Pseudomonas putida for metabolism of aromatics. J. Bacteriol. 176:2517-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marqués, S., M. Manzanera, M. M. González-Perez, R. Ruiz, and J. L. Ramos. 1999. Biodegradation, plasmid-encoded catabolic pathways, host factors and cell metabolism. Environ. Microbiol. 1:103-104. [DOI] [PubMed] [Google Scholar]
- 13.Marqués, S., M.-T. Gallegos, M. Manzanera, A. Holtel, K. N. Timmis, and J. L. Ramos. 1998. Activation and repression of transcription at the double tandem divergent promoters for the xylR and xylS genes of the TOL plasmid of Pseudomonas putida. J. Bacteriol. 180:2889-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 15.Perez-Martin, J., and V. de Lorenzo. 1996. ATP binding to the sigma 54-dependent activator XylR triggers a protein multimerization cycle catalyzed by UAS DNA. Cell 86:331-339. [DOI] [PubMed] [Google Scholar]
- 16.Ramos, J. L., S. Marqués, and K. N. Timmis. 1997. Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid-encoded regulators. Annu. Rev. Microbiol. 51:341-373. [DOI] [PubMed] [Google Scholar]
- 17.Ramos, J. L., N. Mermod, and K. N. Timmis. 1987. Regulatory circuits controlling transcription of TOL plasmid operon encoding meta cleavage pathway for degradation of alkylbenzoates by Pseudomonas. Mol. Microbiol. 1:293-300. [DOI] [PubMed] [Google Scholar]
- 18.Ramos, J. L., A. Stolz, W. Reineke, and K. N. Timmis. 1986. Altered effector specificities in regulators of gene expression: TOL plasmid xylS mutants and their use to engineer expansion of the range of aromatics degraded by bacteria. Proc. Natl. Acad. Sci. USA 83:8467-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silhavy, T. J., and J. R. Beckwith. 1985. Uses of lac fusions for the study of biological problems. Microbiol. Rev. 49:398-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spooner, R. A., K. Lindsay, and F. C. H. Franklin. 1986. Genetic, functional and sequence analysis of the xylR and xylS regulatory genes of the TOL plasmid pWW0. J. Gen. Microbiol. 132:1347-1358. [DOI] [PubMed] [Google Scholar]
- 21.Stormo, G. D., T. D. Schneider, and L. M. Gold. 1982. Characterization of translational initiation sites in E. coli. Nucleic Acids Res. 10:2971-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]