Abstract
Temperate bacteriophages with plasmid prophages are uncommon in nature, and of these only phages N15 and PY54 are known to have a linear plasmid prophage with closed hairpin telomeres. We report here the complete nucleotide sequence of the 51,601-bp Klebsiella oxytoca linear plasmid pKO2, and we demonstrate experimentally that it is also a prophage. We call this bacteriophage φKO2. An analysis of the 64 predicted φKO2 genes indicate that it is a fairly close relative of phage N15; they share a mosaic relationship that is typical of different members of double-stranded DNA tailed-phage groups. Although the head, tail shaft, and lysis genes are not recognizably homologous between these phages, other genes such as the plasmid partitioning, replicase, prophage repressor, and protelomerase genes (and their putative targets) are so similar that we predict that they must have nearly identical DNA binding specificities. The φKO2 virion is unusual in that its phage λ-like tails have an exceptionally long (3,433 amino acids) central tip tail fiber protein. The φKO2 genome also carries putative homologues of bacterial dinI and umuD genes, both of which are involved in the host SOS response. We show that these divergently transcribed genes are regulated by LexA protein binding to a single target site that overlaps both promoters.
Temperate bacteriophages usually physically integrate their prophage DNAs into the host bacterium's chromosome when they establish lysogeny. However, a few prophages, such as those of phages P1, N15, LE1, φ20, and φBB-1, are not integrated but replicate in the lysogen as low-copy-number plasmids (32, 38, 52, 53, 84). Of these, the Escherichia coli double-stranded DNA (dsDNA) tailed-phage N15 has an unusual linear prophage plasmid that has covalently closed hairpin telomeres (67, 68). Although N15 has been studied in some detail (83, 85, 87), few other phages with similar linear DNA prophages have been described. While this paper was being prepared, another linear plasmid prophage, PY54, was reported for Yersinia enterocolitica (48). Furthermore, the linear, hairpin-ended plasmids Lp28-2 and Lp54 in the spirochete Borrelia burgdorferi B31-MI carry sequence similarities to dsDNA phage virion assembly genes (19, 33); however, they have not yet been shown to be bone fide prophages. Linear plasmids are very rare in the γ-Proteobacteria, but it has been reported that Klebsiella oxytoca strain CCUG 15788 (a nickel-resistant bacterium isolated from the mineral oil-water coolant-lubricant emulsion of a metal working machine in Göteborg, Sweden, in 1989 [69]) harbors a linear plasmid, pKO2, of about 50 kbp (95). Since this is close to the size of the N15 linear plasmid prophage, we were prompted to investigate the possibility that this plasmid could be a prophage. We report here the complete nucleotide sequence of pKO2, a linear plasmid prophage that encodes the synthesis of a tailed-phage particle, and the analysis of some of its properties.
MATERIALS AND METHODS
Bacterial strains and plasmid construction.
The bacterial strains used for this study are described in Table 1. All bacterial cultures were grown in Luria-Bertani broth at 37°C. The preparation of K. oxytoca CCUG 15788 DNA in agarose blocks, contour-clamped homogeneous electric field pulsed-field electrophoresis gels, and native and alkaline agarose electrophoresis gels and for Southern blot analysis was performed as previously described (17, 18, 93), and DNA cloning in vitro was performed as previously described (14). Plasmid pBLSK (Stratagene, La Jolla, Calif.) and pET15b (Invitrogen, Carlsbad, Calif.) manipulations were performed by using E. coli NF1829 and XL-1-Blue (Stratagene), respectively, and pET15b expression constructs were induced by IPTG (isopropyl-β-d-thiogalactopyranoside) in E. coli BL21 (DE3). Plasmid F′128 derivatives were constructed by the strategy described by Bunny et al. (11). The P22-lacZ reporter constructs were made as follows. A 365-bp sequence of φKO2 DNA (from bp 25894 through 26258) which included the putative φKO2 P22 promoter and the first 19 codons of the 5′ end of gene 22 was PCR amplified by using the primers 5′-GTTGTAAAACGACGGCCAGTGAATCCGTAATCATGGTCATCTTAGCACCGGCTAATCCCT and 5′-TCAGGCATCGGCGCACGGCTGTATTCGTGATCTCCGGCTCCCATCCTGTAGCGTTTAACCC, whose 40-nucleotide 3′ tails (underlined) are the same as sequences downstream of the mphC gene and at the 5′ end of the lacZ gene of the plasmid carrying F′128 mphC31::Tn10-dTet lacA4643::Tn10d-Cam, respectively (see below and Fig. 2B and C of reference 11). Electroporation of strain TT24565 (Table 1) with this amplified DNA fragment, followed by selection for the loss of tetracycline resistance (7), resulted in a modified F′128 plasmid in which homologous recombination had occurred between the parent plasmid and the PCR fragment's non-φKO2 tails, such that the amplified DNA replaced the plasmid's mph operon and fused the first 19 codons of gene 22 in-frame with the 5′ terminus of the lacZ gene. The inserted DNA was amplified with outside PCR primers, and the DNA product was sequenced directly to confirm the plasmid's structure and to ensure that no PCR-generated sequence changes had occurred. The resulting male strain (UB-1535) was then mated on solid medium for 6 to 8 h with strains TT23771, TT23933, TT24089, and TT24090, and in each case the recipient carrying the modified F′ plasmid was selected by growth in glucose minimal medium in the presence of 30 μg of chloramphenicol/ml. An analogous P23 reporter construct carrying 191 bp of φKO2 DNA with lacZ fused to the first 19 codons of gene 23 was made in a similar manner by using oligonucleotides 5′-GTTGTAAAACGACGGCCAGTGAATCCGTAATCATGGTCATTTCATCAAGCGAAAGCTGCTT-3′ and 5′-TCAGGCATCGGCGCACGGCTGTATTCGTGATCTCCGGCTCCTTAGCACCGGCTAATCCCTC-3′ as PCR primers.
TABLE 1.
Bacterial strains used for this study
| Strain | Genotypea | Source or reference |
|---|---|---|
| K. oxytoca | ||
| CCUG15788 | Wild type | 95 |
| E. coli K-12 | ||
| NF1829 | araD139 Δ(ara ABOIC-leu)7679 galUK Δ(lac)X74 recA+rspL thi/F′ lacIq1 lacZ::Tn5 lacY+ | 91 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1/F′ proAB lacIq lacZ::Tn10 (Tetr) | Stratagene |
| BL21(DE3) | ompT hsdSB (rB− mB−) gal dcm λ (DE3) | 96 |
| S. enterica serovar Typhimurium LT2 | ||
| TT24565 | metA22 metE551 trpD2 ilv-452 leu pro (leaky) hsdLT6 hsdSA29 hsdB strA120/pKD46/F′128 pro+mphC31::Tn10-dTet lac Y 4643::Tn10d-Cam | This study |
| TT23771 | lexA+sulA46::Spc Δ(Fels-2, Gifsy-1, Gifsy-2) proB1657::Tn10 | This study |
| TT23933 | lexA41::Cam(sw) sulA46::Spc Δ(Fels-2, Gifsy-1, Gifsy-2) proB1657::Tn10 | This study |
| TT24089 | lexA33::[Rif lexA3(Ind−)](sw) sulA+ Δ(Fels-2, Gifsy-1, Gifsy-2) proB1657::Tn10 | This study |
| TT24090 | lexA33::[Rif 1exA3(Ind−)](sw) sulA46::Spc Δ(Fels-2, Gifsy-1, Gifsy-2) proB1657::Tn10 | This study |
| UB-1535 | metA22 metE551 trpD2 ilv-452 leu pro (leaky) hsdLT6 hsdSA29 hsdB StrA120/pKD46/F′128 pro+ (KO2-P22::lacZ+) lacY4643:: Tn10d-Cam | This study |
| UB-1547 | lexA33::[Rif lexA3(Ind−)](sw) sulA+ Δ(Fels-2, Gifsy-1, Gifsy-2) proB1657::Tn10/F′128 pro+ (KO2-P22::lacZ+) lacY4643::Tn10d-Cam | This study |
| UB-1548 | lexA+ sulA46::Spc Δ(Fels-2, Gifsy-1, Gifsy-2) proB1657::Tn10/F′128 pro+ (KO2-P22::lacZ+) lacY4643::Tn10d-Cam | This study |
| UB-1549 | lexA33::[Rif lexA3(Ind−)](sw) sulA46::Spc Δ(Fels-2, Gifsy-1, Gifsy-2) proB1657::Tn10/F′128 pro+ (KO2-P22::lacZ+) lacY4643::Tn10d-Cam | This study |
| UB-1550 | lexA41::Cam(sw) sulA46::Spc Δ(Fels-2, Gifsy-1, Gifsy-2) proB1657::Tn10/F′128 pro+ (KO2-P22::lacZ+) lacY4643::Tn10d-Cam | This study |
| UB-1555 | lexA+ sulA46::Spc Δ(Fels-2, Gifsy-1, Gifsy-2) proB1657::Tn10/F′128 pro+ (KO2-P23::lacZ+) lacY4643::Tn10d-Cam | This study |
| UB-1556 | lexA41::Cam(sw) sulA46::Spc Δ(Fels-2, Gifsy-1, Gifsy-2) proB1657::Tn10/F′128 pro+ (KO2-P23::lacZ+) lacY4643::Tn10d-Cam | This study |
| UB-1557 | lexA33::[Rif lexA3(Ind−)](sw) sulA46::Spc Δ(Fels-2, Gifsy-1, Gifsy-2) proB1657::Tn10/F′128 pro+ (KO2-P23::lacZ+) lacY4643::Tn10d-Cam | This study |
| UB-1558 | lexA33::[Rif lexA3(Ind−)](sw) sulA+ Δ(Fels-2, Gifsy-1, Gifsy-2) proB1657::Tn10/F′128 pro+ (KO2-P23::lacZ+) lacY4643::Tn10d-Cam | This study |
The null lexA41::Cam(sw) strain, which has a chloramphenicol resistance gene inserted into the fourth codon of the lexA gene and Δ(Fels-2, Gifsy-1, Gifsy-2) and sulA46::Spc mutations were described by Bunny et al. (11). The noninducible lexA33::[Rif lexA3(Ind−)](sw) allele has the Salmonella lexA gene replaced by an E. coli lexA gene that contains the lexA3 mutation that blocks LexA protein autocleavage (64). The plasmid pKD46 expresses the phage λ recombination gene products, which allow efficient recombinational transformation by DNA with only short terminal homologies with the target DNA (23). KO2-P22::lacZ+ and KO2-P23::lacZ+ indicate the presence of the φKO2 gene 22 and 23 promoters, respectively, driving expression of the F′ lacZ gene (see text).
FIG. 2.
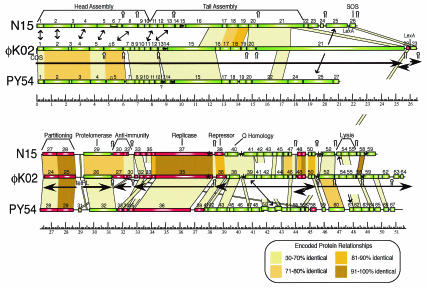
Map of the φKO2 genome. The genomes of phages φKO2, N15, and PY54 are shown with a scale of kilobase pairs for the φKO2 DNA. Predicted genes are indicated by rectangular boxes (those transcribed rightward are green and those transcribed leftward are red). Yellow and brown regions connect homologous regions of the two genomes. The predicted φKO2 transcription units are indicated below its genome. SOS refers to genes that may be involved in modulating and are controlled by the host SOS response. Double-headed arrows connect pairs of genes which have similarities to other genes of similar function but which have no discernible sequence similarity themselves. Asterisks denote putative φKO2 and N15 partitioning sites recognized by gene 27 and 28 proteins, and stem-loops denote predicted rho-independent transcription terminators (above the map for N15 and below the one for φKO2).
Virion production and purification and electron microscopy.
φKO2 virions and DNAs were prepared as follows: K. oxytoca CCUG 15788 was grown in Luria-Bertani broth at 37°C to a density of 2 × 108 cells/ml, mitomycin C (Sigma, St. Louis, Mo.) was added to a final concentration of 0.1 μg/ml, shaking was continued for 2 to 4 h at 37°C, and the culture was shaken with chloroform to complete the lysis. Cell debris was removed by centrifugation for 20 min at 8,000 rpm in a Beckman JA-10 rotor at 4°C, and phage particles were then pelleted by spinning overnight in the same rotor. The pellet was gently resuspended in TM (10 mM TrisCl [pH 7.4], 1 mM MgCl2), and the particles were separated in a CsCl density step gradient as previously described (31) and then dialyzed against TM. About 1 mg of particles was obtained per liter of culture, suggesting a yield of about 50 particles/cell. Electron microscopic analysis showed that both tailed virions and tailless heads were present in this preparation; however, a subsequent sucrose density velocity gradient largely separated these two components (1 ml of about 1-mg/ml particles was layered onto a preformed 10 to 40% sucrose gradient and spun in an SW41 Beckman rotor at 25,000 rpm for 120 min). The visibly opalescent head (faster sedimenting) and virion (slower sedimenting) bands were removed by syringe puncturing of the tubes. Finally, particles were concentrated by pelleting overnight and resuspension in TM as described above. DNAs were isolated from φKO2 virions by sodium dodecyl sulfate (SDS) release and ethanol precipitation. Virions were stained for electron microscopy with 0.5% uranyl formate on a carbon-coated Parlodion film and were examined in a Zeiss EM902 microscope operating at 80 kV.
DNA and protein methods.
Nucleotide sequence determination was done by the dideoxy chain termination method, as previously described (76). Bulk sequencing of a random plasmid library of sheared DNA was performed until an average of more than sevenfold coverage was obtained (768 sequencing runs). Thirty-two primers were used to sequence across weak areas, with φKO2 virion DNA as a template, and PhredPhrap (40) was used to assemble the sequence into one circular contig. The DNA sequence analysis software packages used were DNA Strider (27), GeneMark (8), the Staden programs (94), BLAST (3), BPROM (http://www.softberry.com/berry.phtml?topic=gfindb), and DNA Master (J. Lawrence [http://cobamide2.bio.pitt.edu/]). Polyacrylamide gel electrophoresis of proteins in SDS, staining with Coomassie brilliant blue R-250 (Bio-Rad, Richmond, Va.), and determination of N-terminal amino acid sequences by the University of Utah Protein Facility were performed as previously described (34).
Nucleotide sequence accession number.
The complete nucleotide sequence for the φKO2 virion has been deposited under GenBank no. AY374448.
RESULTS
The φKO2 virion.
Treatment with 0.5 μg of mitomycin C/ml in L broth at 37°C causes K. oxytoca CCUG 15788 cultures to lyse after several hours. Tailed-phage virion-like particles could be purified to near homogeneity from these lysates by differential centrifugation followed by CsCl density step gradient centrifugation (31). The DNA molecules within these particles are monodisperse and about 50 kbp in length, as measured by pulsed-field gel electrophoresis and by summing of the band sizes after cleavage by restriction enzyme HindIII or EcoRI. The isolated virion DNAs hybridized strongly in Southern blot analyses (93) to the pKO2 plasmid DNA displayed in pulsed-field gel electrophoresis gels, showing that the pKO2 plasmid DNA is present in the φKO2 virions (data not shown). When plasmid pKO2 and virion φKO2 DNAs were cleaved by restriction enzymes and the resulting bands were compared by electrophoresis, a number of bands were identical in size; however, each DNA produced several restriction fragments whose sizes were different in the plasmid and virion DNAs. This is explained by the circularly permuted relationship between virion and prophage DNAs, similar to what is seen for other temperate phages. The ends of both DNAs are characterized in more detail below.
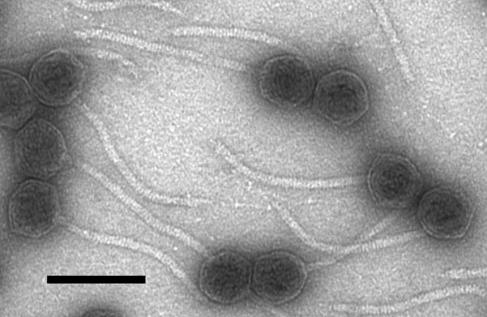
Negative-stain electron microscopy of the φKO2 virions showed heads that are hexagonal in outline (and thus almost certainly icosahedral in shape), with flexible, apparently noncontractile tails that are similar in appearance to the tails of phage λ and many other phages (Fig. 1). The heads are 60 nm in diameter, and the tails are about 9.0 nm wide and 180 nm long and contain about 45 stacked disks (for comparison, these values for λ virions are 60, 18, and 150 nm, respectively, and 32 disks). Tail fibers are not obviously visible, but in a few instances, where the tail tips of two or more phages lie in close proximity, some fibrous material can be seen in the vicinity of the tail tips. This is consistent with but does not prove the hypothesis that φKO2 does have tail fibers but that they are too thin to visualize individually. Although these φKO2 virions appear to be morphologically intact, our attempts to propagate them on several other K. oxytoca strains, as well as several Klebsiella pneumoniae, Salmonella enterica, and E. coli strains, were not successful (we have not yet been able to isolate a K. oxytoca CCUG 15788 strain that is devoid of the pKO2 plasmid for such a test). The strain specificity of most phages means that this propagation failure is not a strong indication that the φKO2 particles are not infectious on the proper host.
FIG. 1.
Electron micrograph of φKO2 virions. Virions were prepared, negatively stained, and examined by electron microscopy as described in Materials and Methods. Bar, 100 nm.
The complete φKO2 nucleotide sequence.
A shotgun library of DNA fragments from virions was used to determine the complete sequence of φKO2 DNA by automated dideoxy methods as described in Materials and Methods. The assembly of 384 shotgun plasmid clones, sequenced in the forward and reverse directions from vector-based primers, and 32 sequences primed by oligonucleotides on φKO2 virion DNA gave a complete sequence of the φKO2 chromosome, which is 51,601 bp in length and 51.9% G+C. This G+C value is very close to the 51.2% value for the related phage N15 but is somewhat lower than the 57.1% G+C value of the K. pneumoniae genome (calculated from publicly available Washington University K. pneumoniae genome project web site data [http://genome.wustl.edu/projects/bacterial/kpneumoniae/]). The φKO2 sequence accurately predicts the sizes of the HindIII, BamHI, and EcoRI fragments, which confirms the correct assembly of the sequence. About 45% of its nucleotide sequence is recognizably similar to and colinearly organized with the phage N15 chromosome; thus, φKO2 is clearly a rather close relative of N15, which indicates that it is a member of the lambdoid phage group. Putative φKO2 genes were identified by using the E. coli codon usage table and the Staden (94) and GeneMark (8) computer programs. Nearly all of the predicted genes have the expected rise in coding potential based on codon usage and a plausible Shine-Dalgarno ribosome binding site. Sixty-four probable protein-encoding genes were located, with 56 having AUG and 8 having GUG translation start codons, occupying 93.4% of the chromosome. These genes, along with their predicted functions and closest homologues, are listed in Table 2, and a physical map of the genome is shown in Fig. 2.
TABLE 2.
Bacteriophage φKO2 gene analysis
| Gene | Predicted protein size (no. of amino- acids) | Predicted function | Phages with best matchesa | Extent of best match (% identity/length of region [no. of aminoacids]) |
|---|---|---|---|---|
| 1 | 143 | Small terminase subunit | PY54, DT1, CP-933C, φST64B | 64/143 |
| 2 | 568 | Large terminase subunit | PY54, ΦP27, SfV, φST64B, φE125 | 71/568 |
| 3 | 59 | Unknown (head assembly?) | PY54f, e14, ΦP27, φST64B, φE125 | 42/59 |
| 4 | 422 | Portalb | PY54, φE125, φST64B, ΦP27 | 75/422 |
| 5 | 306 | Maturation protease/scaffold | PY54, φE125, Gifsy-1, λc | 64/306 |
| 6 | 428 | Major head shell (coat) proteinb | PY54c, φE125 | 76/366 |
| 7 | 152 | No matches (head assembly?) | PY54 | 43/86 |
| 8 | 98 | Unknown (head assembly?) | PY54, SfV, φST64B | 50/98 |
| 9 | 112 | No matches (virion assembly?) | PY54 | 59/112 |
| 10 | 129 | No matches (virion assembly?) | PY54 | 56/129 |
| 11 | 133 | Unknown (tail assembly?) | PY54, D3, λSo, φE125, HK97 | 68/133 |
| 12 | 155 | Major tail shaft proteinb | PY54 | 70/143 |
| 13 | 122 | Tail chaperone? | PY54 | 62/122 |
| 14e | 202 | Frameshifted tail chaperone? | PY54f | 52/202 |
| 15 | 1,118 | Tail “tape measure” | PY54, HK022, TP901-1c | 56/1,118 |
| 16 | 112 | Tail tip structure (λ gpM) | HK97, HK022, φE125, N15, λc | 67/112 |
| 17 | 251 | Tail tip structure (λ gpL) | HK022, N15, HK97, λc | 74/251 |
| 18 | 236 | Tail tip structure (λ gpK) | HK97, N15, HK022, λc | 86/333 |
| 19 | 111 | Moron gene | Xanthomonas prophage XccP1 | 33/99 |
| 20 | 197 | Tail tip structure (λ gpI) | N15, HK022, φE125, λc | 69/194 |
| 21 | 3,433 | Tail tip fiber (λ gpJ) | HK022, N15, λSo, λc | 69/941 |
| 22 | 80 | DinI SOS response regulator | Stm6d, Gifsy-2 | 76/67 |
| 23 | 127 | UmuD′ DNA polymerase subunit | Stm6d, N15, P1c | 68/129 |
| 24 | 323 | Plasmid partitioning (SopB) | N15c, PY54, P1c | 76/320 |
| 25 | 387 | Plasmid partitioning (SopA) | N15c, PY54, P7 | 92/387 |
| 26 | 640 | Protelomerase | N15c, PY54c, VHML | 75/640 |
| 27 | 260 | Auxiliary antirepressor | N15c, CP-933N, φP27, φ11, A118 | 74/262 |
| 28 | 76 | Antirepressor | N15c, PY54c | 78/76 |
| 29 | 54 | Inhibitor of cell division | N15c, PY54, P4c, φR73 | 85/54 |
| 306 | 58 | Control of genes 27 to 29? | φR73, P4, N15 | 60/43 |
| 31 | 75 | No matches | None | |
| 32 | 111 | Unknown | N15, PY54 | 69/116 |
| 33 | 182 | Unknown | P22, PY54 | 35/110 |
| 34 | 82 | Unknown | N15 | 81/82 |
| 35 | 1,328 | Replicase | N15c, PY54 | 92/1,318 |
| 36 | 202 | Prophage repressor | N15c, PY54c | 88/202 |
| 37 | 69 | Cro repressor | N15, PY54c | 87/65 |
| 38 | 245 | Q-like antiterminator | N15, PY54, Gifsy-1 | 70/248 |
| 39 | 202 | Exonuclease III; Pol III ɛ | N15, 186 | 62/197 |
| 40 | 44 | No matches | None | |
| 41 | 110 | No matches | None | |
| 42 | 226 | No matches | None | |
| 43 | 97 | Unknown | N15 | 61/94 |
| 44 | 203 | Unknown | CP-933P, N15, ST64T | 30/109 |
| 45 | 108 | Unknown | N15, ɛ15 plasmid pKM101 | 77/108 |
| 46 | 79 | Unknown | N15, PY54 | 73/73 |
| 47 | 68 | No matches | None | |
| 48 | 95 | Possible Cro-like repressor | N15 | 85/95 |
| 49 | 104 | Unknown | N15, Xylella prophage XfP3 | 93/104 |
| 50 | 73 | Unknown | N15 | 80/73 |
| 51 | 75 | Unknown | N15 | 71/75 |
| 52 | 353 | DNA adenine methylase | PY54, SfV, φP27, VMHL, N15 | 77/353 |
| 53 | 101 | No matches | None | |
| 54 | 163 | No matches | None | |
| 55 | 154 | No matches | None | |
| 56 | 57 | No matches | None | |
| 57 | 32 | No matches | None | |
| 58 | 99 | Unknown | N15, PY54, λSo, Omega, TM4 | 90/99 |
| 59 | 241 | Unknown | φ80 | 66/74 |
| 60 | 120 | Holin? | PY54, φP27, ST64T, SfV, φE125 | 65/120 |
| 61 | 96 | No matches | None | |
| 62 | 146 | Unknown | Salmonella plasmid pHCM2 | 33/130 |
| 63 | 127 | Unknown | S. enterica serovar Typhi prophage Sti1d | 68/127 |
| 64 | 66 | No matches | None |
Matches are listed in descending order of extent of similarity. The matching genes (names not shown) of the indicated phages typically occupy very similar positions on those genomes. Unless denoted by a footnote, they have not been shown by direct experimentation to have the function indicated, but they have sequence similarities to ones that have been so studied.
Presence in φKO2 confirmed by N-terminal amino acid analysis (see text).
This phage's gene has been shown directly to have the indicated function.
The names of S. enterica serovar Typhimurium and Typhi defective prophages Stm6 and Sti1, respectively, are provisional (13).
Note that gene 14 is likely expressed as a frameshifted protein whose N terminus is identical to that of the gene 13 protein but whose C terminus has extra sequence; gene 30 may not be an authentic gene.
Gene not annotated in original genome sequence publication.
Prophage and virion DNA circular permutation.
The overall organization of the φKO2 genome is similar to that of phage N15, with divergently transcribed putative early operons and a λ-like late operon arrangement. Its prophage and virion DNA ends are located at similar positions in the genome sequence as those of N15. After the complete sequence of the φKO2 genome was known, by sequencing of the ends of the virion DNA, we established that φKO2 has 10-nucleotide single-stranded 3′ extensions, 3′-TGCCGCTTCA…-5′ and 5′-…ACGGCGAAGT-3′, at the left and right ends of the mature virion DNA, respectively. The cohesive end site lies 63 bp to the left of the start of gene 1 (below), which encodes the small terminase subunit.
In the pKO2 linear prophage plasmid, the cohesive ends of the virion DNA are covalently joined, and the linear prophage plasmid is formed by opening at a different site, called telRL (for the site of telomere formation), on the opposite side of the circularized genome. Restriction analysis of the linear plasmid DNA showed that its ends are located at positions similar to those of the phage N15 prophage plasmid ends, as determined by its telRL site (data not shown). The φKO2 sequence at this location contains the perfect inverted repeat 5′-CCATTATACGC · GCGTATAATGG-3′ (the dot denotes the center of symmetry), which is identical to the central region of the N15 telRL site (25, 67). We also showed (by a procedure identical to that of Casjens and Huang [17]) that the two strands of terminal HindIII and EcoRI restriction fragments of the linear pKO2 plasmid DNA reanneal extremely rapidly (snapback) compared to internal restriction fragments (data not shown). This strongly indicates that the pKO2 plasmid ends are covalently closed hairpins (also see below).
The predicted late operon genes.
The left portion of the φKO2 virion chromosome is organized in a manner similar to that of the lambdoid phages, with putative head genes near the left end and tail genes clustered immediately to their right. All of these genes are transcribed in a rightward direction. The predicted head and tail shaft (see below) genes 1 through 15 are all quite similar to parallel genes in Yersinia phage PY54 (48), but in neither phage had they been studied in much further detail. Genes 1, 2, 3, 4, 5, 6, and 8 each have matches to putative head genes of other phages, and genes 11 and 16 through 21 match phage tail genes. These matches are all in the order that is expected from the conserved head and tail gene orders found in most dsDNA phage genomes (13, 16). For example, the head gene homologies are as follows: small terminase subunit gene with φKO2 gene 1, large terminase subunit gene with φKO2 gene 2, portal protein gene with φKO2 gene 4, maturation protease gene with φKO2 gene 5, and coat protein gene with φKO2 gene 6. Excluding PY54, the φKO2 head genes most closely resemble those of E. coli phage ΦP27 (86), Shigella flexneri phage SfV (1), and Burkholderia mallei phage φE125 (101) (Table 2). These form a phage head type that is distantly but still convincingly related to phages whose head assembly and DNA packaging have been studied in detail, including E. coli phage HK97 (54). Like φKO2, the cohesive ends of PY54, φE125, and ΦP27 are all 10-nucleotide single-stranded 3′ extensions; however, the sequences of the φKO2 cohesive ends are very different from those of other phages (1, 47, 101). The φKO2 head genes are not recognizably homologous to the λ-like head genes of N15, whose virion DNA has 5′-extended 12-nucleotide cohesive ends.
To begin to confirm the above predictions, we displayed the proteins of whole φKO2 virions and of purified heads by SDS gel electrophoresis (Fig. 3) and excised the virion band gp6 (both tailless heads and tailed virions are produced in large numbers after mitomycin C induction; see Materials and Methods). This must be the major head (capsid) protein, since it is by far the most abundant virion protein in purified heads. Its N-terminal amino acid sequence was determined, and this sequence, NH2-Ala-Ile-Ser-Thr-Ala-Ala-Gly, is present only once in all of the predicted φKO2-encoded proteins, at an internal position in the unprocessed gene 6 protein. The N-terminal alanine is predicted to be encoded by codon 126. The coat proteins of many viruses are proteolytically cleaved during assembly, and those present in mature φP27, SfV, and φSLT virions (three database matches that are cleaved) have been shown to have the N-terminal 115, 115, and 112 amino acids, respectively, removed (2, 73, 86). However, the φKO2 gene 6 protein is only rather weakly similar to these coat proteins, some of its sequence relatives are not cleaved (e.g., the phage A118 coat protein [66]), and it has an in-frame AUG methionine codon 125. These points raise the possibility that translation could begin at this AUG followed by removal of that methionine; however, there is no convincing Shine-Dalgarno ribosome binding sequence upstream of this AUG, whereas there is such a sequence (AGAGAGAA) upstream of the predicted gene 6 start codon. The size of the coat protein measured by SDS-polyacrylamide gel electrophoresis is about 33,000 Da, which is very close to the predicted size of the cleaved gene 6 protein of 32,731 Da. In addition, the φKO2 gene 5 protein is homologous to other phage maturation proteases. These facts combine to make it extremely probable that the gene 6 protein is translated from the whole predicted open reading frame (see GenBank annotation) and is subsequently cleaved between amino acids 125 and 126, most likely during head assembly by analogy with the well-studied phage systems. We suggest that, due to the high degree of similarity in the maturation protease and coat protein genes, a similar situation holds true for phage PY54 (48).
FIG. 3.
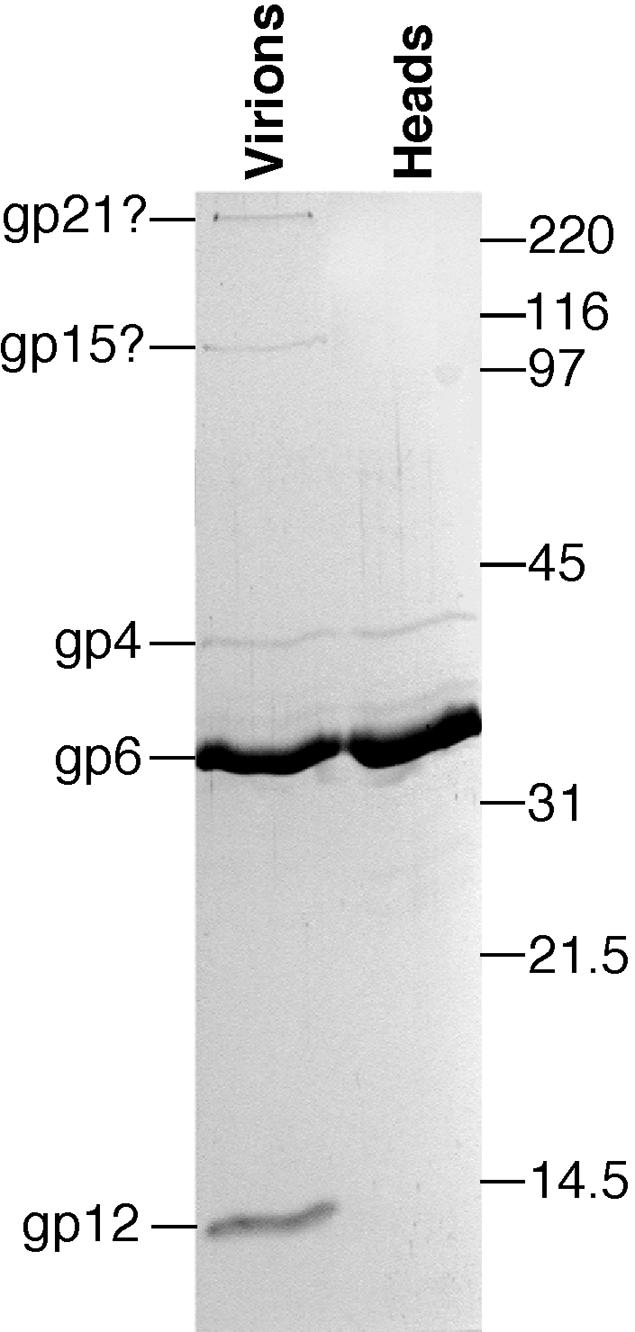
Separation of φKO2 proteins by SDS-polyacrylamide gel electrophoresis. The protein components of purified virions and heads, separated by sucrose gradient centrifugation, were displayed by SDS-10% polyacrylamide gel electrophoresis and were stained with Coomassie brilliant blue. The positions of standard proteins are indicated on the right, and the identities of φKO2 proteins are indicated on the left (gpX indicates the product of gene X).
For a different approach to this question, we asked, prior to determining the N-terminal sequence, whether we could predict the site of the putative cleavage by homology to capsid proteins with characterized cleavage sites. We aligned the gp6 sequence with the numerous homologous sequences in the databases by using the program PSI-BLAST. Of the capsid proteins with known cleavage sites, that of E. coli phage HK97 had the best match to φKO2 in the vicinity of the cleavage site. Despite only 18% overall sequence identity between the two proteins, there was an 18-amino-acid region covering the HK97 cleavage site where the HK97 and φKO2 sequences aligned with six identities, six conservative substitutions, and no gaps, and this positioned the HK97 cleavage site between Met125 and Ala126 of the aligned φKO2 sequence. We believe that this correct prediction of the chemically determined φKO2 N terminus based on homology with the HK97 cleavage site further strengthens the case for cleavage at this site rather than at an internal translational start codon. We noted that the dipeptide defining the cleavage site is not well conserved among phages that do this cleavage: the cleavage site is Lys-Ser in HK97 (28), Arg-Asp in φC31 (92), and Met-Ala in φKO2. In HK97, the cleavage site Lys was changed to Leu and cleavage was still observed, though at a reduced rate, arguing that some of the cleavage specificity must reside in the local conformation of the polypeptide (29).
We also determined the N-terminal sequence of band gp4 (Fig. 2) to be NH2-Met-Phe-Ile-Pro-?-Met (“?” indicates an unidentified amino acid), which matches the predicted N terminus of the φKO2 gene 4 protein. The intensity of this band relative to the coat protein band is consistent with the 12 portal protein molecules per 400 coat protein molecules observed in other T=7 phage heads (16). However, the observed size, as measured by SDS gel electrophoresis, of the predicted 47,354-Da gene 4 protein in φKO2 heads is about 40,000 Da; either it migrates anomalously in such gels or it is proteolytically processed, but not at the N terminus.
Among φKO2 genes 7 through 14, only two similarities to genes in the current database exist outside of phage PY54: gene 8 matches the similarly located phage SfV gene 6 (the proteins they encode are identical in 25 of 76 amino acids), and gene 11 weakly matches similarly located genes in Pseudomonas phage D3 (59), Burkholderia phage φE125 (101), E. coli phage HK97 (54), and the Shewanella prophage λSo (45). The positions of these genes relative to the conserved order of other lambdoid phage genes suggest that gene 8 likely encodes a head completion protein, while gene 11 may encode the protein on tails that binds heads (15, 16, 54). The only gene 12 database match is one to PY54 gene 12, and the latter is present in PY54 virions (48). The N-terminal sequence analysis of the φKO2 major tail protein band (present in virions but not in heads) (Fig. 3) gave the sequence NH2-Ala-Asp-Asn-Gln-Thr. This matches the predicted N terminus of the gene 12 protein perfectly if its N-terminal methionine is removed (which is not uncommon with a penultimate alanine [36]). Its measured molecular weight in SDS-polyacrylamide gel electrophoresis gels of about 14,000 Da is not far from the predicted size of 16,154 Da. The location of gene 12 on the φKO2 map, the large number of molecules in the virion, and their complete absence from heads show that it encodes the building block of the shaft of the tail. The narrowness of the φKO2 tail is likely explained by its smaller shaft subunit (155 amino acids compared to 246 amino acids for λ). A comparison of the φKO2 and PY54 genes in this region showed that PY54 open reading frame 13 is neatly absent from φKO2 (Fig. 2). We noted that for PY54, (i) gene 13 does not have a convincing Shine-Dalgarno sequence, (ii) genes 12 and 13 overlap, and (iii) virions contain two proteins, of 18 and 30 kDa, whose nonidentical peptide mass spectra are very similar. These facts would all be explained if PY54 gene 13 were expressed only as a −1 programmed ribosomal frameshift from gene 12. Any ribosomes that shifted would thus create a gene 12 protein with the gene 13 protein frame amino acids at its C terminus. Since such frameshifts do not occur during every ribosome passage, two sizes of “gene 12 protein” would be made (and the amino acid sequence encoded by the gene 13 frame would be homologous to the C-terminal portion of the HK97 major tail shaft protein). This seems reasonable since in the short overlap between these two reading frames there is a C-CCA-AAG sequence (hyphens separate codons in the gene 12 frame) on which ribosomes can have a −1 shift; this sequence was shown to give a very high level of frameshifting at the IS3 orfA-B frameshift site, even though the tRNA nucleotide paired with the A in the CCA codon should, in theory, pair with a C after the shift (63), and there is a GGGGAA Shine-Dalgarno-like sequence upstream of the putative shift site that might stimulate shifting (60).
The overlapping φKO2 reading frames 13 and 14 appear to be analogous, but not recognizably similar in sequence, to the similarly located G and G-T genes, respectively, of phage λ. In the latter phage, ribosomes initiate at the gene G start codon, but about 4% of them undergo a programmed −1 frameshift and continue on in the G-T frame; no ribosomes initiate in the G-T frame (61). Many other tailed phages have a pair of tail genes with a similar frameshifting relationship (e.g., see references 20 and 102), yet there is often no recognizable similarity among them (J. Xu, R. Duda, R. Hendrix, and S. Casjens, unpublished data). In φKO2, gene 13 has a putative Shine-Dalgarno ribosome binding site, 5′-AAGGA, but the downstream frame has no convincing translation start codon. Furthermore, in the short overlap between the gene 13 and downstream open reading frames, there is a 5′-G-GAA-AAA-AAC-UAA (-Glu-Lys-Asn-stop in the 13 frame) sequence that could potentially program such a shift from the 13 frame into the downstream frame when Glu and Lys tRNAs or Lys and Asn tRNAs are bound to the ribosome, and the stop codon could provide the translational pause that is often associated with such events. The Lys-Asn tRNA shift from the AAA-AAC codons is possible, since a −1 shift when the Lys and Asn tRNAs are bound to the ribosome is identical to the shift that is known to occur during the gag-pol frameshift in mouse mammary tumor virus translation (49) and has been shown to occur in 2.1% of ribosomes traversing a 5′-A-AAA-AAC-UUG-UAA sequence in E. coli (100). We noted that phage PY54 gene 14 and an unannotated open reading frame between it and gene 15 are similar in sequence to these two φKO2 genes, including identical putative shift sites, so the above discussion applies to them as well.
The genes that encode the proteins that build the conical tail tip of phages λ, HK97, and N15 (for example) are similar to φKO2 genes 15, 16, 17, 18, 20, and 21; the latter genes encode predicted proteins that are similar to those made by λ genes H, M, L, K, I, and J, respectively. However, there are two noncanonical features in this cluster of genes. The first is gene 19, which (i) does not match tail assembly genes in other lambdoid phages, (ii) does match (33 of 99 identical amino acids) gene XCC2962 of the P2-like prophage XccP1 of Xanthomonas campestris (22), and (iii) is an island of substantially lower G+C content (39.3%) than the bulk of the φKO2 genome. The XCC2962 gene is not within XccP1's morphogenetic gene cluster, but lies at one end of the prophage. Thus, φKO2 gene 19 should be classified as a “moron” (46, 54) or recent addition in the φKO2 genome, which likely has no function in tail assembly even though it resides in this cluster.
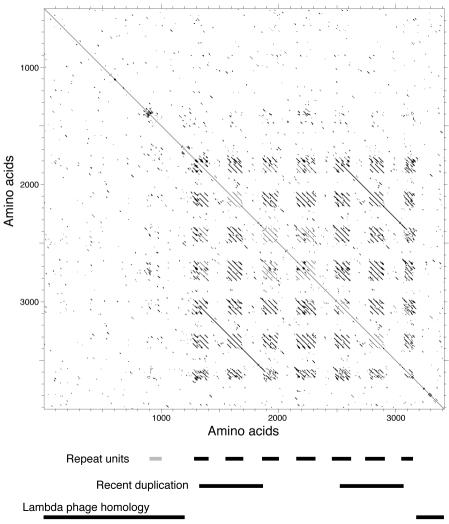
The second unusual feature of the φKO2 tail gene cluster is gene 21. Its prototypic homologue, the phage λ J protein, confers adsorption specificity to the virion and forms a short fiber that protrudes distally from the tail tip and contacts the host's LamB outer membrane protein during adsorption (55, 99). The predicted φKO2 gene 21 protein is exceptionally large, with 3,433 amino acids, compared to its lambdoid phage homologues, which vary in length from 815 amino acids in Salmonella phage Gifsy-2 (70) to 1,421 amino acids in Neisseria phage 2120 (GenBank accession no. AJ278707). A more detailed examination revealed that the gene 21 protein can be viewed as an approximately 1,300-amino-acid λ J-like protein with a large insertion relative to its known homologues (Fig. 4). This insertion is made up of eight imprecise direct repeats of a 130- to 180-amino-acid sequence (the most N-terminal repeat, between about amino acids 830 and 980, is quite different from the others), each of which is separated by about 170 amino acids. The amino acids of the repeats are strongly predicted to form coiled-coil structures, while the remainder of the protein is not. Each of these eight repeats is in turn composed of several shorter 44-amino-acid repeats (Fig. 4). A section of this repeat region that includes part of repeats 2 and 4, all of repeat 3, and the intervening sequences is nearly exactly duplicated in repeat units 6, 7, and 8, suggesting a more recent nontandem duplication of this subregion. The gene 21 repeat region's best match in the database is a λ J-like gene in Xanthomonas phage Xp10 (BLAST E = 6 × 10−8) (GenBank no. AY299121). Other putative tail fiber genes which contain single, several-hundred-amino-acid blocks that are related to the 44-amino-acid repeats in gene 21 are found in phage HK97 (54), Lactococcus lactis phages Tuc2009 (GenBank no. AF109874) and Lj965 (26), Neisseria meningitidis phage 2120 (GenBank no. AJ278707), and a putative prophage in the Bacillus anthracis A2012 genome (GenBank no. NC_003995). Analyses of the tail fiber genes of other phages indicated that the horizontal transfer of sequence blocks within these genes is common (43, 89). SDS-polyacrylamide gel electrophoresis of φKO2 virions showed that a small number of very large protein molecules of about the size predicted for the gene 21 protein are present in virions but not in heads (Fig. 3); the only gene large enough to encode such a large protein is gene 21. It is thus likely that this gene is in fact expressed and that its protein is present as the bacterial receptor adsorption protein of φKO2 tails.
FIG. 4.
Self-comparison of the putative φKO2 gene 21-encoded tail fiber. The gene 21 protein sequence was compared to itself by DNA Strider to produce a dot matrix plot in which each dot indicates seven or more identities in a 23-amino-acid scanning window. The repeat units are indicated below the matrix plot; the leftmost repeat is less conserved than the others and is barely recognizable at this stringency. Homology to the phage λ gene J protein is also indicated.
At the opposite end of the virion chromosome, and at the promoter-proximal end of the φKO2 putative late operon, lie genes 52 through 64, which also appear to be part of the late operon. The putative late promoter, by homology with the parallel region of N15 (83), lies between genes 51 and 52. Of these 13 genes, only 1, gene 52, encodes a protein with convincing matches to proteins of known function, the DNA adenine methylases. Genes 58, 59, 62, and 63 all match genes of unknown functions (but usually similar locations) of other phages, prophages, or plasmids of other bacteria, and gene 60 matches genes in phages φP27, SfV, φE125, and HK97 that in turn have weak similarities to holins. The remaining seven genes in this region are novel. It is notable that no lysis gene cluster is recognizable in the φKO2 late operon. Holins form membrane disruptions through which the cell-wall-degrading enzymes escape to the periplasm, but the weakly holin-like gene 60 is similar to genes of the aforementioned phages which are not part of the lysis gene cluster in those cases; in each case, they lie several genes transcriptionally downstream from the putative lysis gene cluster (1, 54, 86, 101). In phage N15, the lysis genes occupy the positions of φKO2's novel genes 53 through 55; perhaps φKO2 has a lysis module of a type that has not been previously observed (gene 53, which lies at the position of genes that encode holins in other lambdoid phages, does have a hydropathy profile that is quite similar to the phage λ and P22 holins).
The φKO2 early genes.
The φKO2 early region, which is similar to but mosaically related to those of phages N15 and PY54, consists of 43 predicted genes in two large divergent operons and several smaller transcription units. As in other lambdoid phages, the putative φKO2 prophage repressor gene 36 lies between the divergent early left and early right operons. It and the putative Cro repressor (encoded by gene 37) are 88 and 87% identical to the N15 cB repressor and Cro, respectively. This high level of similarity and the fact that their putative three OR and two OL operator binding sites all match the N15 5′-TTATAN6TATAA early operator consensus (83) suggest that N15 and φKO2 have the same repressor target specificity.
The early left region genes 31 through 35 are similar to those of phage N15 and include one encoding a putative replicase (gene 35) and four of unknown function, including a partial homolog of the phage P22 early left operon gene eaF (gene 33). The φKO2 and N15 replicase proteins are 92% identical and their putative replication origin sequences (φKO2 bp 34582 to 34645, inside the replicase gene) have 62 of 64 identical base pairs (82), so it is likely that these replicases have the same origin specificity. The putative early right operon, genes 37 through 51, is also largely homologous to the parallel N15 region. In this region, only φKO2 genes 40, 41, 42, and 47 (all of which have no database matches) do not have N15 or PY54 homologues, and gene 39 is truncated compared to its N15 homologue (Fig. 2). The first three of these genes replace N15 genes 42 and 43, and φKO2 47 is partly similar to sequences between N15 genes 47 and 48, although no gene is predicted there in N15. As in phage N15, only the first gene in this operon is similar to genes of known function, the lambdoid phage gene Q-type transcription antiterminators, although gene 39 is weakly similar to DNA exonuclease III and polymerase ɛ subunits.
Genes 22, 23, 24-25, 26, 27 to 30, and 48-49 appear to form six additional short transcription units (Fig. 2). All but 22 have similarly positioned homologues on the phage N15 chromosome that are expressed in the lysogen and during early lytic growth of that phage (83). Genes 48 and 49 are homologues to N15 genes 48 and 49, but their role is unknown. Genes 24 and 25 encode convincing homologues of the SopB- and SopA-type plasmid partitioning proteins, respectively, and so almost certainly function in this process, as their N15 homologues have been shown to do (41, 79). The φKO2 and N15 SopA and SopB proteins are 92 and 76% identical, respectively, and the φKO2 genome contains four copies of the 5′-GTGCGACCA · CGGTCGCAY-3′ (the dot marks the center of symmetry) inverted repeat at bp 37905, 40345, 45384, and 48618. N15 has very similar or identical sequences at locations that are homologous to the first three. The φKO2 site at bp 48618 does not have a convincing orthologue in N15; however, N15 carries an identical site in the region downstream of its gene 58 which is apparently not homologous to any φKO2 gene (Fig. 2). It thus appears that the φKO2 and N15 partitioning apparatuses have identical or very similar DNA target site specificities, and the conservation of the location of the nonhomologous sites nearest the right end of the virion chromosome suggests that partitioning sites at these locations may be important in spite of their apparent redundancy. We also note that, although phages very often carry DNA target sites near the genes that encode the proteins that bind to them (16), these two phages both carry their multiple putative partitioning sites nearer the center of the linear prophage plasmid, far from the genes that encode the Sop proteins. Perhaps their central location is important for physically moving the DNA molecules during the partitioning process.
Genes 27, 28, and 29 of φKO2 are similar to the anti-immunity operon that is found in this location in N15; operons with a similar function and regulation are also found in phages P4, φR73, P1, and P7 (5, 9, 21, 37, 88). Their putative proteins are 74, 79, and 81% identical to the N15 antB-, antA-, and icd-encoded proteins, respectively; these N15 proteins have auxiliary antirepressor, antirepressor, and inhibition of cell division functions, respectively (81). The 5′ portion of the N15 transcript is processed into a highly folded cA RNA that activates a terminator 5′ to N15 genes 30, 31, and 32. The putative φKO2 cA RNA differs in sequence from that of N15 in 27 of 89 positions, but all differences are either in unpaired regions or are compensated for by changes that preserve the predicted secondary structure (see Fig. 5C in reference 81). This conservation of secondary structure interactions supports the importance of these interactions in cA RNA functioning. A fourth potential φKO2 gene in this region is gene 30; it could, in theory, encode a small protein from the 5′ region of this operon, but experimental analysis has yet to show whether this is true.
FIG. 5.
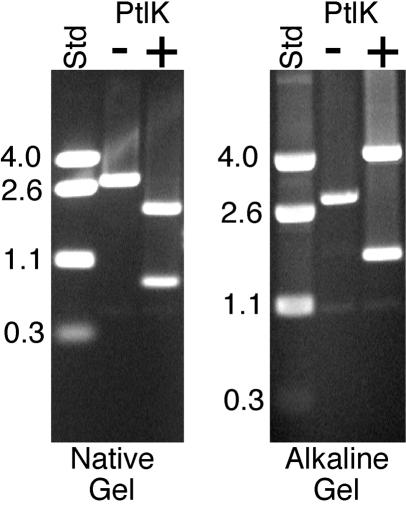
φKO2 gene 26 protein has protelomerase activity. Plasmid pBLSK DNA containing the cloned 56-bp sequence that includes the φKO2 telRL site between its HindIII and BamHI sites was first linearized with restriction endonuclease AluNI. It was then treated with 1 pmol of purified gene 26 protein (PtlK) at 37°C for 30 min in a solution containing 20 mM Tris-Cl (pH 7.5), 50 mM K-glutamate, 1 mM dithiothreitol, and 0.1 mM EDTA. The resulting DNA was separated in native and alkaline agarose gels and stained with ethidium bromide. Size standards (Std) are given in kilobase pairs to the left of each panel.
Gene 26 encodes a protein that is 75 and 42% identical to the phage N15 and PY54 protelomerases (25, 47), respectively, and near its left boundary lies a sequence that is very similar to the N15 telRL site (see above), the site at which the N15 protelomerase creates the ends of the linear prophage plasmid. In order to prove that this φKO2 gene encodes its protelomerase function, we expressed φKO2 gene 26 from the expression plasmid vector pET15b so that the protein made was histidine tagged at its amino terminus. This protein was purified to near homogeneity by nickel-affinity chromatography with Ni-nitrilotriacetic acid agarose (Qiagen, Valencia, Calif.) (51). Figure 5 (left panel) shows that this enzyme cleaves the inserted φKO2 DNA at the position of the putative telRL site; plasmid DNA without the inserted φKO2 telRL site was not cleaved (data not shown). An analysis of the cleaved DNA products in alkaline agarose gels (Fig. 5, right panel) showed that covalently closed hairpin ends are formed during this cleavage, since the reaction products migrate as expected when the two strands are covalently joined. Thus, φKO2 gene 26 encodes a functional protelomerase that no doubt functions to create the plasmid's linear telomeres, like the homologous N15 and PY54 proteins do (51, 80).
Genes 22 and 23 are divergently transcribed and have SOS-related predicted functions; the gene 22 protein is similar to DinI proteins and the gene 23 protein is a UmuD′ homolog (see Discussion). Previous studies with phage N15 indicated that its umuD homologous gene 26 (it has no dinI homolog) is expressed in a lysogen and throughout lytic infection (83), and the same is true of genes surrounding the PY54 gene 56 dinI homolog (48). Interestingly, the sequence 5′-TACTGAATGCATATACAGTA-3′ between φKO2 genes 22 and 23 is an excellent match to the consensus E. coli LexA binding site (5′-taCTGtatatatataCAGta-3′, where uppercase nucleotides are universally present and lowercase nucleotides are not absolutely conserved [35, 62]), and there are potential promoter sequences oriented in both directions that overlap this sequence (we found no other LexA consensus sequences that overlap potential promoters in the φKO2 genome) (Fig. 6). We noted that previously unrecognized potential LexA binding sites lie 14 bp 5′ of N15 gene 26 (5′-TACTGTAAAAAAAAACAGTT-3′) and 20 bp 5′ of PY54 gene 56 (5′-GACTGTATACATATACAGTA-3′). We therefore cloned a 365-bp region of φKO2 DNA that included the putative gene 22 promoter (called P22) and the LexA operator into a low-copy-number F′ plasmid (see Materials and Methods and reference 11), in which gene 22 was fused in-frame to a lacZ reporter gene. This plasmid was then placed into lexA wild-type and mutant Salmonella strains (Table 1; see Materials and Methods), and expression of the gene 22-lacZ fusion protein was measured with and without SOS induction by mitomycin C. Table 3 shows that the P22 promoter drives a substantial level of hybrid gene expression. This lacZ expression was 73-fold higher in the absence of wild-type host LexA protein than in its presence, indicating that active LexA protein greatly lowers transcription from P22. In addition, if P22 is controlled by LexA, repression should be lifted if the host's SOS response is triggered. As predicted by this model, the presence of mitomycin C caused a 25-fold increase in lacZ expression in the host with wild-type lexA. Finally, in a noninducible lexA (Ind−) mutant, in which LexA is not released from its operator by SOS induction signals (64), the level of lacZ expression was not increased by mitomycin C treatment, as was expected if LexA cleavage allows gene 22 transcription. These experiments clearly demonstrate that the transcription of φKO2 gene 22 from the P22 promoter is controlled by the host's LexA repressor. Parallel experiments with the putative gene 23 promoter gave similar results, as mitomycin C caused a 34-fold increase in expression in a lexA+ strain and a lexA null mutation caused a 100-fold increase compared to the lexA+ strain. These experiments also show that under these conditions, promoter P23 is not as strong as P22, since the reporter expression level in the absence of the LexA repressor is about 9- and 14-fold higher for P22 under lexA or mitomycin C treatment regimens, respectively.
FIG. 6.
Region between φKO2 genes 22 and 23. The sequences of both strands of the region between genes 22 and 23 are shown, with a gray background marking the LexA-binding site, whose universally conserved base pairs are underlined. The −10 and −35 regions of putative promoters P22 and P23 are indicated by black and white rectangles, respectively, and arrows indicate the predicted mRNA start sites.
TABLE 3.
Control of the φKO2 gene 22 and 23 promoters by host LexA protein
| Relevant host genotypea | φKO2 promoter | β-Galactosidase activity (Miller units)b
|
|
|---|---|---|---|
| Without mitomycin C | With mitomycin C | ||
| UB-1548 (lexA+ sulA) | P22 | 27 | 672 |
| UB-1550 (lexA null sulA) | P22 | 1,962 | 2,610 |
| UB-1549 (lexA ind sulA) | P22 | 7 | 11 |
| UB-1547 (lexA ind sulA+) | P22 | 8 | 9 |
| UB-1555 (lexA+ sulA) | P23 | 2 | 68 |
| UB-1556 (lexA null sulA) | P23 | 219 | 187 |
| UB-1557 (lexA ind sulA) | P23 | 0.3 | 0.7 |
| UB-1558 (lexA ind sulA+) | P23 | 0.2 | 0.7 |
S. enterica LT2 lexA null mutants are lethal unless a sulA mutation is also present. All of these strains are cured of prophages Fels-2, Gifsy-1, and Gifsy-2, since these prophages can also interfere with the growth of lexA-negative strains (11). The lexA ind sulA+ strains are included to show that the sulA mutation does not cause SOS inducibility.
β-Galactosidase activity units show the change in A420 per minute per milliliter of cells per optical density unit at 600 nm. Cultures were grown with shaking at 37°C to an optical density at 600 nm of 0.3, when mitomycin C was added to 1 μg/ml to one-half of the culture, and growth was allowed to proceed for one additional hour. Cells were chloroform permeabilized and assayed for β-galactosidase as described by Miller (72). Activity values are the averages of several independent determinations.
φKO2 transcription.
Each of the putative transcripts indicated in Fig. 2 has, at its 5′ end, a predicted sigma 70 promoter sequence, and the deduced transcription pattern is very similar to that of N15. As in N15, several regions have multiple closely spaced promoter-like sequences (5′ of genes 38, 29, 30, 37, and 39); however, the significance of these apparently conserved arrangements is unknown. In particular, we noted that none of these promoters, with the exception of the LexA-responsive promoters (see above), has been tested experimentally, and therefore their identification is regarded as tentative. In addition, putative rho-independent (stem-loop) transcription terminators are located between early protein coding regions immediately 3′ of genes 22, 23, 26, 31, 36, 51, 52, 58, and 62 (Fig. 2). All, except those after gene 62, have homologs in N15. The biological function of these terminators is not known. It has been suggested that they may serve to suppress inappropriate transcription from the repressed prophage but are overridden during lytic growth (54). In accord with this, there may be a correlation between phages with multiple terminators scattered throughout their lytic genes and those that have (or appear to have) transcription antitermination functions that are active during lytic growth. An alternative, but not mutually exclusive, view is that the terminators, which may not work at 100% efficiency, serve to fine-tune the levels of transcription in different regions of the genome. In addition, putative rho-independent transcription terminators lie in the short gaps immediately 3′ of genes 5, 6, 12, 19, 20, and 21 in the late operon, and it is interesting that the first three of these occupy positions that are exactly analogous to similar terminators in nonhomologous sequences in the phage N15 late operon. Two of these follow the very highly translated genes 6 and 12, and similarly located terminators have been noted in other phage genomes, including phages as diverse as coliphages λ and T7, but the reason for this conservation is unknown (30, 54).
DISCUSSION
The φKO2 genome.
We have shown that the linear plasmid pKO2 present in K. oxytoca CCUC 15788 is a prophage that is related to E. coli phage N15 and Yersinia phage PY54 and that apparently has a lifestyle that is very similar to theirs. The N15, PY54, and φKO2 genomes are mosaically related. Some regions are sufficiently similar that we argue that the N15 and φKO2 plasmid partitioning proteins, replicase, protelomerases (hairpin end generation), and prophage repressors have the same or extremely similar target site specificities in both phages. The similar overall genome organizations of phages N15, PY54, φKO2, and λ and the close relationship of some of their genes to each other or to those of other canonical lambdoid phages suggest that it is legitimate to include the former three within the lambdoid phage group, but in a subgroup that has a different strategy of lysogeny. The late operon of φKO2 is similar to that of N15 except that it has very different head and tail shaft assembly genes, and its λ J-like tail fiber gene has an ∼7,000 bp insertion relative to known homologues; on the other hand, PY54 and φKO2 have similar head and tail shaft genes but are rather different elsewhere. The φKO2 tail fiber gene encodes a 3,433-amino-acid protein; only the 3,500-amino-acid phage N4 virion RNA polymerase is larger among the known phage-encoded proteins (56). The regions of the φKO2 genome outside the late operon are largely similar to, but are mosaically related to, those of N15.
Control of phage genes by the host's LexA repressor.
We also performed experiments that demonstrate that φKO2 genes 22 and 23, homologues of bacterial dinI and umuD genes, respectively, are regulated by the host's LexA repressor. In other contexts, both dinI- and umuD-like proteins affect the bacterium's SOS response to DNA damage. With E. coli, the DinI protein has been shown to modulate the bacterial SOS response by inhibiting activated RecA-mediated LexA and UmuD protein autocleavage (4, 98, 103, 104). The E. coli protein was originally named damage inducible gene I (105), and dinI homologues have been called msgA, tum, and impC. The virulence gene msgA (macrophage survival gene A) in S. enterica serovar Typhimurium allows increased survival of Salmonella in mouse macrophages (42), the phage 186 tum gene encodes a protein with an antirepressor function and a C-terminal DinI-like domain (10, 90), and impC on Salmonella plasmid pTT110 lies adjacent to the umuD homologue impA, but its function is unknown (65). It is possible that the different phenotypes that result from the loss of these DinI homologues result from the same underlying effect on global SOS control of gene expression (57). Characterized UmuD proteins are subunits of an error-prone DNA polymerase that participates in the repair of DNA damage during the SOS response (97). The E. coli UmuD protein is synthesized in an inactive form, and RecA-mediated autocleavage of its peptide backbone activates it to become a subunit of the UmuC/UmuD′ error-prone DNA polymerase (39).
Both dinI-like and umuD-like genes are found in other temperate phage genomes, in which their roles are not known. Nearly all of the published enterobacterial genome sequences contain multiple dinI-like genes, and the members of this gene family are present in both prophage and apparently non-phage-related contexts in these genomes. DinI homologs that are not obviously phage or prophage encoded are, for example, E. coli K-12 dinI (6) and S. enterica serovar Typhimurium LT2 gene STM1162 (70). Many of the homologs are phage encoded; for example, at least six of the eight homologues in S. enterica LT2 and three of five in both S. enterica serovar Typhi CT18 and E. coli EDL933 lie within prophages. Among the closest homologs to φKO2 gene 22 are S. enterica CT18 gene STY1883, which is not obviously prophage associated (75), and LT2 gene STM2231, which appears to be part of a badly decayed prophage, Stm6 (Stm6 is our name for a previously unnamed defective lambdoid prophage present at genes STM2230 through STM2244 [13, 70]). Homologs of dinI are also found in the genomes of phage 186 (10), the Salmonella LT2 prophages Gifsy-1, Gifsy-2, and Fels-2 (11, 70) as well as prophages Sti1 and Sti5 (genes STY1011 to STY1077 and STY2467 to STY2470, respectively [13], in the genome of S. enterica serovar Typhi CT18 [75]), and prophages CP-933V and CP-933O in E. coli EDL933 (77). Similarly, the following umuD homologs have been found in phage genomes: (i) the φKO2 gene 23 described here, (ii) the phage P1 humD gene (62, 71), (iii) S. enterica serovar Typhimurium gene STM2230 of defective prophage Stm6 (see above), and (iv) X. campestris prophage XccP1 gene Xcc2963 (22). In most proteobacterial genomes umuD homologues are not known to lie in prophage contexts. Curiously, the translation of the phage UmuD homologues (except perhaps that of XccP1) begins at the site where the RecA-mediated activation cleavage of the bacterial protein occurs, suggesting that the phage proteins are made in an active form.
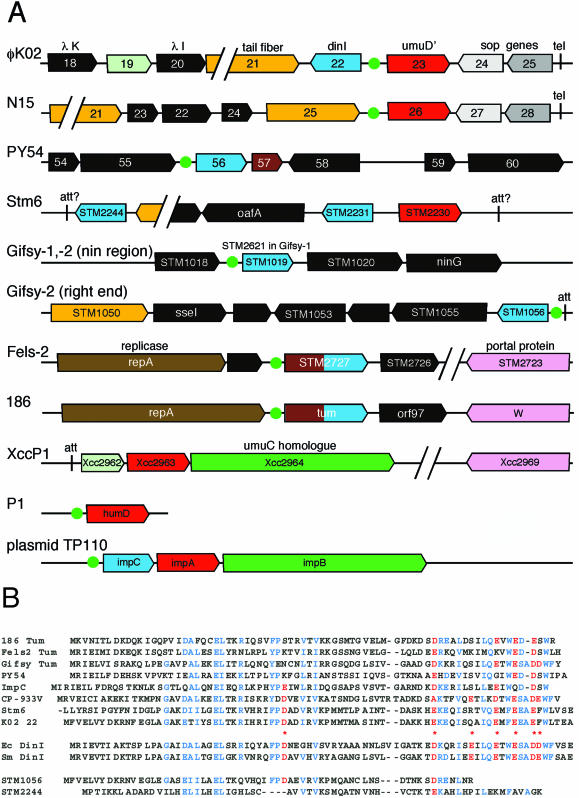
The dinI and umuD homologues lie in a number of strikingly different genomic contexts in different phages (Fig. 7A). The dinI-like genes are present in two different contexts on known lambdoid phages: (i) within the nin region (distal portion of the early right operon) of phages Gifsy-1 and Gifsy-2 and S. enterica CT18 prophage Sti1 (the PY54 gene is in a structurally similar but nonhomologous region) and (ii) between the attachment or tel site and the tail fiber gene in φKO2, Gifsy-2, and the prophages Stm6, Sti1, CP-933V, and CP-933O. It is curious that Gifsy-2, Stm6, and Sti1 each carries two dinI homologs. Homologues of dinI are also found in some P2-like phages (e.g., phage 186), transcriptionally downstream of the replication initiation genes. Finally, φKO2, N15, Stm6, XccP1, and P1 each carries a umuD homologue (Stm6 also has the decaying pseudogene remains of a second umuD homologue between genes STM2231 and STM2232). These are present in three different contexts, those of XccP1, P1, and the other three phages. The facts that dinI- and umuD-like genes lie in different locations in related phage genomes as well as in genomes of phages that are only extremely distantly related and that in several cases they lie next to the attachment site combine to suggest that these genes have been independently (and perhaps recently) acquired by different phages.
FIG. 7.
Phage-borne SOS genes and proteins. (A) Phage-, prophage-, and plasmid-borne dinI and umuD homologues are present in a number of different genetic contexts. Each line represents a section of the genome of the indicated genetic element (see text for details). The directional boxes represent genes and their transcriptional directions. Boxes of the same color are homologous genes, except for the orange boxes, which are all involved in tail fiber assembly but are not all homologues, and the black boxes, each of which has no homologue in the figure. (B) Comparison of 12 DinI family protein amino acid sequences. Seven phage-encoded DinI homologues are shown in the top group of sequences; the two DinI proteins from E. coli and Serratia marcescens, which are known by experimentation to down-regulate the SOS response, are shown in the middle;and two less similar phage homologues are shown below. Above the E. coli protein, asterisks indicate the seven negatively charged surface residues that make up its postulated DNA-mimic surface (78). These seven residues are indicated in red when homologous residues are present, and other highly conserved residues are shown in blue. From the top down, the proteins shown are as follows: (i) the phage 186 Tum protein; (ii) the phage Fels-2 gene STM2727 protein (we believe that a translation start site 25 codons 3′ of that reported by McClelland et al. [70] is the more likely true start for this gene, as STM2727 is identical to the retron 67-encoded Orf6 protein [50]); (iii) the phage Gifsy-2 gene STM1019 protein (identical to the phage Gifsy-1 gene STM2621 and prophage Sti1 STY1032 proteins); (iv) plasmid TP110-encoded ImpC protein; (v) phage PY54 gene 56 protein; (vi) E. coli prophage CP-933V gene Z3305 protein (the Shigella flexneri 301 prophage gene SF1879 protein only differs from Z3305 protein at its third amino acid); (vii) Salmonella defective prophage Stm6 gene STM2231 protein (16 predicted N-terminal amino acids are not shown; its poor Shine-Dalgarno and UUG start codon cast some doubt on the functionality of this gene); (viii) φKO2 gene 22 protein; (ix) E. coli K-12 dinI protein (gene b1061; identical proteins are encoded by the non-prophage-associated E. coli EDL933 gene Z1698 and Shigella flexneri 301 gene SF1067; S. enterica LT2 gene STM1162 and CT18 gene STY1200 proteins are 85 and 83% identical to the b1061 protein, respectively); (x) Serratia marcescens dinI protein; (xi) Gifsy-2 gene STM1056 protein; and (xi) Stm6 gene STM2244 protein.
The three-dimensional structure of the E. coli K-12 dinI protein has been determined, and a negatively charged ridge on its surface that is composed of its C-terminal α-helix is thought to mimic the electrostatic surface character of DNA, enabling it to bind to the single-stranded DNA-binding site of the RecA protein (78, 98). If the phage-encoded dinI homologues were to have this SOS-modulating function, it would be expected that the C-terminal helix, and in particular its negative charges, would be conserved. Figure 7B shows that most, including the φKO2 gene 22 protein, but not all, of the DinI proteins are indeed more highly conserved near the C terminus and that they retain nearly all of the negative charges on the putative DNA-mimic surface, suggesting that they may have the same SOS-modulating function as DinI itself. The several DinI relatives that have not conserved this surface (e.g., Gifsy-2's STM1056) might either have some other function or be nonfunctional dinI pseudogenes.
Interplay between the host and prophage in a lysogen is clearly important, but relatively few cases of direct control of prophage gene expression by host regulatory proteins have been elucidated. Among these are the control of the 186 tum gene promoter (10), λ's Poop promoter (58, 62), and Fels-2, Gifsy-1, and Gifsy-2 (genes STM2731, STM2621, and STM1019, respectively) by the LexA repressor (11); control of the Mu Pmom promoter by the OxyR protein (44); control of the prophage-borne diphtheria toxin gene by the DtxR repressor (74); and control of the PStx1 Shiga toxin promoter of phage H-19B by the iron-regulated Fur repressor (12, 24). We show here that the φKO2 genes 22 and 23 are similarly regulated. We note that several of the other phage-borne umuD and dinI homologues have appropriately located LexA-binding sites (Fig. 7A). The expression of the N15 umuD-like gene from the uninduced prophage (83) suggests that LexA repression there may be incomplete, and the data in Table 3 suggest that in φKO2, LexA repression may also be incomplete on φKO2. The specific roles for phage-encoded DinI and UmuD′ proteins are not yet understood (the P1 UmuD homologue, HumD, can form a functional DNA polymerase complex with E. coli UmuC [71]); however, their common presence suggests that prophages may use them to modulate the host SOS response for their own purposes.
Acknowledgments
We thank Hans Schlegel for the gift of K. oxytoca strain CCUC 15788 and John Roth for many stimulating discussions. We also thank Robert Schackmann for help with N-terminal amino acid sequence determination and Jeffrey Lawrence for help with calculating the K. pneumoniae G+C content.
This work was supported by NSF grant 990526 to S.C. and NIH grant GM51975 to R.H. and G.H.
REFERENCES
- 1.Allison, G. E., D. Angeles, N. Tran-Dinh, and N. K. Verma. 2002. Complete genomic sequence of SfV, a serotype-converting temperate bacteriophage of Shigella flexneri. J. Bacteriol. 184:1974-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, G. E., D. C. Angeles, P. Huan, and N. K. Verma. 2003. Morphology of temperate bacteriophage SfV and characterisation of the DNA packaging and capsid genes: the structural genes evolved from two different phage families. Virology 308:114-127. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkmen, M., and M. J. Benedik. 2002. Multi-copy repression of Serratia marcescens nuclease expression by dinI. Curr. Microbiol. 44:44-48. [DOI] [PubMed] [Google Scholar]
- 5.Biere, A. L., M. Citron, and H. Schuster. 1992. Transcriptional control via translational repression by c4 antisense RNA of bacteriophages P1 and P7. Genes Dev. 6:2409-2416. [DOI] [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 7.Bochner, B. R., H. C. Huang, G. L. Schieven, and B. N. Ames. 1980. Positive selection for loss of tetracycline resistance. J. Bacteriol. 143:926-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borodovsky, M., and J. McInish. 1993. GeneMark: parallel gene recognition for both strands. Comput. Chem. 17:123-133. [Google Scholar]
- 9.Briani, F., D. Ghisotti, and G. Deho. 2000. Antisense RNA-dependent transcription termination sites that modulate lysogenic development of satellite phage P4. Mol. Microbiol. 36:1124-1134. [DOI] [PubMed] [Google Scholar]
- 10.Brumby, A. M., I. Lamont, I. B. Dodd, and J. B. Egan. 1996. Defining the SOS operon of coliphage 186. Virology 219:105-114. [DOI] [PubMed] [Google Scholar]
- 11.Bunny, K., J. Liu, and J. Roth. 2002. Phenotypes of lexA mutations in Salmonella enterica: evidence for a lethal lexA null phenotype due to the Fels-2 prophage. J. Bacteriol. 184:6235-6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calderwood, S. B., and J. J. Mekalanos. 1987. Iron regulation of Shiga-like toxin expression in Escherichia coli is mediated by the fur locus. J. Bacteriol. 169:4759-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casjens, S. 2003. Prophages in bacterial genomics: what have we learned so far? Mol. Microbiol. 249:277-300. [DOI] [PubMed] [Google Scholar]
- 14.Casjens, S., K. Eppler, L. Sampson, R. Parr, and E. Wyckoff. 1991. Fine structure genetic and physical map of the gene 3 to 10 region of the bacteriophage P22 chromosome. Genetics 127:637-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casjens, S., G. Hatfull, and R. Hendrix. 1992. Evolution of dsDNA tailed-bacteriophage genomes. Semin. Virol. 3:383-397. [Google Scholar]
- 16.Casjens, S., and R. Hendrix. 1988. Control mechanisms in dsDNA bacteriophage assembly, p. 15-91. In R. Calendar (ed.), The bacteriophages, vol. 1. Plenum Press, New York, N.Y. [Google Scholar]
- 17.Casjens, S., and W. M. Huang. 1993. Linear chromosomal physical and genetic map of Borrelia burgdorferi, the Lyme disease agent. Mol. Microbiol. 8:967-980. [DOI] [PubMed] [Google Scholar]
- 18.Casjens, S., M. Murphy, M. DeLange, L. Sampson, R. van Vugt, and W. M. Huang. 1997. Telomeres of the linear chromosomes of Lyme disease spirochaetes: nucleotide sequence and possible exchange with linear plasmid telomeres. Mol. Microbiol. 26:581-596. [DOI] [PubMed] [Google Scholar]
- 19.Casjens, S., N. Palmer, R. Van Vugt, W. Mun Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 20.Christie, G. E., L. M. Temple, B. A. Bartlett, and T. S. Goodwin. 2002. Programmed translational frameshift in the bacteriophage P2 FETUD tail gene operon. J. Bacteriol. 184:6522-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Citron, M., and H. Schuster. 1992. The c4 repressor of bacteriophage P1 is a processed 77 base antisense RNA. Nucleic Acids Res. 20:3085-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.da Silva, A. C., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, L. M. Alves, A. M. do Amaral, M. C. Bertolini, L. E. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. Ferreira, R. C. Ferreira, M. I. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. Lemos, M. V. Lemos, E. C. Locali, M. A. Machado, A. M. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. Sena, C. Silva, R. F. de Souza, L. A. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. Tezza, M. Trindade dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 23.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Grandis, S., J. Ginsberg, M. Toone, S. Climie, J. Friesen, and J. Brunton. 1987. Nucleotide sequence and promoter mapping of the Escherichia coli Shiga-like toxin operon of bacteriophage H-19B. J. Bacteriol. 169:4313-4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deneke, J., G. Ziegelin, R. Lurz, and E. Lanka. 2000. The protelomerase of temperate Escherichia coli phage N15 has cleaving-joining activity. Proc. Natl. Acad. Sci. USA 97:7721-7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desiere, F., R. D. Pridmore, and H. Brussow. 2000. Comparative genomics of the late gene cluster from Lactobacillus phages. Virology 275:294-305. [DOI] [PubMed] [Google Scholar]
- 27.Douglas, S. E. 1994. DNA Strider. A Macintosh program for handling protein and nucleic acid sequences. Methods Mol. Biol. 25:181-194. [DOI] [PubMed] [Google Scholar]
- 28.Duda, R., J. Hempel, H. Michel, J. Shabanowitz, D. Hunt, and R. Hendrix. 1995. Structural transitions during bacteriophage HK97 head assembly. J. Mol. Biol. 247:618-635. [DOI] [PubMed] [Google Scholar]
- 29.Duda, R., K. Martincic, Z. Xie, and R. Hendrix. 1995. Bacteriophage HK97 head assembly. FEMS Microbiol. Rev. 17:41-46. [DOI] [PubMed] [Google Scholar]
- 30.Dunn, J., and W. Studier. 1983. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J. Mol. Biol. 166:477-535. [DOI] [PubMed] [Google Scholar]
- 31.Earnshaw, W., S. Casjens, and S. Harrison. 1976. Assembly of the head of bacteriophage P22, X-ray diffraction from heads, proheads and related structures. J. Mol. Biol. 104:387-410. [DOI] [PubMed] [Google Scholar]
- 32.Eggers, C. H., S. Casjens, S. F. Hayes, C. F. Garon, C. J. Damman, D. B. Oliver, and D. S. Samuels. 2000. Bacteriophages of spirochetes. J. Mol. Microbiol. Biotechnol. 2:365-373. [PubMed] [Google Scholar]
- 33.Eggers, C. H., S. Casjens, and D. S. Samuels. 2001. Bacteriophages of Borrelia burgdorferi and other spirochetes, p. 35-44. In M. Saier and J. Garcia-Lara (ed.), The spirochetes. Molecular and cellular biology. Horizon Scientific Press, Wiltshire, United Kingdom.
- 34.Eppler, K., E. Wyckoff, J. Goates, R. Parr, and S. Casjens. 1991. Nucleotide sequence of the bacteriophage P22 genes required for DNA packaging. Virology 183:519-538. [DOI] [PubMed] [Google Scholar]
- 35.Fernandez De Henestrosa, A. R., T. Ogi, S. Aoyagi, D. Chafin, J. J. Hayes, H. Ohmori, and R. Woodgate. 2000. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 35:1560-1572. [DOI] [PubMed] [Google Scholar]
- 36.Flinta, C., B. Persson, H. Jornvall, and G. von Heijne. 1986. Sequence determinants of cytosolic N-terminal protein processing. Eur. J. Biochem. 154:193-196. [DOI] [PubMed] [Google Scholar]
- 37.Forti, F., P. Sabbattini, G. Sironi, S. Zangrossi, G. Deho, and D. Ghisotti. 1995. Immunity determinant of phage-plasmid P4 is a short processed RNA. J. Mol. Biol. 249:869-878. [DOI] [PubMed] [Google Scholar]
- 38.Girons, I. S., P. Bourhy, C. Ottone, M. Picardeau, D. Yelton, R. W. Hendrix, P. Glaser, and N. Charon. 2000. The LE1 bacteriophage replicates as a plasmid within Leptospira biflexa: construction of an L. biflexa-Escherichia coli shuttle vector. J. Bacteriol. 182:5700-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonzalez, M., and R. Woodgate. 2002. The “tale” of UmuD and its role in SOS mutagenesis. Bioessays 24:141-148. [DOI] [PubMed] [Google Scholar]
- 40.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 41.Grigoriev, P. S., and M. B. Lobocka. 2001. Determinants of segregational stability of the linear plasmid-prophage N15 of Escherichia coli. Mol. Microbiol. 42:355-368. [DOI] [PubMed] [Google Scholar]
- 42.Gunn, J. S., C. M. Alpuche-Aranda, W. P. Loomis, W. J. Belden, and S. I. Miller. 1995. Characterization of the Salmonella typhimurium pagC/pagD chromosomal region. J. Bacteriol. 177:5040-5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haggard-Ljungquist, E., C. Halling, and R. Calendar. 1992. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J. Bacteriol. 174:1462-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hattman, S., and W. Sun. 1997. Escherichia coli OxyR modulation of bacteriophage Mu mom expression in dam+ cells can be attributed to its ability to bind hemimethylated Pmom promoter DNA. Nucleic Acids Res. 25:4385-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heidelberg, J. F., I. T. Paulsen, K. E. Nelson, E. J. Gaidos, W. C. Nelson, T. D. Read, J. A. Eisen, R. Seshadri, N. Ward, B. Methe, R. A. Clayton, T. Meyer, A. Tsapin, J. Scott, M. Beanan, L. Brinkac, S. Daugherty, R. T. DeBoy, R. J. Dodson, A. S. Durkin, D. H. Haft, J. F. Kolonay, R. Madupu, J. D. Peterson, L. A. Umayam, O. White, A. M. Wolf, J. Vamathevan, J. Weidman, M. Impraim, K. Lee, K. Berry, C. Lee, J. Mueller, H. Khouri, J. Gill, T. R. Utterback, L. A. McDonald, T. V. Feldblyum, H. O. Smith, J. C. Venter, K. H. Nealson, and C. M. Fraser. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20:1118-1123. [DOI] [PubMed] [Google Scholar]
- 46.Hendrix, R. W., J. G. Lawrence, G. F. Hatfull, and S. Casjens. 2000. The origins and ongoing evolution of viruses. Trends Microbiol. 8:504-508. [DOI] [PubMed] [Google Scholar]
- 47.Hertwig, S., I. Klein, R. Lurz, E. Lanka, and B. Appel. 2003. PY54, a linear plasmid prophage of Yersinia enterocolitica with covalently closed ends. Mol. Microbiol. 48:989-1003. [DOI] [PubMed] [Google Scholar]
- 48.Hertwig, S., I. Klein, V. Schmidt, S. Beck, J. A. Hammerl, and B. Appel. 2003. Sequence analysis of the genome of the temperate Yersinia enterocolitica phage PY54. J. Mol. Biol. 331:605-622. [DOI] [PubMed] [Google Scholar]
- 49.Hizi, A., L. Henderson, T. Copeland, R. R. Sowde, C. Hixson, and S. Oroszlan. 1987. Characterization of mouse mammary tumor virus gag-pro gene products and the ribosomal frameshift site by protein sequencing. Proc. Natl. Acad. Sci. USA 84:7041-7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hsu, M., M. Inouye, and S. Inouye. 1990. Retron for 67-base multicopy single stranded DNA from Escherichia coli: a potential transposable element encoding both reverse transcriptase and Dam methylase function. Proc. Natl. Acad. Sci. USA 87:9454-9458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang, W., L. Joss, T. Hseih, and S. Casjens. Protelomerase uses a Y-recombinase/topoisomerase IB type mechanism to generate DNA hairpin ends. J. Mol. Biol., in press. [DOI] [PubMed]
- 52.Ikeda, H., and J. Tomizowa. 1968. Prophage P1, an extrachromosomal replication unit. Cold Spring Harbor Symp. Quant. Biol. 33:791-798. [DOI] [PubMed] [Google Scholar]
- 53.Inal, J. M., and K. V. Karunakaran. 1996. φ20, a temperate bacteriophage isolated from Bacillus anthracis exists as a plasmidial prophage. Curr. Microbiol. 32:171-175. [DOI] [PubMed] [Google Scholar]
- 54.Juhala, R. J., M. E. Ford, R. L. Duda, A. Youlton, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 299:27-51. [DOI] [PubMed] [Google Scholar]
- 55.Katsura, I. 1983. Tail assembly and injection, p. 331-346. In R. Hendrix, J. Roberts, F. W. Stahl, and R. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 56.Kazmierczak, K. M., E. K. Davydova, A. A. Mustaev, and L. B. Rothman-Denes. 2002. The phage N4 virion RNA polymerase catalytic domain is related to single-subunit RNA polymerases. EMBO J. 21:5815-5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khil, P. P., and R. D. Camerini-Otero. 2002. Over 1000 genes are involved in the DNA damage response of Escherichia coli. Mol. Microbiol. 44:89-105. [DOI] [PubMed] [Google Scholar]
- 58.Krinke, L., M. Mahoney, and D. L. Wulff. 1991. The role of the OOP antisense RNA in coliphage lambda development. Mol. Microbiol. 5:1265-1272. [DOI] [PubMed] [Google Scholar]
- 59.Kropinski, A. M. 2000. Sequence of the genome of the temperate, serotype-converting, Pseudomonas aeruginosa bacteriophage D3. J. Bacteriol. 182:6066-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Larsen, B., N. M. Wills, R. F. Gesteland, and J. F. Atkins. 1994. rRNA-mRNA base pairing stimulates a programmed −1 ribosomal frameshift. J. Bacteriol. 176:6842-6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Levin, M., R. Hendrix, and S. Casjens. 1993. A programmed translational frameshift is required for the synthesis of a bacteriophage lambda tail assembly protein. J. Mol. Biol. 234:124-139. [DOI] [PubMed] [Google Scholar]
- 62.Lewis, L. K., G. R. Harlow, L. A. Gregg-Jolly, and D. W. Mount. 1994. Identification of high affinity binding sites for LexA which define new DNA damage-inducible genes in Escherichia coli. J. Mol. Biol. 241:507-523. [DOI] [PubMed] [Google Scholar]
- 63.Licznar, P., N. Mejlhede, M. Prere, N. Wills, R. Gesteland, J. Atkins, and O. Fayet. 2003. Programmed translational −1 frameshifting on hexanucleotide motifs and the wobble properties of tRNAs. EMBO J. 22:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Little, J. W. 1980. Isolation of recombinant plasmids and phage carrying the lexA gene of Escherichia coli K-12. Gene 10:237-247. [DOI] [PubMed] [Google Scholar]
- 65.Lodwick, D., D. Owen, and P. Strike. 1990. DNA sequence analysis of the imp UV protection and mutation operon of the plasmid TP110: identification of a third gene. Nucleic Acids Res. 18:5045-5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Loessner, M. J., R. B. Inman, P. Lauer, and R. Calendar. 2000. Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes: implications for phage evolution. Mol. Microbiol. 35:324-340. [DOI] [PubMed] [Google Scholar]
- 67.Malinin, A., A. Vostrov, V. Rybchin, and A. Sverchevsky. 1992. Structure of the linear plasmid N15 ends. Mol. Genet. Mikrobiol. Virusol. 1992:19-22 (In Russian.) [PubMed] [Google Scholar]
- 68.Malinin, A. Y., A. L. Vasiliev, G. A. Holodiy, A. N. Vostrov, V. N. Rybchin, and A. N. Sverchevsky. 1992. The structure of the region of the Escherichia coli linear plasmid prophage N15. Mol. Genet. Mikrobiol. Virusol. 1992:22-24 (In Russian.) [Google Scholar]
- 69.Mattsby-Baltzer, I., M. Sandin, B. Ahlstrom, S. Allenmark, M. Edebo, E. Falsen, K. Pedersen, N. Rodin, R. A. Thompson, and L. Edebo. 1989. Microbial growth and accumulation in industrial metal-working fluids. Appl. Environ. Microbiol. 55:2681-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 71.McLenigan, M. P., O. I. Kulaeva, D. G. Ennis, A. S. Levine, and R. Woodgate. 1999. The bacteriophage P1 HumD protein is a functional homolog of the prokaryotic UmuD′-like proteins and facilitates SOS mutagenesis in Escherichia coli. J. Bacteriol. 181:7005-7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 73.Narita, S., J. Kaneko, J. Chiba, Y. Piemont, S. Jarraud, J. Etienne, and Y. Kamio. 2001. Phage conversion of Panton-Valentine leukocidin in Staphylococcus aureus: molecular analysis of a PVL-converting phage, φSLT. Gene 268:195-206. [DOI] [PubMed] [Google Scholar]
- 74.Oram, D. M., A. Avdalovic, and R. K. Holmes. 2002. Construction and characterization of transposon insertion mutations in Corynebacterium diphtheriae that affect expression of the diphtheria toxin repressor (DtxR). J. Bacteriol. 184:5723-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 76.Pedulla, M. L., M. E. Ford, J. M. Houtz, T. Karthikeyan, C. Wadsworth, J. A. Lewis, D. Jacobs-Sera, J. Falbo, J. Gross, N. R. Pannunzio, W. Brucker, V. Kumar, J. Kandasamy, L. Keenan, S. Bardarov, J. Kriakov, J. G. Lawrence, W. R. Jacobs, Jr., R. W. Hendrix, and G. F. Hatfull. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171-182. [DOI] [PubMed] [Google Scholar]
- 77.Perna, N. T., G. Plunkett, III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 78.Ramirez, B., O. Voloshin, R. Camerini-Otero, and A. Bax. 2000. Solution structure of DinI provides insight into its mode of RecA inactivation. Protein Sci. 9:2161-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ravin, N., and D. Lane. 1999. Partition of the linear plasmid N15: interactions of N15 partition functions with the sop locus of the F plasmid. J. Bacteriol. 181:6898-6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ravin, N., T. Strakhova, and V. Kuprianov. 2001. The protelomerase of the phage-plasmid N15 is responsible for its maintenance in linear form. J. Mol. Biol. 312:899-906. [DOI] [PubMed] [Google Scholar]
- 81.Ravin, N., A. Svarchevsky, and G. Deho. 1999. The anti-immunity system of phage-plasmid N15: identification of the antirepressor gene and its control by a small processed RNA. Mol. Microbiol. 34:980-984. [DOI] [PubMed] [Google Scholar]
- 82.Ravin, V., V. Kurprianov, E. B. Gilcrease, and S. Casjens. 2003. Bidirectional replication from an internal ori site of the linear N15 plasmid DNA. Nucleic Acids Res. 31:6552-6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ravin, V., N. Ravin, S. Casjens, M. E. Ford, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequence and analysis of the atypical temperate bacteriophage N15. J. Mol. Biol. 299:53-73. [DOI] [PubMed] [Google Scholar]
- 84.Ravin, V., and M. G. Shulga. 1970. The evidence of extrachromosomal location phage prophage N15. Virology 40:800-805. [DOI] [PubMed] [Google Scholar]
- 85.Ravin, V. K., and E. I. Golub. 1967. A study of phage conversion in Escherichia coli. I. The acquisition of resistance to bacteriophage T1 as a result of lysogenization. Genetika 4:113-121 (In Russian.)
- 86.Recktenwald, J., and H. Schmidt. 2002. The nucleotide sequence of Shiga toxin (Stx) 2e-encoding phage φP27 is not related to other Stx phage genomes, but the modular genetic structure is conserved. Infect. Immun. 70:1896-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rybchin, V. N., and A. N. Svarchevsky. 1999. The plasmid prophage N15: a linear DNA with covalently closed ends. Mol. Microbiol. 33:895-903. [DOI] [PubMed] [Google Scholar]
- 88.Sabbattini, P., F. Forti, D. Ghisotti, and G. Deho. 1995. Control of transcription termination by an RNA factor in bacteriophage P4 immunity: identification of the target sites. J. Bacteriol. 177:1425-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sandmeier, H., S. Iida, and W. Arber. 1992. DNA inversion regions Min of plasmid p15B and Cin of bacteriophage P1: evolution of bacteriophage tail fiber genes. J. Bacteriol. 174:3936-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shearwin, K. E., A. M. Brumby, and J. B. Egan. 1998. The Tum protein of coliphage 186 is an antirepressor. J. Biol. Chem. 273:5708-5715. [DOI] [PubMed] [Google Scholar]
- 91.Shultz, J., T. J. Silhavy, M. L. Berman, N. Fiil, and S. D. Emr. 1982. A previously unidentified gene in the spc operon of Escherichia coli K12 specifies a component of the protein export machinery. Cell 31:227-235. [DOI] [PubMed] [Google Scholar]
- 92.Smith, M. C., R. N. Burns, S. E. Wilson, and M. A. Gregory. 1999. The complete genome sequence of the Streptomyces temperate phage φC31: evolutionary relationships to other viruses. Nucleic Acids Res. 27:2145-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Southern, E. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 94.Staden, R. 1986. The current status and portability of our sequence handling software. Nucleic Acids Res. 14:217-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stoppel, R. D., M. Meyer, and H. G. Schlegel. 1995. The nickel resistance determinant cloned from the enterobacterium Klebsiella oxytoca: conjugational transfer, expression, regulation and DNA homologies to various nickel-resistant bacteria. Biometals 8:70-79. [DOI] [PubMed] [Google Scholar]
- 96.Studier, W., A. Rosenberg, J. Dunn, and J. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 97.Tang, M., X. Shen, E. G. Frank, M. O'Donnell, R. Woodgate, and M. F. Goodman. 1999. UmuD′(2)C is an error-prone DNA polymerase, Escherichia coli pol V. Proc. Natl. Acad. Sci. USA 96:8919-8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Voloshin, O. N., B. E. Ramirez, A. Bax, and R. D. Camerini-Otero. 2001. A model for the abrogation of the SOS response by an SOS protein: a negatively charged helix in DinI mimics DNA in its interaction with RecA. Genes Dev. 15:415-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang, J., M. Hofnung, and A. Charbit. 2000. The C-terminal portion of the tail fiber protein of bacteriophage lambda is responsible for binding to LamB, its receptor at the surface of Escherichia coli K-12. J. Bacteriol. 182:508-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Weiss, R., D. Dunn, M. Shuh, J. Atkins, and R. Gesteland. 1989. E. coli ribosomes re-phase on retroviral frameshift signals at rates ranging from 2 to 5 percent. New Biol. 1:159-169. [PubMed] [Google Scholar]
- 101.Woods, D. E., J. A. Jeddeloh, D. L. Fritz, and D. DeShazer. 2002. Burkholderia thailandensis E125 harbors a temperate bacteriophage specific for Burkholderia mallei. J. Bacteriol. 184:4003-4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xu, J. 2000. A conserved frameshift strategy in dsDNA long tailed phages. Ph.D. University of Pittsburgh, Pittsburgh, Pa.
- 103.Yasuda, T., K. Morimatsu, T. Horii, T. Nagata, and H. Ohmori. 1998. Inhibition of Escherichia coli RecA coprotease activities by DinI. EMBO J. 17:3207-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yasuda, T., K. Morimatsu, R. Kato, J. Usukura, M. Takahashi, and H. Ohmori. 2001. Physical interactions between DinI and RecA nucleoprotein filament for the regulation of SOS mutagenesis. EMBO J. 20:1192-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yasuda, T., T. Nagata, and H. Ohmori. 1996. Multicopy suppressors of the cold-sensitive phenotype of the pcsA68 (dinD68) mutation in Escherichia coli. J. Bacteriol. 178:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]