Abstract
The ability of Legionella pneumophila to grow and cause disease in the host is completely dependent on a type IV secretion system known as the Dot/Icm complex. This membrane-spanning apparatus translocates effector molecules into host cells in a process that is poorly understood but that is known to require the putative ATPase DotB. One possible role for DotB is suggested by its similarity to the PilT family of proteins, which mediate pilus retraction. To better understand the molecular behavior of DotB, we have purified the protein and shown that it forms stable homohexameric rings and hydrolyzes ATP with a specific activity of 6.4 nmol of ATP/min/mg of protein. ATPase activity is critical to the function of DotB, as alteration of the conserved Walker box lysine residue resulted in a mutant protein, DotB K162Q, which failed to bind or hydrolyze ATP and which could not complement a ΔdotB strain for intracellular growth in macrophages. Consistent with the ability of DotB to interact with itself, the dotBK162Q allele exhibited transdominance over wild-type dotB, providing the first example of such a mutation in L. pneumophila. Finally, the DotB K162Q mutant protein had a significantly enhanced membrane localization in L. pneumophila compared to wild-type DotB, suggesting a relationship between nucleotide binding and membrane association. These results are consistent with a model in which DotB cycles between the cytoplasm and the Dot/Icm complex at the membrane, where it hydrolyzes nucleotides to provide energy to the complex.
Legionella pneumophila, an intracellular bacterial pathogen, causes Legionnaires' disease by replicating inside alveolar macrophages (13). This ability is dependent on a set of genes called dot or icm, which encode a specialized type IV secretion system (32, 37). The type IV secretion system family is composed of both classical plasmid transfer systems and a related protein export apparatus used by a wide range of pathogens to export toxins (reviewed in reference 4). For L. pneumophila, the Dot/Icm complex is believed to translocate numerous effector molecules into host cells in order to modify the endocytic pathway. Thus far, two translocated proteins, LidA and RalF, have been identified (6, 19). RalF is an ADP-ribosylating factor that functions to recruit the macrophage protein ARF1 to L. pneumophila-containing phagosomes, while the function of LidA remains unknown. Because neither of these is absolutely required for L. pneumophila intracellular growth, additional Dot/Icm-secreted proteins are believed to exist (6).
The Dot/Icm apparatus is a macromolecular complex that comprises a large number of membrane proteins and several proteins predicted to have cytoplasmic localization. One such cytoplasmic protein, DotB, contains conserved nucleotide binding domains present in many ATPases, as well as domains specific to ATPases found in bacterial type II and type IV secretion systems (23, 27, 39). Though DotB is known to be a critical component of the Dot/Icm complex (37, 38), its molecular function is not yet clear. BLAST searches reveal that DotB is most similar to several uncharacterized putative ATPases found in other type IVB secretion systems including a Coxiella burnetii DotB homologue (BLAST score, e−135) and the TraJ protein of plasmid R64 (BLAST score, e−41). DotB also has limited similarity to the type IVA secretion ATPases HP0525 of Helicobacter pylori (BLAST score, e−4) and VirB11 of Agrobacterium tumefaciens (BLAST score, e−2). Finally, DotB has extensive similarity to the PilT family of proteins (BLAST score, e−38) found in bacterial type II secretion systems. PilT proteins are ATPases that mediate the retraction of adhesive pili, a process essential to bacterial twitching motility and natural transformation (10, 18, 34).
Since PilT proteins are components of type II secretion systems, which are evolutionarily quite divergent from type IV secretion systems (22), the similarity between DotB and PilT is surprising. Pilus retraction is well documented both in type II secretion systems (e.g., type IV pili) and in the conjugal type IV secretion systems (20). However, in the latter case, the mechanism of this retraction is completely unknown; no pilus retraction factors or PilT-like proteins have yet been identified. Now, L. pneumophila, C. burnetii, and plasmid R64 DNA sequences have revealed the existence of three PilT homologues in the type IVB secretion systems. Whether these proteins function as the type IVA secretion ATPases are thought to, as assembly and export chaperones for their respective secretion complexes (14-16), or whether they might mediate retraction of pili elaborated by type IV secretion complexes remains to be determined.
The families of type II and type IV secretion ATPases perform a wide range of functions including assembly of transmembrane complexes, pilus assembly, pilus retraction, and substrate export (3, 10, 14, 15, 28, 29, 36). Remarkably, despite their diverse functions, biochemical characterization of a number of these proteins has revealed that they possess a unifying set of phenotypes. First, in addition to the Walker A and B boxes, these proteins contain conserved domains that cluster them together as a superfamily (23, 27, 39). Second, they exhibit ATPase activity in vitro which is essential to their function (3, 5, 10, 15, 27, 29, 35). Third, they multimerize and most are known to form homohexameric rings (3, 14, 15, 24, 26, 27, 36). Fourth, all are predicted to be hydrophilic but those localized can be found in both the cytoplasm and the membrane fractions of the bacterial cells (3, 10, 25, 27, 30). Finally, several of these purified proteins exhibit increased ATPase activity in the presence of phospholipids, suggesting that the bacterial membrane is the site of their enzymatic activity (3, 15, 27).
To determine the molecular function of the putative ATPase and PilT homologue DotB, we have chosen to purify the protein and examine it biochemically. ATPase activity, multimeric state, and membrane localization have all been addressed. In addition, a mutant form of the protein was engineered and characterized with the goal of understanding the importance of DotB ATPase activity to its function in L. pneumophila pathogenesis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains are listed in Table 1. Strain Lp02 (thyA hsdR rpsL) is a derivative of the L. pneumophila serogroup 1 clinical isolate Philadelphia-1 (2). All L. pneumophila strains were cultured on ACES [N-(2-acetamido)-2-aminoethanesulfonic acid]-buffered charcoal yeast extract agar (CYE) or in ACES-buffered yeast extract broth (AYE) as described previously (7, 9). Lp02 and Lp02 derivatives were cultured on CYE or AYE supplemented with 100 μg of thymidine/ml as needed. Escherichia coli strains C600 and XL1-Blue were cultured in Luria-Bertani broth containing ampicillin at 100 μg/ml for maintenance of plasmid selection.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant properties | Reference or source |
|---|---|---|
| L. pneumophila | ||
| Philadelphia-1 | Clinical isolate, serogroup 1 | CDCa |
| Lp02 | Philadelphia-1 SmR, r−thyA | 2 |
| JV918 | Lp02 ΔdotB | This study |
| JV3079 | Lp02 ΔdotB::Flag coding sequence fused to dotB | This study |
| E. coli | ||
| C600 | e14−(McrA−) supE44 thi-1 thr-1 leuB6 lacY1 tonA21 | Stratagene |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB laclqZΔM15 Tn10(Tetr)] | Stratagene |
| Plasmids | ||
| pQE30 | Expression vector for His-tagged proteins (Ampr) | Qiagen |
| pJB1540 | pQE30 with thrombin site | This study |
| pJB1572 | pJB1540 with dotB | This study |
| pJB2442 | pJB1540 with dotBK162Q | This study |
| pJB908 | RSF1010 cloning vector (AmprtdΔi) | This study |
| pJB1153 | pJB908 with dotB | This study |
| pJB1568 | pJB908 with dotBK162Q | This study |
| pJB1192 | pJB908 with His coding sequence fused to dotB | This study |
| pJB2444 | pJB908 with His coding sequence fused to dotBK162Q | This study |
| pSR47S | R6K suicide vector (KanrsacB) | 17 |
| pJB920 | pSR47S with dotB flanking regions | This study |
| pJB2911 | pSR47S with Flag coding sequence fused to dotB | This study |
CDC, Centers for Disease Control and Prevention (Atlanta, Ga.).
Strain and plasmid construction.
The suicide plasmid pJB920, used to create the ΔdotB strain JV918, was constructed by PCR amplification of 500-bp regions flanking the dotB open reading frame (ORF) from Lp02 chromosomal DNA with primers engineered to have SalI, BamHI, or EagI restriction sites. The resulting fragments were cut with SalI/BamHI or BamHI/EagI, cloned in a three-piece ligation into the SalI/NotI sites of the suicide vector pSR47S (17), and confirmed by sequencing. Next, pJB920 was transformed into Lp02, where it integrated via homologous recombination into the chromosome, generating a dotB/ΔdotB merodiploid. The merodiploid was resolved by counterselection based on sucrose sensitivity, and the resulting colonies were screened for the absence of dotB via PCR, thus generating strain JV918.
The suicide plasmid pJB2911, used to create the Flag-DotB strain JV3079, was constructed as follows. Regions of 500 bp immediately upstream and downstream of the dotB start codon were amplified by PCR from Lp02 chromosomal DNA with primers engineered to have SalI, BamHI, or EagI restriction sites. The resulting fragments were cut with SalI/BamHI or BamHI/EagI, cloned in a three-piece ligation into the SalI/NotI sites of the suicide vector pSR47S (17), and confirmed by sequencing. Next, oligonucleotides encoding the Flag epitope (DYKDDDDK) were inserted into the BamHI site engineered immediately following the dotB start codon, resulting in destruction of the site. Finally, this suicide plasmid (pJB2911) was transformed into Lp02 to generate a dotB/Flag-dotB merodiploid, which was then resolved. Colonies were screened by Western blotting for the presence of Flag-DotB, and strain JV3079, which contains only Flag-DotB, was generated.
The cloning vector pJB908, a derivative of the RSF1010 vector pKB5 (2), was used to create dotB and dotBK162Q complementing clones. To make pJB908, the pKB5 origin of conjugative transfer (oriT), contained on an SfiI/BstZ17I fragment, was replaced with an SfiI/BstZ17I fragment with oriT deleted (8). To make the plasmid pJB1153, which encodes the wild-type DotB protein, the dotB ORF was amplified by PCR from Lp02 chromosomal DNA with primers containing BamHI or SalI restriction sites at their 5 prime ends. The resulting PCR product was digested with BamHI and SalI, cloned into the BamHI/SalI sites of pJB908, and confirmed by sequencing. To make the plasmid pJB1568, which encodes the Walker box mutant protein DotB K162Q, the 5 prime and 3 prime portions of the dotB ORF were amplified separately. Each primer described above was paired with one of two overlapping divergent primers engineered to replace DotB lysine 162 with a glutamine. These primers were designed by using the Primer Generator program (http://www.med.jhu.edu/medcenter/primer/primer.cgi) and resulted in PCR products with altered dotB sequences, creating the K162Q mutation as well as a novel AvaI site. 5′ and 3′ PCR products were digested with BamHI and AvaI or AvaI and SalI, cloned in a three-piece ligation into the BamHI/SalI sites of pJB908, and confirmed by sequencing. Primers were as follows: dotB, 5′CCCGGATCCAGTAAGTTAATGTCTATGG and 3′CCCGTCGACGATTATTGTTGATATTCTTTGG (boldface letters indicate start or stop codons, and restriction sites are underlined); K162Q (dotB internal), 5′CCCCTCGGGCCAATCGACATTGTTAGCTTCC and 3′CCCGGCCCGAGCCGGTAGCGCCTGTAATAAAC (boldface letters indicate bases that differ from the dotB sequence, and restriction sites are underlined).
The expression vectors for His-tagged versions of DotB and DotB K162Q that were used for protein purification were engineered with plasmid pJB1540, a derivative of the Qiagen (Valencia, Calif.) expression vector pQE30. Plasmid pJB1540 encodes a thrombin cleavage site downstream of the sequence encoding the six-His tag and was constructed by cloning overlapping oligonucleotides containing a thrombin cleavage site and BamHI cohesive ends into the BamHI site of pQE30, destroying the upstream BamHI site. The wild-type dotB and dotBK162Q ORFs (minus the ATG start codons) were amplified by PCR from pJB1153 or pJB1568 plasmid DNA with primers containing BamHI or SalI restriction sites at their 5 prime ends. The resulting PCR product was digested with BamHI and SalI, cloned into the BamHI/SalI sites of pJB1540, and confirmed by sequencing. This resulted in the constructs pJB1572 (wt) and pJB2442 (K162Q mutant). Coding sequences for His-tagged versions of wild-type and mutant DotB were subcloned from pJB1572 and pJB2442 into the RSF1010 vector pJB908 for expression in L. pneumophila. These constructs were designated pJB1192 and pJB2444, respectively.
Intracellular growth assays.
The human monocytic cell line U937 was passaged, and cells were differentiated with phorbol 12-myristate 13-acetate (Sigma) as described previously (21). L. pneumophila strains to be used for infection of U937 cells were grown to exponential phase, back-diluted into media containing 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) for overexpression of dotB, and then grown to early stationary phase. Approximately 109 bacteria were pelleted, washed one time in sterile deionized water, resuspended in 1 ml of sterile deionized water, and then diluted 1:4,000 into RPMI 1640 (BioWhittaker) tissue culture medium. This suspension (0.5 ml) was added to a monolayer of differentiated U937 cells (106 per well) in 24-well dishes, and the mixture was coincubated for 1 h at 37°C in 5% CO2. Medium containing extracellular bacteria was then aspirated, and the U937 cell monolayer was washed two times with 0.5 ml of RPMI 1640 containing 1 mM IPTG. Cell monolayers were maintained at 37°C in 5% CO2 for 3 days, and each day bacteria were recovered and plated to determine total CFU in a given well. Because L. pneumophila cannot replicate in RPMI 1640, daily quantitation of CFU allows an accurate representation of bacterial intracellular growth over time.
Antibody generation and affinity purification.
Purified His-DotB was injected into rabbits (Cocalico, Inc.) for generation of a polyclonal DotB antibody, which was subsequently affinity purified as follows. Purified His-DotB (2 mg) was separated via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene fluoride membrane (Millipore), and the His-DotB protein band was cut away from the rest of the membrane. The resulting membrane strip was incubated with 1.5 ml of polyclonal serum diluted in BLOTTO (phosphate-buffered saline [PBS], 5% nonfat dry milk) for 3 h at room temperature, washed in PBS, and incubated with 8 ml of 0.2 M glycine (pH 2.8) for 2 min in order to elute antibody from the membrane. Glycine was neutralized by addition of 2 ml of 1 M Tris (pH 8.8), purified antibody was removed, and a second round of elution was performed. Pooled purified antibody was then concentrated with Ultrafree 15 filter columns with a 10-kDa molecular weight cutoff (Millipore), and buffer was exchanged with PD-10 columns (Amersham Biosciences) into PBS containing 0.02% sodium azide and bovine serum albumin (BSA) at 1 mg/ml.
Electron microscopy.
Proteins were diluted to 16 μg/ml in cold KHMgE buffer (70 mM KCl, 30 mM HEPES [pH 7.4], 1 mM MgCl2, 3 mM EGTA) with or without nucleotides (1 mM) and processed according to published protocols (11, 12). Platinum replicas were viewed in a JEOL transmission electron microscope and photographed in stereo. Micrographs (×68,000) were digitized by video imaging and were transferred to a computer.
Membrane fractionation.
Bacterial cells (100 OD600 [optical density at 600 nm] units) were grown to late stationary phase, harvested, and stored at −20°C until needed. For preparation of membrane fractions, cells were resuspended in 1 ml of cold Tris-sucrose (50 mM Tris [pH 8.0], 500 mM sucrose, 5 mM EDTA); lysozyme was added to 100 μg/ml, and cells were incubated on ice for 1 h to create spheroplasts. These were then pelleted at 5,000 × g (4°C), resuspended in 2.5 ml of cold 20 mM Tris (pH 8.0), and lysed by sonication. Lysate was cleared by 10 min of centrifugation at 10,000 × g and 4°C. Lysate (the total protein fraction) was then subjected to ultracentrifugation for 1 h at 100,000 × g and 4°C to pellet the membrane fractions. The membrane pellet was washed in cold 20 mM Tris (pH 8.0) and then repelleted for 30 min at 100,000 × g (4°C) and resuspended. This fraction represented the total membrane.
Protein purification.
C600 cells containing plasmid pJB1572 were grown in 5 liters of Luria-Bertani broth at 37°C. Cells were induced in mid-log phase to express DotB by adding 200 μM IPTG to the medium and were grown to stationary phase (OD600, ∼5) before harvesting. A cell pellet of approximately 80 g was obtained and was resuspended in 150 ml of cold buffer A (20 mM Tris [pH 7.6], 100 mM NaCl, 10% glycerol, 0.01% Brij 35) with 10 mM imidazole; lysozyme was added to 1 mg/ml, cells were incubated for 30 min on ice and lysed by sonication on ice, and lysate was cleared by 40 min of centrifugation at 20,000 × g and 4°C. Lysate was split into 50-ml screw-cap conical tubes (nine tubes, 40 ml/tube), and 0.5 ml of nickel-nitrilotriacetic acid-agarose (Ni-NTA-agarose) (Qiagen) was added to each. Tubes were incubated, with mixing, for 2 h at 4°C. Three columns (1.5 by 12 cm; Bio-Rad) were packed with the loaded resin. Each column was washed stepwise via gravity flow with cold buffer A containing 10 and 30 mM imidazole, and protein was eluted off the resin in cold buffer A with 100 mM imidazole. Elution fractions were pooled, total protein content was determined by the Bio-Rad protein assay (based on the method of Bradford), and thrombin was added at 16 μg per mg (estimated) of His-DotB in order to remove the N-terminal tag. The cleavage reaction was allowed to proceed for 2 h at 25°C. Proteins were then applied to an 8-ml Source 15Q (Amersham Biosciences) ion-exchange column equilibrated in buffer A; this and subsequent steps in DotB purification were performed with AKTA Explorer fast protein liquid chromatography equipment by Pharmacia. Proteins were eluted with an 80-ml linear gradient from 100 to 500 mM NaCl in buffer A. DotB eluted at 230 mM NaCl. Fractions containing DotB were pooled, and buffer was exchanged with a PD-10 column (Amersham Biosciences) into buffer B (20 mM KPO4, 50 mM KCl, 10% glycerol, 1 mM dithiothreitol [DTT]) and applied to a 3-ml hydroxyapatite (Bio-Rad) column equilibrated in buffer B. Proteins were eluted with a 45-ml linear gradient from 20 to 220 mM KPO4 and 0 to 500 mM NaCl in buffer B. DotB eluted at approximately 350 mM KPO4-150 mM NaCl. Fractions containing DotB were pooled, protein was exchanged into buffer D (buffer A plus 1 mM DTT and 0.1 mM EDTA), and the sample was concentrated to a final volume of 400 μl with Ultrafree 4 filter columns with a 10-kDa molecular mass cutoff (Millipore). Concentrated DotB was then applied in a single injection to a 25-ml Superose 12 column (Amersham Biosciences) equilibrated in buffer D. DotB eluted in single peak at a total elution volume of 9.8 ml. Fractions containing DotB were pooled and brought to a final concentration of 2 mg/ml. This procedure yielded, on average, 3 to 4 mg of highly purified DotB (quantitated 97% pure; AlphaImager software). Purification of DotB K162Q (expressed in C600 with pJB2442) was performed in exactly the same fashion, and preparations were equally pure.
ATPase and ATP binding assays.
ATP hydrolysis reactions were performed with the EnzChek phosphate assay kit (Molecular Probes). Each reaction mixture contained 3 to 24 μM purified protein (molarity was calculated for monomeric DotB) in buffer D with 1 mM ATP, 10 mM MgCl2, 4 mM EnzChek substrate (2-amino-6-mercapto-7-methylpurine riboside [MESG]), and 0.1 U of EnzChek enzyme (purine nucleoside phosphorylase [PNP]). In the presence of inorganic phosphate MESG is converted via PNP to ribose 1-phosphate and 2-amino-6-mercapto-7-methylpurine. This results in an absorbance shift from 330 to 360 nm, thus creating a spectrophotometric readout for the concentration of inorganic phosphate in each reaction. Reactions were performed in 100-μl volumes in 96-well plates and were allowed to proceed for 30 min at 25°C, at which point the A360 was determined with a VERSAmax plate reader (Molecular Devices). Total phosphate concentrations were determined by comparing sample absorbances to those of a phosphate standard curve.
For ATP binding reactions, 20 μg of purified protein was mock preincubated or preincubated with 10 mM ATP for 1 h at 4°C and then added to 400-μl reaction mixtures containing 20 mM MES (morpholineethanesulfonic acid; pH 7.0), 40 μl of ATP- or AMP-agarose (Sigma), and 100 μg of BSA (Sigma). Samples were incubated at 4°C for 90 min with mixing, and nucleotide-agarose was then pelleted by centrifugation and washed three times with 400 μl of cold 20 mM MES (pH 7.0). Bound proteins were eluted by addition of 40 μl of SDS-PAGE sample buffer and 5 min of boiling and then separated by SDS-PAGE for Coomassie staining and visualization. Alternatively, 40 μl of ATP- or AMP-agarose was added to 500 μl of L. pneumophila lysate in buffer E (50 mM Tris [pH 8.0], 150 mM NaCl, 1% Triton X-100) from 3 OD600 equivalents of L. pneumophila cells. Competing ATP was added to a final concentration of 10 mM. Reaction mixtures were incubated at 4°C for 6 h with mixing, and nucleotide-agarose was pelleted by centrifugation and washed three times with 500 μl of cold buffer E. Bound proteins were eluted by addition of 50 μl of SDS-PAGE sample buffer and 5 min of boiling, separated by SDS-PAGE, and transferred to a polyvinylidene fluoride membrane for analysis by Western blotting.
RESULTS
The K162 residue is essential to DotB function.
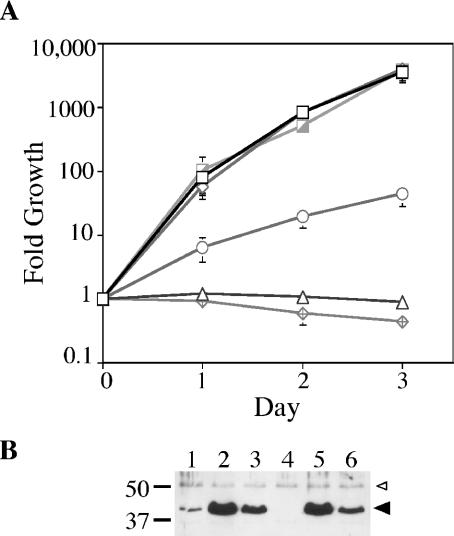
To determine whether the predicted ATPase activity of DotB might be important for its function, a mutation in the DotB Walker A box was engineered, changing the highly conserved lysine to a glutamine. When the DotB K162Q mutant protein was expressed from the Ptac promoter in the L. pneumophila dotB deletion strain JV918, intracellular growth was identical to that of the dotB deletion strain containing the empty expression vector, indicating that the Walker box mutant was not able to function in L. pneumophila (Fig. 1A). One possibility was that the DotB K162Q protein was not stable, despite the fact that a lysine-to-glutamine change should be structurally conservative. To test this, bacterial cells used for U937 infections were subjected to analysis by Western blotting with affinity-purified DotB antibody. Though apparently not as stable as wild-type DotB (Fig. 1B, compare lanes 6 and 5), DotB K162Q was found to be present in greater abundance than chromosomally encoded DotB in the wild-type strain Lp02 (Fig. 1B, compare lanes 6 and 1). Based on this observation, we concluded that the lack of functionality of DotB K162Q could not be attributed to an insufficient amount of protein.
FIG. 1.
Complementation of a dotB mutant for intracellular growth and transdominance of DotB K162Q in wild-type L. pneumophila. (A) Intracellular growth in U937 monocytes. L. pneumophila ΔdotB strain JV918 containing empty vector pJB908 (triangles), the DotB complementing clone pJB1153 (hatched squares), and the DotB K162Q clone pJB1568 (diamonds) was assayed for growth in U937 cells over time. In addition, L. pneumophila wild-type strain Lp02 containing empty vector pJB908 (open squares), the DotB complementing clone pJB1153 (open diamonds), and the DotB K162Q clone pJB1568 (circles) was also assayed for growth in U937 cells. (B) Strains used in panel A were examined by Western blotting for relative DotB levels at the time of infection. Lane 1, Lp02(pJB908); lane 2, Lp02(pJB1153); lane 3, Lp02(pJB1568); lane 4, JV918(pJB908); lane 5, JV918 (pJB1153); lane 6, JV918(pJB1568). The molecular masses of relevant markers (in kilodaltons) are on the left. Large arrowhead, DotB or DotB K162Q band; small arrowhead, cross-reacting band that serves to confirm overall sample equivalence.
In addition to being nonfunctional in a ΔdotB strain, DotB K162Q was found to inhibit the intracellular growth of the wild-type L. pneumophila strain Lp02 about 100-fold (Fig. 1A). This inhibition was not simply due to the increased level of DotB protein in the cells, as the overexpression of wild-type DotB did not affect Lp02 intracellular multiplication (Fig. 1A). Therefore, the Walker box mutant protein is not functional on its own and the mutation, when expressed in high copy number, is dominant negative in nature.
DotB is an ATPase.
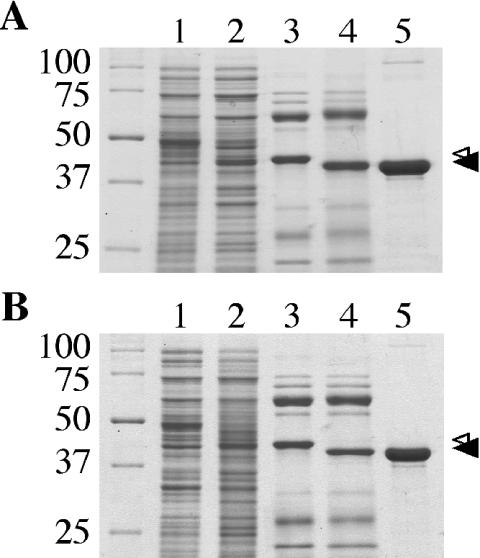
Based on the presence of several conserved nucleotide binding and hydrolysis domains within DotB sequence and on the requirement of the DotB Walker A box residue K162 for function (Fig. 1A), we predicted the DotB protein would exhibit ATP-hydrolyzing activity. To test this prediction, we engineered an N-terminal hexahistidine fusion that was expressed in E. coli and purified by Ni2+ affinity, ion-exchange, hydroxyapatite, and gel filtration chromatography (Fig. 2A). Targeted protease digestion was used to remove the His tag (Fig. 2A, compare lanes 3 and 4), leaving G-S-DotB (a glycine and a serine fused to DotB amino acid residues 2 to 377). The major protein band (42 kDa) was recognized by a polyclonal DotB antibody (data not shown). On average, 5-liter cultures yielded 3 to 4 mg of purified DotB, and the protein preparation was estimated to be approximately 97% pure.
FIG. 2.
Purification of DotB and DotB K162Q from E. coli. (A) Coomassie-stained SDS-PAGE gel showing wild-type DotB purification steps. Lane 1, cell extract from E. coli C600 containing plasmid pJB1572, no induction; lane 2, cell extract from C600(pJB1572), IPTG induced; lane 3, eluate from Ni-NTA-agarose after incubation with His-DotB lysate; lane 4, Ni-NTA eluate after exposure to thrombin; lane 5, purified DotB after three additional separation procedures. (B) Coomassie-stained SDS-PAGE gel showing DotB K162Q purification steps. Lanes are as for panel A, except that DotB K162Q was expressed with C600(pJB2442). (A and B) Small arrowhead, His-DotB or His-DotB K162Q bands; large arrowhead, bands representing DotB or DotB K162Q with the His tags removed. Standards of known molecular masses (kilodaltons) are on the left.
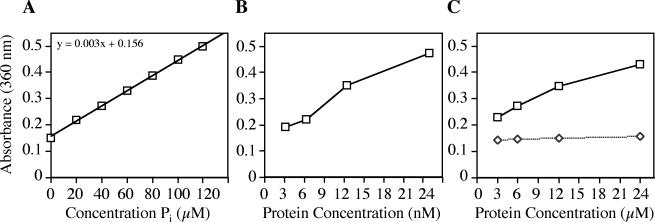
To assay ATPase activity of DotB, we employed a colorimetric assay that measures the presence of inorganic phosphate in solution (EnzChek; Molecular Probes). When free phosphate is present, it is used as a reagent in a chemical reaction that generates a product with maximum absorbance at 360 nm. Thus, the liberation of Pi by an ATPase, such as our positive control apyrase, can be quantitated by comparing A360 to that of a phosphate standard curve. When this simple end point assay was performed, increasing amounts of apyrase corresponded to increased A360 readings, indicating that the enzyme hydrolyzed ATP in proportion to its concentration (Fig. 3B). Positive readings were absolutely dependent on the addition of ATP to the reaction and on the presence of apyrase (data not shown), indicating that the source of Pi was enzymatically hydrolyzed nucleotides.
FIG. 3.
ATPase activity of purified DotB. ATPase activity was determined by measuring the amount of free phosphate in the reaction by a colorimetric assay (EnzChek; Molecular Probes). (A) Standard curve showing absorbance at 360 nm as a function of the concentration of phosphate. (B) ATPase activity of the positive control apyrase. (C) ATPase activity of DotB (squares) and DotB K162Q (diamonds). The specific activities of apyrase and DotB were calculated to be 3.0 μmol/min/mg and 6.4 nmol/min/mg, respectively. Each point represents three replicates.
Likewise, purified DotB liberated Pi from ATP in a concentration-dependent fashion, though to a lesser extent than the very active ATPase apyrase (Fig. 3C). DotB-specific activity was calculated to be 6.4 nmol of ATP/min/mg of protein, while that of apyrase was found to be 3.0 μmol/min/mg. Because the activity of DotB was quite low, we needed to demonstrate that it was due to the presence of DotB rather than a contaminating ATPase that might be present in the preparation in trace amounts. To eliminate this possibility, we employed the nonfunctional DotB K162Q mutant protein, which was predicted to lack enzymatic activity. DotB K162Q was purified in exactly the same fashion as the wild-type protein (Fig. 2B) and was tested by the EnzChek assay. The mutant protein did not generate detectable levels of Pi, indicating that it is enzymatically null (Fig. 3C) and that the activity seen for the wild-type protein is bona fide.
DotB is a hexameric ring.
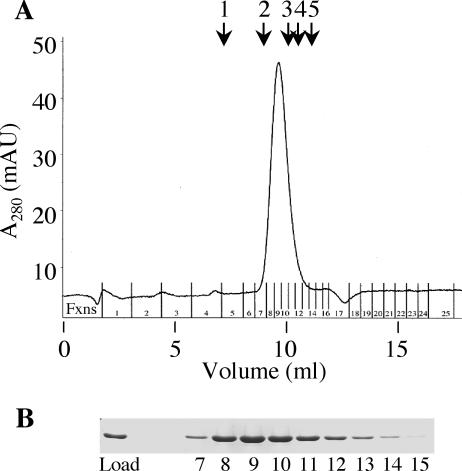
The discovery that several type IVA ATPases exist as homohexameric rings has inspired a model of function wherein the central chamber is the site of potential chaperone activity that is required for translocation and assembly of secretion complex components (14-16, 40). Initial experiments using native PAGE suggested that purified DotB could be found only in a hexameric state (data not shown). To more precisely determine DotB molecular mass, we subjected purified DotB to gel filtration analysis. The protein was found to have a native molecular mass of ∼259 kDa, a value consistent with the predicted molecular mass of a hexamer (252 kDa) (Fig. 4). In addition, the gel filtration elution profile for DotB K162Q was identical to that of wild-type DotB, suggesting that it also can form a hexamer (data not shown).
FIG. 4.
DotB is a hexamer. (A) Superose 12 elution profile for purified wild-type DotB. Milli-absorbance units (mAU) are shown as a function of elution volume. DotB eluted as a single peak corresponding to a molecular mass of 259 kDa. Arrows show elution peaks for standards of known molecular masses (Amersham Biosciences): 1, thyroglobulin (669 kDa); 2, ferritin (440 kDa); 3, catalase (232 kDa); 4, aldolase (158 kDa); 5, BSA (67 kDa). (B) Coomassie-stained SDS-PAGE gel demonstrating that the quantities of DotB in elution fractions (Fxns) 7 to 15 correlate with the single protein elution peak. The elution profile is representative of three experiments and is identical to that for DotB K162Q (data not shown).
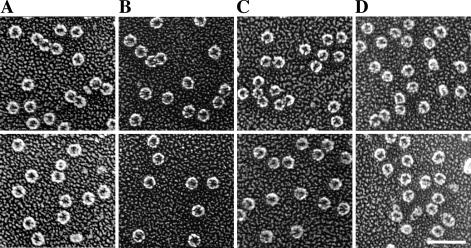
Next, we employed quick-freeze, deep-etch electron microscopy (11, 12), one of the most sensitive and accurate techniques available for visual examination of proteins, to observe the conformation of DotB. Purified protein was diluted in KHMgE buffer and incubated with no nucleotides, with ADP, or with ATPγS and then prepared and visualized with platinum replicas as described in Materials and Methods. Wild-type DotB appeared to exist solely and uniformly as rings of ∼10 nm in diameter, each with an electron-dense center that may represent a completely open hole—as in a doughnut shape—or perhaps the hollow chamber of a bowl-shaped molecule (Fig. 5A). Frequently, these rings were closely juxtaposed, even sometimes appearing to be linked. The protein was found in the same ring conformation regardless of the presence of nucleotides, confirming that nucleotide binding is not needed for hexamerization (Fig. 5B and C). Although changes in individual protein molecules were not discernible, the DotB sample containing ATPγS appeared to contain an increased number of linked rings (data not shown), reminiscent of tube-like structures observed for R388 TrwD (15). A preparation of purified DotB K162Q was also subjected to analysis, with the prediction that its loss of ability to hydrolyze nucleotides might be related to an altered conformation. However the mutant protein, like the wild type, appeared as ∼10-nm rings and was not affected by the presence of nucleotides (Fig. 5D and data not shown).
FIG. 5.
Purified DotB and DotB K162Q proteins form hexameric rings ∼10 nm in diameter. (A to C) Wild-type DotB was visualized in the absence of added nucleotides (A), revealing the putative apo form, as well as in the presence of 1 mM ADP (B) or 1 mM ATPγS (C). (D) DotB K162Q was also visualized in the putative apoprotein (shown here) and nucleotide-bound (not shown) forms. Scale bar, 50 nm. Images are representative of several thousand molecules visualized for each condition.
DotB K162Q forms mixed multimers with wild-type DotB.
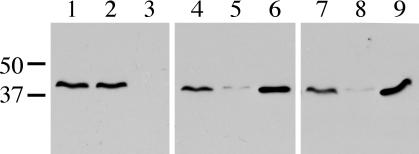
The observation that the dotBK162Q mutation is transdominant suggests that the mutant protein may exert this effect via formation of nonfunctional mixed multimers. To investigate this possibility, the capacity of the DotB K162Q mutant protein to multimerize with wild-type DotB was assayed by using a strain containing two different versions of epitope-tagged DotB. Wild-type DotB with an N-terminal Flag tag was expressed from the L. pneumophila chromosome, while DotB K162Q with an N-terminal His tag was expressed from a plasmid. Both fusion proteins were confirmed to be functional by intracellular growth assays (data not shown). Cell lysates of strains containing Flag-DotB and empty vector, Flag-DotB and the His-DotB complementing clone, or Flag-DotB and the His-DotB K162Q clone were made and passed over Ni-NTA-agarose. As expected, Flag-DotB in the absence of His-DotB did not bind to the Ni2+ matrix and was present in the flowthrough fraction rather than the elution fraction (Fig. 6, compare lanes 2 and 3). However, when Flag-DotB was in the presence of His-DotB, it appeared in the Ni-NTA-agarose elution fractions (Fig. 6, compare lanes 5 and 6), indicating an interaction between Flag- and His-tagged wild-type DotB. Moreover, the same result was obtained when Flag-DotB was in the presence of His-DotB K162Q (Fig. 6, compare lanes 8 and 9). The elution fraction containing Flag-DotB and His-DotB K162Q was further subjected to gel filtration and Western blot analysis to confirm the presence of both proteins in hexameric multimers (data not shown). Thus, the Walker box mutant protein DotB K162Q can interact with wild-type DotB to form mixed hexamers.
FIG. 6.
Detection of mixed DotB/DotB K162Q multimers. Lysate from L. pneumophila Flag-DotB strain JV3079 containing one of three plasmids was subjected to Ni-NTA-agarose. Strains were JV3079 with empty vector pJB908 (lanes 1 to 3), JV3079 with the His-DotB complementing clone pJB1192 (lanes 4 to 6), and JV3079 with the His-DotB K162Q clone pJB2444 (lanes 7 to 9). For each strain, three samples were taken: total protein (lanes 1, 4, and 7), Ni-NTA-agarose flowthrough (lanes 2, 5, and 8), and Ni-NTA-agarose eluate (lanes 3, 6, and 9). Samples were separated via SDS-PAGE, blotted onto a polyvinylidene fluoride membrane, and used in a Flag Western blot to detect the presence of Flag-DotB. The molecular masses of relevant markers (in kilodaltons) are on the left.
DotB K162Q shows enhanced membrane localization specific to L. pneumophila.
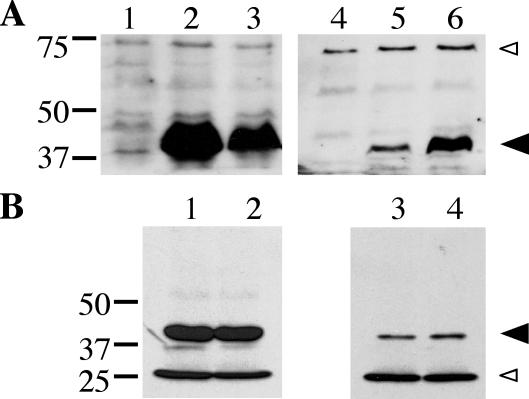
Though the DotB protein lacks any obvious hydrophobic regions and therefore does not appear to have membrane-spanning domains, there is a detectable amount of DotB in L. pneumophila membrane fractions (Fig. 7A, lane 5). This is consistent with observations of numerous secretion ATPases from type II and type IV systems, which partition to various degrees to the cytoplasmic and membrane fractions (10, 25, 27, 29, 30). Interestingly, a Walker A box mutant version of the A. tumefaciens VirB11 protein, VirB11 K175A, displays significantly enhanced membrane association relative to wild-type VirB11 (25). To examine the possibility that DotB K162Q might exhibit an altered localization profile, we prepared cell fractions from the L. pneumophila ΔdotB strain JV918 containing either empty vector, the wild-type DotB complementing clone, or the DotB K162Q clone and compared them by Western blotting to determine DotB and DotB K162Q localization.
FIG. 7.
Subcellular localization of DotB and DotB K162Q. (A) Total protein (lanes 1 to 3) and total membrane protein (lanes 4 to 6) fractions were taken from three L. pneumophila strains and were subjected to DotB Western blotting. Strains included the ΔdotB strain JV918 with empty vector pJB908 (lanes 1 and 4), JV918 with the DotB complementing clone pJB1153 (lanes 2 and 5), and JV918 with the DotB K162Q clone pJB1568 (lanes 3 and 6). The molecular masses of relevant markers (in kilodaltons) are on the left. (B) Total protein (lanes 1 and 2) and total membrane protein (lanes 3 and 4) fractions were taken from two E. coli strains and were used in DotB Western blotting. Strains included XL1-Blue with the DotB complementing clone pJB1153 (lanes 1 and 3) and XL1-Blue with the DotB K162Q clone pJB1565 (lanes 2 and 4). (A and B) Large arrowhead, DotB; small arrowhead, cross-reacting band that confirms overall sample equivalence. The molecular masses of relevant markers (in kilodaltons) are on the left. Results are representative of several experiments.
It appeared that a significant portion of the Walker box mutant protein was associated with L. pneumophila membranes. While soluble protein fractions (whole-cell lysate) contained less total DotB K162Q than wild-type DotB (Fig. 7A, lane 3 versus 2), indicating a relative lack of DotB K162Q stability, fractions composed of only membrane proteins contained more DotB K162Q than wild-type DotB (Fig. 7A, lane 6 versus 5). Analysis of serially diluted cell fractions allowed an estimation that, while approximately 5% of wild-type DotB was membrane associated, 40% of the Walker box mutant protein DotB K162Q was present in the membrane fraction (data not shown). Importantly, when this same experiment was repeated using E. coli as a host strain, equivalent amounts of DotB and DotB K162Q were localized to the membrane fractions (Fig. 7B, compare lanes 3 and 4). Thus, the enhanced membrane localization of the DotB K162Q protein is a Legionella-specific phenomenon and may indicate the presence of a specific receptor for DotB in the L. pneumophila inner membrane.
DotB K162Q cannot bind nucleotides.
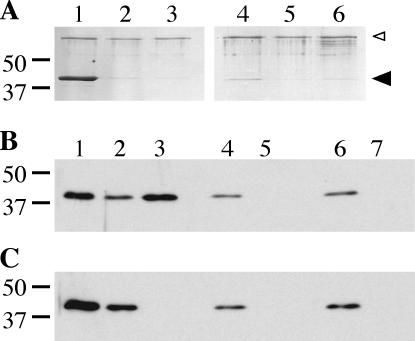
In search of an explanation for the enhanced membrane affinity of DotB K162Q, we wondered whether its association with the membrane might be affected by its capacity to bind nucleotides. To examine this ability, we took two approaches, exposing either purified protein or L. pneumophila whole-cell lysate to ATP- or AMP-agarose beads (Sigma). The beads were washed, and the eluate was analyzed via Coomassie-stained protein gels (Fig. 8A) or DotB Western blotting (Fig. 8B and C). Purified wild-type protein was present in the ATP-agarose eluate and was not present in eluate from AMP-agarose, to which it should not bind, or in eluate from ATP-agarose with competing free ATP (Fig. 8A, compare lanes 1, 2, and 3). Likewise, DotB from L. pneumophila lysate displayed the same pattern in the analogous elution fractions (Fig. 8B, compare lanes 3, 5, and 7). In contrast, DotB K162Q was not present in either ATP-agarose elution fraction (Fig. 8A, lane 4, and C, lane 3), indicating that it could not bind ATP either as a purified protein or when present in L. pneumophila lysate. Thus, DotB K162Q cannot bind ATP, a property which may help to explain its enhanced membrane affinity.
FIG. 8.
Nucleotide binding capacity of DotB and DotB K162Q. (A) Coomassie-stained SDS-PAGE gel showing elution fractions from DotB (lanes 1 to 3) and DotB K162Q (lanes 4 to 6) ATP binding reactions. Purified protein was incubated with ATP-agarose (lanes 1 and 4), AMP-agarose (lanes 2 and 5), or ATP-agarose in the presence of 10 mM competing free nucleotides (lanes 3 and 6), washed, and eluted. The molecular masses of relevant markers (in kilodaltons) are on the left. Large arrowhead, DotB; small arrowhead, BSA added to reactions to prevent nonspecific binding. (B) Western blot of fractions from binding reactions using L. pneumophila lysate containing wild-type DotB (ΔdotB strain JV918 with DotB complementing clone pJB1153). Whole-cell lysate (lane 1) was incubated with ATP-agarose (lanes 2 and 3), AMP-agarose (lanes 4 and 5), or ATP-agarose in the presence of 10 mM competing free nucleotide (lanes 6 and 7), washed, and eluted. Flowthrough (lanes 2, 4, and 6) and elution (lanes 3, 5, and 7) fractions were collected and subjected to DotB Western blotting. (C) Western blot of fractions from binding reactions using L. pneumophila lysate containing DotB K162Q (ΔdotB strain JV918 with DotB K162Q clone pJB1568). Whole-cell lysate (lane 1) was incubated with ATP-agarose (lanes 2 and 3), AMP-agarose (lanes 4 and 5), or ATP-agarose in the presence of 10 mM competing free nucleotides (lanes 6 and 7), washed, and eluted. Flowthrough (lanes 2, 4, and 6) and elution (lanes 3, 5, and 7) fractions were collected and subjected to DotB Western blotting. The molecular masses of relevant markers (in kilodaltons) are on the left.
DISCUSSION
We have purified the L. pneumophila DotB protein, shown that it forms homohexameric rings, and demonstrated for the first time that it exhibits ATPase activity. Moreover, ATPase activity is critical to DotB function, since a Walker A box mutant protein (DotB K162Q) that is deficient in ATP binding and hydrolysis cannot complement a ΔdotB strain for intracellular growth. We have also observed that the dotBK162Q mutation is dominant negative, likely due to the capacity of the mutant protein to form mixed multimers with wild-type DotB. Finally, we have demonstrated that the DotB K162Q protein has enhanced membrane localization in L. pneumophila but not in E. coli, suggesting the existence of a DotB-specific inner membrane receptor.
The ATPase activity measured for DotB is equivalent to that seen for several other secretion ATPases. R64 PilQ, Aquifex aeolicus PilT, R388 TrwD, and Actinobacillus actinomycetemcomitans TadA have similarly modest activities of between 1 and 15 nmol/min/mg (3, 10, 27, 29). Because this level of activity appears to be inconsistent with the idea that type II and type IV secretion ATPases are molecular motors, it has been proposed that they may function as chaperones facilitating the assembly or disassembly of secretion complexes and pili (10). Alternatively, modest activity could be due to the absence of stimulatory cofactors in ATPase reactions. For example, the addition of phospholipids to purified H. pylori HP0525, RP4 TrbB, TrwD, and TadA increased their enzymatic activity 2- to 10-fold (3, 15, 27), suggesting that interactions with the cytoplasmic membrane may regulate their function.
To confirm the importance of DotB ATPase activity, we constructed a mutant version of the protein which was enzymatically null and tested it for functionality in the context of L. pneumophila intracellular growth. Similar to a strain lacking DotB, a strain containing the DotB K162Q Walker A box mutant protein in place of the wild-type protein was completely defective for growth inside macrophages. Thus, DotB ATPase activity appears to be absolutely essential to its function in the Dot/Icm complex. Additionally, we observed that the DotB K162Q protein inhibited the intracellular growth of a wild-type L. pneumophila strain; this is the first description of a dominant negative mutation in Legionella. The DotB K162Q protein should prove to be a useful tool; potential applications include selective inactivation of the Dot/Icm complex at various stages of infection and inactivation of the DotB homologue in C. burnetii. The latter would be especially useful because genetic disruptions are not yet feasible in this system (33, 41, 42).
Other examples of secretion ATPase Walker A box mutations that are dominant negative in character include pilQK238Q and trwDK203Q (27, 29). In these cases, it was proposed that dominance resulted from the formation of wild-type and mutant mixed multimers; however, this remained untested. Here we have successfully demonstrated that DotB K162Q is capable of forming mixed multimers with wild-type DotB in vivo, and this likely explains the dominant negative phenotype of the dotBK162Q mutation. Consistent with this idea, a model derived from several crystal structures of the H. pylori type IVA secretion ATPase HP0525 proposes that the ATPase cycles between at least three conformations via concerted changes that are likely to depend on the functionality of all six subunits (31, 40). Thus, inactivation of even one subunit may be sufficient to render the entire complex nonfunctional, so that mixed multimers of functional and nonfunctional monomers would exhibit a null phenotype.
In addition to its nonfunctional and dominant negative properties, the DotB K162Q mutant protein exhibited enhanced membrane localization. Such augmented membrane localization was likewise observed for the A. tumefaciens Walker box VirB11 K175A mutant protein (25). Though the membrane association of VirB11 does not require other VirB proteins (25, 35), several type II secretion ATPases have been shown to localize to the bacterial membrane only in the presence of specific components of their respective secretion complexes (1, 24, 30). Because the DotB K162Q mutant protein displays enhanced membrane localization only in the context of L. pneumophila and not in E. coli, this suggests that there may be an inner membrane receptor for DotB, perhaps another Dot/Icm protein. Further characterization of DotB K162Q should help to confirm the existence of such a receptor.
It is not clear what dictates the increased membrane localization of the DotB and VirB11 Walker A box mutant proteins. One possibility is that these proteins are locked in an altered conformation that is more conducive to membrane association. Experiments with PilQ have shown that it, like HP0525, changes conformation upon ATP binding. Interestingly, when a Walker A box mutant version of PilQ was examined, it was found to exist only in what appeared to be the ATP-bound conformation (29). To check DotB K162Q for an altered conformation, high-resolution electron microscopy comparing purified wild-type and mutant proteins was performed. We could not detect gross conformational differences between them, either in the presence or absence of nucleotides. Furthermore, limited proteolysis failed to reveal any differences (data not shown). In both cases, such changes may be too subtle to detect. Regardless, it is clear that the mutant protein differs from wild-type DotB in that it is no longer able to bind and hydrolyze nucleotides.
Our work is in accordance with a model where secretion ATPases cycle between cytoplasmic and membrane-linked states by associating and disassociating with secretion complexes at the inner membrane. While at the membrane, secretion ATPases may function as gating molecules, cycling through closed and open forms regulated by ATP binding and hydrolysis. For DotB K162Q, its inability to bind and hydrolyze nucleotides is likely to abolish this mode of action, leaving it trapped in an intermediate step of normal DotB cycling at the L. pneumophila membrane.
In this study, we have demonstrated that the L. pneumophila DotB protein has several characteristics in common with assembly ATPases of type II and type IVA secretion systems and with the retraction ATPase PilT: specifically, DotB is a hydrophilic, membrane-associated ATPase that exists as a hexameric ring, the enzymatic activity of which is critical to its function. The significance of the intriguing homology between DotB and the pilus retraction protein PilT remains to be determined, and a potential role for DotB in pilus retraction awaits identification of a putative Dot/Icm pilus. We have also demonstrated here, based on observations of the mutant protein DotB K162Q, that DotB hexamers likely need to hydrolyze ATP in a concerted or rapidly successive fashion in order to function effectively. These data will serve as the foundation for further analysis of the DotB protein and should lead to a better understanding of its function on molecular and macromolecular levels.
Acknowledgments
We thank Virginia Miller and Katrina Forest for critical analysis of the manuscript and Angel Cantwell and Enrico Di Cera for the generous gift of purified recombinant thrombin.
J.A.S. was supported by Washington University Division of Infectious Disease training grant 5 T32 AI07172-22. This work was supported by NIH grant AI49950-03 to S.J.H. and NIH grant AI048052-03 as well as a grant from the Whittaker Foundation to J.P.V.
REFERENCES
- 1.Ball, G., V. Chapon-Herve, S. Bleves, G. Michel, and M. Bally. 1999. Assembly of XcpR in the cytoplasmic membrane is required for extracellular protein secretion in Pseudomonas aeruginosa. J. Bacteriol. 181:382-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger, K. H., and R. R. Isberg. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7:7-19. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharjee, M. K., S. C. Kachlany, D. H. Fine, and D. H. Figurski. 2001. Nonspecific adherence and fibril biogenesis by Actinobacillus actinomycetemcomitans: TadA protein is an ATPase. J. Bacteriol. 183:5927-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christie, P. J., and J. P. Vogel. 2000. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8:354-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christie, P. J., J. E. Ward, Jr., M. P. Gordon, and E. W. Nester. 1989. A gene required for transfer of T-DNA to plants encodes an ATPase with autophosphorylating activity. Proc. Natl. Acad. Sci. USA 86:9677-9681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conover, G. M., I. Derre, J. P. Vogel, and R. R. Isberg. 2003. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol. Microbiol. 48:305-321. [DOI] [PubMed] [Google Scholar]
- 7.Feeley, J. C., R. J. Gibson, G. W. Gorman, N. C. Langford, J. K. Rasheed, D. C. Mackel, and W. B. Baine. 1979. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J. Clin. Microbiol. 10:437-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frey, J., M. M. Bagdasarian, and M. Bagdasarian. 1992. Replication and copy number control of the broad-host-range plasmid RSF1010. Gene 113:101-106. [DOI] [PubMed] [Google Scholar]
- 9.Gabay, J. E., and M. A. Horwitz. 1985. Isolation and characterization of the cytoplasmic and outer membranes of the Legionnaires' disease bacterium (Legionella pneumophila). J. Exp. Med. 161:409-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herdendorf, T. J., D. R. McCaslin, and K. T. Forest. 2002. Aquifex aeolicus PilT, homologue of a surface motility protein, is a thermostable oligomeric NTPase. J. Bacteriol. 184:6465-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heuser, J. 1989. Protocol for 3-D visualization of molecules on mica via the quick-freeze, deep-etch technique. J. Electron Microsc. Tech. 13:244-263. [DOI] [PubMed] [Google Scholar]
- 12.Heuser, J. E. 1983. Procedure for freeze-drying molecules adsorbed to mica flakes. J. Mol. Biol. 169:155-195. [DOI] [PubMed] [Google Scholar]
- 13.Horwitz, M. A., and S. C. Silverstein. 1980. Legionnaires' disease bacterium (Legionella pneumophila) multiples intracellularly in human monocytes. J. Clin. Investig. 66:441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krause, S., M. Barcena, W. Pansegrau, R. Lurz, J. M. Carazo, and E. Lanka. 2000. Sequence-related protein export NTPases encoded by the conjugative transfer region of RP4 and by the cag pathogenicity island of Helicobacter pylori share similar hexameric ring structures. Proc. Natl. Acad. Sci. USA 97:3067-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krause, S., W. Pansegrau, R. Lurz, F. de la Cruz, and E. Lanka. 2000. Enzymology of type IV macromolecule secretion systems: the conjugative transfer regions of plasmids RP4 and R388 and the cag pathogenicity island of Helicobacter pylori encode structurally and functionally related nucleoside triphosphate hydrolases. J. Bacteriol. 182:2761-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Machon, C., S. Rivas, A. Albert, F. M. Goni, and F. de la Cruz. 2002. TrwD, the hexameric traffic ATPase encoded by plasmid R388, induces membrane destabilization and hemifusion of lipid vesicles. J. Bacteriol. 184:1661-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merriam, J. J., R. Mathur, R. Maxfield-Boumil, and R. R. Isberg. 1997. Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect. Immun. 65:2497-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merz, A. J., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98-102. [DOI] [PubMed] [Google Scholar]
- 19.Nagai, H., J. C. Kagan, X. Zhu, R. A. Kahn, and C. R. Roy. 2002. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295:679-682. [DOI] [PubMed] [Google Scholar]
- 20.Novotny, C. P., and P. Fives-Taylor. 1974. Retraction of F pili. J. Bacteriol. 117:1306-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearlman, E., A. H. Jiwa, N. C. Engleberg, and B. I. Eisenstein. 1988. Growth of Legionella pneumophila in a human macrophage-like (U937) cell line. Microb. Pathog. 5:87-95. [DOI] [PubMed] [Google Scholar]
- 22.Planet, P. J., S. C. Kachlany, R. DeSalle, and D. H. Figurski. 2001. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc. Natl. Acad. Sci. USA 98:2503-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Possot, O., and A. P. Pugsley. 1994. Molecular characterization of PulE, a protein required for pullulanase secretion. Mol. Microbiol. 12:287-299. [DOI] [PubMed] [Google Scholar]
- 24.Py, B., L. Loiseau, and F. Barras. 1999. Assembly of the type II secretion machinery of Erwinia chrysanthemi: direct interaction and associated conformational change between OutE, the putative ATP-binding component and the membrane protein OutL. J. Mol. Biol. 289:659-670. [DOI] [PubMed] [Google Scholar]
- 25.Rashkova, S., G. M. Spudich, and P. J. Christie. 1997. Characterization of membrane and protein interaction determinants of the Agrobacterium tumefaciens VirB11 ATPase. J. Bacteriol. 179:583-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rashkova, S., X. R. Zhou, J. Chen, and P. J. Christie. 2000. Self-assembly of the Agrobacterium tumefaciens VirB11 traffic ATPase. J. Bacteriol. 182:4137-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rivas, S., S. Bolland, E. Cabezon, F. M. Goni, and F. de la Cruz. 1997. TrwD, a protein encoded by the IncW plasmid R388, displays an ATP hydrolase activity essential for bacterial conjugation. J. Biol. Chem. 272:25583-25590. [DOI] [PubMed] [Google Scholar]
- 28.Sagulenko, E., V. Sagulenko, J. Chen, and P. J. Christie. 2001. Role of Agrobacterium VirB11 ATPase in T-pilus assembly and substrate selection. J. Bacteriol. 183:5813-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakai, D., T. Horiuchi, and T. Komano. 2001. ATPase activity and multimer formation of PilQ protein are required for thin pilus biogenesis in plasmid R64. J. Biol. Chem. 276:17968-17975. [DOI] [PubMed] [Google Scholar]
- 30.Sandkvist, M., M. Bagdasarian, S. P. Howard, and V. J. DiRita. 1995. Interaction between the autokinase EpsE and EpsL in the cytoplasmic membrane is required for extracellular secretion in Vibrio cholerae. EMBO J. 14:1664-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savvides, S. N., H. J. Yeo, M. R. Beck, F. Blaesing, R. Lurz, E. Lanka, R. Buhrdorf, W. Fischer, R. Haas, and G. Waksman. 2003. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J. 22:1969-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Segal, G., M. Purcell, and H. A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. USA 95:1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sexton, J. A., and J. P. Vogel. 2002. Type IVB secretion by intracellular pathogens. Traffic 3:178-185. [DOI] [PubMed] [Google Scholar]
- 34.Skerker, J. M., and H. C. Berg. 2001. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. USA 98:6901-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stephens, K. M., C. Roush, and E. Nester. 1995. Agrobacterium tumefaciens VirB11 protein requires a consensus nucleotide-binding site for function in virulence. J. Bacteriol. 177:27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turner, L. R., J. W. Olson, and S. Lory. 1997. The XcpR protein of Pseudomonas aeruginosa dimerizes via its N-terminus. Mol. Microbiol. 26:877-887. [DOI] [PubMed] [Google Scholar]
- 37.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873-876. [DOI] [PubMed] [Google Scholar]
- 38.Vogel, J. P., C. Roy, and R. R. Isberg. 1996. Use of salt to isolate Legionella pneumophila mutants unable to replicate in macrophages. Ann. N. Y. Acad. Sci. 797:271-272. [DOI] [PubMed] [Google Scholar]
- 39.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeo, H. J., S. N. Savvides, A. B. Herr, E. Lanka, and G. Waksman. 2000. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system. Mol. Cell 6:1461-1472. [DOI] [PubMed] [Google Scholar]
- 41.Zamboni, D. S., S. McGrath, M. Rabinovitch, and C. R. Roy. 2003. Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system. Mol. Microbiol. 49:965-976. [DOI] [PubMed] [Google Scholar]
- 42.Zusman, T., G. Yerushalmi, and G. Segal. 2003. Functional similarities between the icm/dot pathogenesis systems of Coxiella burnetii and Legionella pneumophila. Infect. Immun. 71:3714-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]