Abstract
The DcuS-DcuR system of Escherichia coli is a two-component sensor-regulator that controls gene expression in response to external C4-dicarboxylates and citrate. The DcuS protein is particularly interesting since it contains two PAS domains, namely a periplasmic C4-dicarboxylate-sensing PAS domain (PASp) and a cytosolic PAS domain (PASc) of uncertain function. For a study of the role of the PASc domain, three different fragments of DcuS were overproduced and examined: they were PASc-kinase, PASc, and kinase. The two kinase-domain-containing fragments were autophosphorylated by [γ-32P]ATP. The rate was not affected by fumarate or succinate, supporting the role of the PASp domain in C4-dicarboxylate sensing. Both of the phosphorylated DcuS constructs were able to rapidly pass their phosphoryl groups to DcuR, and after phosphorylation, DcuR dephosphorylated rapidly. No prosthetic group or significant quantity of metal was found associated with either of the PASc-containing proteins. The DNA-binding specificity of DcuR was studied by use of the pure protein. It was found to be converted from a monomer to a dimer upon acetylphosphate treatment, and native polyacrylamide gel electrophoresis suggested that it can oligomerize. DcuR specifically bound to the promoters of the three known DcuSR-regulated genes (dctA, dcuB, and frdA), with apparent KDs of 6 to 32 μM for untreated DcuR and ≤1 to 2 μM for the acetylphosphate-treated form. The binding sites were located by DNase I footprinting, allowing a putative DcuR-binding motif [tandemly repeated (T/A)(A/T)(T/C)(A/T)AA sequences] to be identified. The DcuR-binding sites of the dcuB, dctA, and frdA genes were located 27, 94, and 86 bp, respectively, upstream of the corresponding +1 sites, and a new promoter was identified for dcuB that responds to DcuR.
Escherichia coli can use C4-dicarboxylates (succinate, fumarate, and malate) and aspartate as sole carbon and energy sources during both aerobic and anaerobic growth (1). The utilization of C4-dicarboxylates requires various enzymes associated with the citric acid cycle as well as C4-dicarboxylate-specific transport systems (12). There are four known C4-dicarboxylate transporters in E. coli: the DctA system, which functions under aerobic conditions, and the Dcu systems, DcuA, DcuB, and DcuC, which act independently during anaerobic growth (5, 8, 13, 19, 32, 44). Some of the genes that encode the C4-dicarboxylate utilization systems are transcriptionally regulated in response to C4-dicarboxylate availability. This regulation is mediated by a two-component response-regulator designated DcuSR (10, 43). DcuSR induces the transcription of the dcuB-fumB operon (encoding DcuB and the anaerobic fumarase B), the frdABCD operon (encoding the anaerobic fumarate reductase complex), and the dctA gene (encoding DctA) in response to external C4-dicarboxylates and citrate (10, 43). In addition, a recent microarray-based global transcriptional profiling study suggested that the DcuSR regulon could comprise at least 39 other genes, including 13 that are involved in lipopolysaccharide biosynthesis and 3 that are involved in anaerobic nitrate and nitrite respiration (26).
The DcuS and DcuR proteins are members of a subgroup (the CitAB family) of two-component sensor-regulators that includes the citrate-responsive CitAB and CitST systems from Klebsiella pneumoniae and Bacillus subtilis, respectively (3, 42). The sensors within this family consist of three domains, as follows: an N-terminal domain containing two transmembrane helices on either side of an ∼140-amino-acid periplasmic segment containing a periplasmic PAS domain (PASp) which acts as a sensor for extracellular tri- and/or dicarboxylates (17, 27, 28); a second (centrally located) PAS domain in the cytosol (PASc), possibly serving as a redox sensor; and a C-terminal transmitter domain. The response-regulator proteins are composed of two domains, an N-terminal receiver domain and a C-terminal DNA-binding domain (10, 13, 24, 43). Recent studies have shown that DcuS autophosphorylates in vitro, using ATP as a substrate, and this activity is stimulated sixfold by C4-dicarboxylates (14). In addition, DcuR has also been shown to autophosphorylate, using the phosphorylated form of DcuS as a substrate, which converts DcuR into a high-affinity (KD, <1 μM) DNA-binding protein with specificity towards a dcuB promoter fragment (14). Thus, the available evidence suggests that the DcuSR system of E. coli functions as a classical two-component sensor-regulator in controlling gene expression in response to external C4-dicarboxylates (13).
This paper identifies the binding sites for DcuR at three DcuSR-regulated promoters (dcuB, dctA, and frdA) and shows that acetylphosphate treatment of DcuR converts the protein from a monomeric form with a low DNA-binding affinity to a dimeric form with a higher DNA-binding affinity. A second promoter is defined for dcuB which is ∼330 bp upstream of the previously identified promoter and appears, unlike the downstream promoter, to be directly regulated by DcuR.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and general procedures.
The plasmids used in this study are listed in Table 1. Bacterial cultures were grown aerobically at 37°C and 250 rpm in L broth with an appropriate antibiotic, unless otherwise stated. Transformations and DNA manipulations were as described by Sambrook et al. (30), unless otherwise stated.
TABLE 1.
Plasmids
| Plasmid | Relevant features and/or genotype | Source or reference |
|---|---|---|
| pEE1 | pSU18, dcuR | This work |
| pET21a | T7 overexpression vector | Novagen |
| pMalC-2 | malE fusion overexpression vector | New England BioLabs |
| pMA1 | pRS1274 plus dcuB promoter segment (−821 to +220) | This work |
| pMA2 | pRS1274 plus dcuB promoter segment (−721 to +220) | This work |
| pMA3 | pRS1274 plus dcuB promoter segment (−616 to +220) | This work |
| pMA4 | pRS1274 plus dcuB promoter segment (−519 to +220) | This work |
| pMA5 | pRS1274 plus dcuB promoter segment (−419 to +220) | This work |
| pMA6 | pRS1274 plus dcuB promoter segment (−302 to +220) | This work |
| pMA7 | pRS1274 plus dcuB promoter segment (−219 to +220) | This work |
| pMAK8 | pRS1274 plus dcuB promoter segment (−129 to +220) | This work |
| pMA9 | pRS528 plus dcuB promoter segment (−821 to −37) | This work |
| pMA10 | pRS528 plus dcuB promoter segment (−821 to −71) | This work |
| pPG2 | dcuS dcuR | 10 |
| pPG25 | pET-21a, dcuRHis | This work |
| pPG26 | pMalC-2, malE pas dcuS202-325 | This work |
| pPG27 | pMalC-2, malE paskin dcuS202-543 | This work |
| pPG28 | pUC118 plus 709-bp dcuR-dcuB fragment | This work |
| pPG28 | pUC118 plus 709-bp dcuR-dcuB fragment | This work |
| pPG30 | pUC118 plus 365-bp dcuB fragment | This work |
| pPG31 | pMalC-2, malE kin dcuS331-543 | This work |
| pPG32 | pUC118 plus 215-bp dcuR-dcuB fragment | This work |
| pPG32 | pUC118 plus 215-bp dcuB fragment | This work |
| pPG33 | pUC118 plus 376-bp yjeA-frdA fragment | This work |
| pPG34 | pUC118 plus 324-bp f651-dctA fragment | This work |
| pPG36 | pUC118 plus 618-bp rpsV-maeA fragment | This work |
| pPG37 | pSU18, ′dcuR124-234 | This work |
| pPG38 | pUC118 plus 631-bp balA-maeB fragment | This work |
| pPG39 | pET21a plus ′dcuR124-234-His | This work |
| pRS1274 | lacZYA transcriptional fusion vector | 31 |
| pRS528 | lacZYA transcriptional fusion vector | 31 |
| pSU18 | Cloning vector | 2 |
| pT2 | pUC118 plus 402-bp yjdI-dcuS fragment | This work |
| pUC118 | Cloning vector | 36 |
Overexpression constructs.
The following DcuS fragments were overproduced as MalE-DcuS fusions by using the overexpression vector pMal-c2: the His-kinase domain, Kin-DcuS (DcuS residues 331 to 543); the PASc domain, PASc-DcuS (residues 202 to 325); and the PASc and His-kinase domains, PAScKin-DcuS (residues 202 to 543). For this purpose, the corresponding regions of dcuS were PCR amplified by using Pfu DNA polymerase, with pPG2 (10) as the template, and were cloned into the XmnI and HindIII sites of pMal-c2, generating pPG31, pPG26, and pPG27, respectively. DcuR and the C-terminal DNA-binding domain (cDcuR; residues 124 to 234) of DcuR were overproduced and purified as C-terminally His-tagged fusion proteins (the overproduced proteins included two additional amino acids, Leu-Glu, just before the His6 tag) by using pET-21a-derived plasmids (pPG25 and pPG39) containing corresponding PCR-amplified NdeI-XhoI fragments.
Overproduction and purification of DcuRHis, cDcuRHis, MalE-Kin-DcuS, MalE-PASc-DcuS, and MalE-PAScKin-DcuS.
The overproduction of His-tagged proteins was achieved by using corresponding E. coli BL21(λDE3) transformants grown in L broth (with 100 μg of ampicillin/ml and 1 mM IPTG [isopropyl-β-d-thiogalactopyranoside]) at 30°C. The proteins were purified as previously described (30) by using His-bind resin (Novagen) according to the manufacturer's instructions. Purified DcuRHis or cDcuRHis was subsequently transferred into storage buffer by dialysis (20 mM bis-Tris, 50 mM KCl, 5 mM dithiothreitol [DTT], and 10% glycerol, pH 7.5) and maintained at −80°C. The MalE-DcuS fusions were overproduced from DH5α transformants as described above (except for the use of 0.3 mM IPTG) and were purified from soluble extracts (as described above) prepared in binding buffer 1 (20 mM HEPES, 200 mM NaCl, 1 mM EDTA, 1 mM DTT, 1 mM benzamidine, and 10% glycerol, pH 7.4) by use of an amylose resin (New England Biolabs) column according to the manufacturer's instructions. The MalE fusions were further purified on a DEAE-Sepharose (Pharmacia) column, with buffer 2 (20 mM bis-Tris, 1 mM DTT, 10% glycerol, and 0.1% Triton X-100, pH 7.5) as the binding buffer and a 0 to 1 M NaCl gradient in buffer 2 as the elution buffer.
Cloning DNA fragments for DcuR-binding analysis.
A 709-bp fragment containing the dcuR-dcuB intergenic region was PCR amplified by using primers dcuBAf and dcuBAr (5′-ccgaattcGGCTGGTCAACTGCCACATCTTGTT-3′ and 5′-ccgaattcACATGTGTGAACCCTCGCGA-3′, respectively; mismatches are shown in lowercase and restriction sites are shown in bold) and was cloned into pUC118 to generate the construct pPG28. Subfragments of 215 and 365 bp were PCR amplified from pPG28 by using primers dcuBBf and dcuBBr (5′-ccgaattcACCGTAATGCAAAATACCCCTGGC-3′ and 5′-cccccgggACCCATTTGTGGGTAAAAATACACA-3′) or dcuBCf (5′-ccgaattcACAAATGGGTAGATCAGATT-3′) and dcuBAr and were cloned into the corresponding sites of pUC118 to generate pPG32 and pPG30, respectively.
The following other fragments were PCR amplified (with corresponding primers) and cloned (see Table 1) for DcuR-binding analysis: a 376-bp yjeA-frdA intergenic fragment (frdAAf, 5′-ccgaattcAGGATGCCGTTTCGCTCATAGTTAA-3′; frdAAr, 5′-ccgaattcGCTTGAAAGGTTTGCAC-3′), a 324-bp f651-dctA intergenic fragment (dctAf, 5′-ccgaattcAAGGCGTGGAGACTGAAGCA-3; dctAr, 5′-cccccgggAGAGAGGTTTTCATAGGGTGTCC-3′), a 402-bp yjdI-dcuS intergenic segment (dcuSlacZf, 5′-ccggatccCCGTTGCCACGTACGCAATTGCCAGAA-3′; dcuSlacZr, 5′-ccgaattcGCACCGAGAACAGTACCGCACTGACCA-3′), a 618-bp rpsV-maeA intergenic fragment (sfcAf, 5′-ccgaattcTTGTTTTTGGTTCCATGTCAC-3′; sfcAr, 5′-ccgaattcATTCAACAGCAGAACCCGCT-3′), and a 631-bp balA-maeB intergenic segment (maeBf, 5′-ccgaattctGAGTAGGCCAGCGCCAGAT-3′; maeBr, 5′-ccgaattccACCTGTTGTTCCTGGGTCT-3′).
Gel retardation assays.
DNA fragments of the dcuB, dctA, frdA, dcuS, maeA, and maeB operator-promoter regions or control fragments (300-bp regions of the fimZ and b0501 genes) were radiolabeled with [α-32P]dATP by use of the Klenow fragment of DNA polymerase. Unincorporated nucleotides were removed by using Nick Spin columns (Amersham Pharmacia Biotech) or Bio-Rad P-30 columns. Radiolabeled DNA fragments (1 nM) were incubated with various amounts of either DcuR or cDcuR (up to 100 μM) in DNA-binding buffer [50 mM Tris-HCl (pH 7), 5 mM MgCl2, 50 mM KCl, 0.1 mM EDTA, 1 mM DTT, 50 ng of poly(dI-dC), and 10% glycerol] at room temperature for 30 min and then were subjected to electrophoresis in a 6% polyacrylamide gel as previously described (30). The gel was then dried and subjected to autoradiography. The phosphorylation of DcuR with acetylphosphate was achieved by incubation of ∼1 mg of DcuR/ml with 50 mM acetylphosphate at 37°C for 1 h.
DNase I footprint assays.
Double-stranded DNA (∼200 bp) for DNase I footprinting was labeled at the 5′ terminus of one strand only by PCR amplification using two oligonucleotide primers, one of which was 5′ end labeled with [γ-32P]ATP by use of T4 polynucleotide kinase. DNase I footprinting was performed as described previously (30) by using serial dilutions of DcuR incubated with labeled target DNA (∼5 ng) in footprinting buffer [50 mM Tris-HCl (pH 7.5), 50 mM KCl, 10 mM MgCl2, 1 mM EDTA, 5% glycerol, and 1 μg of poly(dI-dC)/ml] in a total volume of 50 μl at room temperature for 30 min.
Dissection of the dcuB operator-promoter region.
Sections of the dcuB operator-promoter region were PCR amplified by using primers designed to generate the fragments indicated in Fig. 1B and C. For cloning purposes, the forward and reverse primers introduced flanking EcoRI and HindIII sites, respectively, where necessary. The resulting fragments were cloned into pRS1274 or pRS528, producing multicopy dcuB-lacZ transcriptional fusions (Table 1 and Fig. 1). The fusion plasmids were transferred into E. coli strain MC4100, and the resulting transformants were grown anaerobically, with or without 40 mM fumarate, in M9 medium supplemented with 0.4% glycerol and 40 mM trimethylamine oxide (TMAO). β-Galactosidase activity was determined as previously described (11).
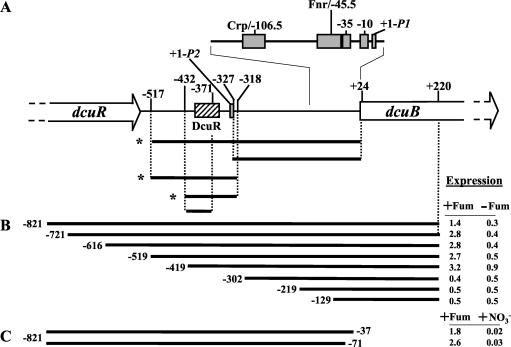
FIG. 1.
DcuR-dependent regulation of the dcuB operator-promoter region. (A) The dcuB operator-promoter region is shown together with a summary of gel retardation experiments performed with DcuR and the indicated DNA fragments. Asterisks indicate those fragments that were specifically retarded by DcuR. The coordinates for each fragment are given with respect to the previously defined +1 site (11). The promoter and predicted Fnr and Crp sites are indicated by gray boxes. The experimentally determined location of the DcuR-binding site is indicated by the hatched box. (B and C) The indicated segments of the dcuB operator-promoter region were used to construct multicopy dcuB-lacZ transcriptional fusions (Table 1; also see Materials and Methods for details). Expression (in micromoles of o-nitrophenyl-β-d-galactopyranoside per minute per gram of protein) was tested anaerobically in M9 medium containing glycerol, Casamino Acids, and TMAO, with (+) or without (−) fumarate (for panel C, the fumarate-free medium included nitrate).
Reverse-transcriptase-mediated primer extension.
The distal transcriptional start site of the dcuB gene was mapped, as previously described (11), by using four ∼25-mer primers (P53, P129, P204, and P278) designed to anneal to the dcuB transcript such that their 5′ termini are positioned 53, 129, 204, and 278 nucleotides, respectively, upstream of the proximal (P1) transcript start site. Total RNAs were prepared by the sodium dodecyl sulfate (SDS)-hot phenol method (1) from MC4100(pMA1) grown anaerobically to log phase in L broth containing 0.4% glycerol and 40 mM fumarate.
DcuS phosphorylation assays.
For phosphorylation assays, purified MalE-Kin-DcuS or MalE-PAScKin-DcuS (5 μM) was incubated at room temperature in kinase buffer (50 mM Tris-HCl [pH 7], 5 mM MgCl2, 50 mM KCl, 0.1 mM EDTA, 1 mM DTT, and 10% glycerol). The autophosphorylation reaction was initiated by the addition of 50 μM [γ-32P]ATP (2,000 mCi/mmol; Amersham Pharmacia Biotech), and samples were withdrawn at regular time intervals, mixed with 2× SDS digestion buffer (124 mM Tris-HCl [pH 7], 20% glycerol, 10% SDS, 0.01% bromophenol blue), and placed on ice. Samples were briefly incubated at 37°C and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and autoradiography.
Phosphotransfer experiments.
MalE-Kin-DcuS or MalE-PAScKin-DcuS (generally 20 μM) was phosphorylated (as described above), and DcuR was added to provide a 1:4 ratio of DcuS to DcuR (unless otherwise stated). Samples were removed at intervals, mixed with 2× SDS digestion buffer, and then electrophoresed. Gels were then dried and autoradiographed.
RESULTS
DcuR specifically binds to a site located 318 to 432 bp upstream of the previously defined dcuB-fumB promoter (P1dcuB).
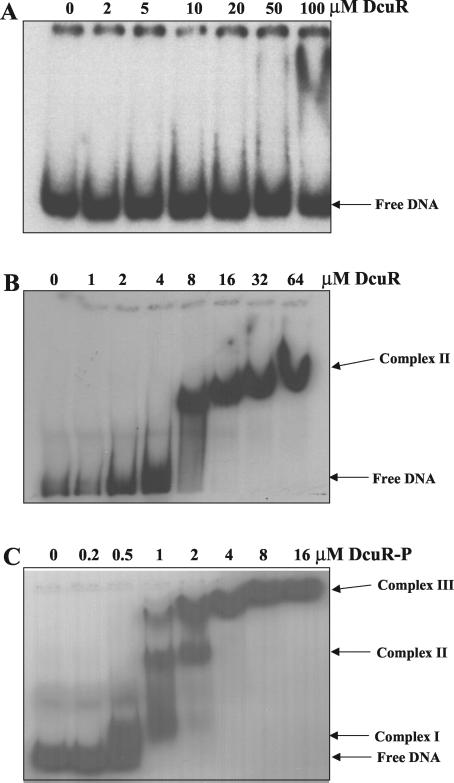
The DcuR protein was overproduced as a C-terminally His6-tagged protein and was purified by Ni2+-affinity chromatography (see Materials and Methods). SDS-PAGE analysis showed that the DcuR polypeptide had the anticipated molecular mass (28.5 kDa), and analytical gel permeation chromatography indicated that the (unphosphorylated) DcuRHis protein is monomeric, since it eluted with an apparent molecular mass of ∼30 kDa (see below). The ability of DcuR to specifically interact with DNA was tested by gel retardation analysis with various fragments of the dcuB promoter region (summarized in Fig. 1A). DcuR was found to specifically bind to a region of DNA located 318 to 432 bp upstream of the known dcuB promoter (P1dcuB) with an apparent affinity of around 6 μM (Fig. 1A and 2). No retardation of negative-control DNA fragments (from the fimZ and b0501 genes) was observed with DcuR concentrations up to 64 μM, indicating that the observed binding of DcuR to the dcuB operator-promoter region is specific. Surprisingly, no specific binding could be detected in the proximal dcuB promoter region (+24 to −327) (Fig. 2A), suggesting that a second dcuB promoter, located upstream of P1dcuB, is required for the DcuSR-dependent induction of dcuB transcription.
FIG. 2.
Gel retardation of the operator-promoter region of dcuB by DcuR. The results of gel retardation analysis of DcuR (A and B) and acetylphosphate-treated DcuR (C) with the −327 to +24 (A) and −517 to −318 (B and C) fragments of the dcuB promoter-operator region are shown. Similar gel retardation studies with −517 to +24, −432 to −318, and −432 to −371 dcuB operator-promoter fragments were performed (data not shown), and the results are summarized in Fig. 1A. The mobility positions of the free DNA and various DcuR-DNA complexes are indicated. For DcuR-P, the concentrations given are for the protein at the beginning of the phosphorylation reaction. The final concentrations of DcuR-P are likely to be lower due to precipitation induced by the acetylphosphate treatment.
DNA-binding affinity of DcuR is enhanced by treatment with acetylphosphate.
A semiquantitative Western blot analysis of DcuR levels in E. coli (results not shown) indicated that the DcuR monomer has a concentration of ∼0.7 to 1.7 μM, which is 4- to 10-fold lower than the apparent affinity of DcuR for the dcuB upstream region. This low affinity presumably results from the monomeric assembly status of unphosphorylated DcuR. In order to raise the affinity of DcuR for its DNA-binding site to physiologically relevant levels, we treated the protein with acetylphosphate, which acts as a substrate for the autophosphorylation of many response regulators (22, 23, 37). Acetylphosphate-treated DcuR (phospho-DcuR) exhibited a marked increase in affinity for the dcuB promoter region (from a KD of 6 μM to a KD of ≤1 μM) (Fig. 2C). This binding affinity is consistent with the estimated concentration of DcuR in vivo, suggesting that the phosphorylation of DcuR is a prerequisite for the induction of dcuB transcription by DcuSR. Retardation of the proximal 351-bp fragment (+24 to −327) required high concentrations (32 μM) of phospho-DcuR (data not shown), indicating that the binding of phospho-DcuR to this fragment is nonspecific. At least three retardation complexes (I to III) were observed with the acetylphosphate-treated protein (Fig. 2C), but only one (complex II) was observed with the untreated protein (Fig. 2B). This difference may reflect the lower binding affinity of untreated DcuR.
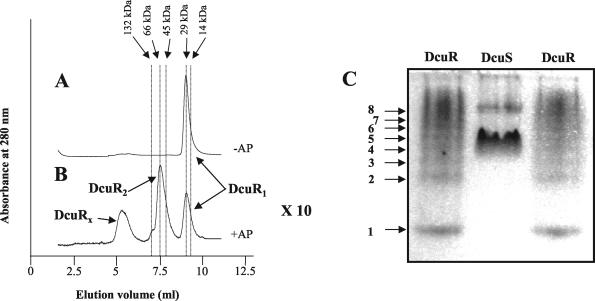
The effect of acetylphosphate treatment on the assembly status of DcuR was investigated by analytical gel permeation chromatography. DcuR incubated in the absence of acetylphosphate had an elution volume that was consistent with a monomeric assembly state (Fig. 3A). Incubation with acetylphosphate resulted in the precipitation of much (∼90%) of the DcuR, as is typically found for response regulators. However, most of the DcuR remaining in solution had an apparent molecular mass of ∼60 to 70 kDa, consistent with a homodimer (Fig. 3B). This suggests that acetylphosphate causes DcuR multimerization. The loss of DcuR solubility caused by acetylphosphate treatment suggests that the estimated KD of ≤1 μM for phospho-DcuR is likely to be higher than the true value. Interestingly, DcuR appeared to oligomerize upon native PAGE (Fig. 3C), suggesting that DcuR has the capacity to form higher order oligomers under certain conditions.
FIG. 3.
Assembly status of DcuR. Elution profiles of acetylphosphate-treated (B) and untreated (A) DcuR during analytical gel permeation chromatography through a BioSelect-125 column (Bio-Rad). The eluant was phosphorylation buffer (see Materials and Methods), and the flow rate was 0.5 ml/min. The arrows at the top indicate the elution volumes of the native molecular mass markers (range, 14,000 to 5,000,000 Da; Sigma) that were used to standardize the column. Monomeric, dimeric, and high-molecular-weight (DcuRx) forms of DcuR are indicated. Note that acetylphosphate treatment resulted in the precipitation of ∼90% of the DcuR, so the scale of the corresponding elution profile has been increased 10-fold for panel B with respect to panel A (×10). (C) Native PAGE (10%) of DcuR (and DcuS). The gel was stained with Coomassie blue. Arrows indicate different oligomeric forms of DcuR (1 to 8). The high-mobility form (1) is presumed to be a monomer.
The cis element conferring DcuSR dependence on dcuB expression is positioned 302 to 419 bp upstream of P1dcuB.
For confirmation that the observed DcuR-binding site is responsible for DcuSR-dependent regulation of the dcuB gene, eight multicopy dcuB-lacZ transcriptional fusions containing truncations at the distal end of the dcuB operator-promoter region were constructed (Fig. 1B). The effect of fumarate on the expression of the dcuB-lacZ fusions was measured after growth under anaerobic conditions in minimal medium containing glycerol, Casamino Acids, and TMAO (Fig. 1B). Those fusions that included the dcuB operator-promoter region downstream of position −419 were induced approximately fivefold by fumarate (similar levels of fumarate induction for chromosomally integrated dcuB-lacZ were observed previously [11], whereas those fusions that lacked the region upstream of position −302 were not affected by fumarate. This indicates that the cis element conferring the response to fumarate (and DcuR) is located in the −419 to −302 region of the dcuB promoter. This position is consistent with that determined by the gel retardation studies, and the data localize the DcuR-binding site to a 102-bp region 318 to 419 bp upstream of the defined dcuB promoter.
Identification of a second dcuB transcriptional start site.
The presence of an additional promoter, located upstream of P1dcuB, was tested by the production of two further multicopy dcuB-lacZ fusions. These fusions contain the −821 to −37 and −821 to −71 regions of the dcuB operator-promoter and therefore lack P1dcuB (Fig. 1C). The absence of P1dcuB had no major effect on the expression of the dcuB-lacZ fusions, and the expression levels of the fusions remained high in the presence of fumarate, confirming that the dcuB gene possesses an additional transcriptional start site positioned upstream of P1dcuB and under DcuSR-dependent control. The lower level of expression in the absence of fumarate shown in Fig. 2C than that shown in Fig. 2B is due to the inclusion of nitrate in place of fumarate and is consistent with previous results (11).
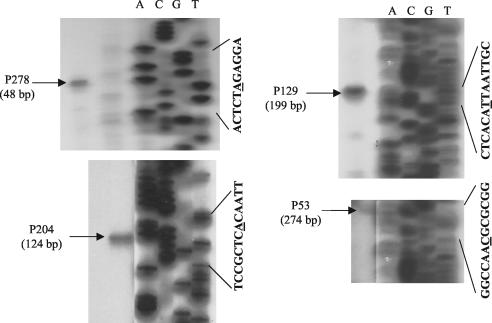
For determination of the position of the upstream promoter (P2dcuB), the transcriptional start site was mapped by reverse-transcriptase-mediated primer extension. Four primers were used for this purpose (see Materials and Methods). All four primers gave extension products that correspond to a distal transcript start site (P2dcuB) located 327 nucleotides upstream of the previously mapped proximal site (Fig. 4; also Fig. 1A and 5A). This +1 site is associated with an appropriately positioned −10 box that matches the consensus sequence well (at four of six nucleotides; Fig. 5A). However, only a relatively poor −35 site, located within the DcuR-binding site, was found for the distal promoter (described below). This suggests that the DcuR-binding site may compensate for the weak −35 site, allowing the otherwise weak promoter to be highly activated by DcuR. This might explain the relatively strong DcuR dependence of dcuB transcription (induced up to 160-fold) (10).
FIG. 4.
Determination of the distal transcriptional start site (P2dcuB) of dcuB by reverse-transcriptase-mediated primer extension. The reverse-transcriptase-mediated primer extension products were generated by using primers (P53, P129, P204, and P278) specific for the dcuB transcript (see Materials and Methods for details). Sequencing ladders (lanes A, C, G, and T) were generated by using the M13 universal primer and M13mp18 as the template. The relevant M13 nucleotide sequences (the underlined residues are those whose corresponding reaction products matched the size of the reverse transcription product) and the sizes of the primer extension products are shown.
FIG. 5.
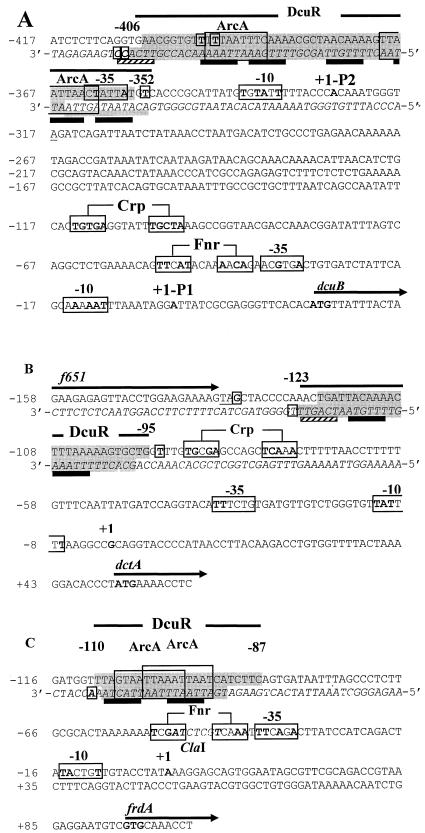
Nucleotide sequences of the dcuB (A), dctA (B), and frdA (C) promoter-operator regions. The DcuR-protected regions are highlighted in dark gray (sequences for both strands are presented, with the noncoding strand in italics), with a weakly protected region underscored with a gray bar and adjacent areas that are potentially protected but were not subject to DNase I cleavage highlighted in light gray. Hypersensitive sites are shown in bold and enclosed in small boxes. Tandemly repeated (T/A)(A/T)(T/C)(A/T)AA sequences possibly corresponding to a DcuR-binding motif are indicated with black bars or, for weak matches, striped bars. Transcription and translation start sites are in bold, and promoters and potential binding sites for Crp and Fnr are in large boxes with residues matching the Crp and Fnr consensus sequences shown in bold. The f651 stop codon is underlined. Sequences within the DcuR-binding regions closely resembling the ArcA-binding consensus [(A/T)TAATTAAC(A/T)] are boxed. Numbering corresponds to distances from the transcriptional start site. A relevant restriction site is indicated in italics.
Definition of the DcuR-binding site in the dcuB operator-promoter region by DNase I footprinting.
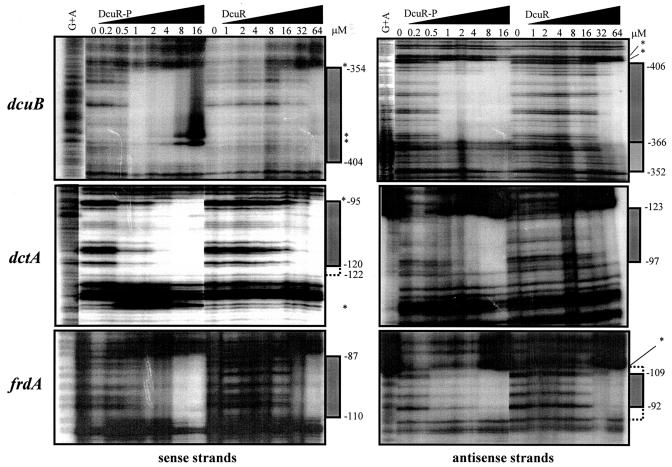
The precise location of the DcuR-binding site in the dcuB operator-promoter region was determined by use of a DNase I footprint assay. Both phospho-DcuR and non-acetylphosphate-treated DcuR protected the same well-defined region of DNA on both the coding and noncoding strands (Fig. 5 and 6). As anticipated, relatively low concentrations (1 μM) of phospho-DcuR were required to achieve protection, whereas much higher concentrations (32 μM) of non-phospho-DcuR were necessary.
FIG. 6.
DNase I footprint analysis of the dcuB, dctA, and frdA promoter-operator regions with phosphorylated and unphosphorylated DcuR. The corresponding DNA fragments were PCR amplified by using a labeled forward or reverse primer to enable radiolabeling of the sense and antisense strands, respectively. The concentrations of DcuR subunit employed are shown, as are the lanes containing the G+A ladder. The regions of protection are indicated by bars on the right side of the autoradiographs (the lightly shaded bar indicates a region that is partially protected), and hypersensitive bands are highlighted with asterisks. Broken bars indicate regions that may be protected but were not cleaved by DNase I. The corresponding coordinates are distances (in base pairs) from the proximal transcription start site (see Fig. 5). For an explanation of DcuR-P concentrations, see the legend for Fig. 2.
The protected region spans positions −406 to −352, encompassing 55 bp of DNA (Fig. 5A). This location is consistent with that indicated by gel retardation and dcuB-lacZ analyses (−419 to −318) (Fig. 1A). The −366 to −352 region on the antisense strand appeared to be less well protected than the corresponding region on the opposite strand (Fig. 6). This may result from weaker binding of DcuR to this region. High concentrations of phospho-DcuR or non-phospho-DcuR caused the appearance of five DNase I hypersensitive sites, with two adjacent sites within the protected region and three sites just outside the DcuR-binding site (Fig. 5 and 6). This suggests that DcuR binding causes a distortion of the DNA duplex at these sites.
Interaction of DcuR with the dctA promoter region.
The expression of the dctA gene is also DcuSR dependent, so the possibility that this gene, like dcuB, is under direct DcuSR control was also examined by gel retardation and DNase I footprint analyses. Gel retardation studies showed that a 324-bp EcoRI-ClaI fragment that includes the entire f651-dctA intergenic region upstream of dctA is specifically retarded by DcuR (not shown). Retardation studies using a smaller, 207-bp fragment (−196 to +11) from within the 324-bp fragment showed that non-phospho-DcuR and phospho-DcuR bind specifically to the dctA operator-promoter region, with apparent affinities of ∼6 and ∼1 μM, respectively (not shown). These values match those obtained with dcuB, indicating that these two promoters have similar affinities for DcuR. DcuR protected a 29-bp region of the dctA promoter at corresponding positions on the sense (−120 to −95) and antisense (−123 to −97) strands (Fig. 5B and 6). This binding site is about half the size of that determined for the dcuB promoter (55 bp), suggesting that the dcuB promoter may bind approximately twice as much DcuR as the dctA promoter.
Phospho-DcuR provided partial protection at 0.5 to 4 μM, with full protection being achieved at ≥8 μM. Non-acetylphosphate-treated DcuR was required at a higher concentration (64 μM) for full protection. As for the dcuB promoter, DcuR binding resulted in the formation of DNase I hypersensitive sites at positions flanking the region of protection (Fig. 5B and 6).
Interaction of DcuR with the frdABCD operator-promoter region.
The only other known DcuSR-controlled promoter in E. coli is that of the frdABCD operon. A gel retardation analysis showed that DcuR specifically binds to the 366-bp EcoRI frdA operator-promoter fragment containing the entire yieA-frdA intergenic region (not shown). When this fragment was divided into two subfragments, the promoter-distal 203-bp EcoRI-ClaI (−245 to −44) fragment bound DcuR specifically, whereas the 163-bp ClaI-EcoRI (−47 to +116) fragment carrying the frdA promoter did not (not shown). A 236-bp fragment (−206 to +30) containing the frdA promoter, the ClaI site, and part of the 203-bp EcoRI-ClaI fragment (containing the DcuR-binding site) was retarded by both non-phospho-DcuR and phospho-DcuR, with approximate KD values of 6 and 1 μM (not shown), as was observed for the dcuB and dctA promoters. The DcuR-binding site is therefore located between positions −206 and −44 in the frdA promoter.
In DNase I footprinting experiments, non-phospho- and phospho-DcuR protected the same 24-bp region of DNA (−110 to −87) on both the coding and noncoding strands. Phospho-DcuR provided partial protection at a concentration of 0.5 μM and full protection at ≥8 μM (Fig. 6). Unphosphorylated DcuR showed no sign of protection until concentrations were ≥32 μM. DcuR binding induced the formation of an apparent DNase I hypersensitive site at position −111 on the antisense strand, just outside the protected region.
The C-terminal domain of DcuR binds to the dcuB, dctA, and frdA operator-promoter regions.
To determine whether the C-terminal domain of DcuR confers DNA-binding ability (as anticipated), a DcuR fragment (cDcuR) containing just the C-terminal domain (residues 124 to 234) was produced (see Materials and Methods) and tested by gel retardation analysis. cDcuR specifically retarded DNA fragments containing the dcuB, dctA, and frdA operator-promoter regions, with apparent binding affinities similar to those obtained for the full-length unphosphorylated DcuR protein (data not shown). This finding confirms that the C-terminal domain provides the DNA-binding capability exhibited by DcuR and is thus consistent with previous work on the isolated C-terminal domain of CitB (24).
DcuR does not specifically interact with the dcuS, maeA, and maeB operator-promoter regions.
The possibilities that the dcuS-dcuR genes are autoregulated and that the genes (maeA and maeB) encoding the two malic enzymes (interconverting malate and pyruvate) are transcriptionally controlled by DcuSR were tested by gel retardation analysis (the malic enzyme gene, ywkA/maeA, of B. subtilis is known to be malate induced [6]). However, the ∼400-, 620-, and 630-bp intergenic regions upstream of dcuS, maeA, and maeB (see Materials and Methods) failed to interact specifically with the DcuR protein (data not shown), suggesting that these genes are unlikely to be directly controlled by the DcuSR system. We note that the global transcriptional profiling data of Oshima et al. (26) also indicate that the maeA and maeB genes are not subject to DcuSR regulation.
Autophosphorylation of DcuS and phosphotransfer to DcuR.
Three versions of DcuS were overproduced as MalE N-terminal fusions: the PASc kinase domain (MalE-PAScKin), the kinase domain (MalE-Kin), and the PASc domain (MalE-PASc). All had molecular masses close to those anticipated (64, 72, and 55 kDa), as estimated by SDS-PAGE. Analytical gel permeation chromatography indicated that the MalE-PAScKin protein had a native molecular mass of ∼130 kDa, suggesting that the protein is a dimer (not shown), as was found for other sensor kinases (33). No evidence for any prosthetic group or metal associated with any of the DcuS subproteins was obtained by either UV visible spectroscopy or inductive coupled plasma mass spectroscopic analysis.
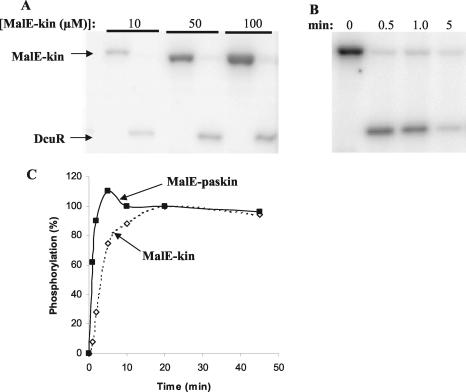
The MalE-PAScKin and MalE-Kin proteins both exhibited autophosphorylation activity upon incubation with [γ-32P]ATP. MalE-PAScKin had an approximately fourfold higher apparent autophosphorylation activity level (initial rate) than the MalE-Kin protein, suggesting that the presence of the PASc domain enhances autophosphorylation activity or possibly causes a lower level of autophosphatase activity (Fig. 7). However, unlike that of the full-length DcuS protein (14), the activity of neither MalE-Kin nor MalE-PAScKin was significantly affected by the presence of succinate or fumarate (data not shown), suggesting that the C4-dicarboxylate responsiveness of DcuS is conferred by the N-terminal periplasmic domain. The phosphorylated forms of the MalE-PAScKin and MalE-Kin proteins were able to act as phosphorus donors for the autophosphorylation of DcuR (Fig. 7 and data not shown). The rate of phospho-group transfer was very high (virtually complete within 30 s). However, after autophosphorylation, DcuR underwent rapid dephosphorylation and was mostly (>90%) dephosphorylated within 5 min (Fig. 7). This apparent phosphatase activity may be due to the intrinsic instability of the presumed aspartylphosphate residue (20, 39) or could be mediated by the dephosphorylated form of DcuS, as observed for other transmitter kinases (25, 35).
FIG. 7.
Autophosphorylation of the DcuS-kinase domain and phosphotransfer to DcuR. (A and B) Phosphorimager-generated autoradiographs of phosphorylated DcuS (MalE-Kin) and DcuR separated by SDS-PAGE (reactions performed at 25°C). DcuS was phosphorylated with [γ-32P]ATP prior to the addition of DcuR. (A) DcuR (5 μM) was added to phosphorylated DcuS at time zero, resulting in the complete dephosphorylation of DcuS within 1 min and concomitant phosphotransfer to DcuR. The level of DcuR phosphorylation increased with increasing concentrations of phosphorylated DcuS. (B) DcuR (5 μM) was added to 20 μM phosphorylated DcuS at time zero. After the initial rapid phosphotransfer from DcuS to DcuR, the phosphorylated form of DcuR dephosphorylated, resulting in the almost complete dephosphorylation of DcuR after a 5-min incubation. (C) Graph comparing the relative rates of autophosphorylation for MalE-Kin and MalE-PAScKin at 25°C. Reactions for the two DcuS proteins were performed in the same tube, and the degree of phosphorylation was determined (as a percentage of the level achieved at 20 min) after the separation of the two proteins by SDS-PAGE. Note that the two proteins incorporated closely similar amounts of 32P after 20 and 45 min of incubation.
DISCUSSION
The results presented here indicate that the DcuSR two-component sensor-regulator system directly induces the transcription of the dcuB-fumB, dctA, and frdABCD genes of E. coli through specific interactions of DcuR with the corresponding operator regions. The results are in general agreement with those of Janausch et al. (14), who showed that the phosphorylated form of DcuR specifically binds to a 646-bp fragment of the dcuB promoter at a protein concentration of 0.7 μM and that the unphosphorylated protein fails to bind at concentrations below 1.5 μM. We found that the treatment of DcuR with acetylphosphate results in the conversion of DcuR from a monomer with a low affinity for its DNA-binding site into a homodimer with a relatively high DNA-binding affinity. It is likely that acetylphosphate acts as an autophosphorylation substrate for DcuR, as reported for other response regulators (21) and for the closely related CitB protein (24). The increase in affinity achieved upon acetylphosphate treatment ranged from 6- to 32-fold, depending on the promoter examined and the method (retardation or footprinting assay) employed. It is likely that the true increase in affinity achieved upon DcuR phosphorylation is higher than this since DcuR may not be fully phosphorylated by acetylphosphate and acetylphosphate treatment resulted in a loss of DcuR due to precipitation.
In vivo, the DcuSR-dependent induction of gene expression is likely to require the phosphorylation and consequent dimerization of DcuR in order to raise the binding affinity of DcuR to physiologically relevant levels. Some response regulators oligomerize upon phosphorylation and then bind to DNA in their oligomerized forms (e.g., NtrC and UhpA) (38, 40, 41). DcuR is also able to form oligomers, as was observed by native PAGE. However, it is unclear whether such oligomers are relevant to the interaction of DcuR with its DNA-binding site.
The three DcuR-binding sites are located 27, 96, and 87 bp upstream of their corresponding dcuB, dctA, and frdA promoters, respectively. The locations in the dctA and frdA promoters are typical for transcriptional activators that promote transcription through interactions with the flexible C-terminal domain of the α-subunit of RNA polymerase (29), as has been shown to be the case for the response-regulator OmpR (18). Alternatively, DcuR could function as an activator at these promoters by interacting with other activators, such as Crp at the dctA promoter or Fnr at the frdA promoter. For dcuB, the DcuR-binding site extends into the anticipated position for the −35 box of the corresponding P2 promoter. This suggests that DcuR may act as a class II activator at the dcuB P2 promoter whereby the DcuR-binding site replaces the −35 region. The expression of dcuB is strongly dependent on Fnr as well as DcuSR (11). However, it is unclear how the Fnr effect is mediated, since although a potential Fnr site was observed for the P1 promoter (11), given the apparent lack of contribution of the P1 promoter to dcuB-lacZ expression (Fig. 1C) revealed here, its relevance is now unclear.
All three DcuR-binding sites are highly enriched in A+T residues (average A+T content of 77%, in contrast to the ∼50% average ratio for the whole genome). In this respect, the binding sites match those of the closely homologous CitB and CitT (76 and 82% A+T-rich, respectively) proteins from K. pneumoniae and B. subtilis (24, 42). The high-affinity CitT binding site contains a tandemly repeated (A/T)(A/T)CAAA hexanucleotide motif (42) which appears six times in the 50-bp DNase I protected region, with individual repeats separated by a spacing of 2 to 4 bp. A similar tandem hexanucleotide repeat, (A/C)ATAAA, can be observed in the 47- and 35-bp binding sites of CitB, which are repeated eight times in total and also have a 2- to 4-bp space between repeats (derived from reference 24). Tandemly repeated hexanucleotide sequences have also been observed in the binding sites of other response-regulators, such as PhoP of B. subtilis, in which these sequences have been shown to be essential for PhoP-dependent regulation (7). An inspection of the three DcuR-binding sites showed a similar directly repeated hexameric sequence, (T/A)(A/T)(T/C)(A/T)AA (preferred bases are shown in bold; see Fig. 5), which could act as a binding motif for DcuR. The spacing is not as regular as for the CitT and CitB motifs, ranging from 1 to 6 bp.
Sequences closely matching the ArcA-binding site consensus sequence are present in two (dcuB and frdA) of the three sites. Although the corresponding genes are anaerobically induced, this induction is reported to be mainly Fnr, rather than ArcA, dependent (11, 15). ArcA represses anaerobic gene expression in E. coli (9). It is unclear whether the ArcA-binding sequences in the DcuR-protected regions are a consequence of similarities between the consensus binding sequences for ArcA and DcuR or whether there could be competition between ArcA and DcuR at these sites.
The stoichiometry of DcuR binding to DNA at the three sites studied here is unclear. Gel retardation analysis resolved two to three DcuR-DNA complexes for each site, but there was no obvious correlation between the number of complexes observed and the size of the DNase I protected region. The closely related CitB protein also forms a series of retardation complexes (four in total) during the interaction of the phosphorylated form with two adjacent binding sites of 82 bp (24), but the stoichiometry of these complexes is also unknown. Other response-regulators have minimum DNase I footprints of about 30 bp. For instance, PhoP protects a 29-bp region (7). This protection is thought to be achieved by the binding of a single PhoP dimer. It would therefore appear reasonable to infer that the smaller DcuR-binding sites (of 24 and 29 bp) occupy a single DcuR dimer, whereas the 55-bp site would contain the equivalent of two DcuR dimers. Each dimer would be anticipated to bind to two adjacent hexanucleotide direct repeats, although other features, such as base composition (A+T richness) and/or the conformation of the DNA duplex, may contribute to the DcuR-binding site.
The isolated kinase and PAS-kinase domains of DcuS possess autokinase activity and are able to act as substrates for the rapid transphosphorylation of DcuR. These findings are consistent with the study of Janausch et al. (14) showing that the complete DcuS protein (including the N-terminal periplasmic sensor domain) autophosphorylates when it is reconstituted in liposomes. However, unlike the complete DcuS protein, the autokinase activity of the isolated kinase and PAS-kinase domains was not enhanced by C4-dicarboxylates. This supports the notion that the N-terminal domain of DcuS is the C4-dicarboxylate sensor domain and is consistent with studies of the periplasmic domain of CitA showing that it directly interacts with citrate (16, 17). Given the rapid autophosphorylation activity of the isolated kinase and PAS-kinase domains, it is probable that in the intact DcuS protein the periplasmic domain acts to inhibit the activity of the autokinase domain in the absence of C4-dicarboxylates. Other sensor kinases also exhibit constitutive autokinase activity when they are N-terminally truncated to remove their sensor domains (4).
The role of the PASc domain has not been resolved, although it does appear to improve the activity of the autokinase domain. Other PAS domains are involved in sensing signals such as light or redox. This capacity is enabled by an association with prosthetic moieties (e.g., 4-hydroxy cinnamoyl, flavin adenine dinucleotide, or heme [34]), although no evidence for any prosthetic group that might provide a clue for the role of the PASc domain was obtained with the overproduced PASc and PASc-kinase domains. It has been suggested that the PASc domain of DcuS may be involved in the modification of the activity of DcuS in response to cellular redox status (10) or intracellular metabolites such as C4-dicarboxylates, although no evidence for this was uncovered here. Since the PASc-kinase construct had a higher level of autokinase activity than the isolated kinase domain, it appears that the PAS domain does not inhibit the activity of the associated kinase domain.
Acknowledgments
We thank the BBSRC for a project grant to S.C.A. and D.J.K., the Egyptian Government for a studentship to A.E.A., and the Royal Society for financial support.
We also thank A. G. Cox and S. Thorpe (Chemistry Department, University of Sheffield) for ICP-MS analysis.
REFERENCES
- 1.Aiba, H., S. Adkya, and B. de Crombrugghe. 1981. Evidence for a functional gal promoter in intact Escherichia coli cells. J. Biol. Chem. 256:11905-11910. [PubMed] [Google Scholar]
- 2.Bartolomé, B., Y. Jubete, E. Martinez, and F. Delacruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 3.Bott, M., M. Meyer, and P. Dimroth. 1995. Regulation of anaerobic citrate metabolism in Klebsiella pneumoniae. Mol. Microbiol. 18:533-546. [DOI] [PubMed] [Google Scholar]
- 4.Bourret, R. B., K. A. Borkovich, and M. I. Simon. 1991. Signal transduction pathways involving protein phosphorylation in prokaryotes. Annu. Rev. Biochem. 60:401-441. [DOI] [PubMed] [Google Scholar]
- 5.Davies, S. J., P. Golby, D. Omrani, S. A Broad, V. L. Harrington, J. R. Guest, D. J. Kelly, and S. C. Andrews. 1999. Inactivation and regulation of the aerobic C4-dicarboxylate transport (dctA) gene of Escherichia coli. J. Bacteriol. 181:5624-5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doan, T., P. Servant, S. Tojo, H. Yamaguchi, G. Lerondel, K.-I. Yoshida, Y. Fujita, and S. Aymerich. 2003. The Bacillus subtilis ywkA gene encodes a maolic enzyme and its transcription is activated by the YufL/YufM two-component system in response to malate. Microbiology 149:2331-2343. [DOI] [PubMed] [Google Scholar]
- 7.Eder, S., W. Liu, and F. M. Hulett. 1999. Mutational analysis of the phoD promoter in Bacillus subtilis: implication for PhoP binding and promoter activation of the Pho regulon promoters. J. Bacteriol. 181:2017-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engel, P., R. Kramer, and G. Unden. 1992. Anaerobic fumarate transport in Escherichia coli by an fnr-dependent dicarboxylate uptake system which is different from aerobic dicarboxylate uptake. J. Bacteriol. 174:5533-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Georgellis, D., O. Kwon, and E. C. C. Lin. 2001. Quinones as the redox signal for the Arc two-component system of bacteria. Science 292:2314-2316. [DOI] [PubMed] [Google Scholar]
- 10.Golby, P., S. Davies, D. J. Kelly, J. R. Guest, and S. C. Andrews. 1999. Identification and characterization of a two-component sensor-kinase and response-regulator system (DcuS-DcuR) controlling gene expression in response to C4-dicarboxylates in Escherichia coli. J. Bacteriol. 181:1238-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golby, P., D. J. Kelly, J. R. Guest, and S. C. Andrews. 1998. Transcriptional regulation and organization of the dcuA and dcuB genes encoding homologous anaerobic C4-dicarboxylate transporters in Escherichia coli. J. Bacteriol. 180:6586-6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guest, J. R. 1992. Oxygen-regulated gene expression in Escherichia coli. J. Gen. Microbiol. 138:2253-2263. [DOI] [PubMed] [Google Scholar]
- 13.Janausch, I. G., E. Zientz, Q. H. Tran, A. Kroger, and G. Unden. 2002. C4-dicarboxylate carriers and sensors in bacteria. Biochim. Biophys. Acta 1553:39-56. [DOI] [PubMed] [Google Scholar]
- 14.Janausch, I. G., I. Garcia-Moreno, and G. Unden. 2002. Function of DcuS from Escherichia coli as a fumarate-stimulated histidine protein kinase in vitro. J. Biol. Chem. 277:39809-39814. [DOI] [PubMed] [Google Scholar]
- 15.Jones, H. M., and R. P. Gunsalus. 1987. Regulation of Escherichia coli fumarate reductase (frdABCD) operon expression by respiratory electron acceptors and the fnr gene product. J. Bacteriol. 169:3340-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaspar, S., and M. Bott. 2002. The sensor kinase CitA (DpiB) of Escherichia coli functions as a high-affinity citrate receptor. Arch. Microbiol. 177:313-321. [DOI] [PubMed] [Google Scholar]
- 17.Kaspar, S., R. Perozzo, S. Reinelt, M. Meyer, K. Pfister, L. Scapozza, and M. Bott. 1999. The periplasmic domain of the histidine autokinase CitA functions as a highly specific citrate receptor. Mol. Microbiol. 33:858-872. [DOI] [PubMed] [Google Scholar]
- 18.Kato, N., H. Aiba, and T. Mizuno. 1996. Suppressor mutations in alpha-subunit of RNA polymerase for a mutant of the positive regulator, OmpR, in Escherichia coli. FEMS Microbiol. Lett. 139:175-180. [DOI] [PubMed] [Google Scholar]
- 19.Kay, W. W., and H. L. Kornberg. 1971. The uptake of C4 dicarboxylic acids by Escherichia coli. Eur. J. Biochem. 18:274-281. [DOI] [PubMed] [Google Scholar]
- 20.Keener, J., and S. Kustu. 1988. Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NtrB and NtrC of enteric bacteria: roles of the conserved N-terminal domain of NtrC. Proc. Natl. Acad. Sci. USA 85:4976-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lukat, G. S., W. R. McCleary, A. M. Stock, and J. B. Stock. 1992. Phosphorylation of bacteria response regulator proteins by low molecular weight phospho-donors. Proc. Natl. Acad. Sci. USA 89:718-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCleary, W. R., and J. B. Stock. 1994. Acetyl phosphate and the activation of 2 component response regulators. J. Biol. Chem. 269:31567-31572. [PubMed] [Google Scholar]
- 23.McCleary, W. R., J. B. Stock, and A. J. Ninfa. 1993. Is acetyl phosphate a global signal in Escherichia coli? J. Bacteriol. 175:2793-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer, M., P. Dimroth, and M. Bott. 1997. In vitro binding of the response regulator CitB and of its carboxy-terminal domain to A+T-rich DNA target sequences in the control region of the divergent citC and citS operons of Klebsiella pneumoniae. J. Mol. Biol. 269:719-731. [DOI] [PubMed] [Google Scholar]
- 25.Ninfa, A. J., and B. Magasanik. 1986. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc. Natl. Acad. Sci. USA 83:5909-5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oshima, T., H. Aiba, Y. Masuda, S. Kannay, M. Sugiura, B. L. Wanner, H. Mori, and T. Mizuno. 2002. Transcriptional analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol. Microbiol. 46:281-291. [DOI] [PubMed] [Google Scholar]
- 27.Pappalardo, L., I. G. Janausch, V. Vijayan, E. Zientz, J. Junker, W. Peti, M. Zweckstetter, G. Unden, and C. Griesinger. 2003. The NMR structure of the sensory domain of the membranous two-component fumarate sensor (histidine protein kinase) DcuS of Escherichia coli. J. Biol. Chem. 278:39185-39188. [DOI] [PubMed] [Google Scholar]
- 28.Reinelt, S., E. Hofmann, T. Gerharz, M. Bott, and D. R. Madden. 2003. The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain. J. Biol. Chem. 278:39189-39196. [DOI] [PubMed] [Google Scholar]
- 29.Rhodius, V. A., and S. J. W. Busby. 1998. Positive activation of gene expression. Curr. Opin. Microbiol. 1:152-159. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 32.Six, S., S. C. Andrews, G. Unden, and J. R. Guest. 1994. Escherichia coli possesses two homologous anaerobic C4-dicarboxylate membrane transporters (DcuA and DcuB) distinct from the aerobic dicarboxylate transport system (Dct). J. Bacteriol. 176:6470-6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 34.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tokishita, S., H. Yamada, H. Aida, and T. Mizuno. 1990. Transmembrane signal transduction and osmoregulation in Escherichia coli. 2. The osmotic sensor, EnvZ, located in the isolated cytoplasmic membrane displays its phosphorylation and dephosphorylation abilities as to the activator, OmpR. J. Biochem. (Tokyo) 108:488-493. [DOI] [PubMed] [Google Scholar]
- 36.Vieira, J., and J. Messing. 1987. Production of single-stranded plasmid DNA. Methods Enzymol. 153:3-34. [DOI] [PubMed] [Google Scholar]
- 37.Wanner, B. L. 1995. Signal transduction and cross regulation in the Escherichia coli phosphate regulon by PhoR, CreC, and acetyl phosphate, p. 203-221. In J. Hoch and T. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, D.C.
- 38.Webber, C. A., and R. J. Kadner. 1997. Involvement of the amino-terminal phosphorylation module of UhpA in activation of uhpT transcription in Escherichia coli. Mol. Microbiol. 24:1039-1048. [DOI] [PubMed] [Google Scholar]
- 39.Weiss, V., and B. Magasanik. 1988. Phosphorylation of nitrogen I (NRI) of Escherichia coli. Proc. Natl. Acad. Sci. USA 85:8919-8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weiss, V., F. Claverie-Martin, and B. Magasanik. 1992. Phosphorylation of nitrogen regulator-I of Escherichia coli induces strong cooperative binding to DNA essential for activation. Proc. Natl. Acad. Sci. USA 89:5088-5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wyman, C., I. Rombel, A. K. North, C. Bustamente, and S. Kustu. 1997. Unusual oligomerization required for activity of NtrC, a bacterial enhancer-binding protein. Science 275:1658-1661. [DOI] [PubMed] [Google Scholar]
- 42.Yamamoto, H., M. Murata, and J. Sekiguchi. 2000. The CitST two-component system regulates the expression of the Mg-citrate transporter in Bacillus subtilis. Mol. Microbiol. 37:898-912. [DOI] [PubMed] [Google Scholar]
- 43.Zientz, E., J. Bongaerts, and G. Unden. 1998. Fumarate regulation of gene expression in Escherichia coli by DcuSR (dcuSR genes) two-component regulatory system. J. Bacteriol. 180:5421-5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zientz, E., S. Six, and G. Unden. 1996. Identification of a third secondary carrier (DcuC) for anaerobic C4-dicarboxylate transport in Escherichia coli: roles of the three Dcu carriers in uptake and exchange. J. Bacteriol. 178:7241-7247. [DOI] [PMC free article] [PubMed] [Google Scholar]