Abstract
Sugar fermentation was long considered the sole means of energy metabolism available to lactic acid bacteria. We recently showed that metabolism of Lactococcus lactis shifts progressively from fermentation to respiration during growth when oxygen and heme are available. To provide insights into this phenomenon, we compared the proteomic profiles of L. lactis under fermentative and respiratory growth conditions in rich medium. We identified 21 proteins whose levels differed significantly between these conditions. Two major groups of proteins were distinguished, one involved in carbon metabolism and the second in nitrogen metabolism. Unexpectedly, enzymes of the proteolytic system (PepO1 and PepC) which are repressed in rich medium in fermentation growth were induced under respiratory conditions despite the availability of free amino acids. A triple mutant (dtpT dtpP oppA) deficient in oligopeptide transport displayed normal respiration, showing that increased proteolytic activity is not an absolute requirement for respiratory metabolism. Transcriptional analysis confirmed that pepO1 is induced under respiration-permissive conditions. This induction was independent of CodY, the major regulator of proteolytic functions in L. lactis. We also observed that pepO1 induction is redox sensitive. In a codY mutant, pepO1 expression was increased twofold in aeration and eightfold in respiration-permissive conditions compared to static conditions. These observations suggest that new regulators activate proteolysis in L. lactis, which help to maintain the energetic needs of L. lactis during respiration.
The lactic acid bacteria are mainly studied for their capacities to ferment carbon sources, which results in acidification of the medium. It was therefore surprising to discover that some bacteria, including Lactococcus lactis, had the capacity for respiration-based growth when a heme source and oxygen were present in the medium (12, 16, 24, 47).
In L. lactis, respiration growth results in improved biomass, higher pH, and markedly greater long-term survival of lactococci (12, 16). However, the mechanisms involved in this stimulation are not yet understood. One reason is that only a limited number of genetic components involved in respiration growth have been identified (12). In contrast, bacteria described as respiratory (e.g., Escherichia coli and Bacillus subtilis) are equipped with redundant functions that ensure the production of electron sources and electron transport chains under different conditions of growth (40). The latter contain several quinone and cytochrome oxidases that facilitate oxygen reduction to water.
L. lactis contains a complete respiratory chain. The cydAB operon (encoding cytochrome bd oxidase) is required for respiration growth on oxygen plus heme, indicating that no other cytochrome oxidases are expressed (12). A quinone biosynthesis pathway is functional in several lactic acid bacteria, and the necessary biosynthesis genes have been deduced from genome sequences (5, 16, 32). Gene products involved in providing electrons from cytoplasmic compounds to the respiratory chain have also been deduced in L. lactis, and some of their functions have been confirmed (16; L. Rezaiki, A. Gruss, and P. Gaudu, unpublished data). As observed in aerobic bacteria, NADH is also used as an electron source for respiration in L. lactis (4). Interestingly, during growth on oxygen plus heme, the respiratory chain expulses protons, allowing growth rescue of an ATPase mutant (4). Respiration may thus protect cells against a decrease in intracellular pH during growth.
The clear distinctions between L. lactis and respiratory bacteria like E. coli and B. subtilis concern the NADH pool and heme sources. (i) The tricarboxylic acid cycle, which recycles the NADH pool for the respiratory chain, is incomplete (26), suggesting that L. lactis has developed other pathways to provide NADH. (ii) L. lactis is not equipped with an intact heme biosynthesis pathway, although some genes are present and functional. For example, ferrochelatase (encoded by hemZ), which catalyses iron incorporation into the heme precursor protoporphyrin IX, is present and active in L. lactis and allows respiration growth in the presence of protoporphyrin IX instead of heme (12). To overcome the absence of heme biosynthesis, L. lactis has heme uptake functions (15). Interestingly, Enterococcus faecalis and Leuconostoc mesenteroides also use environmental heme for respiration (22, 47) and appear to lack complete tricarboxylic acid cycles.
Studies of respiration regulation in aerobic bacteria such as E. coli and B. subtilis have revealed the roles of two-component systems and [4Fe/4S] FNR-like proteins (34, 55). In E. coli, the two-component system ArcA/ArcB governs genes involved in the tricarboxylic acid cycle and respiratory chain, while FNR acts on fermentation and anaerobic respiration. In B. subtilis, analogs of the above plus two other regulators, CcpA and PhoP/PhoR, have been identified. The catabolite control protein CcpA controls expression of glycolytic genes and tricarboxylic acid cycle and respiratory chain enzymes, while PhoP/PhoR controls the ArcA/ArcB analogs (called ResE/ResD) (3, 54). In L. lactis, an FNR homologue and two-component systems were previously identified, but their roles with respect to oxygen physiology were not clearly determined (17, 37).
In contrast to this situation, the regulation of heme-requiring respiration remains to be elucidated. We reported recently that respiration-permissive growth of L. lactis on glucose as a carbon source is diauxic. Cells grow initially via fermentation. As sugar is depleted from the medium, respiration growth takes over and lactate is consumed. CcpA, the catabolic response regulator, plays a key role in coordinating the shift from fermentation to respiration (15). In the presence of glucose, CcpA represses heme uptake, thus favoring fermentation during the first phase of growth. As sugar is consumed, CcpA relieves its suppression of heme uptake, allowing respiration to proceed (15). These results indicated that respiration is a highly regulated event in time in L. lactis that depends on the induction of pathways responding to the addition of heme and oxygen to the medium.
In view of the novelty of respiration growth in L. lactis compared to other bacteria, we decided to use a global proteomic approach to identify new proteins that are altered by respiration growth. This technique has been used successfully to visualize variations of protein levels as well as posttranslational modifications (21). Our studies reveal that, compared to fermentation, respiration affects the level as well as the posttranslational processing of certain proteins. Unexpectedly, several enzymes of the proteolytic system are induced, although this induction did not correspond to nitrogen starvation. Our results rather suggest that their induction is triggered by a modification of the cell redox state. This study represents a first approach to understanding the physiological changes associated with respiration in a bacterium requiring exogenous heme for respiration.
MATERIALS AND METHODS
Strains and growth conditions.
The strains (Table 1) were grown in M17 medium (Difco) supplemented with 1% glucose (GM17). Erythromycin (2.5 μg/ml) was added as appropriate. Heme was added for respiration growth to a final concentration of 10 μg/ml, as described previously (12). Generally, a 1/250 dilution of a fresh overnight L. lactis culture was used as the inoculum. Cells were cultured at 30°C under static, aerated, and aerated plus heme (referred to as respiration-permissive) conditions as described previously (12). Note that metabolism is fermentative under both static and aerated conditions.
TABLE 1.
Strains and fusions used in this study
| Strain | Relevant characteristic(s) | Reference |
|---|---|---|
| MG1363 | Plasmid-free derivative of NCDO0712 | 14 |
| YS5F | MG1363 Δopp ΔpepO1 ΔdtpT dppF::pINTdppF Eryr | 45 |
| JIM7506 | MG1363 bearing pepO1::lacZ fusion integrated in opp-pepO1 locus, Eryr | 20 |
| JIM8106 | JIM7506 codY::ISS1 Eryr | 20 |
Bidimensional electrophoresis. (i) Sample preparation.
Cells were harvested in late exponential growth phase at an optical density at 600 nm (OD600) of 1.6 (for cells grown via fermentation or aeration) or at an OD600 of 3.5 (for cells grown under respiration-permissive conditions). Cells were disrupted by pressure (2.5 kbar) with a cell disrupter (Constant System Ltd.). Extracts were centrifuged at 10,000 rpm for 20 min at 4°C. Cell walls and membranes were eliminated by a second centrifugation at 50,000 rpm for 30 min at 4°C, and the cytosolic fraction was harvested. Protein concentrations were measured according to the Bradford procedure with bovine serum albumin as the standard (6).
(ii) Electrophoresis.
Cytoplasmic proteins (250 μg per sample) were subjected to benzonase treatment and then precipitated with the methanol-chloroform method as clearly described previously (21). The pellet was resuspended in a solution of 7 M urea, 2 M thiourea, 4% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 100 mM dithiothreitol, 4.7 mM Tris-HCl, pH 8.8, and 0.45% IPG buffer, pH 4 to 7 (Amersham). The first dimension was performed with 17-cm IPG strips, pH 4 to 7 (Bio-Rad), followed by the second dimension on a 12% polyacrylamide gel. Gels were stained with BioSafe colloidal Coomassie blue (Bio-Rad) for at least 1 h and then destained with deionized water. Although this method is less sensitive than radioactive labeling (which follows protein expression during a time interval) or other staining procedures, it facilitates studies in rich medium, in which radiolabeling is laborious, and allows direct analysis of excised protein spots by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis.
Protein identification.
Two-dimensional gels were tested between four and six times on two independent sample preparations. Stained proteins displaying significant and reproducible intensities that differed between the tested growth conditions were excised from the gels. Excised gel plugs were washed in 50% CH3CN plus 0.15% (vol/vol) trifluoroacetic acid and then transferred to Eppendorf tubes. Digestions were performed in 50 mM NH4CO3 (pH 8.0) and 2 μl of 0.25-μg/μl modified trypsin (Promega, sequencing grade) for 18 h in a Thermomixer (Eppendorf) at 37°C with vortexing. Supernatants (0.5 μl) were spotted directly onto the MALDI plate. Samples were then dried at room temperature before 0.5 μl of matrix solution (α-cyano-4-hydroxycinnamic acid in 50% [vol/vol] acetonitrile and 0.1% [vol/vol] trifluoroacetic acid, freshly prepared) was added. Mass spectra were determined on a Voyager-DE-STR time-of-flight mass spectrometer (Applied Biosystems, Framingham, Mass.) equipped with a nitrogen laser emitting at a λ of 337 nm (Laser Science, Franklin, Mass.). Spectra were recorded in positive reflector mode with 20 V as the accelerating voltage, a delayed extraction time of 130 ns, and a 62% grid voltage. Monoisotopic mass lists were searched against an L. lactis database with the MS-Fit program (http://prospector.ucsf.edu, version 3.2.1). A minimum of four matching peptides and a sequence coverage above 20% were required before considering positive hits. For spots with low coverage (less than 20%), the N-terminal sequences were determined as clearly described previously (21).
Free amino acid composition.
Samples were taken in late exponential phase (at the same OD600 as for two-dimensional electrophoresis) and from stationary-phase cultures. Peptides from the medium were precipitated with 5-sulfosalicylic acid (3% final concentration) at 4°C for 1 h and then centrifuged at 15,000 rpm for 5 min. Supernatants were diluted 20-fold into 0.1 M lithium citrate, pH 2.2, and filtered on 0.45-μm filters. Free amino acid contents were determined by ion-exchange chromatography and postcolumn ninhydrin derivatization with a Biotronik LC3000 automatic analyzer (Maintal, Germany). Free amino acid concentrations in M17 were as follows: aspartic acid, 0.76 mM; threonine, 0.76 mM; serine, 1.1 mM; glutamic acid, 2.3 mM; glutamine, not detected; proline, 0.5 mM; glycine, 1.43 mM, alanine, 2.24 mM; valine, 1.4 mM; cysteine, not detected; methionine, 0.47 mM; isoleucine, 1 mM; leucine, 2.8 mM; tyrosine, 0.67 mM; phenylalanine, 1.36 mM; lysine, 1.8 mM; and arginine, 1.2 mM. Measurements of asparagine, histidine, and tryptophan are not presented due to limits in test sensitivity and instability of these amino acids. Analyses were performed on two independent culture samples.
β-Galactosidase essay.
pepO1 expression was determined by measuring β-galactosidase activity (30) in the codY mutant (JIM8106) and its isogenic control strain (JIM7605), both carrying the pepO1′-lacZ fusion (20).
RESULTS
Alterations in protein expression under respiratory-permissive compared to fermentation conditions.
To identify proteins involved in respiratory metabolism, we compared proteomic profiles of L. lactis strain MG1363 cultured under static, aerated, and respiration-permissive conditions. Proteins were extracted from cells harvested during late log phase in fermentation (static or aerated conditions, OD600 = 1.6) and in respiration (OD600 = 3.5). At the time cultures were taken, the pHs of the cultures were all around 5.5. However, in contrast to growth under fermentation conditions, respiration growth leads to a subsequent rise in pH (12).
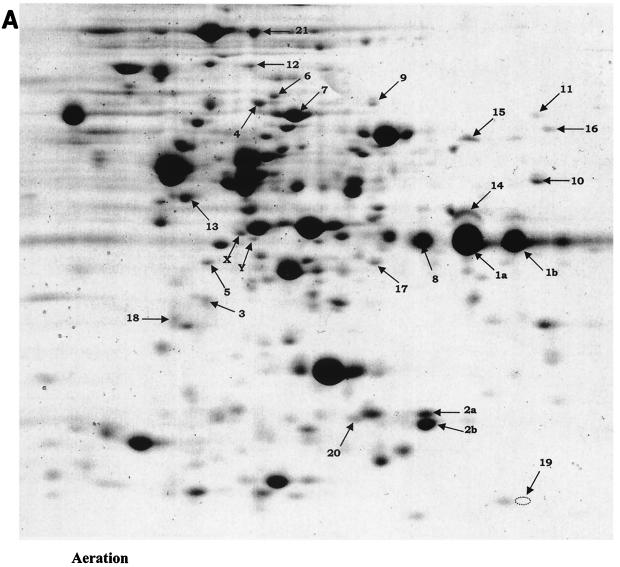
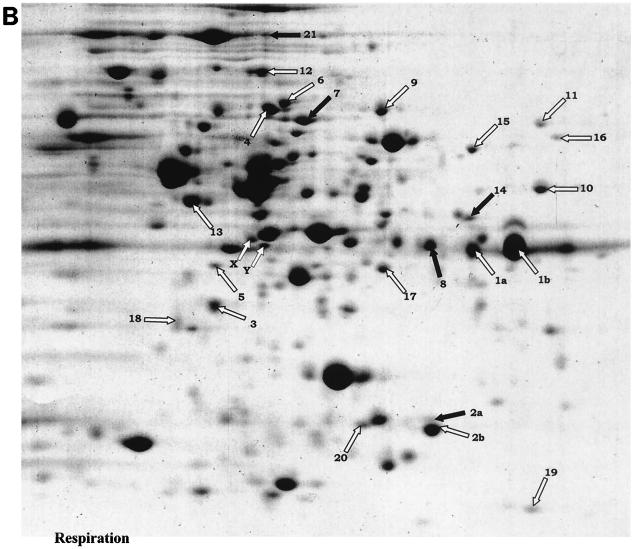
The proteome profiles of static and aeration cultures were indistinguishable in our growth conditions (data not shown). This similarity was not surprising, given the minor effects of aeration on L. lactis MG1363 metabolism (12, 23, 35). Lactic acid is still the major end product of glucose degradation, although traces of acetate and acetoin are detected in aeration growth. It is also likely that Coomassie blue staining limits the detection of poorly expressed proteins. We therefore compared profiles of aeration and respiration growth in the studies presented below (Fig. 1A and B). We observed that the levels of 21 proteins out of ≈250 detected on the two-dimensional gels were significantly altered in respiration-permissive cultures of L. lactis MG1363 compared to fermentation cultures (Fig. 1A and B; Table 2). These proteins were classified into two main groups, corresponding to those involved in carbon metabolism and nitrogen metabolism, plus one group comprising four unclassified proteins.
FIG. 1.
Proteomic analyses of cytosolic proteins of L. lactis MG1363 under respiration versus aeration growth. Protein extracts (250 μg of protein per sample) of late-log-phase cultures (see Materials and Methods) were analyzed by two-dimensional gel electrophoresis (pH gradient 4 to 7). Gels were stained with colloidal Coomassie blue. (A) Profile of aerated culture extract (OD600 = 1.6). Note that aeration and fermentation culture extract profiles were indistinguishable (data not shown). Fine arrows indicate protein spots whose intensities were reproducibly and significantly different under the two conditions (see Table 2). (B) Profile of respiration culture extract (OD600 = 3.5). Black arrows show spots whose amounts are decreased in respiration growth; white arrows show spots whose amounts are increased in respiration growth. a and b correspond to posttranslational protein events. X and Y both correspond to RpoA protein, as does the larger adjacent spot to their upper right; however, protein levels were not reproducibly different. Gels were repeated at least four times for each condition, with two independently prepared samples.
TABLE 2.
List of proteins modified during heme-dependent respiration versus fermentation growtha
| Group | Spot no. | Name | Protein | Effect | Method(s) |
|---|---|---|---|---|---|
| 1: Carbon metabolism | 1a, 1b | GapdhB | Glyceraldehyde-3-phosphate dehydrogenase; a and b isoforms | Post-trans. | PMF, N |
| 2a, 2b | Pmg | Phosphoglycerate mutase; a and b proteins | Post-trans. | PMF, N | |
| 3 | PdhE1β | Pyruvate dehydrogenase E1β subunit | ++ | PMF, N | |
| 4 | PdhD | Pyruvate dehydrogenase D subunit | ++ | PMF, N | |
| 5 | LlpL | Lipoate-protein ligase | ++ | PMF | |
| 6 | Als | α-Acetolactate synthase | ++ | PMF | |
| 7 | BglA | Phospho-β-glucosidase | −− | PMF | |
| 8 | RmlB | dTDP-glucose 4,6-dehydratase | −− | PMF | |
| 9 | PurH | Bifunctional purine biosynthesis protein | ++ | PMF | |
| 10 | GlyA | Serine hydroxymethyltransferase | ++ | PMF | |
| 11 | Fhs | Formyltetrahydrofolate synthetase | ++ | PMF, N | |
| 2: Nitrogen metabolism | 12 | PepO1 | Neutral endopeptidase | ++ | PMF |
| 13 | PepC | Aminopeptidase C | ++ | PMF | |
| 14 | PepA | Glutamyl-aminopeptidase | − | PMF, N | |
| 15 | GatA | Glu-tRNA amidotransferase subunit A | ++ | PMF | |
| 16 | ThrC | Threonine synthase | ++ | PMF, N | |
| 17 | MetC | Cystathionine β-lyase | ++ | PMF | |
| 3: Unclassified | 18 | TrxB1 | Thioredoxin reductase | + | PMF |
| 19 | YgfC | Potential transcriptional regulator | ++++ | PMF, N | |
| 20 | YchH | Potential acetyltransferase | ++ | PMF | |
| 21 | TypA | GTP-binding protein | −− | PMF |
Post-trans., posttranslational events; PMF, peptide mass fingerprinting; N, N-terminal sequencing. Greater and lesser expression under respiration compared to aeration fermentation conditions is evaluated as + to ++++ and − to −−, respectively. The number of symbols reflects the intensity of the difference observed.
Group 1: carbon metabolism. (i) Fermentation pathway.
Glyceraldehyde-3-phosphate dehydrogenase B (GapB), encoded by gapB, catalyzes the conversion of 3-phosphoglycerate into 1,3-diphosphoglycerate and concomitantly reduces NAD+. L. lactis encodes a second glyceraldehyde dehydrogenase, GapA (5), which is poorly, if at all, produced and migrates at distinct positions on two-dimensional gels (56). GapB appeared as two major isoforms, a and b, in agreement with previous results (25, 56) (Fig. 1; spots 1a and 1b). Under fermentation conditions, the amounts of each isoform are approximately equal. In contrast, their relative amounts are drastically modified during growth under respiration-permissive conditions. The amounts of the a isoform are strongly decreased, whereas the amounts of the b isoform remain unchanged. GapB appears to be the major source of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) activity in L. lactis (56). To our knowledge, this is the first demonstration that GAPDH isoform levels vary during growth.
Phosphoglycerate mutase (Pmg), encoded by pmg, catalyzes the conversion of 3-phosphoglycerate to 2-phosphoglycerate. It appeared as two spots (a and b) having different molecular weights (Fig. 1; spots 2a and 2b), suggesting that the b form may result from maturation or degradation of the a form. Amounts of the a Pmg form are markedly lower under respiration-permissive conditions. Two subunits of pyruvate dehydrogenase (PdhE1β, encoded by pdhB, and PdhD, encoded by pdhD) and lipoate-protein ligase (LplL, encoded by lplL), were overproduced under respiration-permissive conditions compared to fermentative conditions (Fig. 1; spots 3, 4, and 5, respectively). These proteins contribute to the activity of the pyruvate dehydrogenase complex (50), which catalyzes the conversion of pyruvate into acetyl-coenzyme A. Pyruvate dehydrogenase complex production results in a concomitant reduction of NAD+. Furthermore, acetyl-coenzyme A is used in fatty acid, ethanol, and acetate production. LplL is a homologue of the Escherichia coli LplA protein, which catalyzes ATP-dependent attachment of lipoic acid to pyruvate dehydrogenase complex proteins (33). In L. lactis IL-1403, lplL is adjacent to the pdh genes, consistent with a role in pyruvate dehydrogenase complex activity (5). The greater expression of these proteins is consistent with the large amounts of acetate produced under oxygen and heme conditions (12, 24).
The amounts of α-acetolactate synthase (Als, encoded by als) are greatly increased by respiration growth (Fig. 1; spot 6). Als catalyzes the conversion of pyruvate into α-acetolactate, which is then degraded to acetoin (39, 50). A second acetolactate synthase is encoded by ilvBN from L. lactis IL1403 (53) and forms part of the operon involved in branched-chain amino acid biosynthesis (Ile, Leu, and Val) (18). These amino acids repress ilvBN transcription. We observed that millimolar quantities of these branched-chain amino acids remain in the medium after growth (see below), suggesting that ilvBN transcription is shut off. We thus consider that the abundant amounts of acetoin produced during respiration growth reflect high activity of als rather than of ilvBN (12, 24).
The amounts of phospho-β-glucosidase (BglA), encoded by bglA, are reduced during respiration growth compared to fermentation (Fig. 1; spot 7). BglA putatively hydrolyzes C6-phosphorylated β-glucosides, releasing glucose 6-phosphate. One such enzyme is the β-galactosidase present in strains growing in milk, where lactose is the main carbon source. Other phospho-β-glucosidase activities have also been identified in lactic acid bacteria (31, 48). This activity is usually detected in cells growing on β-glucosides like salicin, arbutin, and cellobiose but not on glucose, which suggests that the corresponding gene is controlled by carbon catabolite repression (31, 48). The presence of BglA under glucose fermentation growth but not in respiration conditions suggests that (i) BglA is probably not under carbon catabolite repression and (ii) BglA might have a role other than to degrade these sugars. Interestingly, a bglA mutant with the UV sensitivity phenotype has been isolated (11). As UV is known to break DNA, which contains a sugar backbone, BglA might be involved in DNA repair during fermentation growth, during which DNA damage is more frequent (L. Rezaiki, B. Cesselin, and A. Gruss, unpublished data).
(ii) Cell wall biosynthesis.
We observed decreased amounts of a dTDP-α-glucose-4,6-dehydratase (RmlB), encoded by rmlB, during respiration growth (Fig. 1; spot 8). RmlB from L. lactis is likely involved in cell wall synthesis, as (i) rmlB is located within a genomic locus involved in cell wall structure (5) and (ii) in E. coli and many other bacteria, its homologs are implicated in cell wall synthesis (7, 27). We propose that decreased RmlB production results in a cell wall modification in the respiration state of L. lactis.
(iii) Carbon/nitrogen metabolism.
Bifunctional purine biosynthesis protein (PurH), serine hydroxymethyl transferase (GlyA), and formyltetrahydrofolate synthetase (Fhs) all participate in de novo purine biosynthesis (GTP, ATP) and are all produced in increased amounts in respiration growth (Fig. 1; spots 9, 10, and 11, respectively). Their coding genes, purH, glyA, and fhs, respectively, belong to the purine regulon controlled by the regulator PurR, which responds to purine starvation (2). Overproduction of these enzymes during respiration growth may reflect a depletion of the purine pool during the first phase of growth (via fermentation). However, as GlyA and Fhs also participate in other pathways (e.g., amino acid biosynthesis) (13), we cannot rule out that their induction serves other roles during respiration.
Group 2: nitrogen metabolism. (i) Translation.
The amounts of Glu-tRNA amidotransferase subunit A (GatA) are increased by respiration growth (Fig. 1; spot 15). The gatA gene is presumably the last gene of the gatCgatBybgDgatA operon in L. lactis IL1403 (5); the Gat complex catalyzes the conversion of glutamic acid-tRNA to glutamine-tRNA; glutamic acid and glutamine are both essential amino acids in L. lactis (43).
(ii) Anabolism.
Cystathionine β-lyase (MetC) (Fig. 1, spot 17) is involved in cysteine biosynthesis. Serine hydroxymethyltransferase (GlyA) (Fig. 1, spot 10, see above) may also be implicated in this pathway. Threonine synthase (ThrC) (Fig. 1, spot 16) is needed for threonine biosynthesis. These proteins are all increased in respiration growth. metC (called metB2 in L. lactis IL1403) is organized in an operon with cysK (encoding cysteine synthase) in L. lactis MG1363 (13), possibly suggesting that the amounts of the latter are also increased in respiration growth. Unexpectedly, increased production of ThrC in respiration suggests that threonine biosynthesis genes are subject to regulation, in contrast to what was reported previously (28).
(iii) Peptide catabolism.
Expression of the exopeptidase PepA, which hydrolyzes oligopeptides from an N-terminal glutamate, serine, or aspartic acid end, is reduced under respiration conditions (Fig. 1, spot 14). The pepA gene is constitutively expressed, although weak induction was observed under nitrogen starvation (19). We therefore considered that PepA plays a minor role in proteolysis compared to other peptidases. Lower PepA levels during respiration are consistent with a secondary role for this peptidase under conditions of rapid growth.
Unlike PepA, the amounts of PepO1 (endopeptidase) and PepC (exopeptidase), which also hydrolyze oligopeptides, are markedly increased in respiration growth (Fig. 1, spots 12 and 13, respectively). Interestingly, these peptidases are strongly induced by nitrogen starvation (19). Whereas pepC is monocistronic, pepO1 belongs to the oppDFBCApepO1 operon in L. lactis MG1363, which also encodes the oligopeptide transporter Opp (19). Differences in oppDFBCA gene products under respiration growth were not detected by proteomic analyses, possibly because they are membrane associated and, as such, eliminated in the conditions we used for sample preparation.
Group 3: unclassified proteins.
The amounts of NAD(P)H:thioredoxin oxidoreductase (encoded by trxB1) are slightly increased in respiration growth (Fig. 1, spot 18). In other bacteria, this enzyme plays an important role in defense against oxidative stress and acts as an electron carrier for key enzymes like ribonucleotide reductase (52). YgfC, a putative regulator, and YchH, classified as a putative acetyltransferase, are induced in respiration growth, whereas TypA, possibly a GTP-binding protein, shows lower expression in respiration (Fig. 1, spots 19, 20, and 21, respectively).
Respiration does not lead to nitrogen starvation.
Proteolysis in lactococci involves peptide transporters such as Opp, DtpT, and DtpP, aminotransferases (AraT, BcaT) and endo- and exopeptidases such as PepO1 and PepC (9, 19). It is repressed in medium rich in free amino acids, such as GM17, but not in milk or in chemically defined medium. In the last, addition of dipeptides containing leucine represses the expression of proteolysis-related functions, indicating that the proteolysis system is indeed regulated. Furthermore, once the medium is exhausted of its free amino acid supply, it shifts to the use of peptides (9). The observed induction of PepO1 and PepC during respiratory growth could thus reflect nitrogen starvation. Two approaches were used to test this hypothesis. We compared the levels of free amino acids in the medium when cells were cultured under fermentation and respiration conditions. We also studied the behavior of a mutant deficient in oligopeptide transport under both conditions.
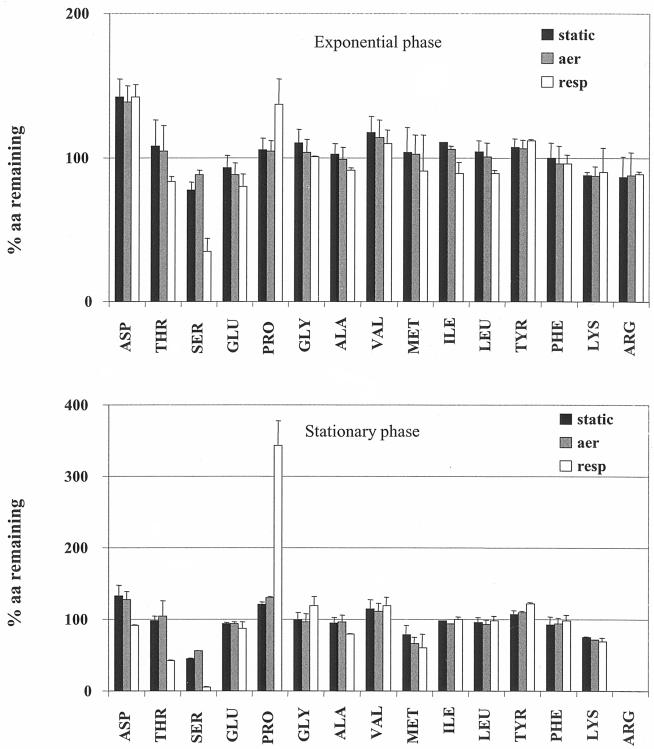
Amino acid composition was analyzed from spent log- and stationary-phase medium after growth under fermentation (static and aerated) and respiration conditions (Fig. 2). For the majority of amino acids, availability varied surprisingly little as a function of growth mode or growth phase. In some cases (e.g., for threonine and serine), amino acid accumulation was respiration specific.
FIG. 2.
Free amino acid (aa) composition of spent medium from L. lactis MG1363 fermentation, aeration, and respiration cultures. Late-log- and stationary-phase supernatant samples of MG1363 cultures were analyzed (see Materials and Methods). Glutamine and cysteine were present at below detectable levels (not shown). Asparagine, histidine, and tryptophan determinations are not presented due to limits in test sensitivity and/or instability of these amino acids. The deviation of amino acid concentrations was less than 15% between independent cultures and determinations.
Here we comment on the two more significant observations. One concerns arginine uptake in the stationary phase of all cultures. The second concerns considerable proline expulsion, which is specific to respiration cultures. The basic amino acid arginine is used for protein synthesis (it constitutes 4% of total L. lactis amino acids in cell composition) (36, 43). It is also involved in resistance to acid stress via the arginine deiminase pathway (42). This pathway generates energy (ATP) and ammonia, which limits acidification of the cytoplasm. The complete exhaustion of arginine from GM17 medium of stationary-phase cultures indicated that this pathway was functional in all growth conditions. Moreover, we also detected citrulline and orthinine, both end products of arginine catabolism via arginine deiminase, in the supernatant; these compounds accounted for more than 80% of the total arginine initially present in the medium (data not shown).
Surprisingly, free proline accumulated in stationary-phase supernatants of respiration cultures despite its abundant presence (about 5% total L. lactis amino acids in cell composition). This indicated that intracellular proline came from de novo synthesis or from oligopeptide uptake during respiration. Interestingly, proline-containing dipeptides (in particular Leu-Pro) are more efficiently incorporated than free amino acids in some L. lactis strains (49). Proline expulsion in L. lactis is specifically associated with respiration growth (Fig. 2), with proline concentrations ≈12-fold higher (reaching 1.2 mM) than those present in stationary-phase supernatants of fermentation cultures. To our knowledge, this is the first reported observation of proline expulsion in lactic acid bacteria.
Oligopeptides are not essential for respiration growth.
We reasoned that if peptides played a role in respiration growth, a mutant defective in known oligopeptide transporters (Opp, DtpT, and DtpP) might be impaired in respiration. Surprisingly, we found that wild-type and Δopp dtpT dtpP triple mutant strains had similar growth capacities (final OD600: fermentation, 2.5 and 2.2, respectively; respiration, 5.5 and 5, respectively). These results indicate that oligopeptides present in the complex GM17 medium are not essential for respiration growth in L. lactis. They further suggest that increased PepO1 and PepC expression in respiration cultures is not triggered by starvation of specific peptides.
Induction of oppA-pepO1 transcription under aeration conditions.
In L. lactis MG1363, expression of components of the proteolytic system responds to the concentrations of available carbon and nitrogen (19). For the lactococcal peptidase genes pepO1 and pepC, transaminases and oligotransporters, the nitrogen source has clear effects on protein levels. CodY is a major regulator responding to nitrogen source availability in controlling proteolysis in L. lactis (20).
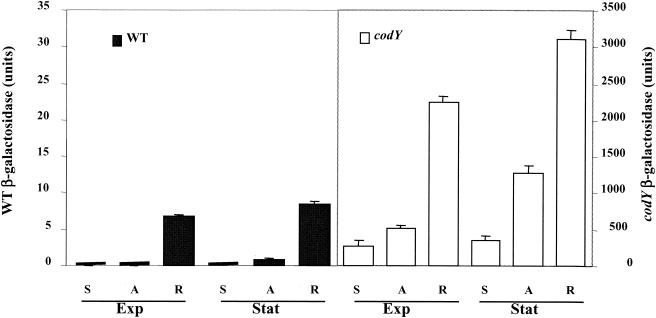
As PepO1 and PepC induction in respiration growth seemed to be independent of amino acid availability (see above), we asked whether CodY is implicated in this induction. To test this, we followed pepO1 expression via an oppA-lacZ transcriptional fusion in wild-type and codY strains under fermentation (static or aerated conditions) and respiration growth (Fig. 3). In agreement with the two-dimensional gel observations, pepO1 expression was induced in respiration growth, in both wild-type and codY mutant strains and in both exponential- and stationary-phase cells (Fig. 3). We also noted a significant stimulatory effect of oxygen on pepO1 expression in the codY mutant. These results indicate the existence of another regulatory pathway for peptidase control in L. lactis that seems to be dependent on oxygen and/or respiration growth and that is independent of the previously reported main nitrogen regulator, CodY.
FIG. 3.
Induction of pepO1 by respiration growth is independent of CodY. JIM7506 (codY and containing a pepO1-lacZ fusion) and the control wild-type strain (containing a pepO1-lacZ fusion) were cultured under static, aeration, or respiration-permissive conditions. β-Galactosidase specific activities were measured in late exponential phase (Exp, OD600 = 1.6 for fermentation and 3.5 for respiration) and in stationary phase (Stat, OD600 = 2.8 for static or aeration growth and 4.5 for respiration growth). Note that β-galactosidase expression scales for the wild-type and the codY mutant strains differ by 100-fold. S, static growth; A, aerated growth; R, respiration growth. Curves correspond to an average of two independent determinations performed on independent cultures. The deviation between measurements was less than 10%.
DISCUSSION
L. lactis has been examined extensively for its properties as a fermenting microorganism, but essentially nothing is known of its life style during respiration. Our data provide the first description of cytoplasmic changes due to heme-dependent respiration, as recently identified in the gram-positive bacterium L. lactis (12, 16). We distinguished ≈250 of the ≈2,300 putative proteins predicted from the whole genome sequence of L. lactis (5). As numerous proteins involved in respiration are predictably membrane associated (e.g., those involved in the electron transport chain), we suspect that the proportion of respiration-related functions in L. lactis is greater than what we can estimate from this work. Our proteome analyses show that L. lactis uses several proteins already present in fermentation conditions to shift to respiration growth. We identified 11 proteins that are involved in the glycolysis and purine pathways and six involved in nitrogen metabolism whose expression levels are altered by respiration. For one protein, YgfC, expression seems to be respiration specific (Fig. 1, spot 19). YgfC might belong to the TetR/AcrR family of transcriptional regulators.
Some proteins, such as superoxide dismutase (encoded by sodA), were previously reported as being overproduced in the presence of oxygen (44). However, we did not see significant differences in SodA levels under static versus aerated conditions. Possibly, sodA transcription may already be induced under acidifying fermentation conditions, which could mask the response to oxygen; indeed, sodA expression was reportedly induced in nonbuffered compared to buffered medium (44).
Posttranslational modification due to respiration.
L. lactis respiration growth resulted in significant posttranslational modifications in GapB, as well as in Pmg. Interestingly, GAPDH modification as reported here has not been observed in the aerobic bacterium Bacillus subtilis (54). In L. lactis, GapB is a key enzyme in the glycolytic pathway (41, 51), and recently Solem et al. reported that GapB amounts were fourfold greater than required for the needs of the cell (51). This may explain why the observed changes in GapB did not result in growth rate differences between respiration and fermentation cultures. The modifications provoked by respiration growth therefore suggest a new role for GapB in L. lactis physiology other than in glycolysis. In this regard, GAPDH has recently been implicated as having a secondary role on the cell surface in bacterial adhesion (1, 10). Moreover, several isoforms of this enzyme were already reported in other Streptococcaceae related to L. lactis, e.g., Streptococcus pneumoniae (8), Enterococcus faecalis (A. Hartke, personal communication), and Streptococcus pyogenes (38). Identification of the nature of the GAPDH modification in L. lactis will be valuable for understanding the physiological role played by these isoforms in respiration growth.
Energy production and respiration.
Blank et al. reported a proton expulsion activity in L. lactis when cells were cultured in the presence of heme plus oxygen (4), as observed in E. coli. Interestingly, although L. lactis does not encode a complete tricarboxylic acid cycle (26), this activity depends on the NADH pool (4). Consequently, it must replenish its NADH pool via other pathways. Greater production of pyruvate dehydrogenase complex components during respiration growth suggests that this complex may participate in NADH recycling. The pyruvate dehydrogenase complex catalyzes the reduction of NAD+ from pyruvate oxidation. This pathway is further confirmed by the accumulation of acetate (12, 24). Moreover the acetate pathway produces one ATP molecule, which constitutes an energetic gain to the cell compared to lactic acid production. Similarly, arginine catabolism may be a means of producing energy. This amino acid is strongly consumed via the arginine deiminase pathway, even during respiration, where, unlike fermentation, the medium pH first drops and then increases late in growth.
Respiration and starvation.
Our results indicate that respiration led to starvation of the purine and sulfur amino acid pools. In the case of purines, PurH, GlyA, and Fhs levels are all increased during respiration. The genes encoding these proteins belong to the PurH regulon, whose expression responds to the purine pool (2). In the case of sulfur amino acids, we noted an increase in MetC levels during respiration. As the metC-cysK operon is governed by CmbR, which responds to cysteine starvation (13), we consider it likely that MetC is induced in response to a depletion in this amino acid. In L. lactis, we showed that cysteine can protect cells against the oxidative stress provoked by heme (15). In contrast to the need for additional purines and sulfur amino acids, depletion during respiration of amino acids (other than arginine) is not so obvious. It is therefore perplexing that PepO1 and PepC were overproduced in respiration growth. We consider it possible that some amino acids are preferentially assimilated as peptides rather than as free amino acids. For example, lactococci preferentially utilize peptides as a proline source (49). Nevertheless, no differences in respiration growth were observed between peptide transport-deficient versus peptide transport-proficient strains.
We observed that PepO1 expression was upregulated by both aeration and respiration growth in the wild-type strain as well as in a codY mutant. This suggests that novel regulators control the expression of proteolysis-related genes independently of CodY, both in an oxidative environment and in respiration. In L. lactis, the only reported potential redox-responsive factors are the FNR-like regulators FlpA and FlpB, although no clear function had been reported (17, 46). Interestingly, flpB is adjacent to the opp-pepO1 operon, so it is tempting to speculate that FlpB has an effect on opp-pepO1 expression. This is consistent with previous findings showing that Flp regulators control zinc uptake (17); zinc is an important cofactor for numerous enzymes, including PepO1 (29).
In summary, several phenomena previously unknown in lactococci were revealed by proteomic analyses of respiration growth. Posttranslational modifications of GapB protein were detected and were respiration growth specific. Proline expulsion occurs in respiring but not fermenting lactococci. Finally, codY-independent regulation of proteolysis during respiration was revealed.
Acknowledgments
We are grateful to C. Henry and J.-C. Huet for mass spectrometry and N-terminal sequencing of proteins and to Y. Sanz and E. Guédon for providing the strains used in this study. We thank our colleagues from the URLGA laboratory and S. Iversen and H. Møllgaard (Chr. Hansen A/S, Denmark) for stimulating discussion during the course of this work.
This work is supported by a research grant (Chr. Hansen A/S, Denmark).
REFERENCES
- 1.Alvarez, R. A., M. W. Blaylock, and J. B. Baseman. 2003. Surface localized glyceraldehyde-3-phosphate dehydrogenase of Mycoplasma genitalium binds mucin. Mol. Microbiol. 48:1417-1427. [DOI] [PubMed] [Google Scholar]
- 2.Beyer, N. H., P. Roepstorff, K. Hammer, and M. Kilstrup. 2003. Proteome analysis of the purine stimulon from Lactococcus lactis. Proteomics 3:786-797. [DOI] [PubMed] [Google Scholar]
- 3.Birkey, S. M., W. Liu, X. Zhang, M. F. Duggan, and F. M. Hulett. 1998. Pho signal transduction network reveals direct transcriptional regulation of one two-component system by another two-component regulator: Bacillus subtilis PhoP directly regulates production of ResD. Mol. Microbiol. 30:943-953. [DOI] [PubMed] [Google Scholar]
- 4.Blank, L. M., B. J. Koebmann, O. Michelsen, L. K. Nielsen, and P. R. Jensen. 2001. Hemin reconstitutes proton extrusion in an H+-ATPase-negative mutant of Lactococcus lactis. J. Bacteriol. 183:6707-6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Breedveld, M., K. Bonting, and L. Dijkhuizen. 1998. Mutational analysis of exopolysaccharide biosynthesis by Lactobacillus sakei 0-1. FEMS Microbiol. Lett. 169:241-249. [DOI] [PubMed] [Google Scholar]
- 8.Cash, P., E. Argo, L. Ford, L. Lawrie, and H. McKenzie. 1999. A proteomic analysis of erythromycin resistance in Streptococcus pneumoniae. Electrophoresis 20:2259-2268. [DOI] [PubMed] [Google Scholar]
- 9.Chambellon, E., and M. Yvon. 2003. CodY-regulated aminotransferases AraT and BcaT play a major role in the growth of Lactococcus lactis in milk by regulating the intracellular pool of amino acids. Appl. Environ. Microbiol. 69:3061-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D’Costa, S. S., T. G. Romer, and M. D. Boyle. 2000. Analysis of expression of a cytosolic enzyme on the surface of Streptococcus pyogenes. Biochem. Biophys. Res. Commun. 278:826-832. [DOI] [PubMed] [Google Scholar]
- 11.Duwat, P., A. Cochu, S. D. Ehrlich, and A. Gruss. 1997. Characterization of Lactococcus lactis UV-sensitive mutants obtained by ISS1 transposition. J. Bacteriol. 179:4473-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duwat, P., S. Sourice, B. Cesselin, G. Lamberet, K. Vido, P. Gaudu, Y. Le Loir, F. Violet, P. Loubiere, and A. Gruss. 2001. Respiration capacity of the fermenting bacterium Lactococcus lactis and its positive effects on growth and survival. J. Bacteriol. 183:4509-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandez, M., M. Kleerebezem, O. P. Kuipers, R. J. Siezen, and R. van Kranenburg. 2002. Regulation of the metC-cysK operon, involved in sulfur metabolism in Lactococcus lactis. J. Bacteriol. 184:82-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaudu, P., G. Lamberet, S. Poncet, and A. Gruss. 2003. CcpA regulation of aerobic and respiration growth in Lactococcus lactis. Mol. Microbiol. 50:183-192. [DOI] [PubMed] [Google Scholar]
- 16.Gaudu, P., K. Vido, B. Cesselin, S. Kulakauskas, J. Tremblay, L. Rezaiki, G. Lamberet, S. Sourice, P. Duwat, and A. Gruss. 2002. Respiration capacity and consequences in Lactococcus lactis. Antonie Leeuwenhoek 82:263-269. [PubMed] [Google Scholar]
- 17.Gostick, D. O., H. G. Griffin, C. A. Shearman, C. Scott, J. Green, M. J. Gasson, and J. R. Guest. 1999. Two operons that encode FNR-like proteins in Lactococcus lactis. Mol. Microbiol. 31:1523-1535. [DOI] [PubMed] [Google Scholar]
- 18.Goupil-Feuillerat, N., M. Cocaign-Bousquet, J. J. Godon, S. D. Ehrlich, and P. Renault. 1997. Dual role of alpha-acetolactate decarboxylase in Lactococcus lactis subsp. lactis. J. Bacteriol. 179:6285-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guedon, E., P. Renault, S. D. Ehrlich, and C. Delorme. 2001. Transcriptional pattern of genes coding for the proteolytic system of Lactococcus lactis and evidence for coordinated regulation of key enzymes by peptide supply. J. Bacteriol. 183:3614-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guedon, E., P. Serror, S. D. Ehrlich, P. Renault, and C. Delorme. 2001. Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in Lactococcus lactis. Mol. Microbiol. 40:1227-1239. [DOI] [PubMed] [Google Scholar]
- 21.Guillot, A., C. Gitton, P. Anglade, and M. Y. Mistou. 2003. Proteomic analysis of Lactococcus lactis, a lactic acid bacterium. Proteomics 3:337-354. [DOI] [PubMed] [Google Scholar]
- 22.Huycke, M. M., D. Moore, W. Joyce, P. Wise, L. Shepard, Y. Kotake, and M. S. Gilmore. 2001. Extracellular superoxide production by Enterococcus faecalis requires demethylmenaquinone and is attenuated by functional terminal quinol oxidases. Mol. Microbiol. 42:729-740. [DOI] [PubMed] [Google Scholar]
- 23.Jensen, N. B., C. R. Melchiorsen, K. V. Jokumsen, and J. Villadsen. 2001. Metabolic behavior of Lactococcus lactis MG1363 in microaerobic continuous cultivation at a low dilution rate. Appl. Environ. Microbiol. 67:2677-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaneko, T., M. Tagahashi, and H. Suzuki. 1990. Acetoin fermentation by citrate-positive Lactococcus lactis subsp. lactis 3022 grown aerobically in the presence of hemin or Cu2+. Appl. Environ. Microbiol. 56:2644-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kilstrup, M., S. Jacobsen, K. Hammer, and F. K. Vogensen. 1997. Induction of heat shock proteins DnaK, GroEL, and GroES by salt stress in Lactococcus lactis. Appl. Environ. Microbiol. 63:1826-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lapujade, P., M. Cocaign-Bousquet, and P. Loubiere. 1998. Glutamate biosynthesis in Lactococcus lactis subsp. lactis NCDO 2118. Appl. Environ. Microbiol. 64:2485-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma, Y., R. J. Stern, M. S. Scherman, V. D. Vissa, W. Yan, V. C. Jones, F. Zhang, S. G. Franzblau, W. H. Lewis, and M. R. McNeil. 2001. Drug targeting Mycobacterium tuberculosis cell wall synthesis: genetics of dTDP-rhamnose synthetic enzymes and development of a microtiter plate-based screen for inhibitors of conversion of dTDP-glucose to dTDP-rhamnose. Antimicrob. Agents Chemother. 45:1407-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madsen, S. M., B. Albrechtsen, E. B. Hansen, and H. Israelsen. 1996. Cloning and transcriptional analysis of two threonine biosynthetic genes from Lactococcus lactis MG1614. J. Bacteriol. 178:3689-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mierau, I., S. T. T. Paris, J. Alfred, A. J. Haandrikman, J. Kok, K. J. Leenhouts, W. N. Konings, and G. Venema. 1993. Cloning and sequencing of the gene for a lactococcal endopeptidase, an enzyme with sequence similarity to mammalian enkephalinase. J. Bacteriol. 175:2087-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, J. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Monedero, V., O. P. Kuipers, E. Jamet, and J. Deutscher. 2001. Regulatory functions of serine-46-phosphorylated HPr in Lactococcus lactis. J. Bacteriol. 183:3391-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morishita, T., N. Tamura, T. Makino, and S. Kudo. 1999. Production of menaquinones by lactic acid bacteria. J. Dairy Sci. 82:1897-1903. [DOI] [PubMed] [Google Scholar]
- 33.Morris, T. W., K. E. Reed, and J. E. Cronan, Jr. 1994. Identification of the gene encoding lipoate-protein ligase A of Escherichia coli. Molecular cloning and characterization of the lplA gene and gene product. J. Biol. Chem. 269:16091-16100. [PubMed] [Google Scholar]
- 34.Nakano, M. M., and P. Zuber. 1998. Anaerobic growth of a “strict aerobe” (Bacillus subtilis). Annu. Rev. Microbiol. 52:165-190. [DOI] [PubMed] [Google Scholar]
- 35.Neves, A. R., A. Ramos, H. Costa, I. I. van Swam, J. Hugenholtz, M. Kleerebezem, W. de Vos, and H. Santos. 2002. Effect of different NADH oxidase levels on glucose metabolism by Lactococcus lactis: kinetics of intracellular metabolite pools determined by in vivo nuclear magnetic resonance. Appl. Environ. Microbiol. 68:6332-6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novák, L., and P. Loubiere. 2000. The metabolic network of Lactococcus lactis: distribution of 14C-labeled substrates between catabolic and anabolic pathways. J. Bacteriol. 182:1136-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Connell-Motherway, M., D. van Sinderen, F. Morel-Deville, G. F. Fitzgerald, S. D. Ehrlich, and P. Morel. 2000. Six putative two-component regulatory systems isolated from Lactococcus lactis subsp. cremoris MG1363. Microbiology 146:935-947. [DOI] [PubMed] [Google Scholar]
- 38.Pancholi, V., and V. A. Fischetti. 1993. Glyceraldehyde-3-phosphate dehydrogenase on the surface of group A streptococci is also an ADP-ribosylating enzyme. Proc. Natl. Acad. Sci. USA 90:8154-8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Platteeuw, C., J. Hugenholtz, M. Starrenburg, I. van Alen-Boerrigter, and W. M. de Vos. 1995. Metabolic engineering of Lactococcus lactis: influence of the overproduction of alpha-acetolactate synthase in strains deficient in lactate dehydrogenase as a function of culture conditions. Appl. Environ. Microbiol. 61:3967-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poole, R. K., and G. M. Cook. 2000. Redundancy of aerobic respiratory chains in bacteria? Routes, reasons and regulation. Adv. Microb. Physiol. 43:165-224. [DOI] [PubMed] [Google Scholar]
- 41.Poolman, B., B. Bosman, J. Kiers, and W. N. Konings. 1987. Control of glycolysis by glyceraldehyde-3-phosphate dehydrogenase in Streptococcus cremoris and Streptococcus lactis. J. Bacteriol. 169:5887-5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poolman, B., A. J. Driessen, and W. N. Konings. 1987. Regulation of arginine-ornithine exchange and the arginine deiminase pathway in Streptococcus lactis. J. Bacteriol. 169:5597-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poolman, B., and W. N. Konings. 1988. Relation of growth of Streptococcus lactis and Streptococcus cremoris to amino acid transport. J. Bacteriol. 170:700-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanders, J. W., K. J. Leenhouts, A. J. Haandrikman, G. Venema, and J. Kok. 1995. Stress response in Lactococcus lactis: cloning, expression analysis, and mutation of the lactococcal superoxide dismutase gene. J. Bacteriol. 177:5254-5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanz, Y., F. C. Lanfermeijer, P. Renault, A. Bolotin, W. N. Konings, and B. Poolman. 2001. Genetic and functional characterization of dpp genes encoding a dipeptide transport system in Lactococcus lactis. Arch. Microbiol. 175:334-343. [DOI] [PubMed] [Google Scholar]
- 46.Scott, C., J. R. Guest, and J. Green. 2000. Characterization of the Lactococcus lactis transcription factor FlpA and demonstration of an in vitro switch. Mol. Microbiol. 35:1383-1393. [DOI] [PubMed] [Google Scholar]
- 47.Sijpesteijn, A. K. 1970. Induction of cytochrome formation and stimulation of oxidative dissimilation by hemin in Streptococcus lactis and Leuconostoc mesenteroides. Antonie Leeuwenhoek 36:335-348. [DOI] [PubMed] [Google Scholar]
- 48.Simons, G., M. Nijhuis, and W. M. de Vos. 1993. Integration and gene replacement in the Lactococcus lactis lac operon: induction of a cryptic phospho-β-glucosidase in lacG-deficient strains. J. Bacteriol. 175:5168-5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smid, E. J., and W. N. Konings. 1990. Relationship between utilization of proline and proline-containing peptides and growth of Lactococcus lactis. J. Bacteriol. 172:5286-5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Snoep, J. L., M. J. Teixeira de Mattos, M. J. Starrenburg, and J. Hugenholtz. 1992. Isolation, characterization, and physiological role of the pyruvate dehydrogenase complex and alpha-acetolactate synthase of Lactococcus lactis subsp. lactis bv. diacetylactis. J. Bacteriol. 174:4838-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Solem, C., B. J. Koebmann, and P. R. Jensen. 2003. Glyceraldehyde-3-phosphate dehydrogenase has no control over glycolytic flux in Lactococcus lactis MG1363. J. Bacteriol. 185:1564-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stewart, E. J., F. Aslund, and J. Beckwith. 1998. Disulfide bond formation in the Escherichia coli cytoplasm: an in vivo role reversal for the thioredoxins. EMBO J. 17:5543-5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swindell, S. R., K. H. Benson, H. G. Griffin, P. Renault, S. D. Ehrlich, and M. J. Gasson. 1996. Genetic manipulation of the pathway for diacetyl metabolism in Lactococcus lactis. Appl. Environ. Microbiol. 62:2641-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tobisch, S., D. Zuhlke, J. Bernhardt, J. Stulke, and M. Hecker. 1999. Role of CcpA in regulation of the central pathways of carbon catabolism in Bacillus subtilis. J. Bacteriol. 181:6996-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Unden, G., and J. Bongaerts. 1997. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1320:217-234. [DOI] [PubMed] [Google Scholar]
- 56.Willemoes, M., M. Kilstrup, P. Roepstorff, and K. Hammer. 2002. Proteome analysis of a Lactococcus lactis strain overexpressing gapA suggests that the gene product is an auxiliary glyceraldehyde-3-phosphate dehydrogenase. Proteomics 2:1041-1046. [DOI] [PubMed] [Google Scholar]