Abstract
An operon encoding enzymes of the agmatine deiminase system (AgDS) has been identified in the cariogenic bacterium Streptococcus mutans UA159. The AgDS is regulated by agmatine induction and carbohydrate catabolite repression. Ammonia is produced from agmatine at low pH, suggesting that the AgDS could augment acid tolerance.
Streptococcus mutans is the etiological agent of dental caries (12). A key virulence attribute of this organism is acid tolerance, since catabolism of dietary carbohydrates lowers the pH of dental plaque to values below 4.0 (5). A primary determinant of acid tolerance is the membrane-bound F1F0-ATPase, although reduction in the proton permeability of the cell membrane and induction of DNA repair pathways and stress proteins also contribute to the acid tolerance of S. mutans (17). Another widely used mechanism for acid resistance in less acid-tolerant oral streptococci is the generation of ammonia by the arginine deiminase system (ADS) (6).
S. mutans metabolizes a wider variety of carbohydrates than many gram-positive bacteria, but the ADS is not present in this organism (1, 15). Consequently, when a cluster of genes with similarity to ADS genes (1) was identified in the S. mutans UA159 genome, it was proposed that the genes encoded enzymes of the agmatine deiminase system (AgDS). The AgDS is analogous to the ADS but is involved in catabolism of agmatine (Fig. 1) (1), a decarboxylated derivative of arginine (19). In lactic acid bacteria and Enterococcus faecalis, the AgDS resembles the ADS. However, in Pseudomonas aeruginosa and other organisms, agmatine is produced from arginine via an intracellular arginine decarboxylase, and putrescine is converted to spermidine or broken down to succinate. The identification of an ADS-like gene cluster in S. mutans is significant because agmatine catabolism may contribute to acid tolerance. Production of alkali from agmatine may have critical implications for the pathogenicity of S. mutans and may profoundly affect the ecology of oral biofilms. Thus, we have initiated an analysis of the AgDS gene cluster.
FIG. 1.
Comparison of the ADS and AgDS pathways. Agmatine can be produced from arginine by arginine decarboxylase. AD, arginine deiminase; CK, carbamate kinase.
Analysis of the sequence of the AgD gene cluster.
The first gene in the operon, otcA, encodes a putative putrescine carbamoyltransferase (PTC) and has been redesignated aguB. AguB is 80% identical to ArgF-2, annotated as an ornithine carbamoyltransferase (OTC) in E. faecalis V583 (Fig. 2) (14). ArgF-2 is one of two putative OTCases encoded by the E. faecalis genome (14), although argF-2 is located in an operon encoding an apparent AgD, carbamate kinase, and antiporter. The other OTC gene is in the ADS operon (4), so ArgF-2 may be a PTC (18). The PTC amino acid sequence is not available, but the enzyme is well characterized and distinct from OTC (21).
FIG. 2.
Putative AgDS gene cluster in S. mutans UA159 and percent identity to similarly organized clusters in other bacteria, based on NCBI Blast using sequences obtained from GenBank (National Center for Biotechnology Information). αPort, agmatine:putrescine antiporter; CK, carbamate kinase; N/A, xasA sequence is not available.
According to Prosite (http://www.expasy.org/prosite), the carbamoyltransferase consensus is F-X-(E/K)-X-S-(G/T)-R-T, with the third residue differentiating aspartate (E) and ornithine (K) carbamoyltransferases. This consensus is present in the amino acid sequence of the S. mutans aguB product, as well as E. faecalis ArgF-2, Lactobacillus lactis subsp. lactis OtcA, Listeria monocytogenes LMO0036, and Lactobacillus sakei ArgF, with a highly conserved Q in the third position, perhaps reflecting a preference for putrescine. The HPTQ residues at positions 143 to 146, which are involved in carbamoylphosphate binding, are present in AguB, as are the HCLP residues at positions 281 to 284, which facilitate ornithine binding (11). Since the structures of ornithine and putrescine are identical at the position of cleavage, this conservation is not surprising. The second gene in the operon, SMU.263, encodes a putative amino acid antiporter and has been redesignated aguD. Similar to the case of other amino acid antiporters, 11 transmembrane helices were predicted for the amino acid sequence of AguD by using the “DAS” - Transmembrane Prediction server at http://www.sbc.su.se/∼miklos/DAS (9). The third gene, SMU.264, encodes AgD and has been redesignated aguA. The GGGNIHCITQQ sequence (13) was identified at the C terminus of AguA. The final gene, arcC, encodes a carbamate kinase and has been redesignated aguC to reflect its association with the AgDS.
In addition to aguBDAC, a transcriptional regulator of the LuxR family is located 239 bp upstream of aguB and transcribed in the opposite direction. LuxR-type proteins belong to the FixJ-NarL superfamily, mainly composed of two-component response regulators (10). Homologs of this regulatory protein were identified 399 and 234 bp upstream of the AgDS gene clusters in E. faecalis and L. lactis subsp. lactis, respectively. Involvement of this protein in AgDS regulation is under investigation.
Cotranscription of aguBDAC.
No terminator-like sequences were identified in the agu intergenic regions. To demonstrate cotranscription of aguBDAC, a polar mutation in aguB was constructed by insertion of an Ωkan cassette harboring strong transcription/translation termination signals (16). Northern blotting was performed (3) with 10 μg of total RNA extracted (8) from wild-type and aguB mutant cells grown in low-carbohydrate tryptone-vitamin-based (TV) broth (7) supplemented with 0.5% galactose and 10 mM agmatine. The RNA was hybridized to an aguB probe and labeled by using the Bright Star labeling kit (Ambion Inc., Austin, Tex.). The probe hybridized to a 4.6-kb wild-type mRNA, consistent with the size of the operon (Fig. 3). No transcript was detected in the aguB mutant strain.
FIG. 3.
Northern blot analysis of total RNA isolated from wild-type and aguB mutant S. mutans strains grown in TV broth containing 0.5% galactose and 10 mM agmatine. RNA (10 μg) was hybridized to an aguB DNA probe.
AgDS expression in S. mutans.
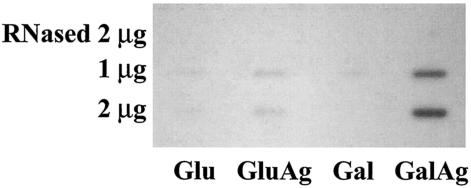
To quantify mRNA under different growth conditions, slot blot analysis was used. Total RNA extracted from cells grown in TV broth plus 0.5% glucose or 0.5% galactose, with or without 10 mM agmatine, was transferred to a 0.45-μm nitrocellulose membrane. The RNA was hybridized to an aguB probe. AgDS-specific mRNAs were detected under all conditions, although expression was severalfold higher when cells were grown in medium containing agmatine (Fig. 4).
FIG. 4.
Slot blot analysis of mRNA isolated from wild-type S. mutans grown in TV broth containing 0.5% glucose (Glu), 0.5% glucose and 10 mM agmatine (GluAg), 0.5% galactose (Gal), or 0.5% galactose and 10 mM agmatine (GalAg). The mRNA was transferred to a nitrocellulose membrane and hybridized to an aguB DNA probe. “RNased” indicates a control in which 2 μg of RNA was treated with RNase prior to application to the membrane.
AgD activity in permeabilized cells grown under different growth conditions was measured by colorimetric determination of N-carbamoylputrescine production from agmatine (Fig. 5A) (2). In wild-type S. mutans, peak AgD activity was observed in cells grown in TV containing the non-catabolite-repressing sugar galactose and agmatine, whereas activity was 65% lower in cells grown in the repressing sugar glucose and agmatine. Thus, the operon is inducible by agmatine and under the control of carbohydrate catabolite repression. No AgD activity was measurable in the strain carrying the polar insertion in aguB, confirming that the operon encodes the AgD enzyme and transcription of the genes occurs from a promoter 5′ to aguB.
FIG. 5.
AgD activity (A) and production of ammonia from agmatine (B) in wild-type (WT) S. mutans grown in TV containing 0.5% glucose (Glu), 0.5% glucose and 10 mM agmatine (GluAg), 0.5% galactose (Gal), or 0.5% galactose and 10 mM agmatine (GalAg). Activity of a polar mutant (AguB−) of S. mutans grown in TV containing 0.5% galactose and 10 mM agmatine was measured.
Ammonia production was measured after cells were incubated with buffer and 10 mM agmatine, using an ammonia detection kit (Diagnostic Chemicals Limited, Charlottetown, Canada). Consistent with AgD activity, ammonia production was observed only in cells grown with agmatine (Fig. 5B). Ammonia production increased two-thirds in cells grown in galactose and agmatine, compared to glucose and agmatine. The optimum pH for ammonia production from agmatine was 4 (Fig. 6).
FIG. 6.
Ammonia production from agmatine at various pHs by wild-type S. mutans grown in TV broth containing 0.5% galactose and 10 mM agmatine.
Summary.
AgDS of S. mutans is functional and under tight genetic control. The system is expressed at a low level relative to urease and arginine deiminase of other oral streptococci, and it is unlikely that agmatine catabolism would cause significant environmental alkalinization. However, ammonia production by the AgDS at low pH would increase ΔpH and provide ATP, which could be used for growth or to extrude protons, and the system is capable of functioning at pH values that are considered extreme in dental plaque (20). Therefore, the AgDS may represent a significant contributor to acid tolerance and thus to the virulence of S. mutans in vivo.
Nucleotide sequence accession number.
The sequences of these genes have been deposited with GenBank and bear accession number BK004003.
Acknowledgments
This research was supported by Public Health Service grant DE10362 from the National Institute of Dental and Craniofacial Research.
REFERENCES
- 1.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 99:14434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archibald, R. M. 1944. Determination of citrulline and allantoin and demonstration of citrulline in blood plasma. J. Biol. Chem. 156:121-142. [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1989. Current protocols in molecular biology. J. Wiley and Sons, New York, N.Y.
- 4.Barcelona-Andres, B., A. Marina, and V. Rubio. 2002. Gene structure, organization, expression and potential regulatory mechanisms of arginine catabolism in Enterococcus faecalis. J. Bacteriol. 184:6289-6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bender, G. R., S. V. W. Sutton, and R. E. Marquis. 1986. Acid tolerance, proton permeabilities and membrane ATPases of oral streptococci. Infect. Immun. 53:331-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burne, R. A., and R. E. Marquis. 2000. Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol. Lett. 193:1-6. [DOI] [PubMed] [Google Scholar]
- 7.Burne, R. A., Z. T. Wen, Y. Y. Chen, and J. E. Penders. 1999. Regulation of expression of the fructan hydrolase gene of Streptococcus mutans GS-5 by induction and carbon catabolite repression. J. Bacteriol. 181:2863-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, Y. Y., and R. A. Burne. 1996. Analysis of Streptococcus salivarius urease expression using continuous chemostat culture. FEMS Microbiol. Lett. 135:223-229. [DOI] [PubMed] [Google Scholar]
- 9.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10:673-676. [DOI] [PubMed] [Google Scholar]
- 10.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35:439-468. [DOI] [PubMed] [Google Scholar]
- 11.Houghton, J. E., D. A. Bencini, G. A. O'Donovan, and J. R. Wild. 1984. Protein differentiation: a comparison of aspartate transcarbamoylase and ornithine transcarbamoylase from Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 81:4864-4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuramitsu, H. K. 1993. Virulence factors of mutans streptococci: role of molecular genetics. Crit. Rev. Oral Biol. Med. 4:159-176. [DOI] [PubMed] [Google Scholar]
- 13.Nakada, Y., Y. Jiang, T. Nishijyo, Y. Itoh, and C. D. Lu. 2001. Molecular characterization and regulation of the aguBA operon, responsible for agmatine utilization in Pseudomonas aeruginosa PAO1. J. Bacteriol. 183:6517-6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paulsen, I. T., L. Banerjei, G. S. A. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Trann, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071-2074. [DOI] [PubMed] [Google Scholar]
- 15.Perch, B., E. Kjems, and T. Ravn. 1974. Biochemical and serological properties of Streptococcus mutans from various human and animal sources. Acta Pathol. Microbiol. Scand. B 82:357-370. [DOI] [PubMed] [Google Scholar]
- 16.Perez-Casal, J., M. G. Caparon, and J. R. Scott. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J. Bacteriol. 173:2617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quivey, R. G., W. L. Kuhnert, and K. Hahn. 2001. Genetics of acid adaptation in oral streptococci. Crit. Rev. Oral Biol. Med. 12:301-314. [DOI] [PubMed] [Google Scholar]
- 18.Roon, R. J., and H. A. Barker. 1972. Fermentation of agmatine in Streptococcus faecalis: occurrence of putrescine transcarbamoylase. J. Bacteriol. 109:44-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simon, J. P., and V. Stalon. 1982. Enzymes of agmatine degradation and the control of their synthesis in Streptococcus faecalis. J. Bacteriol. 152:676-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stephan, R. M. 1940. Changes in hydrogen-ion concentration on tooth surfaces and in carious lesions. Am. J. Dent. 27:718-723. [Google Scholar]
- 21.Wargnies, B., N. Lauwers, and V. Stalon. 1979. Structure and properties of the putrescine carbamoyltransferase of Streptococcus faecalis. Eur. J. Biochem. 101:143-152. [DOI] [PubMed] [Google Scholar]