Abstract
The Bacillus subtilis ResD-ResE two-component regulatory system activates genes involved in nitrate respiration in response to oxygen limitation or nitric oxide (NO). The sensor kinase ResE activates the response regulator ResD through phosphorylation, which then binds to the regulatory region of genes involved in anaerobiosis to activate their transcription. ResE is composed of an N-terminal signal input domain and a C-terminal catalytic domain. The N-terminal domain contains two transmembrane subdomains and a large extracytoplasmic loop. It also has a cytoplasmic PAS subdomain between the HAMP linker and C-terminal kinase domain. In an attempt to identify the signal-sensing subdomain of ResE, a series of deletions and amino acid substitutions were generated in the N-terminal domain. The results indicated that cytoplasmic ResE lacking the transmembrane segments and the extracytoplasmic loop retains the ability to sense oxygen limitation and NO, which leads to transcriptional activation of ResDE-dependent genes. This activity was eliminated by the deletion of the PAS subdomain, demonstrating that the PAS subdomain participates in signal reception. The study also raised the possibility that the extracytoplasmic region may serve as a second signal-sensing subdomain. This suggests that the extracytoplasmic region could contribute to amplification of ResE activity leading to the robust activation of genes required for anaerobic metabolism in B. subtilis.
Two-component regulatory systems in the prokaryote and lower eukaryote play a major role in the adaptation of cells to diverse changes in environmental conditions. A soil gram-positive bacterium, Bacillus subtilis, copes with oxygen limitation by switching to anaerobic respiration using nitrate as a terminal electron acceptor (for a review, see reference 21). Expression of the genes required for the alteration to anaerobic metabolism is activated by the ResD-ResE signal transduction system, which is also involved in aerobic respiration (22, 33). ResE, a membrane-bound histidine sensor kinase, perceives a signal (or signals) related to oxygen limitation and undergoes autophosphorylation at a conserved histidine residue. The level of phosphorylation of ResD is determined by the balance of the two activities possessed by ResE, a phosphate donor for ResD and a phosphatase of ResD-phosphate. Since the activity of ResD as a transcriptional activator is modulated by the level of phosphorylation, the output of signal transduction is determined by switches of ResE activity between kinase-biased and phosphatase-biased forms. A previous study strongly suggested that the kinase and phosphatase activities of ResE are reciprocally regulated by oxygen level (19). Two important questions remain unsolved regarding how ResE responds to oxygen limitation. First, what is the ligand for ResE which reflects oxygen limitation if it is not oxygen per se? Second, which part of ResE is involved in signal sensing?
ResE is predicted to consist of two transmembrane helices (TM1 and TM2) and a long extracytoplasmic (periplasmic) region flanked by the transmembrane subdomains (Fig. 1). The second transmembrane region is followed by a HAMP (histidine kinase, adenylyl cyclase, methyl-accepting chemotaxis protein, and phosphatase) linker and a PAS subdomain. The N-terminal signal input domain, composed of these subdomains, is connected to a conserved kinase or transmitter domain. The periplasmic subdomain and the PAS subdomain of various sensor kinases are reported to be involved in sensing signals, and the HAMP linker is known to transmit signals between the input and output domains. Transmembrane sensor kinases often utilize an extracytoplasmic sensor subdomain to receive external stimuli. A B. subtilis sensor histidine kinase, ComP, senses an extracellular pheromone, ComX, leading to the initiation of competence development for genetic transformation. The deletion of the second extracytoplasmic loop of ComP conferred ComX-independent kinase activity (25). The periplasmic region of PhoQ kinase of the PhoP-PhoQ two-component regulatory system in Salmonella enterica and Escherichia coli specifically binds Mg2+ in vitro and is essential in vivo to activate PhoP-dependent transcription (4, 36, 37). The periplasmic region of CitA kinase, which is required for the expression of the citrate fermentation genes in Klebsiella pneumoniae, binds citrate (11). A recent study showed that the crystal structure of the CitA periplasmic subdomain in complex with citrate reveals a PAS fold (27).
FIG. 1.
Schematic diagram of the putative subdomain organization of ResE and of mutant ResE constructs. The pDG148-derived plasmids carrying resE are listed. All resE expression systems utilize the IPTG-inducible Pspac promoter and the resE SD sequence (E) or the vector-derived SD sequence (V). Numbers in parentheses represent start and end points of amino acid deletions. Solid lines show regions cloned into plasmid pDG148, and dotted lines show deleted regions. The sites and identities of amino acid substitutions in the HAMP subdomain are indicated (X). The top panel shows the conserved histidine-374 (H) and ATPase subdomains in the kinase domain and the two transmembrane subdomains (TM1 and TM2), HAMP, and PAS subdomains in the signal input domain.
PAS subdomains, known to be important for signal sensing like the periplasmic subdomain, have been identified both in prokaryotic and eukaryotic proteins as being involved in sensing light, oxygen, and the redox state of the cells. Ligands interacting with PAS subdomains include flavin adenine dinucleotide, heme, 2Fe-2S cluster, and 4-hydroxycinnamoyl (for reviews, see references 35 and 43). The best-studied kinase that possesses a PAS subdomain for signal sensing is FixL, which is required for nitrogen fixation in response to oxygen limitation (10, 14). The FixL heme subdomain has a PAS fold, and oxygen binds to the ferrous iron of heme, which inhibits kinase activity. In some cases PAS subdomains are involved in protein-protein interaction as well as ligand binding. Kinase A of the B. subtilis phosphorelay that controls sporulation initiation has three PAS subdomains (A, B, and C); PAS-B and PAS-C have been implicated in dimerization, and PAS-A has been shown to be important for autophosphorylation activity and is required for either signal sensing or structural integrity or both (39). A further study showed that the PAS-A subdomain binds ATP and catalyzes the exchange of phosphate between ATP and nucleoside diphosphates (31). ATP hydrolysis in the PAS subdomain was proposed to drive the conformational changes that activate or deactivate the kinase in response to signal-ligand binding.
Although the HAMP subdomain is not directly involved in signal sensing, it plays an important role in signal transmission. Many membrane-bound sensors, including E. coli NarX, have a HAMP linker immediately adjacent to a transmembrane region on the inside of the cytoplasmic membrane. HAMP linkers are predicted to consist of two amphipathic α-helices (AS-1 and AS-2) (for a review, see reference 41). The HAMP subdomain of NarX transmits the signal received by the P box of the periplasmic region (3, 6) to the output domain (1). Similarly, mutational analyses suggested that the periplasmic subdomain of E. coli EnvZ is required for sensing osmolarity signals (40), and its HAMP linker region serves to transmit the signal to the kinase domain (24).
Despite the importance of the ResD-ResE signal transduction system in aerobic and anaerobic respiration, no study has been undertaken to determine how ResE senses a signal. Oxygen itself is unlikely to be the signal for ResE because ResDE is also required for aerobic respiration (33). The presence of the long extracytoplasmic loop of ResE may suggest that the region is involved in sensing an extracellular signal as has been shown in kinases of similar architecture. Alternatively, the PAS subdomain may be the signal-sensing region and the membrane anchoring could simply be needed for efficient interaction between the PAS subdomain and the signal generated from some membrane-associated source. In this study deletion and amino acid substitution analysis of each putative signal input subdomain of ResE were carried out to determine which region is important for signal reception.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
All B. subtilis strains used in this study are derivatives of JH642 (trpC2 pheA1) (Table 1). E. coli DH5α was used to propagate pDG148-derived plasmids, and transformants were selected on Luria broth agar supplemented with 25 μg of ampicillin per ml. The multicopy plasmid pDG148 (32) is a shuttle vector between E. coli and B. subtilis and carries the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Pspac promoter that was used in this study to produce wild-type and mutant ResE. The plasmids were introduced into a B. subtilis resE mutant (LAB2234) (16), and transformants were selected on Difco sporulation agar (17) supplemented with 5 μg of neomycin per ml and 75 μg of spectinomycin per ml. Two ResDE-dependent promoters (nasD and hmp promoters) fused to a lacZ reporter gene were used to evaluate the effects of deletions and amino acid substitutions of ResE. SPβ phage lysate bearing nasD-lacZ (18) or hmp-lacZ (20) was used to transduce resE mutants that carry plasmids encoding various ResE proteins, and transductants were selected for chloramphenicol (Cm) resistance (5 μg/ml). Strain ORB4724 (narGH resE) was generated by transforming LAB2408 (narGH::ble) with chromosomal DNA prepared from LAB2234. Strain ORB4724 was transformed with various resE-carrying plasmids, and each transformant was lysogenized with SPβ bearing hmp-lacZ as described above.
TABLE 1.
B. subtilis strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| JH642 | trpC2 pheA1 | J. A. Hoch |
| LAB2234 | trpC2 pheA1 ΔresE::spc | 16 |
| LAB2408 | trpC2 pheA1 ΔnarGH::ble | 16 |
| ORB4327 | trpC2 pheA1 ΔresE::spc pMMN525 | This study |
| ORB4332 | trpC2 pheA1 ΔresE::spc pMMN525 SPβc2del2::Tn917::pMMN392 | This study |
| ORB4333 | trpC2 pheA1 ΔresE::spc pMMN525 SPβc2del2::Tn917::pML107 | This study |
| ORB4356 | trpC2 pheA1 ΔresE::spc pAB3 | This study |
| ORB4371 | trpC2 pheA1 ΔresE::spc pAB3 SPβc2del2::Tn917::pMMN392 | This study |
| ORB4372 | trpC2 pheA1 ΔresE::spc pAB3 SPβc2del2::Tn917::pML107 | This study |
| ORB4387 | trpC2 pheA1 ΔresE::spc pAB4 | This study |
| ORB4410 | trpC2 pheA1 ΔresE::spc pAB4 SPβc2del2::Tn917::pMMN392 | This study |
| ORB4411 | trpC2 pheA1 ΔresE::spc pAB4 SPβc2del2::Tn917::pML107 | This study |
| ORB4441 | trpC2 pheA1 ΔresE::spc pMMN534 | This study |
| ORB4446 | trpC2 pheA1 ΔresE::spc pMMN534 SPβc2del2::Tn917::pMMN392 | This study |
| ORB4447 | trpC2 pheA1 ΔresE::spc pMMN534 SPβc2del2::Tn917::pML107 | This study |
| ORB4476 | trpC2 pheA1 ΔresE::spc pMMN536 | This study |
| ORB4477 | trpC2 pheA1 ΔresE::spc pMMN537 | This study |
| ORB4481 | trpC2 pheA1 ΔresE::spc pMMN536 SPβc2del2::Tn917::pMMN392 | This study |
| ORB4482 | trpC2 pheA1 ΔresE::spc pMMN536 SPβc2del2::Tn917::pML107 | This study |
| ORB4484 | trpC2 pheA1 ΔresE::spc pMMN537 SPβc2del2::Tn917::pMMN392 | This study |
| ORB4485 | trpC2 pheA1 ΔresE::spc pMMN537 SPβc2del2::Tn917::pML107 | This study |
| ORB4620 | trpC2 pheA1 ΔresE::spc pMMN544 | This study |
| ORB4622 | trpC2 pheA1 ΔresE::spc pMMN544 SPβc2del2::Tn917::pMMN392 | This study |
| ORB4623 | trpC2 pheA1 ΔresE::spc pMMN544 SPβc2del2::Tn917::pML107 | This study |
| ORB4711 | trpC2 pheA1 ΔresE::spc pMMN559 | This study |
| ORB4717 | trpC2 pheA1 ΔresE::spc pMMN559 SPβc2del2::Tn917::pMMN392 | This study |
| ORB4718 | trpC2 pheA1 ΔresE::spc pMMN559 SPβc2del2::Tn917::pML107 | This study |
| ORB4724 | trpC2 pheA1 ΔresE::spc ΔnarGH::ble | This study |
| ORB4755 | trpC2 pheA1 ΔresE::spc pMMN563 | This study |
| ORB4756 | trpC2 pheA1 ΔresE::spc pMMN564 | This study |
| ORB4760 | trpC2 pheA1 ΔresE::spc pMMN563 SPβc2del2::Tn917::pMMN392 | This study |
| ORB4761 | trpC2 pheA1 ΔresE::spc pMMN563 SPβc2del2::Tn917::pML107 | This study |
| ORB4763 | trpC2 pheA1 ΔresE::spc pMMN564 SPβc2del2::Tn917::pMMN392 | This study |
| ORB4764 | trpC2 pheA1 ΔresE::spc pMMN564 SPβc2del2::Tn917::pML107 | This study |
| ORB4765 | trpC2 pheA1 ΔresE::spc pMMN565 | This study |
| ORB4773 | trpC2 pheA1 ΔresE::spc pMMN565 SPβc2del2::Tn917::pMMN392 | This study |
| ORB4774 | trpC2 pheA1 ΔresE::spc pMMN565 SPβc2del2::Tn917::pML107 | This study |
| ORB4782 | trpC2 pheA1 ΔresE::spc ΔnarGH::ble pMMN525 SPβc2del2::Tn917::pML107 | This study |
| ORB4783 | trpC2 pheA1 ΔresE::spc ΔnarGH::ble pMMN563 SPβc2del2::Tn917::pML107 | This study |
| ORB4784 | trpC2 pheA1 ΔresE::spc ΔnarGH::ble pMMN565 SPβc2del2::Tn917::pML107 | This study |
| ORB4790 | trpC2 pheA1 ΔresE::spc ΔnarGH::ble pMMN534 SPβc2del2::Tn917::pML107 | This study |
| ORB4791 | trpC2 pheA1 ΔresE::spc ΔnarGH::ble pMMN544 SPβc2del2::Tn917::pML107 | This study |
| ORB4792 | trpC2 pheA1 ΔresE::spc ΔnarGH::ble pMMN564 SPβc2del2::Tn917::pML107 | This study |
| Plasmids | ||
| pDG148 | E. coli/B. subtilis shuttle plasmid; Ampr Neor | 32 |
| pAB3 | pDG148 encoding cytoplasmic ResE without HAMP subdomain | This study |
| pAB4 | pDG148 encoding cytoplasmic ResE without HAMP and PAS subdomains | This study |
| pMMN525 | pDG148 encoding full-length ResE (own SD sequence) | This study |
| pMMN534 | pDG148 encoding cytoplasmic ResE with transmembrane subdomain 2 | This study |
| pMMN536 | pDG148 encoding full-length ResE with a mutation in HAMP (A207V) | This study |
| pMMN537 | pDG148 encoding full-length ResE with a mutation in HAMP (E226D) | This study |
| pMMN544 | pDG148 encoding full-length ResE without HAMP subdomain | This study |
| pMMN559 | pDG148 encoding full-length ResE without PAS subdomain | This study |
| pMMN563 | pDG148 encoding full-length ResE (vector SD sequence) | This study |
| pMMN564 | pDG148 encoding cytoplasmic ResE without PAS subdomain | This study |
| pMMN565 | pDG148 encoding cytoplasmic ResE (cytoplasmic region) | This study |
Construction of plasmids carrying resE and resE derivatives.
The resE gene was amplified by PCR using the two primers oMN00-109 and oMN00-110 and chromosomal DNA prepared from JH642 as a template. The resE gene, first cloned into an integration plasmid, was isolated by digesting with HindIII and SphI and subsequently cloned into the plasmid pDG148 digested with the same enzymes to generate the plasmid pMMN525. These multiple steps are in accordance with the original plan in which various resE genes were integrated at the amyE locus under control of the Pspac promoter; however, this strategy was not feasible because truncated cytoplasmic ResE proteins were observed to be less stable compared to the full-length ResE (see Results). Therefore, a multicopy pDG148 plasmid was used instead to overproduce ResE to allow the evaluation of phenotypes of the less stable mutant ResE.
The plasmid pMMN525 has a full-length intact resE gene including its own Shine-Dalgarno (SD) sequence downstream of the Pspac promoter. The following pDG148-derived plasmids carrying four mutant resE genes were constructed from pMMN525: pMMN544, pMMN536, pMMN537, and pMMN559. The plasmid pMMN544, which is identical to pMMN525 except with a deletion in the HAMP subdomain, was constructed by two-step PCR. Two overlapped PCR products were generated from pMMN525 with the primers oPspacup/oMN03-229 and oPspacdn/oMN03-228. The PCR products were used as templates for the second PCR using the primers oPspacup and oPspacdn. The resultant PCR fragment, after digestion with HindIII and SphI, was cloned into the plasmid pDG148. The plasmids pMMN536 and pMMN537 each have an amino acid substitution in the HAMP subdomain (Ala-207 to Val and Glu-226 to Asp, respectively); they were constructed in a similar way involving two-step PCR except that the primer pairs oPspacup/oMN03-225 and oPsapcdn/oMN03-224 (for pMMN536) and oPspacup/oMN03-227 and oPspacdn/oMN03-226 (for pMMN537) were used in the first PCR. The plasmid pMMN559 lacks the PAS subdomain, which was constructed by replacing a BssHII-SphI fragment which contains the PAS subdomain by a BssHII-SphI fragment of pMMN564. The plasmid pMMN564 harbors a resE gene encoding a truncated ResE which lacks the PAS subdomain (see below).
Other sets of resE constructs were made in which the pDG148-derived SD sequence was utilized instead of the resE SD sequence. The plasmid pMMN563 carrying a full-length intact resE was constructed by cloning the PCR product amplified with the primers oMN03-242 and oMN03-214 into the plasmid pDG148 digested with SalI and SphI. The resE gene, other than the SD sequence, in pMMN563 was identical to that in the plasmid pMMN525. pMMN565 was generated by cloning the PCR product amplified with the primers oMN03-243 and oMN02-214 and encodes a truncated cytoplasmic ResE lacking two transmembrane subdomains and the extracytoplasmic region. ResE proteins produced by the plasmids pAB3 and pAB4 are the truncated ResE that lack the HAMP and the HAMP plus PAS subdomains, respectively. The resE genes in plasmids pAB3 and pAB4 were generated by PCR using the primers oMN02-215/oMN02-214 and oMN02-216/0MN02-214, respectively. A truncated resE gene lacking a region encoding the PAS subdomain was generated by two-step PCR as described above, with the primer pairs oPspacup/oMN02-218 and oPspacdn/oMN02-217 used in the first reactions and oPspacup/oPspacdn used in the second. The resE gene was cloned into pDG148, which was cleaved with SalI and SphI, to generate pMMN564.
The oligonucleotide primers used for construction of the plasmids are listed in Table 2, and the resE sequences in the plasmids were confirmed by DNA sequence analysis. The location and nature of the resE deletions and mutations are schematically presented in Fig. 1.
TABLE 2.
Oligonucleotide primers used in this study
| Name | Primer sequence (5′ to 3′)a |
|---|---|
| oPspacup | GACTTTATCTACAAGGTGTG |
| oPspacdn | AAATGATGACCTCGTTTCCA |
| oMN00-109 | GAACAAGGTGGATCCGGAAGCTTCGAAAAAAAT TGTCACCGTA |
| oMN00-110 | CTTATGGAACAGGATCCTTCGGCAAATTCAGA |
| oMN02-214 | CGCACCGCATGCAAATTCAGACTCGATTTT |
| oMN02-215 | GGAAGCGTCGACATGCTCAATCAAGAAAAA |
| oMN02-216 | GCATTCGTCGACATGATTGAGATGACGCTT |
| oMN02-217 | CTCAATCAAGAAAAACAGCAAATGATTGAGATG |
| oMN02-218 | CATCTCAATCATTTGCTGTTTTTCTTGATTGAG |
| oMN02-219 | GGAATGGTCGACATGGAGTATATTTTTCTTGCCGCT |
| oMN03-224 | AAAAATGAGAGAAGGCGTGCAGGATTTGGCGAA |
| oMN03-225 | TTCGCCAAATCCTGCACGCCTTCTCTCATTTTT |
| oMN03-226 | ATTTTAACTCAGGATGATATCGGTGAACTGGC |
| oMN03-227 | GCCAGTTCACCGATATCATCCTGAGTTAAAAT |
| oMN03-228 | CGCATTCTTTTTATCAAGCCTCAATCAAGAAAAAGAGC |
| oMN03-229 | GCTCTTTTTCTTGATTGAGGCTTGATAAAAAGAATGCG |
| oMN03-242 | GGCGGAGTCGACATGAAATTTTGGAAAAGC |
| oMN03-243 | AATAATGTCGACATGGTCACGTACCCTCTC |
Restriction enzyme sites are underlined.
Cell fractionation.
B. subtilis cells were cultured in 35 ml of 2× yeast-tryptone (YT) medium (17) with or without 1 mM IPTG. At the mid-log phase of growth (optical density at 600 nm [OD600] of around 0.5), cells were harvested and resuspended with 5 ml of 20 mM potassium phosphate buffer (pH 7.5)-15 mM MgCl2-20% sucrose. Lysozyme (1 mg/ml) was added, and the suspension was incubated by gentle shaking at 37°C for 30 min. The protoplasts were collected by centrifugation at 7,000 × g for 5 min. The sedimented protoplasts were lysed by resuspension in 5 ml of lysis buffer (30 mM Tris-HCl [pH 8.0]-1 mM EDTA). The cells were completely lysed by being passed three times through a thin needle, yielding the whole-cell extract. The whole-cell extract was centrifuged at 27,000 × g for 30 min at 4°C in a Ti70 rotor (Beckman) to recover supernatant as cytoplasmic fraction. The pellet was carefully resuspended in 5 ml of lysis buffer containing 0.5% Triton X-100. After centrifugation at 27,000 × g for 30 min at 4°C the supernatant was kept as the membrane fraction.
Western blot analysis.
The protein concentration of whole-cell extract, cytoplasmic fraction, and membrane fraction prepared above was determined using the Bio-Rad protein assay solution. Equal amounts of protein (6 μg for whole-cell extract, 5 μg for cytoplasmic fraction, and 2.5 μg for membrane fraction) were loaded onto 12% sodium dodecyl sulfate (SDS) polyacrylamide gels. After electrophoresis, the proteins were electrotransferred to a nitrocellulose filter and were probed with anti-ResE antibody as previously described (19). In order to verify the accuracy of cell fractionation, immunoblot analysis was carried out with antibodies against cytochrome aa3 quinol oxidase subunit II (QoxA, a membrane protein) and phosphoglycerate mutase (Pgm, a cytoplasmic protein). Anti-QoxA and anti-Pgm sera were kindly provided by Claes von Wachenfeldt and Peter Setlow, respectively.
Measurement of β-galactosidase activity.
Cells were grown aerobically and anaerobically in 2× YT supplemented with 1% glucose, 0.2% potassium nitrate, and appropriate antibiotics. IPTG (0.02 or 1 mM) was added as indicated. The anaerobic cultures were performed by filling the cell suspension to the top of tubes as previously described (22). Cells were inoculated from cultures grown overnight on Difco sporulation agar medium (starting OD600 of 0.02). Samples were withdrawn at time intervals and β-galactosidase activity was determined as previously described (17) and shown as Miller units (13).
The effect of nitric oxide (NO) on hmp-lacZ expression was examined in narGH resE cells carrying various resE-containing plasmids. Since the narGH mutant lacks respiratory nitrate reductase activity, the effect of endogenous NO produced through nitrate reduction could be eliminated (15). Cells were grown anaerobically in serum bottles filled with 2× YT supplemented with 0.5% glucose, 0.5% pyruvate, appropriate antibiotics, and 1 mM (or 0.02 mM when indicated) IPTG. Cells at the mid-log phase of growth (OD600 of approximately 0.3) were treated with 10 μM NO (using 1.8 mM saturated solution prepared from NO gas) as described previously (15). Cells were harvested for measurement of β-galactosidase activity after incubation for 30 min in the presence and absence of NO.
RESULTS
Construction and expression of resE carrying subdomain deletion and mutation.
To localize regions of ResE that are important for oxygen-dependent regulation, a series of deletions from the 5′ end of resE as well as internal in-frame deletions in the putative signal input subdomains were constructed as described in Materials and Methods (Fig. 1). In addition, two amino acid substitutions in the HAMP subdomain were generated. All resE genes were cloned into a multicopy plasmid, pDG148, so that expression of resE was controlled by the addition of IPTG. As positive controls two constructs were made carrying a full-length resE with its own SD sequence (pMMN525) or pDG148-derived SD sequence (pMMN563). All other resE constructs carrying transmembrane subdomains and the extracytoplasmic loop (called membrane-bound ResE) had the resE SD sequence. Truncated cytoplasmic resE constructs (cytoplasmic ResE) and resE with TM2 carry the pDG148-derived SD sequence.
Before examining the effect of the resE subdomain mutations on ResDE-dependent gene expression, we determined whether the mutant proteins were produced at a similar level as that of the full-length protein by Western analysis with anti-ResE antibody (Fig. 2A). Comparison of ResE produced by the two full-length, intact resE constructs (encoded by pMMN563 and pMMN525) indicates that the vector-derived SD sequence is stronger than the native SD sequence (Fig. 2A, compare lanes l and 2). We then compared the concentration of ResE in these resE plasmid-carrying strains to that in wild-type cells. Figure 2B shows that a much lower amount of ResE was produced in aerobic wild-type cultures compared to anaerobic cultures (lane 1) as expected from the previous finding that the resE gene is primarily transcribed from a ResDE-dependent promoter (33). In contrast, ResE production was insensitive to the oxygen level when the resE gene was expressed from the IPTG-inducible Pspac promoter (Fig. 2B, lanes 2 and 3). Consequently, the level of ResE during aerobic growth is much higher in cells carrying resE on the multicopy plasmid than in wild type. Under anaerobic conditions ResE was slightly more abundant in cells carrying pMMN525 (resE SD sequence) but much more abundant in cells bearing pMMN563 (vector SD sequence) than in wild-type cells containing a single copy of resE. These results indicated that ResE was overproduced in the cells carrying multicopy resE under the control of the Pspac promoter. The system is appropriate to assess the ability of ResE to sense oxygen limitation because the expression of resE is relieved from oxygen-dependent transcriptional regulation.
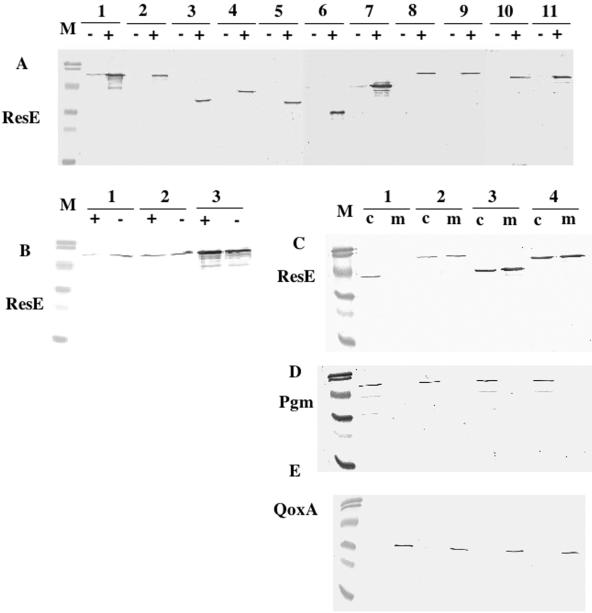
FIG. 2.
Production of the wild-type and mutant ResE proteins. (A) resE mutant cells carrying various resE-carrying plasmids shown in Fig. 1 were grown aerobically in 2× YT in the absence (−) or presence (+) of 1 mM IPTG. Equal amounts (6 μg) of total protein from each whole-cell lysate were resolved by 12% SDS-polyacrylamide gel electrophoresis and probed by using anti-ResE antibody. Lane 1, cells carrying pMMN563; lane 2, pMMN525; lane 3, pMMN564; lane 4, pMMN565; lane 5, pAB3; lane 6, pAB4; lane 7, pMMN534; lane 8, pMMN536; lane 9, pMMN537; lane 10, pMMN544; and lane 11, pMMN559. (B) Whole-cell lysate was prepared from aerobic (+) and anaerobic (−) cultures of wild-type JH642 (lane 1) or resE mutants carrying the plasmids pMMN525 (lane 2) and pMMN563 (lane 3). Western analysis was done as described for panel A. (C to E) Whole-cell lysate from IPTG-induced cultures was further separated into cytoplasmic (c) and membrane (m) fractions. Equal protein samples (5 μg from the cytoplasmic fraction and 2.5 μg from the membrane fraction) were loaded onto a 12% SDS-polyacrylamide gel. Western analysis was done by using anti-ResE antibody, anti-Pgm antibody, and anti-QoxA antibody. Pgm and QoxA localize to the cytoplasm and membrane, respectively. Lanes 1, cells carrying pMMN565; lanes 2, pMMN525; lanes 3, pMMN534; and lanes 4, pMMN563. Sizes of molecular mass markers (M) are 108.0, 90.0, 50.7, 35.5, 28.6, and 21.2 kDa.
Although other truncation mutants utilize the vector SD sequence (Fig. 2A, lanes 3 to 7), only the strain carrying the plasmid pMMN534 that encodes ResE with TM2 produces ResE at a level similar to that of the wild-type ResE translated from the same SD sequence (Fig. 2A, lanes 1 and 7. This result suggests that the cytoplasmic ResE variants are less stable because of a difference in mRNA stability, altered protein folding, or lack of membrane anchoring. Membrane-bound ResE mutants carrying deletions or mutations (lanes 8 to 11) produced equal amounts of proteins as wild-type ResE (lane 2). All of the proteins correspond to the predicted molecular weights and are produced only in the presence of IPTG, except for the full-length ResE (lane 1) and ResE with TM2 (lane 7), both of which utilized the vector SD sequence. This result implied that small amounts of protein produced by the leaky Pspac promoter were stabilized by membrane anchoring.
Because one of the questions we want to answer is whether the membrane anchoring of ResE is important for signal sensing, localization of four representative ResE proteins was examined by cell fractionation. To verify the effectiveness of the cell fractionation method, antibodies against a cytoplasmic protein, Pgm, and a membrane protein, QoxA, were used in Western analysis of cytoplasmic and membrane fractions (Fig. 2D and E). The result showed that cytoplasmic and membrane fractions were properly separated in each sample. Figure 2C shows that truncated ResE localized exclusively in the cytoplasmic fraction (lane 1), while the two full-length intact ResE proteins (lanes 2 and 4) and ResE with TM2 localized to both the membrane and cytoplasm (lane 3). Some proportions of full-length and TM2 ResE remained in the cytoplasm probably because of the proteins' overproduction. Since the localization patterns were similar between the full-length ResE protein and the TM2-bearing ResE, we concluded that TM2 is sufficient to anchor ResE to the membrane.
Effect of ResE levels on the expression of nasD and hmp.
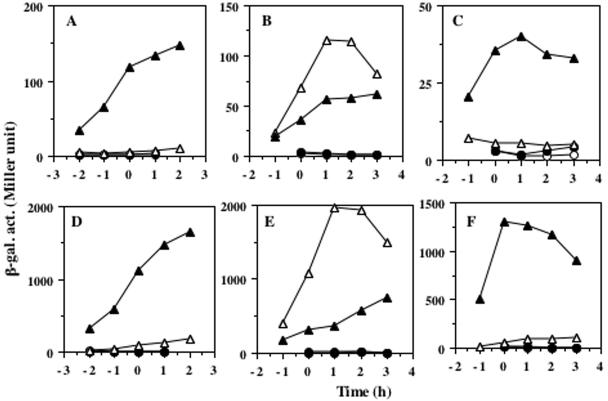
Purification of membrane-bound sensor kinase has been difficult, and most of the in vitro work has been carried out by using truncated soluble kinases, with a few exceptions (12, 26, 30, 38). This is an obstacle to studying the signal-sensing mechanisms of sensor kinases in vitro. Therefore, we first investigated whether a truncated soluble ResE still senses and responds to oxygen limitation by measuring the ResDE-dependent expression of nasD-lacZ and hmp-lacZ under aerobic and anaerobic conditions. Cells carrying the full-length resE (with the resE SD sequence) expressed nasD and hmp only under oxygen-limited conditions, and the expression was largely IPTG dependent (Fig. 3A and D). The expression of nasD and hmp in the strain producing the truncated cytoplasmic ResE was also induced under anaerobic conditions with IPTG, although the levels of expression were lower than those in cells with the full-length ResE (Fig. 3C and F). This result clearly demonstrated that the cytoplasmic region of ResE is at least partly, if not solely, responsible for perceiving the signal derived from oxygen limitation. The full-length ResE with the vector-derived SD sequence also responds to oxygen-limitation; however, the expression was unexpectedly higher in the absence of IPTG than in the presence of IPTG (Fig. 3B and E).
FIG. 3.
Expression of nasD-lacZ and hmp-lacZ in cells grown under aerobic and anaerobic conditions. resE strains carrying the plasmid pMMN525 (full-length resE with resE SD sequence) (A and D), pMMN563 (full-length resE with vector SD sequence) (B and E), and pMMN565 (cytoplasmic resE with vector SD sequence) (C and F) were grown in 2× YT supplemented with 1% glucose and 0.2% potassium nitrate and in the absence or presence of IPTG. β-Galactosidase activities (β-gal. act.) of nasD-lacZ (A to C) and hmp-lacZ (D to F) were measured at time intervals. ○, aerobic growth in the absence of IPTG; •, aerobic growth in the presence of 1 mM IPTG; ▵, anaerobic growth in the absence of IPTG: ▴, anaerobic growth in the presence of 1 mM IPTG. Time zero indicates the end of the exponential growth.
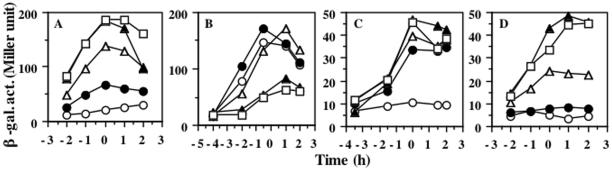
One possible reason that the full-length ResE produced from the vector-derived SD sequence has a negative effect on nasD-lacZ and hmp-lacZ expression when the expression was increased by the addition of IPTG is that the overproduced ResE may be inhibitory to ResD activity. Overproduction of the histidine kinase domain of UhpB is known to inhibit action of the cognate response regulator UhpA in E. coli (42). A similar negative effect by overproduction of kinase on expression of its target genes was observed in PhoR, which is involved in expression of the phosphate (Pho) regulon in B. subtilis (28). If this is the case, one could imagine that another ResE variant carrying only TM2, which was shown to be overproduced (Fig. 2), would have a similar negative effect on nasD expression when IPTG was added in the culture medium. This hypothesis was tested in the experiment whose results are shown in Fig. 4. Four strains carrying various resE genes were grown anaerobically in the absence and presence of different concentrations of IPTG. The amount of ResE proteins increased in parallel to the concentration of IPTG (data not shown). The expression of nasD in the strains that produce the full-length ResE (resE SD sequence) (Fig. 4A) and the cytoplasmic ResE (Fig. 4D) increases in proportion to the concentration of IPTG. In contrast, nasD expression in the strain producing the full-length ResE (vector SD sequence) was fully activated in the absence or presence of lower levels of IPTG, but increasing the amount of IPTG to 0.2 mM and above dramatically reduced expression of nasD-lacZ (Fig. 4B), which was in keeping with the result presented above (Fig. 3). Increasing the amount of IPTG did not result in reduced nasD expression in cells producing TM2-carrying ResE; instead, no induction of nasD-lacZ was observed in the absence of IPTG (Fig. 4C). This result suggests that overexpression of only the cytoplasmic portion of ResE does not cause a negative effect on ResDE-dependent gene expression, unlike the case of overproduction of UhpB. Rather, the result can be explained by an inhibitory effect caused by the overproduction of the extracytoplasmic region.
FIG. 4.
Effect of IPTG concentration on nasD-lacZ expression. resE strains carrying the plasmid pMMN525 (full-length resE with resE SD sequence) (A), pMMN563 (full-length resE with vector SD sequence) (B), pMMN534 (TM2-carrying resE with vector SD sequence) (C), and pMMN565 (cytoplasmic resE with vector SD sequence) (D) were grown as described in the legend of Fig. 3 in the absence of IPTG (○) or in 0.02 mM IPTG (•), 0.05 mM IPTG (▵), 0.2 mM IPTG (▴), or 1 mM IPTG (□). Time zero indicates the end of exponential growth. β-Galactosidase activity (β-gal. act.) is shown in Miller units.
Effect of ResE subdomain mutations on the aerobic and anaerobic expression of nasD and hmp.
Aerobic and anaerobic expression of nasD and hmp was examined in cells carrying plasmids that encode ResE variants (Table 3). IPTG was added at a concentration of 1 mM for all strains except the strain carrying the plasmid pMMN563, to which 0.02 mM was added in accordance with the result described above and to maintain nearly equal intracellular ResE concentrations, based on immunoblot analysis (data not shown). Two amino acid substitutions in the HAMP subdomain were constructed in the membrane-bound ResE. These substitutions were chosen because the corresponding mutations (Ala-207 to Val in AS-1 and Glu-226 to Asp in the C-terminal end of the connector) in NarX result in impaired and constitutive phenotypes, respectively (1). These mutations, unlike those of NarX, had no significant effect on nasD and hmp regulation except that an overall higher level of activity was detected in cells carrying resE (E226D). The aerobic expression of both genes was partially derepressed by the complete deletion of the HAMP subdomain (pMMN544). These results indicate that the HAMP subdomain has only a minor role in transmitting the signal from the extracytoplasmic region to the output domain. On the contrary, the in-frame deletion of the PAS subdomain almost completely abolished gene expression, indicating that the PAS subdomain is essential for the activity of ResE.
TABLE 3.
Effect of deletions and amino acid substitutions in ResE on ResDE-dependent gene expression under aerobic and anaerobic conditions
| Plasmid | Relevant characteristic (subdomain)b | β-Galactosidase activity (Miller units)a
|
|||
|---|---|---|---|---|---|
|
nasD-lacZ
|
hmp-lacZ
|
||||
| +O2 | −O2 | +O2 | −O2 | ||
| pMMN525 | Full | 1.0 ± 0.3 | 163 ± 21 | 9.0 ± 0.5 | 1965 ± 354 |
| pMMN536 | Full (HAMP A207V) | 1.0 ± 0.0 | 122 ± 6.0 | 7.0 ± 2.0 | 1834 ± 185 |
| pMMN537 | Full (HAMP E226D) | 2.5 ± 0.5 | 354 ± 34 | 17 ± 2.0 | 2507 ± 211 |
| pMMN544 | Full (− HAMP) | 8.0 ± 1.7 | 259 ± 56 | 41 ± 5.0 | 2730 ± 118 |
| pMMN559 | Full (− PAS) | 1.5 ± 0.3 | 3.0 ± 0.0 | 12 ± 6.0 | 110 ± 17 |
| pMMN563 | Full | 1.0 ± 0.2 | 177 ± 5.0 | 12 ± 4.0 | 1856 ± 302 |
| pMMN534 | Cytoplasmic (+ TM2) | 1.0 ± 0.0 | 37 ± 7.0 | 10 ± 3.0 | 654 ± 26 |
| pMMN565 | Cytoplasmic | 1.0 ± 0.3 | 39 ± 6.0 | 8.0 ± 0.5 | 858 ± 167 |
| pMMN564 | Cytoplasmic (− PAS) | 1.0 ± 0.0 | 2.0 ± 0.0 | 6.0 ± 1.0 | 59 ± 2.0 |
| pAB3 | Cytoplasmic (− HAMP) | 1.0 ± 0.0 | 11 ± 1.0 | 6.0 ± 1.5 | 259 ± 33 |
| pAB4 | Cytoplasmic (− HAMP, − PAS) | 1.0 ± 0.0 | 2.0 ± 0.0 | 12 ± 6.0 | 95 ± 18 |
Cells were grown in 2× YT supplemented with 1% glucose, 0.2% potassium nitrate, and 1 mM IPTG (0.02 mM IPTG in cells carrying pMMN563). Samples were taken at time intervals for measurement of β-galactosidase activity under aerobic (+O2) and anaerobic (−O2) conditions. The maximal levels of activity are given, and all values are the averages of three to four independent experiments with standard deviations.
Full, full-length ResE; +, with the indicated subdomain; −, without the indicated subdomain. Mutations in HAMP are also indicated.
As shown above, the cytoplasmic ResE retained activity that is responsive to oxygen limitation, although the expression of nasD and hmp was reduced by two- to fourfold compared to cells carrying the full-length ResE. One possible explanation for the reduced activity displayed by the cytoplasmic ResE is that multiple signals are sensed by different subdomains of ResE; that is, the cytoplasmic region senses a signal, and the extracytoplasmic region receives another signal. The other possibility is that the cytoplasmic region is the only signal-sensing region but that membrane anchoring is needed for efficient perception of the signal. To examine the alternative possibilities, the TM2 region was added back to the cytoplasmic ResE. As shown in Fig. 2, ResE produced in cells carrying the plasmid pMMN534 localizes membrane as efficiently as the full-length ResE protein. However, the expression of nasD and hmp in the strain with TM2-ResE was similar to that in cells producing the cytoplasmic ResE, indicating that the membrane anchoring apparently did not result in full activation of nasD and hmp and, rather, suggesting the possibility that the extracytoplasmic region serves as a second sensing subdomain. The same HAMP deletion in pMMN544 was introduced in the cytoplasmic ResE (pAB3). The deletion caused reduced gene expression, contrary to the result for membrane-bound ResE lacking the HAMP subdomain. Although we cannot eliminate the possibility that the HAMP linker has some role in the cytoplasmic forms of ResE, the reduced activity of cytoplasmic ResE lacking the HAMP linker might, instead, be caused by improper folding of the protein. Again the deletion of the PAS subdomain severely affects the function of ResE. These results revealed that the PAS subdomain has a pivotal role in ResE activity.
Effect of ResE subdomain deletions on induction of hmp expression by NO.
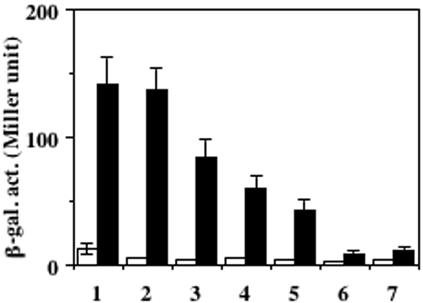
We have reported previously that the expression of ResDE-dependent genes was induced by NO during either aerobic or anaerobic growth (15). B. subtilis cells grown anaerobically with nitrate as an electron acceptor generate a low amount of NO nonenzymatically from nitrite, which is the product of the reduction catalyzed by nitrate reductase. Therefore, a mutation in narG (nitrate reductase gene) was introduced into the resE mutant to prevent endogenous NO production, and the resultant strain was transformed with pDG148 derivatives carrying resE. The narG mutant grew well anaerobically by fermentation in the medium supplemented with glucose and pyruvate (16). As reported previously (15) and shown in Fig. 5, oxygen limitation is not sufficient to activate hmp transcription, and the expression was induced over 10-fold with 10 μM NO in cells carrying the full-length resE genes (lane 1). The inhibitory effect by overproduction of the full-length ResE protein was also observed (compare lanes 2 and 3). The hmp expression was moderately increased by the addition of NO in the case of cytoplasmic ResE (lane 5) and TM2-carrying ResE (lane 4). On the contrary, the deletion of the PAS subdomain (lane 6) led to severe reduction in the stimulatory effect by NO. The residual induction observed in the PAS-lacking ResE was due to ResDE-independent activation of hmp with NO as previously reported (15), because a low level of induction was detected in cells without a resE-carrying plasmid (lane 7). These results indicate that the PAS subdomain is also critical to sense an NO-derived signal and that the extracytoplasmic region is needed for full activation of ResE.
FIG. 5.
Induction of hmp-lacZ with NO. narGH resE strains carrying the plasmid pMMN525 (full-length resE with resE SD) (column 1), pMMN563 (full-length resE with vector SD sequence) (columns 2 and 3), pMMN534 (TM2-carrying resE with vector SD sequence) (column 4), pMMN565 (cytoplasmic resE with vector SD sequence) (column 5), pMMN564 (cytoplasmic resE lacking PAS with vector SD sequence) (column 6), and without plasmid (column 7) were grown anaerobically in 2× YT supplemented with 0.5% glucose, 0.5% pyruvate, and 1 mM IPTG (0.02 mM IPTG for the culture shown in column 2). At the mid-log phase of growth, cells were incubated for 30 min without (open columns) or with (filled columns) 10 μM NO. β-Galactosidase activity (β-gal. act.) is shown in Miller units. The data are the averages of three to five experiments with standard deviations.
DISCUSSION
The study described herein aimed at identifying subdomains of ResE involved in sensing signal(s) derived from oxygen limitation. The results showing that the expression of nasD-lacZ and hmp-lacZ was regulated by oxygen levels in cells carrying the truncated resE clearly demonstrated that one sensing subdomain resides in the cytoplasmic region. ResE is involved in sensing signals related to the redox state of the cells. Disulfide bond formation of cysteines plays an important role for redox sensing as shown in RegB (see below). However, no cysteine residue is present in ResE, suggesting that there is another mechanism for sensing the redox state. The PAS subdomain is an attractive target of redox control and the most probable candidate for the cytoplasmic ligand-binding site because PAS subdomains are known to function in sensing various signals including oxygen concentration and the redox state of cells (35, 43). Our deletion analysis strongly supports this hypothesis. It is worth noting that the PAS subdomains in some sensor kinases are not important for responding to signals. PhoR of B. subtilis, like ResE, belongs to the class IIIA family of kinases according to a classification based on sequence similarities in the vicinity of the phosphorylated histidine (7). PhoR also has a large extracytoplasmic subdomain along with a cytoplasmic PAS subdomain; however, unlike the result for ResE, deletion of the entire N-terminal region including the PAS subdomain did not alter activation of the Pho response. The C-terminal kinase domain is sufficient for Pho regulation, implicating some unknown mechanisms for phosphate sensing (28). ArcB, a membrane-bound sensor kinase of E. coli, also responds to the redox conditions of cells, and an in vitro study using a truncated cytoplasmic protein showed that the oxidized forms of quinones inhibit autophosphorylation (8). This result suggests that the cytoplasmic region is sufficient to respond to the signal and that membrane anchoring is necessary in vivo for efficient signal reception. E. coli ArcB contains the PAS subdomain; however, the role of the PAS subdomain for sensing the redox state of quinones has not been reported. Interestingly, Haemophilus influenzae ArcB, which lacks the PAS subdomain, is able to activate the Arc regulon in E. coli responding to the redox condition of the growth, suggesting that, at least in H. influenzae, ArcB senses the redox-related signal through a PAS-independent mechanism (9). Whether the autokinase activity of H. influenzae ArcB is directly inhibited by oxidized quinones has not been tested.
We raised the possibility that the extracytoplasmic region serves as a second sensing subdomain based on the following observations. First, the expression of nasD-lacZ and hmp-lacZ was reproducibly lower with the cytoplasmic ResE than with the full-length ResE protein. Second, the lower activity of the cytoplasmic ResE is not attributed to the lack of membrane anchoring since the TM2-carrying ResE, which localizes to the membrane, exhibited a similar level of activity as the cytoplasmic protein. Third, the overproduction of the full-length ResE reduces ResDE-dependent gene expression. A likely explanation for this inhibitory effect is as follows. The default state of the intact ResE might be in the phosphatase-dominant mode (aerobic conditions), and it is shifted to the kinase-dominant state when the extracytoplasmic region binds the ligand (anaerobic conditions). Overproduction of the full-length ResE leaves larger populations of ResE in the ligand-unbound state, that is, the phosphatase-positive mode. Consistent with this hypothesis, TM2-carrying ResE did not display the inhibitory effect of overproduction.
Although we favor the idea that the negative effect of the overproduced full-length ResE protein is attributed to titration of the signal molecule by the extracytoplasmic region, we cannot exclude other possibilities. For example, the overproduced protein may be misfolded, which results in the formation of inactive dimers. However, this improper folding appears to occur only with the full-length ResE and not with the overproduced TM2-ResE, which is certainly possible though not probable. There is no evidence that the full-length ResE protein is more prone to misfolding; on the contrary, equally efficient membrane localization of each protein suggested that both proteins undergo proper folding. Interestingly, the inhibitory effect of PhoR overproduction on the PhoP activity described above was only observed with the full-length PhoR. Cells overproducing PhoR that lacked most of the extracytoplasmic region exhibited increased expression of the Pho regulon, a phenomenon similar to that observed in ResE. It has been suggested that the effect is due to the shift of the ratio of PhoP to PhoR (28). No matter what the exact mechanism is, the negative effect from overproduction of ResE and PhoR kinase is evidently exerted only if the protein carries the extracytoplasmic region.
Our study showed that the PAS subdomain is indispensable for the activity of membrane-bound ResE as well as the cytoplasmic form. This result might suggest that the PAS subdomain is needed for an additional role besides signal sensing. Alternatively, the deletion of the PAS subdomain may cause an alteration of the protein structure, which affects signal transmission from the extracytoplasmic subdomain to the kinase region. The last possibility is that the signal sensed by the extracytoplasmic region acts synergistically with the signal received by the PAS subdomain. The signal reception by the extracytoplasmic region might enhance the effect of the PAS subdomain. Such a synergistic effect was reported for VirA as described below.
A well-studied precedent for sensor kinases sensing multiple signal ligands is VirA, which is required for tumorigenicity in Agrobacterium. The periplasmic subdomain is essential for sensing monosaccharides, and the linker subdomain is required for sensing plant-derived phenolic compounds and acidity. Interestingly, VirA activates the cognate response regulator VirG in response to high levels of phenolic compounds, but the periplasmic sensing allows VirA to respond to low levels of phenolic compounds (2, 5, 29). Another example is PrrB (RegB) that may have two sensing regions. The PrrBA two-component system of Rhodobacter sphaeroides (RegBA in Rhodobacter capsulatus) is required for the expression of genes involved in photosynthesis as well as those that function in CO2 and N2 fixation. PrrB senses a signal generated by electron flow through the cbb3 oxidase, which inhibits the default kinase-positive mode of PrrB. PrrB has six membrane-spanning regions, and the central portion of the transmembrane subdomain including the second periplasmic loop was shown to play an important role in sensing and transducing the signal (23). Recently a conserved cysteine in the kinase domain of R. capsulatus RegB was shown to play a role in redox sensing. RegB undergoes a conformational change from active dimer to inactive tetramer in vitro through intermolecular disulfide bond formation under oxidizing conditions (34). Since R. sphaeroides PrrB also possesses the redox box that bears the conserved cysteine, it would be interesting to see how the sensing by the transmembrane subdomain and by the redox box in the kinase domain are coordinated in redox-sensing signal transduction in the purple phototrophic bacteria.
Signal transduction of ResE activated by NO is similar to the response to oxygen limitation as evident by the results of the deletion analysis, which showed that the same deletions of ResE affect similarly the response to both oxygen limitation and NO. These findings support our previous hypothesis that ResDE-dependent gene expression induced by NO is a property of anaerobically grown cells (15). Future studies will be focused on finding the signal ligand(s) sensed by the PAS subdomain and possibly by the extracytoplasmic region as well as on determining how these signals are involved in activation of ResE.
Acknowledgments
We thank Peter Setlow and Claes von Wachenfeldt for the gift of anti-PGM and anti-QoxA sera, respectively. We also thank Harold Tjalsma for the protocol of cell fractionation. We are grateful to Shunji Nakano and Peter Zuber for their valuable discussion and critical reading of the manuscript.
This work was supported by a grant from the National Science Foundation (grant MCB0110513).
REFERENCES
- 1.Appleman, J. A., and V. Stewart. 2003. Mutational analysis of a conserved signal-transducing element: the HAMP linker of the Escherichia coli nitrate sensor NarX. J. Bacteriol. 185:89-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cangelosi, G. A., R. C. Ankenbauer, and E. W. Nester. 1990. Sugars induce the Agrobacterium virulence genes through a periplasmic binding protein and a transmembrane signal protein. Proc. Natl. Acad. Sci. USA 87:6708-6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavicchioli, R., R. C. Chiang, L. V. Kalman, and R. P. Gunsalus. 1996. Role of the periplasmic domain of the Escherichia coli NarX sensor-transmitter protein in nitrate-dependent signal transduction and gene regulation. Mol. Microbiol. 21:901-911. [DOI] [PubMed] [Google Scholar]
- 4.Chamnongpol, S., M. Cromie, and E. A. Groisman. 2003. Mg2+ sensing by the Mg2+ sensor PhoQ of Salmonella enterica. J. Mol. Biol. 325:795-807. [DOI] [PubMed] [Google Scholar]
- 5.Chang, C.-H., and S. C. Winans. 1992. Functional roles assigned to the periplasmic, linker, and receiver domains of the Agrobacterium tumefaciens VirA protein. J. Bacteriol. 174:7033-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiang, R. C., R. Cavicchioli, and R. P. Gunsalus. 1992. Identification and characterization of narQ, a second nitrate sensor for nitrate-dependent gene regulation in Escherichia coli. Mol. Microbiol. 6:1913-1923. [DOI] [PubMed] [Google Scholar]
- 7.Fabret, C., V. A. Feher, and J. A. Hoch. 1999. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J. Bacteriol. 181:1975-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Georgellis, D., O. Kwon, and E. C. C. Lin. 2001. Quinones as the redox signal for the arc two-component system of bacteria. Science 292:2314-2316. [DOI] [PubMed] [Google Scholar]
- 9.Georgellis, D., O. Kwon, E. C. C. Lin, S. M. Wong, and B. J. Akerley. 2001. Redox signal transduction by the ArcB sensor kinase of Haemophilus influenzae lacking the PAS domain. J. Bacteriol. 183:7206-7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong, W., B. Hao, S. S. Mansy, G. Gonzalez, M. A. Gilles-Gonzalez, and M. K. Chan. 1998. Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction. Proc. Natl. Acad. Sci. USA 95:15177-15182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaspar, S., R. Perozzo, S. Reinelt, M. Meyer, K. Pfister, L. Scapozza, and M. Bott. 1999. The periplasmic domain of the histidine autokinase CitA functions as a highly specific citrate receptor. Mol. Microbiol. 33:858-872. [DOI] [PubMed] [Google Scholar]
- 12.Lee, A. I., A. Delgado, and R. P. Gunsalus. 1999. Signal-dependent phosphorylation of the membrane-bound NarX two-component sensor-transmitter protein of Escherichia coli: nitrate elicits a superior anion ligand response compared to nitrite. J. Bacteriol. 181:5309-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 14.Monson, E. K., M. Weinstein, G. S. Ditta, and D. R. Helinski. 1992. The FixL protein of Rhizobium meliloti can be separated into a heme-binding oxygen-sensing domain and a functional C-terminal kinase domain. Proc. Natl. Acad. Sci. USA 89:4280-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakano, M. M. 2002. Induction of ResDE-dependent gene expression in Bacillus subtilis in response to nitric oxide and nitrosative stress. J. Bacteriol. 184:1783-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakano, M. M., Y. P. Dailly, P. Zuber, and D. P. Clark. 1997. Characterization of anaerobic fermentative growth in Bacillus subtilis: identification of fermentation end products and genes required for the growth. J. Bacteriol. 179:6749-6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakano, M. M., M. A. Marahiel, and P. Zuber. 1988. Identification of a genetic locus required for biosynthesis of the lipopeptide antibiotic surfactin in Bacillus subtilis. J. Bacteriol. 170:5662-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakano, M. M., F. Yang, P. Hardin, and P. Zuber. 1995. Nitrogen regulation of nasA and the nasB operon, which encode genes required for nitrate assimilation in Bacillus subtilis. J. Bacteriol. 177:573-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakano, M. M., and Y. Zhu. 2001. Involvement of the ResE phosphatase activity in down-regulation of ResD-controlled genes in Bacillus subtilis during aerobic growth. J. Bacteriol. 183:1938-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakano, M. M., Y. Zhu, M. LaCelle, X. Zhang, and F. M. Hulett. 2000. Interaction of ResD with regulatory regions of anaerobically induced genes in Bacillus subtilis. Mol. Microbiol. 37:1198-1207. [DOI] [PubMed] [Google Scholar]
- 21.Nakano, M. M., and P. Zuber. 1998. Anaerobic growth of a “strict aerobe” (Bacillus subtilis). Annu. Rev. Microbiol. 52:165-190. [DOI] [PubMed] [Google Scholar]
- 22.Nakano, M. M., P. Zuber, P. Glaser, A. Danchin, and F. M. Hulett. 1996. Two-component regulatory proteins ResD-ResE are required for transcriptional activation of fnr upon oxygen limitation in Bacillus subtilis. J. Bacteriol. 178:3796-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oh, J. I., I. J. Ko, and S. Kaplan. 2001. The default state of the membrane-localized histidine kinase PrrB of Rhodobacter sphaeroides 2.4.1 is in the kinase-positive mode. J. Bacteriol. 183:6807-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park, H., and M. Inouye. 1997. Mutational analysis of the linker region of EnvZ, an osmosensor in Escherichia coli. J. Bacteriol. 179:4382-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piazza, F., P. Tortosa, and D. Dubnau. 1999. Mutational analysis and membrane topology of ComP, a quorum-sensing histidine kinase of Bacillus subtilis controlling competence development. J. Bacteriol. 181:4540-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Potter, C. A., A. Ward, C. Laguri, M. P. Williamson, P. J. Henderson, and M. K. Phillips-Jones. 2002. Expression, purification and characterisation of full-length histidine protein kinase RegB from Rhodobacter sphaeroides. J. Mol. Biol. 320:201-213. [DOI] [PubMed] [Google Scholar]
- 27.Reinelt, S., E. Hofmann, T. Gerharz, M. Bott, and D. R. Madden. 2003. The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain. J. Biol. Chem. 278:39189-39196. [DOI] [PubMed] [Google Scholar]
- 28.Shi, L., and F. M. Hulett. 1999. The cytoplasmic kinase domain of PhoR is sufficient for the low phosphate-inducible expression of pho regulon genes in Bacillus subtilis. Mol. Microbiol. 31:211-222. [DOI] [PubMed] [Google Scholar]
- 29.Shimoda, N., A. Toyoda-Yamamoto, J. Nagamine, S. Usami, M. Katayama, Y. Sakagami, and Y. Machida. 1990. Control of expression of Agrobacterium vir genes by synergistic actions of phenolic signal molecules and monosaccharides. Proc. Natl. Acad. Sci. USA 87:6684-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stallkamp, I., W. Dowhan, K. Altendorf, and K. Jung. 1999. Negatively charged phospholipids influence the activity of the sensor kinase KdpD of Escherichia coli. Arch. Microbiol. 172:295-302. [DOI] [PubMed] [Google Scholar]
- 31.Stephenson, K., and J. A. Hoch. 2001. PAS-A domain of phosphorelay sensor kinase A: a catalytic ATP-binding domain involved in the initiation of development in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98:15251-15256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stragier, P., C. Bonamy, and C. Karmazyn-Campelli. 1988. Processing of a sporulation factor in Bacillus subtilis: how morphological structure could control gene expression. Cell 52:697-704. [DOI] [PubMed] [Google Scholar]
- 33.Sun, G., E. Sharkova, R. Chesnut, S. Birkey, M. F. Duggan, A. Sorokin, P. Pujic, S. D. Ehrlich, and F. M. Hulett. 1996. Regulators of aerobic and anaerobic respiration in Bacillus subtilis. J. Bacteriol. 178:1374-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swem, L. R., B. J. Kraft, D. L. Swem, A. T. Setterdahl, S. Masuda, D. B. Knaff, J. M. Zaleski, and C. E. Bauer. 2003. Signal transduction by the global regulator RegB is mediated by a redox-active cysteine. EMBO J. 22:4699-4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vescovi, E. G., Y. M. Ayala, E. Di Cera, and E. A. Groisman. 1997. Characterization of the bacterial sensor protein PhoQ. Evidence for distinct binding sites for Mg2+ and Ca2+. J. Biol. Chem. 272:1440-1443. [DOI] [PubMed] [Google Scholar]
- 37.Waldburger, C. D., and R. T. Sauer. 1996. Signal detection by the PhoQ sensor-transmitter. Characterization of the sensor domain and a response-impaired mutant that identifies ligand-binding determinants. J. Biol. Chem. 271:26630-26636. [DOI] [PubMed] [Google Scholar]
- 38.Walker, M., and J. A. DeMoss. 1993. Phosphorylation and dephosphorylation catalyzed in vitro by purified components of the nitrate sensing system, NarX and NarL. J. Biol. Chem. 268:8391-8393. [PubMed] [Google Scholar]
- 39.Wang, L., C. Fabret, K. Kanamaru, K. Stephenson, V. Dartois, M. Perego, and J. A. Hoch. 2001. Dissection of the functional and structural domains of phosphorelay histidine kinase A of Bacillus subtilis. J. Bacteriol. 183:2795-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waukau, J., and S. Forst. 1999. Identification of a conserved N-terminal sequence involved in transmembrane signal transduction in EnvZ. J. Bacteriol. 181:5534-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams, S. B., and V. Stewart. 1999. Functional similarities among two-component sensors and methyl-accepting chemotaxis proteins suggest a role for linker region amphipathic helices in transmembrane signal transduction. Mol. Microbiol. 33:1093-1102. [DOI] [PubMed] [Google Scholar]
- 42.Wright, J. S., I. N. Olekhnovich, G. Touchie, and R. J. Kadner. 2000. The histidine kinase domain of UhpB inhibits UhpA action at the Escherichia coli uhpT promoter. J. Bacteriol. 182:6279-6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhulin, I. B., B. L. Taylor, and R. Dixon. 1997. PAS domain S-boxes in Archaea, Bacteria and sensors for oxygen and redox. Trends Biochem. Sci. 22:331-333. [DOI] [PubMed] [Google Scholar]