Abstract
The gram-positive human pathogen Staphylococcus aureus is often isolated with media containing potassium tellurite, to which it has a higher level of resistance than Escherichia coli. The S. aureus cysM gene was isolated in a screen for genes that would increase the level of tellurite resistance of E. coli DH5α. The protein encoded by S. aureus cysM is sequentially and functionally homologous to the O-acetylserine (thiol)-lyase B family of cysteine synthase proteins. An S. aureus cysM knockout mutant grows poorly in cysteine-limiting conditions, and analysis of the thiol content in cell extracts showed that the cysM mutant produced significantly less cysteine than wild-type S. aureus SH1000. S. aureus SH1000 cannot use sulfate, sulfite, or sulfonates as the source of sulfur in cysteine biosynthesis, which is explained by the absence of genes required for the uptake and reduction of these compounds in the S. aureus genome. S. aureus SH1000, however, can utilize thiosulfate, sulfide, or glutathione as the sole source of sulfur. Mutation of cysM caused increased sensitivity of S. aureus to tellurite, hydrogen peroxide, acid, and diamide and also significantly reduced the ability of S. aureus to recover from starvation in amino acid- or phosphate-limiting conditions, indicating a role for cysteine in the S. aureus stress response and survival mechanisms.
Cysteine is an essential amino acid that performs vital functions in the catalytic activity and structure of many proteins. Cysteine residues are required for essential and ubiquitous proteins with iron-sulfur (Fe-S) clusters, including cytochromes and aconitase (4, 21). In many organisms, the cysteine-containing molecules glutathione and thioredoxin play a major role in maintaining an intracellular reducing environment and protection against oxidative stress (6, 14, 23, 51). The formation of disulfide bonds between cysteine residues is the critical step in the activation of bacterial transcriptional regulators such as OxyR (8, 71) and the molecular chaperone Hsp33 (2, 34). Disulfide bonds are also needed for proper folding and stability of some proteins, particularly those found in extracytoplasmic compartments (68).
In bacteria, cysteine is synthesized from serine by incorporation of sulfide or thiosulfate. Sulfide is obtained from the transport and reduction of inorganic sulfate or from organic sulfonate compounds such as taurine (61, 62). The final step in cysteine biosynthesis is catalyzed by either O-acetylserine (thiol)-lyase A or O-acetylserine (thiol)-lyase B, encoded by the genes cysK and cysM, respectively (19, 30, 31). The CysK and CysM proteins from Escherichia coli are 43% identical. CysK synthesizes cysteine from O-acetylserine and sulfide, while the CysM protein differs in that it can also utilize thiosulfate instead of sulfide. The reaction between O-acetylserine and thiosulfate produces S-sulfocysteine, which is converted into cysteine by an as yet uncharacterized mechanism (44).
It has been proposed that the O-acetylserine (thiol)-lyase B isozyme is preferentially used during growth in anaerobic growth conditions (37). In E. coli, cysteine can be used to donate the sulfur moiety for methionine biosynthesis in a set of reactions known as the trans-sulfuration pathway. This pathway can be reversed in Bacillus subtilis, which can therefore use methionine as its sole source of sulfur (24). The genes involved in cysteine biosynthesis and sulfur assimilation in E. coli and Salmonella enterica serovar Typhimurium have been well characterized (reviewed in reference 37). More recently, cysteine biosynthesis has been studied in the gram-positive bacteria B. subtilis (24, 63) and Lactococcus lactis (18), and also in the archaeon genus Methanosarcina (5, 36). In contrast, cysteine biosynthesis and sulfur assimilation in the gram-positive Staphylococcus aureus have not been well studied.
S. aureus is a medically important human pathogen capable of causing a variety of infections, ranging from minor skin and wound infections to life-threatening diseases (41). Staphylococci are often isolated and identified with growth media containing potassium tellurite (K2TeO3), to which they have a higher level of resistance than many other bacteria. Although tellurite has been used in health laboratories and the food industry for over 80 years as a selective agent for the isolation of pathogens, the mechanism of tellurite resistance in staphylococci is poorly understood (3, 42, 69, 70).
We initially isolated a cysteine synthase homologue in a screen for S. aureus genes that would confer increased tellurite resistance on E. coli. In this paper we show that this locus is functionally homologous to CysM and is involved in stress resistance. In addition, we investigated the cysteine biosynthetic pathways of S. aureus and its ability to grow with different sources of sulfur.
MATERIALS AND METHODS
Media and growth conditions.
S. aureus and E. coli strains, plasmids, and oligonucleotides are listed in Table 1. E. coli was grown in Luria-Bertani (LB) medium at 37°C. S. aureus was grown at 25°C or 37°C with shaking at 250 rpm in brain heart infusion (BHI) (Oxoid), tryptic soy broth (TSB) (Difco), or chemically defined medium (CDM) (32, 66). For growth on solid media, 1% wt/vol agar was added. To measure the sulfur requirements of S. aureus strains on CDM agar plates, CDM was prepared without l-cysteine. This medium contains 2 mM MgSO4. l-Cystine, glutathione, sodium sulfate, 4-nitrocatechol sulfate, sodium sulfite, sodium sulfide, sodium thiosulfate, ethanedisulfonate, or benzenesulfonate was added at a concentration of 500 μM. For anaerobic growth, plates were incubated in an anaerobic growth cabinet (MK3 Anaerobic Incubator, Don Whitley Systems). For liquid growth tests, l-cysteine or sodium thiosulfate was added at concentrations up to 500 μM, and cultures were inoculated from an overnight preculture to an optical density at 600 nm (OD600) of 0.005. When included, antibiotics were added at the following concentrations: ampicillin, 100 mg liter−1; chloramphenicol, 5 mg liter −1, erythromycin, 5 mg liter−1; lincomycin, 5 mg liter −1; kanamycin, 50 mg liter −1; and tetracycline, 5 mg liter −1.
TABLE 1.
Strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or primer | Genotype or description | Reference or source |
|---|---|---|
| E. coli | ||
| DH5α | φ80 Δ(lacZ)M15 Δ(argF-lac)U169 endA1 recA1 hsdR17 (rK− mK+) deoR thi-1 supE44 gyrA96 relA1 | 52 |
| XLOLR | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 recA1 gyrA96 thi-1 relA1 [F′ proAB lacIqZΔM15 Tn10 (Tetr)] λr Su− | Stratagene |
| S. aureus | ||
| 8325-4 | Wild-type strain cured of prophages | Lab stock |
| RN4220 | Restriction-deficient transformation recipient | Lab stock |
| SH1000 | Functional rsbU+ derivative of 8325-4 | 26 |
| J96 | SH1000 cysM::tet | This study |
| SMH2052 | SH1000 cysJ::Tn917 | 28 |
| J62 | SH1000 cysM-lacZ | This study |
| J106 | J96/pJIM80 | This study |
| J108 | J96/pMK4 | This study |
| J116 | SH1000/pMK4 | This study |
| Plasmids | ||
| pOB | pGEM3Zf(+) cloning vector | 26 |
| pAZ106 | Promoterless lacZ eryr insertion vector | 35 |
| pDG1513 | Tetr cassette vector | 25 |
| pBK-CMV | Kanr phagemid recircularised from ZAP Express vector | Stratagene |
| pMK4 | E. coli — S. aureus catr shuttle vector | 55 |
| pJIM27 | pBK-CMV with S. aureus cysM (543551-545725)a | This study |
| pJIM28 | pBK-CMV with S. aureus cysM (545746-543385) | This study |
| pJIM29 | pBK-CMV with S. aureus cysM (545725-543551) | This study |
| pJIM30 | pBK-CMV with S. aureus cysM (545725-543994) | This study |
| pJIM31 | pBK-CMV containing random 2-kb fragment of S. aureus DNA | This study |
| pJIM62 | 4.8-kb fragment containing cysM in pOB | This study |
| pJIM64 | pJIM62 containing cysM::tet cassette insertion | This study |
| pJIM44 | 6-kb fragment in pAZ106 forming cysM-lacZ fusion | This study |
| pJIM80 | cysM locus in pMK4 for complementation | This study |
| Primersb | ||
| JKL48 | GTCGTTGAATTCTTATAGCAG | |
| JKL49 | CCAGCGAATTCCTGAACGTG | |
| JKL7 | CTTGGATCCAACGTCAAACAGAATCTAGACGCGG | |
| JKL22 | CGCTCTTGGCGAATTCTTTCGGG | |
| T3 | ATTAACCCTCACTAAAG | |
| T7 | AATACGACTCACTATAG |
Coordinates of fragment in S. aureus N315 genome database (38).
Restriction sites are underlined.
Screen for tellurite-resistant clones.
A phagemid library of S. aureus 8325-4 genomic DNA was prepared by mass excision from a λZAP Express (Stratagene) library made previously (20). The pBK-CMV phagemids are recircularized in E. coli XLOLR cells, have a kanamycin resistance marker, and contain random inserts of up to 12 kb of partially Sau3A-digested S. aureus DNA. To determine the MIC of tellurite, an overnight culture was diluted in phosphate-buffered saline (PBS), and a 10-μl inoculum containing approximately 5 × 104 bacteria was spotted onto LB agar containing different concentrations of K2TeO3. The MIC of tellurite was defined as the lowest concentration of K2TeO3 at which there was no bacterial growth. The MIC of tellurite for E. coli XLOLR and DH5α was found to be 200 nM and 1 μM, respectively; 105 random XLOLR clones were plated onto LB plates containing kanamycin and 1 μM K2TeO3. Several kanamycin- and tellurite-resistant colonies were obtained. Phagemid DNA was purified from these colonies and transformed into E. coli DH5α to verify that the increased tellurite resistance was plasmid linked.
To determine the maximum level of tellurite resistance, the clones were streaked out on LB containing up to 1 mM K2TeO3. The inserts of plasmids conferring tellurite resistance were sequenced with the Big Dye dideoxy terminator cycle sequencing kit and an ABI 373A DNA sequencer according to the manufacturer's instructions (Applied Biosystems) with oligonucleotide primers complementary to the T3 and T7 promoter sequences of pBK-CMV. Sequences were compared to the S. aureus N315 genomic DNA sequence (38) and investigated with the NCBI-BLAST homology search program (http://www.ncbi.nlm.nih.gov/BLAST/) (1).
Construction of strains and plasmids.
DNA manipulations and gel electrophoresis were carried out according to methods described in Sambrook et al. (52). To construct the S. aureus cysM::tet knockout, a 4.8-kb PCR fragment that is flanked by EcoRI restriction sites was amplified with primers JKL48 and JKL49. This fragment was restricted with EcoRI and ligated with EcoRI-cut pOB (27), creating pJIM62. A unique ClaI site 508 bp downstream from the putative start codon of the cysM open reading frame was used to insert a 2.1-kb ClaI fragment containing a tetracycline resistance cassette from pDG1513 (25), creating pJIM64.
To construct the single-crossover chromosomal cysM-lacZ fusion, a 6-kb PCR fragment was amplified with primers JKL7 and JKL22, which contain BamHI and EcoRI restriction sites, respectively. The cut fragment was ligated into the lacZ fusion vector pAZ106 (35) digested with BamHI and EcoRI, creating pJIM44, in which the lacZ gene is fused 300 bp downstream of the putative cysM start codon. To complement the cysM::tet mutant, a 3-kb EcoRI-PstI fragment from pJIM62 was ligated into the shuttle vector pMK4 (55) cut with EcoRI and PstI, creating pJIM80. The insert begins 108 bp upstream from the start codon of the hsp33 gene upstream of cysM and ends 682 bp downstream from the start codon of the folP gene downstream of cysM. Transformation into S. aureus RN4220 was performed as described by Schenk and Ladagga (53), selecting for tetracycline resistance, erythromycin resistance, and lincomycin resistance (for pJIM64), erythromycin resistance and lincomycin resistance (for pJIM44), or chloramphenicol resistance (for pJIM80 and pMK4) colonies.
Phage transduction into recipient strains was performed as described by Novick (48) with φ11 as the transducing phage. J96 (SH1000 cysM::tet) was isolated after transduction of an integrated RN4220 transformant of pJIM64 into S. aureus strain SH1000 (26), selecting for tetracycline-resistant, erythromycin-sensitive colonies. J62 (SH1000 cysM-lacZ) was isolated as an erythromycin-resistant, lincomycin-resistant colony after transduction of an integrated transformant of pJIM44. Southern blotting was used to verify the location and correct integration of DNA at chromosomal loci. J106 (J96/pJIM80), J108 (J96/pMK4), and J116 (SH1000/pMK4) were isolated as chloramphenicol-resistant colonies after transduction into SH1000 from RN4220 transformed with pJIM80 or pMK4. The presence of pJIM80 or pMK4 was confirmed by PCR with forward and reverse universal primers which are complementary to regions that flank the pMK4 polylinker. Strain SMH2052 was obtained from a random mutagenesis study (28) and contains a Tn917 transposon insertion 655 bp downstream of the putative cysJ start codon.
Analysis of thiols from S. aureus and E. coli.
S. aureus and E. coli cultures were grown in TSB medium for thiol analysis. E. coli carrying pJIM31 was isolated as a kanamycin-resistant, tellurite-sensitive colony and used as a neutral control in comparison with pJIM27 carrying S. aureus cysM. Washed cell pellets (100 to 250 mg) were resuspended in 1 ml of 50% vol/vol acetonitrile in Tris-HCl buffer (20 mM, pH 8.0), containing 2 mM monobromobimane (Calbiochem) and incubated at 60°C for 15 min in the dark. Control samples were treated with 5 mM N-ethylmaleimide for 10 min under the same conditions before the addition of monobromobimane (to 2 mM). The cellular debris was removed by centrifugation, and the samples were diluted in 10 mM aqueous methane sulfonic acid for reverse-phase high-pressure liquid chromatography (HPLC) analysis or to be kept frozen for future analyses. Thiol standards were prepared as described (17). The amount of thiol in the supernatant fraction is expressed on the basis of the dry weight of the residual cell pellet from each extract, which was determined by drying the cell pellet in an oven (80°C) until a constant weight was obtained. These residual dry weights were found to be 70 to 80% of the dry weight obtained for cells that had not been extracted.
HPLC analysis of thiol-bimane derivatives.
Duplicate samples of cell extracts were routinely analyzed for thiols as their bimane derivatives by at least two different HPLC protocols (45). The chromatographic conditions used in these protocols, the sources for reagents, the preparation of thiol-bimane standards, and the HPLC equipment used have been described in detail elsewhere (17). Briefly, in the HPLC trifluoroacetic acid-methanol method (45), a reverse-phase column with trifluoroacetic acid-water and methanol gradients was used to separate most low-molecular-weight thiol derivatives normally encountered in biological extracts but not those of highly charged thiols such as coenzyme A. To confirm the identity and amounts of thiols found by the trifluoroacetic acid method, the coenzyme A method was used, which constituted a tetrabutylammonium phosphate (TBAP) ion-pairing protocol designed for the separation of coenzyme A-bimane derivatives (17). This method used a C8 RP column (C8 Symmetry, 3.9 by 150 mm; Waters) at a flow rate of 1.0 ml min−1. The chromatographic protocol employed solvents and gradients as follows: solvent A, 10% (vol/vol) methanol, 0.25% (vol/vol) acetic acid, and 10 mM TBAP, pH 3.4; solvent B, 90% (vol/vol) methanol, 0.25% (vol/vol) acetic acid, and 10 mM TBAP. At time zero, 10% B; 15 min at 25% B; 30 min at 50% B; 40 min at 75% B; 45 min at 100% B, wash, equilibrate, and reinject.
Stress resistance and starvation survival assays.
Determination of the MIC of tellurite for S. aureus strains was done in the same way as for E. coli strains (see above) except with BHI medium instead of LB. Hydrogen peroxide resistance assays were carried out as described by Watson et al. (66), with the following modifications: cells were grown in amino acid-limiting CDM (1%, wt/vol, glucose) to exponential phase (OD600 = 0.1). Following the addition of H2O2 to a final concentration of 10 mM and incubation, cells were serially diluted in PBS containing catalase at 10 mg ml−1, and viability was assessed by overnight growth on BHI agar. Liquid tellurite resistance assays were performed in the same way except with K2TeO3 at a final concentration of 200 mM instead of H2O2 and serial dilution in PBS.
Acid resistance assays were performed by growing cells to exponential phase in BHI, harvesting, and resuspension in BHI acidified to pH 2 with HCl. Cells were serially diluted in 4× PBS and viability was determined on BHI agar. Disk diffusion assays were performed as follows: 5 ml of BHI top agar (0.7%, wt/vol) was seeded with 5 μl of an exponential-phase S. aureus BHI culture (OD600 = 0.2), and used as an overlay on a BHI agar plate. Sterile 13-mm antibiotic disks were placed on top of the overlay, and either 20 μl of 500 mM diamide, 35 μl of 2 M methyl viologen, or 20 μl of K2TeO3 was added to the disk. Zones of growth inhibition were measured after 24 h of incubation at 37°C. Starvation survival experiments were performed in amino acid-limiting, glucose-limiting, or phosphate-limiting CDM (32, 66); 50-ml cultures were grown for 24 h with shaking at 37°C, then kept static at 25°C. Samples were serially diluted and viability was assessed by growth on BHI agar. The results presented here are representative of three independent experiments that showed less than 10% variability.
β-Galactosidase assays.
Expression of cysM-lacZ in S. aureus was measured in BHI cultures of J62 grown with shaking at 37°C. Cultures were inoculated from exponential-phase precultures to an OD600 of 0.001. To test for induction of cysM-lacZ, subinhibitory concentrations of diamide (200 μM), methyl viologen (25 μM), or K2TeO3 (5 μM) were added after 2 h of growth. Levels of β-galactosidase activity were measured as described previously (27) with 4-methylumbelliferyl-β-d-galactosidase as the substrate. Assays were performed in duplicate, and the values were averaged. The results presented here are representative of two independent experiments that showed less than 10% variability.
RESULTS
S. aureus cysteine synthase gene confers increased tellurite resistance on E. coli.
The MIC of tellurite for S. aureus 8325-4 and S. aureus SH1000 (an rsbU+ derivative of 8325-4) (26) was 6 mM and 7 mM, respectively. In contrast, the MIC of tellurite for E. coli XLOLR and DH5α was 200 nM and 1 μM, respectively. A phagemid library of S. aureus 8325-4 DNA was used to screen for genes conferring increased tellurite resistance on E. coli. The library was prepared by mass excision from a λZAP Express library made previously by Foster (20); 105 clones were screened on the basis of their ability to enable growth of E. coli XLOLR on LB agar containing 1 μM K2TeO3.
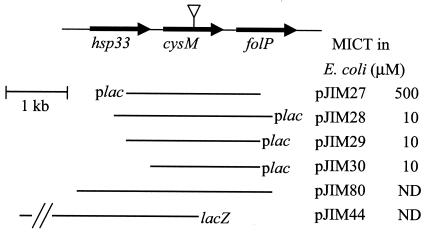
Phagemid DNA was purified from nine apparently tellurite-resistant colonies and transformed into E. coli DH5α to verify that the increased tellurite resistance was plasmid linked. Four clones conferred increased tellurite resistance on E. coli DH5α (pJIM27 to pJIM30, Fig. 1). All four clones contained an overlapping region with only one intact open reading frame (SA0471), encoding a putative protein of 310 amino acid residues, with 45% amino acid identity to CysK of E. coli and 35% amino acid identity to CysM of E. coli. Downstream of SA0471 is a putative Rho-independent terminator (17 to 47 bases from the stop codon). Plasmids pJIM28 to pJIM30 enabled E. coli DH5α to grow on LB medium containing up to 10 μM tellurite. pJIM27, which carries the same insert but in the opposite orientation, so that the cysM gene is downstream of the pBK-CMV lac promoter, conferred resistance up to 500 μM tellurite.
FIG. 1.
S. aureus cysM region and plasmids conferring tellurite resistance in E. coli. The inserts in pJIM27, pJIM28, pJIM29, and pJIM30 are shown in relation to the lac promoter on the pBK-CMV vector. The MIC of tellurite (MICT) of each clone is shown. The MIC of tellurite for E. coli DH5α is 1 μM. The inverted triangle shows the position of the tetracycline resistance cassette insertion in S. aureus J96 (cysM::tet). Plasmids pJIM80, used for complementation, and pJIM44, used to make S. aureus J62 (cysM-lacZ), are shown. ND, not determined.
High levels of glutathione are produced by E. coli carrying an S. aureus cysteine synthase.
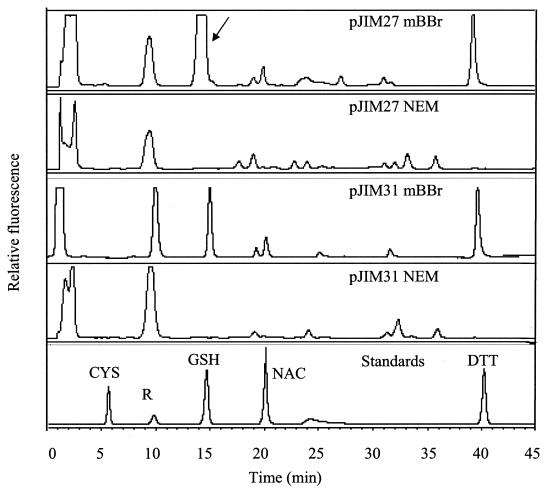
The thiol content of cultures of E. coli carrying pJIM27 was analyzed by separating the bimane derivatives of low-molecular-weight thiols in cell extracts by reverse-phase HPLC. E. coli containing pJIM31, a pBK-CMV derivative that did not confer increased tellurite resistance, was used as a control. Extracts from both strains gave a peak corresponding to glutathione (Fig. 2). The amount of glutathione in these cultures was quantified, and in E. coli/pJIM27 cultures the amount of glutathione was significantly higher throughout growth compared to the control strain. A culture of E. coli/pJIM31 at an OD600 of 3.9 contained 1.1 nmol of glutathione per mg of dry cell weight. In contrast, a culture of E. coli/pJIM27 at a similar OD600 of 3.5 produced 9.93 nmol of glutathione per mg of dry cell weight.
FIG. 2.
Reverse-phase HPLC of E. coli thiols. Traces shown are from extracts of E. coli/pJIM27 and E. coli/pJIM31 (control) cultures grown in TSB to an OD600 of 1.7 and 2.0, respectively. Arrow points to the glutathione peak. CYS, cysteine; GSH, glutathione; NAC, N-acetylcysteine; DTT, dithiothreitol; R, peak produced by chemical reagents; NEM, N-ethylmaleimide; mBBr, monobromobimane.
Cysteine synthase proteins and molecules containing cysteine, including glutathione, have previously been shown to be involved in tellurite resistance in E. coli (15, 49, 59, 60). Tellurite resistance in S. aureus is uncharacterized, and furthermore, little is known about cysteine biosynthesis and its importance in this bacterium. It was therefore of interest to investigate the role of this cysteine synthase locus in S. aureus.
Search for cysteine biosynthetic genes in the S. aureus genome.
The process of sulfur assimilation has been shown to involve many genes and enzymatic reactions in bacteria such as E. coli and B. subtilis (reviewed in references 24 and 37). All of the published S. aureus genome sequences contain cysteine synthase homologues (38; http://www.genome.ou.edu/staph.html, http://www.tigr.org), but auxotrophy for cysteine has been reported in some S. aureus strains (16).
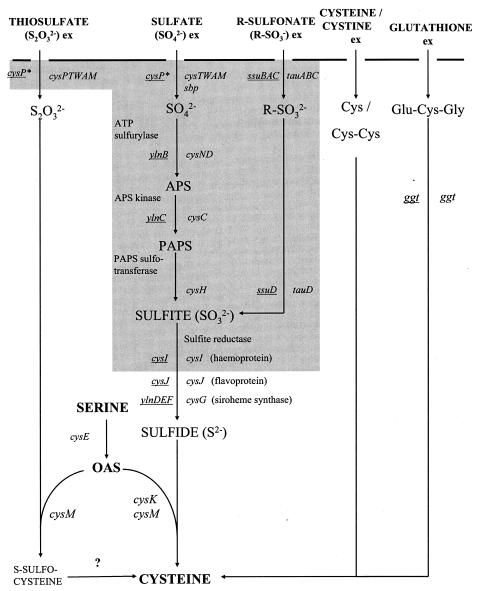
A systematic search of the S. aureus 8325 and S. aureus N315 genome sequences was performed, looking for homologues of genes known or thought to be involved in bacterial cysteine biosynthesis and sulfur assimilation. Neither sequence contained any homologues of the E. coli or B. subtilis genes required for sulfate or thiosulfate uptake, reduction of sulfate to sulfite, or uptake and reduction of organic sulfonates (Fig. 3). The S. aureus N315 genome encodes homologues of the B. subtilis proteins CysE (SA0487, 64% amino acid identity); CysJ (SA2413, 50% identity); YlnD (SA2186, 42% identity); YlnE (SA2189, 21% identity); YlnF (SA2186, 35% identity); and the γ-glutamyl peptidase encoded by ggt (SA0202, 37% identity). The S. aureus N315 genome also has three cysteine synthase gene homologues (Table 2), including SA0471, which corresponds to the gene conferring increased tellurite resistance carried on pJIM27 to pJIM30.
FIG. 3.
Sulfur assimilation and cysteine biosynthesis in E. coli and B. subtilis. Genes involved in uptake and assimilation of inorganic and organic sulfur sources in E. coli and B. subtilis are shown. B. subtilis genes are underlined. The shaded region covers genes for which no apparent homologue can be found in the S. aureus N315 genome sequence. Note that E. coli, B. subtilis, and S. aureus all encode cysE, cysK, and cysM homologues. Ex, external sulfur source. Cysteine, cystine, and glutathione may be used as organic sources of sulfur. APS, adenosine 5′-phosphosulfate; PAPS, 3′-phosphoadenosine 5′-phosphosulfate; OAS, O-acetylserine. cysP*, the B. subtilis CysP sulfate/thiosulfate transporter is distinct from the E. coli CysP thiosulfate-binding protein.
TABLE 2.
Cysteine synthase homologues in S. aureus N315
| S. aureus N315 gene no. | N315 coordinates (no. of amino acids) | % identitya to CysK/CysM | Open reading framesb
|
|
|---|---|---|---|---|
| Upstream | Downstream | |||
| SA0112 | 128772-129752 (327) | 33/35 | sirABC (Fe3+ uptake) | Putative ornithine cyclodeaminase |
| SA0418 | 479707-480612 (302) | 36/33 | metB (cystathionine γ-synthase) | Putative Na+-dependent transporter |
| SA0471 | 544265-545197 (311) | 45/35 | hsp33 (redox-sensitive chaperone) | folPBK (folic acid biosynthesis) |
Growth of S. aureus with different sulfur sources.
The genomic data suggested that S. aureus potentially can use sulfide or thiosulfate to produce cysteine, but does not have any of the recognized bacterial systems for the uptake and reduction of sulfate or sulfonates. A strain carrying a knockout mutation in the SA0471 cysteine synthase locus was constructed (J96). In addition, strain SMH2052, which has a transposon insertion in the cysJ homologue, was isolated in a random mutagenesis study (28). To investigate cysteine biosynthesis and the role of these two genes in S. aureus, the ability of SH1000, J96 (cysM), and SMH2052 (cysJ) to grow with different sources of sulfur was tested (Table 3). None of the three strains could grow on chemically defined medium (CDM) agar plates lacking cysteine (note that CDM contains MgSO4 and methionine at final concentrations of 2 mM and 200 μM, respectively) or with the addition of sulfate, sulfite, ethanedisulfonate, or benzenesulfonate. All three strains could grow with cysteine, cystine, or glutathione as the sole sulfur source. In the presence of sodium sulfide, strains SH1000 and SMH2052 grew normally, while J96 (cysM) colonies grew poorly, only appearing after 3 days of incubation. In anaerobic conditions, the results for each strain were the same as were found in aerobic conditions (data not shown). In addition, a number of other S. aureus strains, including Newman and COL, were unable to utilize sulfate as a sole sulfur source (data not shown).
TABLE 3.
Growth of S. aureus strains on different sulfur sourcesa
| Sulfur source added | Growth
|
||
|---|---|---|---|
| SH1000 | J96 (cysM) | SMH2052 (cysJ) | |
| None (SO42− + methionine) | − | − | − |
| l-Cystine | + | + | + |
| l-Cysteine | + | + | + |
| Glutathione | + | + | + |
| 4-Nitrocatechol sulfate | − | − | − |
| Sodium sulfate | − | − | − |
| Sodium sulfide | + | +/− | + |
| Ethanedisulfonate | − | − | − |
| Benzenesulfonate | − | − | − |
| Sodium thiosulfate | + | − | + |
Strains were grown on CDM agar containing 2 mM MgSO4 and 200 μM methionine. Different sulfur sources were added at 500 μM. +, normal growth (colony diameter, 2 to 3 mm) after 24 h at 37°C; −, no growth; +/−, poor growth (colony diameter up to 1 mm).
S. aureus SH1000 requires cysM to utilize thiosulfate as the sole sulfur source.
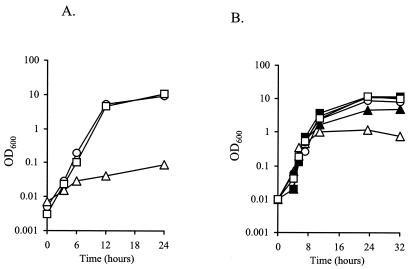
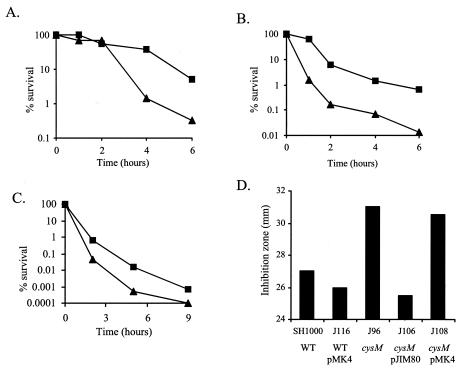
Significantly, SH1000 and SMH2052 (cysJ), but not J96 (cysM), could grow with sodium thiosulfate as the sole sulfur source. Plasmid pJIM80, which carries the S. aureus cysM gene in the shuttle vector pMK4, was constructed. The introduction of pJIM80 into J96 (cysM) complemented the cysM mutation, enabling growth to wild-type levels in CDM broth containing 500 μM thiosulfate, whereas the addition of pMK4 into J96 (cysM) did not (Fig. 4A). cysM mutants of E. coli are unable to grow on thiosulfate, as CysK cannot utilize thiosulfate as a substrate. The growth and complementation data demonstrated that the gene inactivated in S. aureus J96 encodes a cysteine synthase functionally homologous to E. coli CysM.
FIG. 4.
(A) Growth of S. aureus strains in CDM broth containing 500 μM sodium thiosulfate as the sole sulfur source. Squares, J116 (SH1000 wild-type carrying pMK4); circles, J106 [J96 (cysM) carrying pJIM80]; triangles, J106 [J96 (cysM) carrying pMK4]. (B) Growth of wild-type S. aureus SH1000 (solid symbols) and J96 (cysM) (open symbols) in CDM broth with various cysteine concentrations. Squares, 200 μM cysteine; circles, 50 μM cysteine; triangles, 10 μM cysteine. Cultures were grown with shaking at 37°C. Results are representative of at least two independent experiments.
S. aureus J96 (cysM) shows reduced growth in cysteine-limiting conditions.
After 32 h of growth in CDM broth containing 200 μM cysteine, the OD600 of SH1000 and J96 (cysM) cultures was 12 and 10.5, respectively (Fig. 4B). However, at lower cysteine concentrations, J96 (cysM) cultures grew to a lower density than wild-type SH1000. After 32 h of growth in the presence of 10 μM cysteine, the OD600 of SH1000 and J96 (cysM) cultures was 4.9 and 0.8, respectively. A similar growth defect could be seen with cystine as the sulfur source (data not shown).
Analysis of the thiol content of S. aureus.
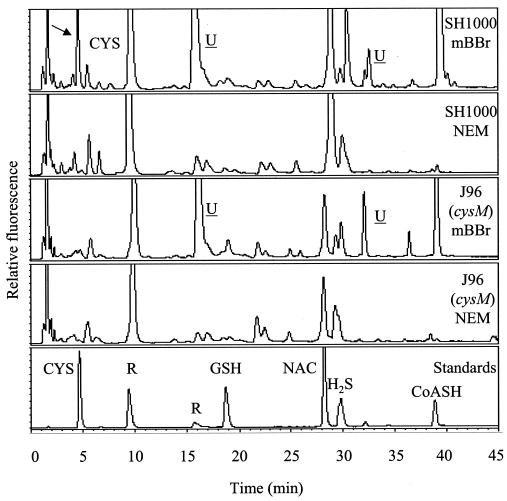
The thiol contents of S. aureus SH1000 and J96 (cysM) were analyzed (Fig. 5). Analysis was performed on extracts of cultures at stages throughout the growth phase. SH1000 wild-type extracts gave peaks corresponding to cysteine and coenzyme A and also peaks at around 16 min and 33 min which represent uncharacterized S. aureus thiol components that did not correspond to any of the known standards. HPLC traces from J96 (cysM) cultures were essentially the same except for the reduction in the size of the peak corresponding to cysteine. When quantified, it was found that, whereas the cysteine concentrations in SH1000 extracts are maintained at around 0.5 nmol per mg of dry cell weight, in J96 (cysM) cultures the cysteine concentration falls to undetectable levels at an OD600 of 1.4 and above (Fig. 5 and data not shown). In contrast, the uncharacterized thiol-containing compounds remained constant.
FIG. 5.
Reverse-phase HPLC of S. aureus thiols. Extracts of S. aureus SH1000 and J96 (cysM) cultures grown in TSB medium to an OD600 of 1.5 were reacted with monobromobimane (mBBr) and separated by reverse-phase HPLC. These extracts are representative of all stages of growth. Control samples were reacted with N-ethylmaleimide (NEM) and monobromobimane. To identify peaks, bimane derivatives of standard thiols were used. CYS, cysteine; GSH, glutathione; NAC, N-acetylcysteine; CoASH, reduced coenzyme A; U, unidentified components; R, peaks produced by reagents. The arrow points to the cysteine peak found in SH1000 but absent in J96 (cysM) samples.
The cysM mutant is sensitive to tellurite, oxidative, and disulfide stress.
Since the S. aureus cysM gene conferred increased tellurite resistance in E. coli, its effect on the same in S. aureus was tested. The MIC of tellurite for J96 was 5 mM, lower than that of SH1000 (7 mM). In liquid assays, when exponential-phase cells were challenged with 200 mM tellurite, J96 (cysM) cultures showed a 15-fold reduction in viability compared to SH1000 after 6 h (Fig. 6A).
FIG. 6.
Stress resistance assays. (A, B, and C) Viability of S. aureus SH1000 (squares) and J96 (cysM) (triangles) exponential-phase cells after challenge with (A) 200 mM K2TeO3, (B) 10 mM H2O2, and (C) hydrochloric acid to pH 2. (D) Disk diffusion assays with 1 M diamide. The sensitivity of J96 (cysM) can be complemented with pJIM80 but not the control pMK4 plasmid.
S. aureus survives a diverse range of stresses during its life cycle (9), and the role of cysM in the response to other stresses was investigated. The viability of J96 (cysM) was reduced 45-fold in comparison to SH1000 6 h after the addition of 10 mM H2O2 (Fig. 6B). J96 (cysM) was also more sensitive to acid stress, its viability reduced 30-fold after 4 h at pH 2 (Fig. 6C). In disk diffusion assays, growth of J96 (cysM) was significantly more inhibited than that of SH1000 by 1 M diamide, a specific thiol oxidant that causes disulfide stress. This diamide sensitivity could also be complemented by pJIM80 (Fig. 6D). However, J96 (cysM) was no more sensitive to methyl viologen than SH1000 (data not shown).
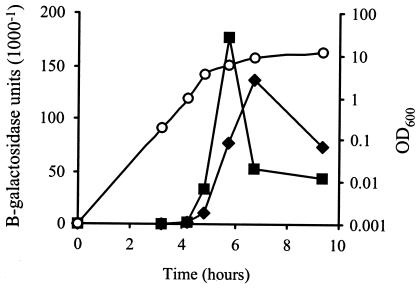
To determine whether the S. aureus cysM gene was induced by stress, cysM-lacZ fusion strain J62 was constructed. Expression of cysM-lacZ reached a sharp peak in the postexponential/early stationary phase of growth in BHI medium (Fig. 7). The addition of diamide, methyl viologen, hydrogen peroxide, or tellurite during the early exponential phase of growth (OD = 0.05) did not lead to early induction of cysM-lacZ expression (data not shown). A slight increase in the level of expression was observed in stationary phase following addition of 5 μM tellurite (Fig. 7).
FIG. 7.
Expression of cysM-lacZ. J62 (SH1000 cysM-lacZ) cultures were grown at 37°C with shaking in BHI medium. Squares, cysM-lacZ expression, no additions; diamonds, cysM-lacZ expression with addition of 5 μM K2TeO3 at 2 h; circles, OD600 of representative culture.
The cysM locus affects starvation survival but not exoprotein production and pathogenicity.
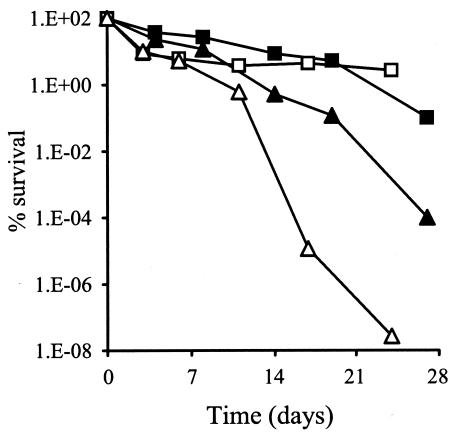
Genes involved in stress resistance have also been shown to affect the viability of S. aureus during long-term starvation conditions (10, 11, 66, 67). In amino acid-limiting CDM containing 1% (wt/vol) glucose, the viability of J96 (cysM) was reduced 1,000-fold in comparison to SH1000 after 27 days, and in phosphate-limiting CDM, J96 (cysM) lost all viability after 21 days (Fig. 8). There was no difference in the viability of J96 and SH1000 grown in glucose-limiting CDM (data not shown).
FIG. 8.
Starvation survival recovery. Cultures of SH1000 (squares) and J96 (triangles) were grown with shaking for 24 h at 37°C in either amino acid-limiting (solid symbols) or phosphate-limiting (open symbols) CDM, and then kept static at 25°C. Samples were removed and serially diluted, and viability was assessed by growth on BHI plates.
The ability of S. aureus to respond to and survive various stresses is vital for its ability to successfully colonize tissues and evade host defense mechanisms, but despite being sensitive to the various stress conditions described above, J96 (cysM) was not attenuated in a mouse lesion model of pathogenicity (7), was no different from SH1000 in hemolytic activity on rabbit or sheep blood plates, and produced the same profile on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels of exoprotein extracts (data not shown).
DISCUSSION
Role of cysM in tellurite resistance in E. coli and S. aureus.
The use of tellurite in selective media for S. aureus was first described by Ludlam in 1949 (42). In Baird-Parker medium, a mixture of egg yolk and tellurite is used to isolate and identify coagulase-positive staphylococci from food (3). S. aureus colonies are distinguished by a zone of clearing in the egg yolk due to lecithinase activity and a shiny black appearance. The black color of bacteria grown on tellurite media is caused by deposits of mostly elemental tellurium produced by the reduction of tellurite (58). The toxicity of tellurite is thought to come from its strong oxidizing ability (58, 60). Genes associated with tellurite resistance are found in many pathogenic bacteria and can be plasmid borne, such as the klaABC operon found on IncP-type plasmids (50).
CysK from Bacillus stearothermophilus and Rhodobacter sphaeroides confers tellurite resistance when introduced into E. coli and Paracoccus denitrificans, respectively (49, 64). The exact mechanism of CysK-mediated tellurite resistance is not known. Cysteine residues are known to be important in the function of many metal-binding proteins (22). The E. coli tellurite resistance determinants TehA and TehB each contain three cysteine residues, and replacement of these cysteines with alanine residues by site-directed mutagenesis leads to a decrease in tellurite resistance (15). In addition, the thiol redox enzymes (glutathione reductase and thioredoxin reductase) and their metabolites (glutathione, glutaredoxin, and thioredoxin), which all contain cysteine residues, have been shown to be involved in tellurite resistance (59, 60). Turner et al. (60) suggested that reduced thiols cause reduction of TeO32− to Te0, possibly via an intermediate telluro-ether disulfide bond (RS-Te-SR), and that this, in combination with the action of other components, contributes to tellurite resistance.
E. coli cells carrying the S. aureus cysM gene produce a large amount of glutathione (γ-glutamyl-cysteinyl-glycine), although the level of cysteine is not greatly higher than in the control cultures. Glutathione protects many organisms from oxidative toxicity by functioning as a slowly autooxidizing reserve of cysteine and as a cofactor in the detoxification of products formed from oxygen reactions (6, 14, 51). The production of glutathione is limited by the availability of cysteine, and therefore it is likely that expression of the S. aureus cysM leads to increased levels of cysteine, which in turn is incorporated into glutathione by E. coli glutathione synthetase.
In pJIM27, the cysM gene is downstream of the pBK-CMV lac promoter. Enhanced expression of the S. aureus cysM gene from the lac promoter may explain the much higher level of glutathione production and tellurite resistance conferred by pJIM27. It is possible that the increased tellurite resistance is because of a direct reaction of glutathione with TeO32− or due to glutathione's reversing the effects of tellurite oxidation on other thiols in the cell. When grown on LB medium containing tellurite, colonies of E. coli carrying pJIM27 also had a grey-black appearance. S. aureus colonies grown on tellurite medium have a characteristic black color, suggesting that the S. aureus cells reduce tellurite. However, glutathione cannot be responsible for tellurite reduction in S. aureus, as it does not synthesize this compound (46). The S. aureus cysM mutant shows increased sensitivity to tellurite, suggesting that cysteine or a compound that requires cysteine for its synthesis other than glutathione contributes to tellurite resistance.
S. aureus SH1000 cultures were shown to contain cysteine, reduced coenzyme A, H2S, and at least two more uncharacterized thiol compounds. In contrast to the wild type, cysteine levels were not maintained throughout growth in the cysM mutant. The level of the other thiols, however, was not significantly altered in the cysM mutant. Reduced coenzyme A has been shown to be the predominant low-molecular-weight thiol in S. aureus (12, 13), and its synthesis does not involve cysteine. It is possible that the cysteine molecule itself is involved in direct reduction of tellurite. Alternatively, protein or peptide synthesis may be affected by the absence of cysteine and that other specific tellurite-reducing peptides or proteins are reduced. Also, it has recently been proposed that important proteins containing iron-sulfur clusters are the indirect targets of tellurite via the production of superoxide (57). Cysteine is important as a source of sulfur for the repair of oxidatively damaged iron-sulfur cluster proteins with crucial roles in metabolism (57).
Role of cysM in S. aureus stress resistance and starvation survival.
The observation that plasmid pJIM80 could complement the sensitivity of J96 (cysM) to diamide, a specific thiol oxidant, suggests that the increased sensitivity of J96 (cysM) is not due to any polar effects on genes downstream of cysM. Tellurite, hydrogen peroxide, acid, and diamide are all substances that can cause imbalance in the thiol redox status of the cytoplasm, or oxidative stress (6, 34, 40, 58). Cysteine residues in the cytoplasm are normally kept in a reduced state, but under oxidative conditions they form disulfide bonds, causing misfolding and inactivation of proteins (2).
In S. aureus, free cysteine, reduced coenzyme A, and other thiols may help maintain the thiol redox balance as well as thioredoxin and thioredoxin reductases, which are encoded by the trxAB genes. A defect in cysteine biosynthesis would also impair protein synthesis, which has been shown to be important in starvation survival of S. aureus (66). This could explain the more pronounced loss of viability in cultures of J96 (cysM) in phosphate- or amino acid-limiting conditions. Glucose limitation leads to the development of a stable survival state in 0.1 to 1% of S. aureus cells (66), and in these conditions, J96 (cysM) apparently obtains enough cysteine from the medium to enable it to remain viable. A recent study has shown that a number of B. subtilis genes are induced by diamide stress, including cysK (40). Although the S. aureus cysM gene is involved in resistance to diamide and other stresses, expression of cysM was not affected by addition of diamide, methyl viologen, or hydrogen peroxide, indicating differences in the stress responses of B. subtilis and S. aureus.
Cysteine biosynthesis and sulfur assimilation in S. aureus.
In cysteine-limiting conditions, the growth of the S. aureus cysM mutant is significantly impaired compared to the wild-type SH1000. This suggests that CysM is the major cysteine synthase in S. aureus, since neither of the other two cysteine synthase homologues in the S. aureus genome can compensate for the mutation of cysM. It has been suggested that CysM-type cysteine synthases are preferentially used during anaerobic growth (37). In S. aureus it appears that CysM is required for growth with thiosulfate in both aerobic and anaerobic conditions.
In other bacteria, including E. coli and B. subtilis, sulfate or organic sulfonates are reduced to sulfide, a substrate for both CysK- and CysM-type cysteine synthases. It is likely that S. aureus cannot utilize sulfate or sulfonates as a sulfur source because its genome does not encode any of the genes required for the uptake and subsequent reduction of such compounds. Recent studies have shown that the genomes of the gram-positive lactic acid bacterium Lactococcus lactis and the gram-negative pathogen Haemophilus influenzae do not encode genes for sulfate uptake and reduction but do have cysteine synthase homologues (18, 65), just as in the genomes of several S. aureus strains. In addition, it has been shown that both L. lactis and H. influenzae are unable to grow with sulfate as the only sulfur source (18, 65). S. aureus can, however, use both thiosulfate and sulfide as a sulfur source. The use of thiosulfate is dependent on cysM, while growth on sulfide is impaired but not totally abolished in the J96 (cysM) mutant, suggesting that one or both of the other cysteine synthases can use sulfide as a substrate, but not as effectively as CysM.
S. aureus CysM is clearly an O-acetylserine (thiol)-lyase B-type protein, despite its having greater sequence homology to E. coli CysK, an O-acetylserine (thiol)-lyase A. In E. coli, thiosulfate is transported with the cysTWA permease in conjunction with a thiosulfate binding protein, CysP (21), but no homologues of these are present in the S. aureus genome. The ability of S. aureus SH1000 to use thiosulfate indicates the presence of an unknown thiosulfate uptake mechanism.
The evidence presented here suggests that many S. aureus strains can assimilate thiosulfate and sulfide but not sulfate. The physiological significance of this is unknown. Sulfate and sulfide are the major available forms of sulfur in soil and water, and both are also present in humans, although sulfide levels are presumably kept low due to its toxicity (47). Thiosulfate is present at least transiently in the environment and is present in humans, although usually at low concentrations (33, 39). It is likely that while in a human host, S. aureus gains most of its sulfur from organic sources and infrequently requires assimilation of inorganic sulfur. The inability of some S. aureus strains to use sulfate or sulfonates could be linked to the scarcity of these compounds in the environment in which S. aureus has evolved. It is interesting that S. aureus SH1000 can utilize the organic compounds cysteine, cystine, and glutathione, which are readily available in the mammalian cell environment (43).
S. aureus SH1000 cannot grow in CDM containing methionine and sulfate as the only sources of sulfur, showing that it is unable to perform the trans-sulfuration reaction which converts methionine into cysteine. Growth on cysteine, cystine, and glutathione suggests that S. aureus has, first, a transport mechanism for these molecules, and second, an ability to break down cystine (Cys-Cys) or glutathione to yield cysteine. No specific cysteine permease has been described, although the S. aureus genome sequence encodes many putative ABC-type transporters, one of which could transport cysteine, and several putative oligopeptide transporters, which could transport cystine or glutathione. In E. coli, γ-glutamyl peptidase cleaves glutathione to cysteinyl-glycine during the transport of an external amino acid, followed by breakdown of cysteinyl-glycine to yield cysteine (56). Although it does not synthesize glutathione, S. aureus can apparently import and metabolize glutathione, as has been shown for Streptococcus mutans (54) and H. influenzae (65). Interestingly, it has recently been reported that imported glutathione forms part of the oxidative stress resistance mechanism of H. influenzae (65). It is theoretically possible that glutathione may be used in a similar way by S. aureus to resist oxidative attack during infection of the host.
This study has shown that a link between metabolism and stress resistance is part of the overall complex physiology of S. aureus which allows it to inhabit so many different niches and be such a successful pathogen. Elucidation of the metabolic capabilities of S. aureus and an understanding of those important for growth of the organism in vivo may lead to the development of novel intervention strategies.
Acknowledgments
This work was funded by the BBSRC.
We thank Michaela Yanku for technical assistance.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aslund, F., and J. Beckwith. 1999. Bridge over troubled waters: sensing stress by disulfide bond formation. Cell 96:751-753. [DOI] [PubMed] [Google Scholar]
- 3.Baird-Parker, A. 1962. An improved diagnostic and selective medium for isolating coagulase positive staphylococci. J. Appl. Bacteriol. 25:12-19. [Google Scholar]
- 4.Beinert, H., and P. J. Kiley. 1999. Fe-S proteins in sensing and regulatory functions. Curr. Opin. Chem. Biol. 3:152-157. [DOI] [PubMed] [Google Scholar]
- 5.Borup, B., and J. G. Ferry. 2000. O-Acetylserine sulfhydrylase from Methanosarcina thermophila. J. Bacteriol. 182:45-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carmel-Harel, O., and G. Storz. 2000. Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu. Rev. Microbiol. 54:439-461. [DOI] [PubMed] [Google Scholar]
- 7.Chan, P. F., S. J. Foster, E. Ingham, and M. O. Clements. 1998. The Staphylococcus aureus alternative sigma factor σB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J. Bacteriol. 180:6082-6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christman, M. F., G. Storz, and B. N. Ames. 1989. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc. Natl. Acad. Sci. USA 86:3484-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clements, M., and S. Foster. 1999. Stress resistance in Staphylococcus aureus. Trends Microbiol. 7:458-462. [DOI] [PubMed] [Google Scholar]
- 10.Clements, M., S. Watson, and S. Foster. 1999. Characterization of the major superoxide dismutase of Staphylococcus aureus and its role in starvation survival, stress resistance, and pathogenicity. J. Bacteriol. 181:3898-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clements, M., S. Watson, R. Poole, and S. Foster. 1999. CtaA of Staphylococcus aureus is required for starvation survival, recovery, and cytochrome biosynthesis. J. Bacteriol. 181:501-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.del Cardayré, S. B., and J. E. Davies. 1998. Staphylococcus aureus coenzyme A disulfide reductase, a new subfamily of pyridine nucleotide-disulfide oxidoreductase. Sequence, expression, and analysis of cdr. J. Biol. Chem. 273:5752-5757. [DOI] [PubMed] [Google Scholar]
- 13.del Cardayré, S. B., K. P. Stock, G. L. Newton, R. C. Fahey, and J. E. Davies. 1998. Coenzyme A disulfide reductase, the primary low molecular weight disulfide reductase from Staphylococcus aureus. Purification and characterization of the native enzyme. J. Biol. Chem. 273:5744-5751. [DOI] [PubMed] [Google Scholar]
- 14.Dickinson, D. A., and H. J. Forman. 2002. Cellular glutathione and thiols metabolism. Biochem. Pharmacol. 64:1019-1026. [DOI] [PubMed] [Google Scholar]
- 15.Dyllick-Brenzinger, M., M. Liu, T. Winstone, D. Taylor, and R. Turner. 2000. The role of cysteine residues in tellurite resistance mediated by the TehAB determinant. Biochem. Biophys. Res. Commun. 277:394-400. [DOI] [PubMed] [Google Scholar]
- 16.Emmet, M., and W. E. Kloos. 1975. Amino acid requirements of staphylococci isolated from human skin. Can. J. Microbiol. 21:729-733. [DOI] [PubMed] [Google Scholar]
- 17.Fahey, R. C., and G. L. Newton. 1987. Determination of low-molecular-weight thiols with monobromobimane fluorescent labeling and high-performance liquid chromatography. Methods Enzymol. 143:85-96. [DOI] [PubMed] [Google Scholar]
- 18.Fernandez, M., M. Kleerebezem, O. P. Kuipers, R. J. Siezen, and R. van Kranenburg. 2002. Regulation of the metC-cysK operon, involved in sulfur metabolism in Lactococcus lactis. J. Bacteriol. 184:82-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fimmel, A. L., and R. E. Loughlin. 1977. Isolation and characterization of cysK mutants of Escherichia coli K12. J. Gen. Microbiol. 103:37-43. [DOI] [PubMed] [Google Scholar]
- 20.Foster, S. 1995. Molecular characterization and functional analysis of the major autolysin of Staphylococcus aureus 8325/4. J. Bacteriol. 177:5723-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frazzon, J., J. R. Fick, and D. R. Dean. 2002. Biosynthesis of iron-sulphur clusters is a complex and highly conserved process. Biochem. Soc. Trans. 30:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Giles, N. M., G. I. Giles, and C. Jacob. 2003. Multiple roles of cysteine in biocatalysis. Biochem. Biophys. Res. Commun. 300:1-4. [DOI] [PubMed] [Google Scholar]
- 23.Gleason, F. K., and A. Holmgren. 1988. Thioredoxin and related proteins in procaryotes. FEMS Microbiol. Rev. 4:271-297. [DOI] [PubMed] [Google Scholar]
- 24.Grundy, F. J., and T. M. Henkin. 2002. Synthesis of serine, glycine, cysteine and methionine, p. 245-254. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 25.Guérout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 26.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. σB Modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horsburgh, M. J., S. J. Wharton, A. G. Cox, E. Ingham, S. Peacock, and S. J. Foster. 2002. MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol. Microbiol. 44:1269-1286. [DOI] [PubMed] [Google Scholar]
- 28.Horsburgh, S. M. 2002. Identification of novel regulators of virulence determinant production in Staphylococcus aureus. Ph.D. thesis. University of Sheffield, Sheffield, England.
- 29.Hryniewicz, M., A. Sirko, A. Palucha, A. Bock, and D. Hulanicka. 1990. Sulfate and thiosulfate transport in Escherichia coli K-12: identification of a gene encoding a novel protein involved in thiosulfate binding. J. Bacteriol. 172:3358-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hulanicka, M. D., N. M. Kredich, and D. M. Treiman. 1974. The structural gene for O-acetylserine sulfhydrylase A in Salmonella typhimurium. Identity with the trzA locus. J. Biol. Chem. 249:867-872. [PubMed] [Google Scholar]
- 31.Hulanicka, M. D., C. Garrett, G. Jagura-Burdzy, and N. M. Kredich. 1986. Cloning and characterization of the cysAMK region of Salmonella typhimurium. J. Bacteriol. 168:322-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hussain, M., J. G. Hastings, and P. J. White. 1991. A chemically defined medium for slime production by coagulase-negative staphylococci. J. Med. Microbiol. 34:143-147. [DOI] [PubMed] [Google Scholar]
- 33.Ivankovich, A. D., B. Braverman, T. S. Stephens, M. Shulman, and H. J. Heyman. 1983. Sodium thiosulfate disposition in humans: relation to sodium nitroprusside toxicity. Anesthesiology 58:11-17. [DOI] [PubMed] [Google Scholar]
- 34.Jakob, U., W. Muse, M. Eser, and J. C. Bardwell. 1999. Chaperone activity with a redox switch. Cell 96:341-352. [DOI] [PubMed] [Google Scholar]
- 35.Kemp, E. H., R. L. Sammons, A. Moir, D. Sun, and P. Setlow. 1991. Analysis of transcriptional control of the gerD spore germination gene of Bacillus subtilis 168. J. Bacteriol. 173:4646-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitabatake, M., M. W. So, D. L. Tumbula, and D. Soll. 2000. Cysteine biosynthesis pathway in the archaeon Methanosarcina barkeri encoded by acquired bacterial genes? J. Bacteriol. 182:143-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kredich, N. M. 1996. Biosynthesis of cysteine, p. 514-527. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, M. Riley, M. Schaechter, and E. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 1. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 38.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 39.Le Faou, A., B. S. Rajagopal, L. Daniels, and G. Fauque. 1990. Thiosulfate, polythionates and elemental sulfur assimilation and reduction in the bacterial world. FEMS Microbiol. Rev. 6:351-381. [DOI] [PubMed] [Google Scholar]
- 40.Leichert, L. I., C. Scharf, and M. Hecker. 2003. Global characterization of disulfide stress in Bacillus subtilis. J. Bacteriol. 185:1967-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 42.Ludlam, G. 1949. A selective medium for the isolation of Staph. aureus from heavily contaminated material. Monthly Bull. Ministry Health 8:15-20. [PubMed] [Google Scholar]
- 43.Lyons, J., A. Rauh-Pfeiffer, Y. M. Yu, X. M. Lu, D. Zurakowski, R. G. Tompkins, A. M. Ajami, V. R. Young, and L. Castillo. 2000. Blood glutathione synthesis rates in healthy adults receiving a sulfur amino acid-free diet. Proc. Natl. Acad. Sci. USA 97:5071-5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakamura, T., H. Iwahashi, and Y. Eguchi. 1984. Enzymatic proof for the identity of the S-sulfocysteine synthase and cysteine synthase B of Salmonella typhimurium. J. Bacteriol. 158:1122-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Newton, G. L., and R. C. Fahey. 1995. Determination of biothiols by bromobimane labeling and high-performance liquid chromatography. Methods Enzymol. 251:148-166. [DOI] [PubMed] [Google Scholar]
- 46.Newton, G. L., K. Arnold, M. S. Price, C. Sherrill, S. B. delCardayre, Y. Aharonowitz, G. Cohen, J. Davies, R. C. Fahey, and C. Davis. 1996. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J. Bacteriol. 178:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicholls, P., and J. K. Kim. 1982. Sulphide as an inhibitor and electron donor for the cytochrome c oxidase system. Can. J. Biochem. 60:613-623. [DOI] [PubMed] [Google Scholar]
- 48.Novick, R. P. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587-636. [DOI] [PubMed] [Google Scholar]
- 49.O'Gara, J., M. Gomelsky, and S. Kaplan. 1997. Identification and molecular genetic analysis of multiple loci contributing to high-level tellurite resistance in Rhodobacter sphaeroides 2.4.1. Appl. Environ. Microbiol. 63:4713-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pansegrau, W., E. Lanka, P. T. Barth, D. H. Figurski, D. G. Guiney, D. Haas, D. R. Helinski, H. Schwab, V. A. Stanisich, and C. M. Thomas. 1994. Complete nucleotide sequence of Birmingham IncP alpha plasmids. Compilation and comparative analysis. J. Mol. Biol. 239:623-663. [DOI] [PubMed] [Google Scholar]
- 51.Penninckx, M. J., and M. T. Elskens. 1993. Metabolism and functions of glutathione in micro-organisms. Adv. Microbiol. Physiol. 34:239-301. [DOI] [PubMed] [Google Scholar]
- 52.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Laboratory Press, Cold Spring Harbor, N.Y.
- 53.Schenk, S., and R. A. Laddaga. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73:133-138. [DOI] [PubMed] [Google Scholar]
- 54.Sherrill, C., and R. C. Fahey. 1998. Import and metabolism of glutathione by Streptococcus mutans. J. Bacteriol. 180:1454-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sullivan, M. A., R. E. Yasbin, and F. E. Young. 1984. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene 29:21-26. [DOI] [PubMed] [Google Scholar]
- 56.Suzuki, H., W. Hashimoto, and H. Kumagai. 1993. Escherichia coli K-12 can utilize an exogenous gamma-glutamyl peptide as an amino acid source, for which gamma-glutamyltranspeptidase is essential. J. Bacteriol. 175:6038-6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tantalean, J. C., M. A. Araya, C. P. Saavedra, D. E. Fuentes, J. M. Perez, I. L. Calderon, P. Youderian, and C. C. Vasquez. 2003. The Geobacillus stearothermophilus V iscS gene, encoding cysteine desulfurase, confers resistance to potassium tellurite in Escherichia coli K-12. J. Bacteriol. 185:5831-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor, D. 1999. Bacterial tellurite resistance. Trends Microbiol. 7:111-115. [DOI] [PubMed] [Google Scholar]
- 59.Turner, R. J., J. H. Weiner, and D. E. Taylor. 1995. The tellurite resistance determinants tehAtehB and klaAklaBtelB have different biochemical requirements. Microbiology 141:3133-3140. [DOI] [PubMed] [Google Scholar]
- 60.Turner, R., J. Weiner, and D. Taylor. 1999. Tellurite-mediated thiol oxidation in Escherichia coli. Microbiology 145:2549-2557. [DOI] [PubMed] [Google Scholar]
- 61.van der Ploeg, J. R., M. A. Weiss, E. Saller, H. Nashimoto, N. Saito, M. A. Kertesz, and T. Leisinger. 1996. Identification of sulfate starvation-regulated genes in Escherichia coli: a gene cluster involved in the utilization of taurine as a sulfur source. J. Bacteriol. 178:5438-5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van der Ploeg, J. R., N. J. Cummings, T. Leisinger, and I. F. Connerton. 1998. Bacillus subtilis genes for the utilization of sulfur from aliphatic sulfonates. Microbiology 144:2555-2561. [DOI] [PubMed] [Google Scholar]
- 63.van der Ploeg, J. R., M. Barone, and T. Leisinger. 2001. Functional analysis of the Bacillus subtilis cysK and cysJI genes. FEMS Microbiol. Lett. 201:29-35. [DOI] [PubMed] [Google Scholar]
- 64.Vasquez, C. C., C. P. Saavedra, C. A. Loyola, M. A. Araya, and S. Pichuantes. 2001. The product of the cysK gene of Bacillus stearothermophilus V mediates potassium tellurite resistance in Escherichia coli. Curr. Microbiol. 43:418-423. [DOI] [PubMed] [Google Scholar]
- 65.Vergauwen, B., F. Pauwels, M. Vaneechoutte, and J. J. Van Beeumen. 2003. Exogenous glutathione completes the defense against oxidative stress in Haemophilus influenzae. J. Bacteriol. 185:1572-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Watson, S., M. Clements, and S. Foster. 1998. Characterization of the starvation-survival response of Staphylococcus aureus. J. Bacteriol. 180:1750-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Watson, S., M. Antonio, and S. Foster. 1998. Isolation and characterization of Staphylococcus aureus starvation-induced, stationary-phase mutants defective in survival or recovery. Microbiology 144:3159-3169. [DOI] [PubMed] [Google Scholar]
- 68.Wedemeyer, W. J., E. Welker, M. Narayan, and H. A. Scheraga. 2000. Disulfide bonds and protein folding. Biochemistry 39:4207-1426. [DOI] [PubMed] [Google Scholar]
- 69.Zadik, P. M., S. Davies, S. Whittaker, and C. Mason. 2001. Evaluation of a new selective medium for methicillin-resistant Staphylococcus aureus. J. Med. Microbiol. 50:476-479. [DOI] [PubMed] [Google Scholar]
- 70.Zebovitz, E., J. B. Evans, and C. F. Niven. 1955. Tellurite-glycine agar: a selective plating medium for the quantitative detection of coagulase-positive staphylococci. J. Bacteriol. 70:686-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zheng, M., F. Aslund, and G. Storz. 1998. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279:1718-1721. [DOI] [PubMed] [Google Scholar]