Abstract
Interleukin (IL)-13 genetic polymorphisms have shown adverse effects on respiratory health. However, few studies have explored the interactive effects between IL-13 haplotypes and environmental exposures on childhood asthma. The aims of our study are to evaluate the effects of IL-13 genetic variants on asthma phenotypes, and explore the potential interaction between IL-13 and household environmental exposures among Taiwanese children. We investigated 3,577 children in the Taiwan Children Health Study from 14 Taiwanese communities. Data regarding children's exposure and disease status were obtained from parents using a structured questionnaire. Four SNPs were tagged accounting for 100% of the variations in IL-13. Multiple logistic regression models with false-discovery rate (FDR) adjustments were fitted to estimate the effects of IL-13 variants on asthma phenotypes. SNP rs1800925, SNP rs20541 and SNP rs848 were significantly associated with increased risks on childhood wheeze with FDR of 0.03, 0.04 and 0.04, respectively. Children carrying two copies of h1011 haplotype showed increased susceptibility to wheeze. Compared to those without carpet use and h1011 haplotype, children carrying h1011 haplotype and using carpet at home had significantly synergistic risks of wheeze (OR, 2.5; 95% CI, 1.4–4.4; p for interaction, 0.01) and late-onset asthma (OR, 4.7; 95% CI, 2.0–10.9; p for interaction, 0.02). In conclusions, IL-13 genetic variants showed significant adverse effects on asthma phenotypes among children. The results also suggested that asthma pathogenesis might be mediated by household carpet use.
Introduction
Asthma not only results in morbidity or school absence in school children [1], [2] but also leads to raising medical costs and social burden [3]. The prevalence of childhood asthma/wheeze has been reported as increasing globally [4], [5]. Diverse genetic and environmental components have been noted for this complex disease. Recent studies have suggested that many genetic variants were associated with childhood asthma-related diseases that occur when environmental factors trigger immune responses [6], [7].
Interleukin (IL)-13 is an important T-helper type 2 (Th2) cytokine involved in the inflammation of asthmatic airways [8]. In animal models, pulmonary expression of IL-13 was reported to include eosinophilic tissue inflammation, subepithelial fibrosis, mucus hypersecretion and airway hyperresponsiveness (AHR) to methacholine [9], [10]. Many epidemiological studies also revealed that the variants in the IL-13 gene were associated with total IgE level, increased eosinophil count, atopy and asthma among children [11], [12], [13], [14].
The most important risk factor in the development of allergic diseases such as asthma is induction of IgE against indoor allergens, and imbalance between T-helper type 1 (Th1) and Th2 cytokine responses for skewing to Th2 response [15]. Household carpet use is known to be a reservoir of major indoor allergens [16], which have been suggested to increase airway inflammation and asthma in children [17], [18]. Previous studies have shown that household environmental tobacco smoke (ETS) and IL-13 genetic variants may have interactive effects on asthma phenotypes [19], [20]. However, there is no related literature concerning the association between childhood exposure to household carpet use and IL-13 genetic polymorphisms that might be involved in asthma susceptibility. Haplotype analyses of IL-13 were also unclear among the Chinese population.
Taiwan Children Health Study (TCHS) is a population-based study representing a wide range of environmental factors and genetic susceptibility. TCHS offers an opportunity to investigate the gene-environment interactive effects on respiratory health. In present study, we explored the associations between IL-13 genotypes/haplotypes, household carpet use and asthma phenotypes among children.
Materials and Methods
Study population
We conducted a population-based survey for children's health in 2007 and the study protocol has been described in detail previously [21], [22]. Briefly, TCHS recruited 5,082 middle-school children from 14 diverse communities in Taiwan. The parents or guardians of each participating student provided written informed consent at study entry. In this analysis, we randomly selected 3,577 seventh-grade children who provided their buccal cells as the DNA resource for genotyping, and there were no differences in the main characteristics between genotyped and non-genotyped subjects. The study protocol was approved by the Institutional Review Board.
Questionnaire of asthma phenotypes
The standard questionnaire for childhood exposures and health status was taken home by students and answered by parents or guardians. Children were considered to have asthma if there was a positive answer to the question “Has a doctor ever diagnosed this child as having asthma?” Wheeze was defined as any occurrence of the child's chest sounding wheezy or whistling. Early-onset asthma was defined as age of onset for asthma before 5 years of age. Late-onset asthma was onset after 5 years of age.
Exposure assessment and covariates
Household carpet use exposure was determined from the question “Have you ever used carpets in the living room, children's bedroom or other bedrooms in your house?” Basic demographic data and possible confounding variables were also collected, including sex, age, community, personal/family history of asthma or atopic diseases, in utero exposures to maternal smoking, ETS, dampness, incense burning, and pet ownership at home.
SNPs selection
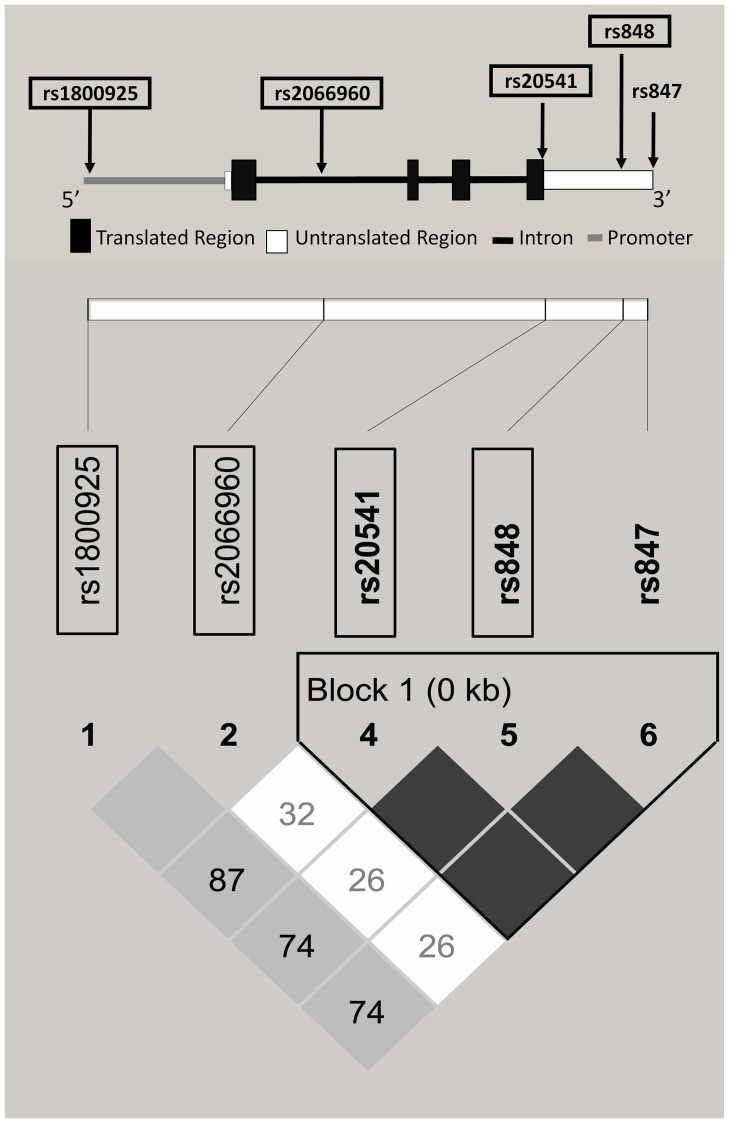
A list of IL-13 SNPs was provided by the Han Chinese in Beijing genome panel (CHB_GENO_PANEL) of Environmental Genome Project of National Institute of Environmental Health Sciences (NIEHS) (http://egp.gs.washington.edu/). We studied the genomic region of the IL-13 and the region 2000 bp upstream of the gene. All SNPs were used as input files for the Haploview v4.1 (http://www.broadinstitute.org/mpg/haploview) to select tag SNPs and to investigate the linkage disequilibrium (LD) patterns for IL-13. One of the six SNPs was excluded due to minor allele frequency (MAF) less than 0.05 [23], [24]. As shown in Fig. 1, one haplotype block in strong LD was defined by the confidence intervals of D' (where the upper CI limit was 0.98 and the lower CI limit was 0.70) [25]. Tagging SNPs were selected because of pairwise tagging tests at the prescribed squared correlation (r2) value ≧0.95 [24]. Pairwise tagging tests simply apply tag SNP represented to all the other SNPs, due to their highly correspondence [26]. The four tag SNPs of IL-13 were SNP rs1800925, SNP rs2066960, SNP rs20541 and SNP rs848, and they captured 100% of allele's variations in IL-13.
Figure 1. The location and LD of IL-13 SNPs were plotted.
D' is pairwise linkage disequilibrium using the Haploview program. The block structure of IL-13 is defined by the confidence intervals of D'. D' values are displayed in the lozenge-shaped cells, and empty cells indicate D' = 1. Four SNPs (squared) are selected and they accounted for 100% of the variations in IL-13.
DNA collection and genotyping
Genomic DNA was isolated from cotton swabs containing oral mucosa using phenol/chloroform extraction method [27]. The TaqMan assays were performed using a TaqMan PCR Core Reagent kit (Applied Biosystems, Foster City, CA) according to manufacturer's instructions. PCR amplification using 1.0∼5.0 ng of genomic DNA was performed with an initial step of 95°C for 5 min followed by 40 cycles of 95°C for 30 s and 60°C for 30 s. The fluorescence profile of each well was measured in an ABI 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA) and the results analyzed with Sequence Detection Software (Applied Biosystems, Foster City, CA). Experimental samples were compared with 36 controls to identify the three genotypes at each locus. Any samples that were outside the parameters defined by the controls were identified as non-informative and were retested. We also repeated 15% of randomly selected DNA samples to verify and confirm the results of genotyping for four IL-13 SNPs. The details of primer and probe sequences were presented in table S1. The genotype completion rates were between 97% and 99% for all loci.
Statistical analysis
Unconditional multiple logistic regression models were fitted to estimate the individual effects of IL-13 on asthma phenotypes. Models are adjusted for age, sex, parental history of asthma, parental history of atopy, in utero exposures to maternal smoking, ETS, dampness, incense burning, pet ownership at home and community. When considering the effects about IL-13 additive genetic model was utilized. Selection of confounders that were included in the model was based on a priori consideration and the standard statistical procedure of 10% change in point estimates [28]. Subjects with missing covariate information were included in the model using missing indicators [29]. Estimates of measurements of LD and r2 in TCHS participants were obtained from Haploview.
Haplotype frequencies were estimated from the IL-13 genotype data using the EM algorithm by TagSNPs [30]. The likelihood ratio test (LRT) was used to detect the global association of four variants of IL-13 haplotypes with asthma phenotypes. We collapsed rare haplotypes (frequency <0.05) into a category in the log additive haplotype analyses. The numbers of copies of each haplotype a person carries and appropriate confidence intervals were estimated using a single imputation procedure [30]. Logistic regression models were based on the co-dominant (two copies of the haplotype vs. no copy and one copy of the haplotype vs. no copy) and dominant (at least one copy of the haplotype vs. no copy) inheritance model. We estimated the false-discovery rate (FDR) for multiple comparisons for main effect associations [31], and used FDR <0.05 as a criterion.
The interaction between genotype or haplotype and household carpet use was assessed by adding an interactive term in the logistic regression model and a likelihood ratio test was used to test its significance. All analyses were conducted using SAS software version 9.1 (SAS Institute, Cary, NC, USA).
Results
A total of 3,577 children with genotyping data were enrolled in current study. The mean age of participants was 12.3±0.5 years and all of them were of ethnic Han Chinese origin (Table 1). Among these children, 8.7% live in households with carpets, and 43.4% have ETS exposure at home. The prevalence of asthma was 8.0% and wheeze occurred in 12.1% of participants.
Table 1. Selected characteristics for participants in Taiwan Children Health Study.
| With genotyping | All eligible participants | |||
| (N = 3577) | (N = 5082) | |||
| N | % | N | % | |
| Demographic information | ||||
| Sex | ||||
| Boy | 1755 | 49.1 | 2464 | 48.5 |
| Girl | 1822 | 50.9 | 2618 | 51.5 |
| Age, yr (Mean ± SD) | 12.26±0.50 | 12.42±0.65 | ||
| Parental asthma† | ||||
| No | 3335 | 96.9 | 4739 | 97.1 |
| Yes | 108 | 3.1 | 140 | 2.9 |
| Parental atopy† | ||||
| No | 2536 | 73.7 | 3617 | 74.1 |
| Yes | 907 | 26.3 | 1262 | 25.9 |
| Home exposures † | ||||
| Carpet use | 308 | 8.7 | 442 | 8.7 |
| In utero exposures to maternal smoking | 142 | 4.0 | 198 | 3.9 |
| ETS at home | 1544 | 43.4 | 2265 | 44.9 |
| Respiratory Outcomes † | ||||
| Asthma | 284 | 8.0 | 375 | 7.4 |
| Wheeze | 431 | 12.1 | 586 | 11.6 |
| Early-onset asthma‡ | 184 | 5.3 | 239 | 4.9 |
| Late-onset asthma§ | 92 | 2.7 | 123 | 2.6 |
Number of subjects do not add up to total N because of missing data.
Early-onset: asthma diagnosed ≦5 yr of age.
Late-onset: asthma diagnosed >5 yr of age.
The genotype frequencies of IL-13 were showed in Table 2 and distributions of the four selected SNPs were in Hardy-Weinberg equilibrium (p value cutoff >10−4) [23]. The pairwise measures of LD for IL-13 were presented in Table S2 and we could find SNP rs20541 and SNP rs848 in a strong LD.
Table 2. Genotype frequencies of IL-13 in this study participants.
| N | % | |
| IL-13 genotypes | ||
| SNP rs1800925 | ||
| CC | 2750 | 73.6 |
| CT | 877 | 23.5 |
| TT | 109 | 2.9 |
| Minor allele frequency | 0.15 | |
| HWE | 8×10−4 | |
| SNP rs2066960 | ||
| CC | 1428 | 37.6 |
| CA | 1725 | 45.4 |
| AA | 645 | 17.0 |
| Minor allele frequency | 0.40 | |
| HWE | 0.01 | |
| SNP rs20541 | ||
| CC | 1774 | 46.5 |
| CT | 1653 | 43.3 |
| TT | 391 | 10.2 |
| Minor allele frequency | 0.33 | |
| HWE | 0.86 | |
| SNP rs848 | ||
| GG | 1718 | 45.9 |
| GT | 1599 | 42.7 |
| TT | 427 | 11.4 |
| Minor allele frequency | 0.33 | |
| HWE | 0.08 | |
HWE: Hardy-Weinberg equilibrium.
After adjustment for age, sex, parental history of asthma, parental history of atopy, in utero exposures to maternal smoking, ETS, dampness, incense burning, pet ownership at home and community, IL-13 SNPs showed statistical significance for the occurrence of wheeze (FDR<0.05) (Table 3 and Table S3). Children carrying T allele of SNP rs1800925 were associated with increased risks on wheeze (OR, 1.3; 95% CI, 1.1–1.5; FDR, 0.03). The variant allele of SNP rs2066960 was protective for wheeze (OR, 0.9; 95% CI, 0.7–1.0; FDR, 0.04). SNP rs20541 and SNP rs848 revealed a similar associated pattern with asthma phenotypes, the increased risks of wheeze were related to children with T allele in additive models.
Table 3. Association of IL-13 with asthma phenotypes, by additive genetic model.
| SNP | Asthma | Wheeze | Early-onset asthma† | Late-onset asthma‡ | ||||||||||||
| OR | 95% CI | P value | FDR | OR | 95% CI | P value | FDR | OR | 95% CI | P value | FDR | OR | 95% CI | P value | FDR | |
| rs1800925 | 1.2 | (0.9,1.5) | 0.21 | 0.41 | 1.3 | (1.1,1.5) | 0.01 | 0.03 | 1.2 | (0.9,1.5) | 0.23 | 0.60 | 1.2 | (0.8,1.7) | 0.41 | 0.76 |
| rs2066960 | 0.9 | (0.7,1.1) | 0.12 | 0.41 | 0.9 | (0.7,1.0) | 0.02 | 0.04 | 1.0 | (0.8,1.2) | 0.60 | 0.60 | 0.7 | (0.5,1.0) | 0.03 | 0.13 |
| rs20541 | 1.1 | (0.9,1.3) | 0.45 | 0.49 | 1.2 | (1.0,1.4) | 0.04 | 0.04 | 1.1 | (0.9,1.4) | 0.44 | 0.60 | 1.1 | (0.8,1.5) | 0.66 | 0.76 |
| rs848 | 1.1 | (0.9,1.3) | 0.49 | 0.49 | 1.2 | (1.0,1.4) | 0.04 | 0.04 | 1.1 | (0.9,1.3) | 0.54 | 0.60 | 1.0 | (0.8,1.4) | 0.76 | 0.76 |
Models are adjusted for age, sex, parental history of asthma, parental history of atopy, in utero exposures to maternal smoking, ETS, dampness, incense burning, pet ownership at home and community.
Early-onset: asthma diagnosed ≦5 yr of age.
Late-onset: asthma diagnosed >5 yr of age.
Haplotype frequencies of the four SNPs were presented in Table 4, and the h0100 was the most common haplotype. The globe haplotype association using likelihood ratio test with 4 degrees of freedom was only significant for wheeze (Table 5). Compared with h0100 haplotype, children with h1011 haplotype had a significantly increased risk of wheeze (OR, 1.5; 95% CI, 1.2–1.8). The h0000 haplotype was correlated with late-onset asthma (OR, 1.5; 95% CI, 1.0–2.2).
Table 4. Haplotype frequencies of IL-13 in this study participants.
| Haplotypea | frequency |
| h0100 | 0.3314 |
| h0000 | 0.3088 |
| H0011 | 0.1356 |
| H1011 | 0.1133 |
| H0111b | 0.0470 |
| H0001b | 0.0173 |
| H1000b | 0.0135 |
| H1100b | 0.0083 |
| H0010b | 0.0079 |
| H1111b | 0.0057 |
| H1010b | 0.0039 |
| H0101b | 0.0033 |
| H0110b | 0.0014 |
| H1001b | 0.0012 |
| H1101b | 0.0010 |
| H1110b | 0.0004 |
0: common allele and 1: minor allele, by the order of SNP1 (rs1800925): C/T; SNP2 (rs2066960): C/A; SNP3 (rs20541): C/T; SNP4 (rs848): G/T.
Haplotypes were collapsed into a category in the haplotype analyses.
Table 5. Association of IL-13 haplotypes with asthma phenotypes among children.
| Asthma | Wheeze | Early-onset asthma† | Late-onset asthma‡ | |||||
| OR | 95%CI | OR | 95%CI | OR | 95%CI | OR | 95%CI | |
| h0100a | 1 | 1 | 1 | 1 | ||||
| h0000a | 1.1 | (0.9,1.4) | 1.1 | (0.9,1.4) | 1.0 | (0.7,1.3) | 1.5 | (1.0,2.2) |
| h0011a | 1.2 | (0.9,1.6) | 1.2 | (0.9,1.5) | 1.2 | (0.8,1.6) | 1.3 | (0.8,2.1) |
| h1011a | 1.2 | (0.9,1.7) | 1.5 | (1.2,1.8) | 1.2 | (0.8,1.7) | 1.5 | (0.9,2.4) |
| Others | 0.8 | (0.6,1.2) | 1.0 | (0.8,1.4) | 0.7 | (0.5,1.2) | 1.0 | (0.5,1.9) |
| globe p value | 0.22 | 0.03 | 0.34 | 0.21 | ||||
Models are adjusted for age, sex, parental history of asthma, parental history of atopy, in utero exposures to maternal smoking, ETS, dampness, incense burning, pet ownership at home and community.
Early-onset: asthma diagnosed ≦5 yr of age.
Late-onset: asthma diagnosed >5 yr of age.
0: common allele and 1: minor allele, by the order of SNP1 (rs1800925): C/T; SNP2 (rs2066960): C/A; SNP3 (rs20541): C/T; SNP4 (rs848): G/T.
The IL-13 haplotype copy numbers analyses were presented in Table 6. Compared with children without h1011 haplotype, those with two copies of h1011 haplotype showed a significantly 2.4-fold increased risk of wheeze (95% CI, 1.3–4.5; FDR, 0.04). It also showed a dose-response relationship on the increased risks, with OR 1.3 (95% CI, 1.0–1.6; FDR, 0.23) for those with one copy of h1011 haplotype, and OR 2.4 (95% CI, 1.3–4.5; FDR, 0.04) for those with two copies.
Table 6. Association of IL-13 haplotype copy numbers with wheeze among children.
| OR | 95%CI | P value | FDR | OR | 95%CI | P value | FDR | ||
| h0100a | h0011a | ||||||||
| 0 copies | 1 | 0 copies | 1 | ||||||
| 1 copy | 0.9 | (0.7,1.1) | 0.19 | 0.39 | 1 copy | 1.1 | (0.9,1.5) | 0.41 | 0.65 |
| 2 copies | 0.7 | (0.5,1.0) | 0.04 | 0.19 | 2 copies | 1.1 | (0.6,2.2) | 0.75 | 0.89 |
| ≧1 copy | 0.8 | (0.7,1.0) | 0.07 | 0.14 | ≧1 copy | 1.1 | (0.9,1.5) | 0.39 | 0.52 |
| h0000a | h1011a | ||||||||
| 0 copies | 1 | 0 copies | 1 | ||||||
| 1 copy | 1.0 | (0.8,1.3) | 0.86 | 0.89 | 1 copy | 1.3 | (1.0,1.6) | 0.08 | 0.23 |
| 2 copies | 1.0 | (0.7,1.4) | 0.89 | 0.90 | 2 copies | 2.4 | (1.3,4.5) | 0.01 | 0.04 |
| ≧1 copy | 1.0 | (0.8,1.3) | 0.92 | 0.92 | ≧1 copy | 1.4 | (1.1,1.7) | 0.02 | 0.07 |
Models are adjusted for age, sex, parental history of asthma, parental history of atopy, in utero exposures to maternal smoking, ETS, dampness, incense burning, pet ownership at home and community.
0: common allele and 1: minor allele, by the order of SNP1 (rs1800925): C/T; SNP2 (rs2066960): C/A; SNP3 (rs20541): C/T; SNP4 (rs848): G/T.
Some household environmental factors were investigated in the TCHS (Table S4). We found no substantial differences on the effect of the IL-13 genetic variants among children in relation to exposure to the other environmental factors, such as ETS, dampness, incense burning, and pet ownership at home. The association of household carpet use and ETS with asthma phenotypes was shown in Table S5. Children in households with carpet use had an increased risk on late-onset asthma (OR, 1.8; 95% CI, 1.0–3.3; FDR, 0.07). To further investigate the interactions of household factors and IL-13 variants on children's respiratory health, we examined the relationships of household carpet use with asthma phenotypes (Table 7). Joint exposure appeared to increase the individual effects of SNP rs1800925 and household carpet use on wheeze (OR, 2.0; 95% CI, 1.2–3.6; p for interaction, 0.03) and late-onset asthma (OR, 3.9; 95% CI, 1.7–9.1; p for interaction, 0.04). SNP rs20541 and SNP rs848 and household carpet use showed synergistic effects on late-onset asthma, with the OR 2.5 (95% CI, 1.2–5.2; p for interaction, 0.03) for those with T allele in SNP rs20541 and household carpet use, and with the OR 2.6 (95% CI, 1.2–5.3; p for interaction, 0.02) for those with T allele in SNP rs848 and household carpet use.
Table 7. Joint effects of carpet use and IL-13 genotypes on asthma phenotypes among children.
| Asthma | Wheeze | Early-onset asthma† | Late-onset asthma‡ | ||||||
| Carpet use | OR | 95%CI | OR | 95%CI | OR | 95%CI | OR | 95%CI | |
| SNP rs1800925 | |||||||||
| CC | No | 1 | 1 | 1 | 1 | ||||
| CC | Yes | 0.7 | (0.4,1.2) | 0.7 | (0.5,1.2) | 0.5 | (0.2,1.2) | 1.1 | (0.5,2.6) |
| CT or TT | No | 1.1 | (0.8,1.5) | 1.2 | (0.9,1.5) | 1.2 | (0.8,1.7) | 1.0 | (0.6,1.7) |
| CT or TT | Yes | 1.4 | (0.7,3.0) | 2.0 | (1.2,3.6) | 0.5 | (0.1,1.9) | 3.9 | (1.7,9.1) |
| P for interaction | 0.17 | 0.03 | 0.75 | 0.04 | |||||
| SNP rs2066960 | |||||||||
| CC | No | 1 | 1 | 1 | 1 | ||||
| CC | Yes | 0.8 | (0.3,1.9) | 1.3 | (0.7,2.3) | 0.6 | (0.2,2.1) | 1.1 | (0.3,3.7) |
| CA or AA | No | 1.0 | (0.7,1.2) | 0.9 | (0.7,1.1) | 1.1 | (0.8,1.6) | 0.7 | (0.4,1.0) |
| CA or AA | Yes | 0.8 | (0.5,1.5) | 0.8 | (0.5,1.3) | 0.4 | (0.2,1.1) | 1.5 | (0.7,3.1) |
| P for interaction | 0.84 | 0.30 | 0.52 | 0.28 | |||||
| SNP rs20541 | |||||||||
| CC | No | 1 | 1 | 1 | 1 | ||||
| CC | Yes | 0.6 | (0.3,1.3) | 0.7 | (0.4,1.3) | 0.6 | (0.3,1.6) | 0.7 | (0.2,2.4) |
| CT or TT | No | 1.0 | (0.8,1.3) | 1.1 | (0.9,1.4) | 1.2 | (0.9,1.6) | 0.8 | (0.5,1.3) |
| CT or TT | Yes | 1.1 | (0.6,2.0) | 1.4 | (0.9,2.3) | 0.4 | (0.1,1.2) | 2.5 | (1.2,5.2) |
| P for interaction | 0.30 | 0.15 | 0.36 | 0.03 | |||||
| SNP rs848 | |||||||||
| GG | No | 1 | 1 | 1 | 1 | ||||
| GG | Yes | 0.6 | (0.3,1.3) | 0.7 | (0.4,1.3) | 0.6 | (0.2,1.6) | 0.7 | (0.2,2.2) |
| GT or TT | No | 1.0 | (0.8,1.3) | 1.2 | (0.9,1.4) | 1.2 | (0.9,1.6) | 0.8 | (0.5,1.2) |
| GT or TT | Yes | 1.1 | (0.6,2.0) | 1.5 | (0.9,2.4) | 0.4 | (0.1,1.2) | 2.6 | (1.2,5.3) |
| P for interaction | 0.26 | 0.13 | 0.39 | 0.02 | |||||
Models are adjusted for age, sex, parental history of asthma, parental history of atopy, in utero exposures to maternal smoking, ETS, dampness, incense burning, pet ownership at home and community.
Early-onset: asthma diagnosed ≦5 yr of age.
Late-onset: asthma diagnosed >5 yr of age.
Because the effect of h1011 was greater than other haplotypes (Table 5), we investigated the association between h1011 haplotype, household carpet use and asthma phenotypes. Compared to those without household carpet use and h1011 haplotype, children carrying h1011 haplotype and living in homes with carpets had increased risks of wheeze (OR, 2.5; 95% CI, 1.4–4.4; p for interaction, 0.01) and late-onset asthma (OR, 4.7; 95% CI, 2.0–10.9; p for interaction, 0.02) (Table 8). However, we could not find the similar pattern for ETS at home on the effects of h1011 haplotype to asthma phenotypes in childhood (Table S6).
Table 8. Joint effects of carpet use and IL-13 haplotype h1011 on asthma phenotypes among children.
| h1011a | ||||||
| No | Yes | |||||
| Carpet use | OR | 95%CI | OR | 95%CI | P for interaction | |
| Asthma | No | 1 | 1.1 | (0.8,1.4) | 0.06 | |
| Yes | 0.6 | (0.3,1.1) | 1.7 | (0.8,3.6) | ||
| Wheeze | No | 1 | 1.2 | (0.9,1.5) | 0.01 | |
| Yes | 0.7 | (0.4,1.1) | 2.5 | (1.4,4.4) | ||
| Early-onset asthma† | No | 1 | 1.1 | (0.8,1.6) | 0.98 | |
| Yes | 0.5 | (0.2,1.1) | 0.5 | (0.1,2.2) | ||
| Late-onset asthma‡ | No | 1 | 1.0 | (0.6,1.7) | 0.02 | |
| Yes | 1.0 | (0.4,2.4) | 4.7 | (2.0,10.9) | ||
Models are adjusted for age, sex, parental history of asthma, parental history of atopy, in utero exposures to maternal smoking, ETS, dampness, incense burning, pet ownership at home and community.
Early-onset: asthma diagnosed ≦5 yr of age.
Late-onset: asthma diagnosed >5 yr of age.
0: common allele and 1: minor allele, by the order of SNP1 (rs1800925): C/T; SNP2 (rs2066960): C/A; SNP3 (rs20541): C/T; SNP4 (rs848): G/T.
Discussion
To the best of our knowledge, this is the first study concerning the potential interactive associations between IL-13, household carpet use and childhood asthma. In our data, IL-13 genetic variants showed significant adverse effects on asthma phenotypes. We found that children carrying h1011 haplotype have increased risks of the occurrence of wheeze. Household carpet use appears to modify the effects of IL-13 gene on wheeze and late-onset asthma.
The IL-13 gene is located on chromosome 5q, which has been suggested to be associated with the risk of elevated serum IgE levels, eosinophilia, airway hyper-sensitiveness and the occurrence of childhood asthma [13], [14]. In our study, children with variant alleles of SNP rs1800925, SNP rs20541 and SNP rs848 have significantly higher risks for wheeze (Table 3). Previous studies have shown that promoter variant in IL-13 (SNP rs1800925) could enhance IL-13 transcription [32] and was associated with atopy, asthma and increased bronchial hyper-reactivity [13], [33], [34]. Another polymorphism in exon4 (SNP rs20541, Arg130Gln) also resulted in the occurrence of asthma, reduced lung function and increased serum IgE [13], [35], [36]. Functional and association studies both showed that 130Gln was related to higher IL-13 levels and a stronger Th2 immune response than 130Arg [37]. Few studies focused on rs848 polymorphism with asthma. Hunninghake et al. have indicated that SNP rs848 was in strong linkage disequilibrium with SNP rs20541 [13], which is consistent with our findings (Fig. 1 and Table S2). However, the effects of SNP rs2066960 variants in previous studies showed that minor allele (A allele) was associated with elevated serum IgE and early-transient wheeze [19], [38]. In our data, we found that SNP rs2066960 A allele was associated with decreased risks of asthma phenotypes (Table 3). The different genetic association on asthma might be attributed to genotype frequencies in different ethnic populations. Based on the haplotype analyses in the present study, we found that three variants of 4 SNPs in IL-13 gene might significantly affect risks of asthma phenotypes in children. Compared with the common haplotype, children carrying h1011 haplotype were more susceptible to development of wheeze (OR, 1.5; 95% CI, 1.2-1.8) (Table 5). Furthermore, the more copy numbers of the h1011 haplotype children carry, the higher risk of wheeze they would possess (Table 6).
IL-13, independent from IL-4, plays a central role for the development of asthma-related symptoms in animal models and human studies [10], [39]. IL-13 was primarily produced by Th2 CD4+ T cells after allergen irritation, and it may induce the entire pathogenic pathway of asthma independently of traditional cells, such as mast cells and eosinophils [8]. Genetic variants in IL-13 have been found to be associated with elevated serum levels of IL-13 [37]. Household carpet use is a significant reservoir of allergens, including house dust mite, dog and cat dander, and fungal concentrations [17], [40]. House dust mites, Dermatophagoides pteronyssinus allergen 1 (Der p 1) and Dermatophagoides farinae (Der f 1), might play important roles in allergic sensitization, as well as in the development of asthma and asthma deterioration in children [41], [42], [43]. In our data, the interactive effects are consistent with IL-13 genotypes and household carpet use on asthma phenotypes (Table 7). Especially in SNP rs1800925, joint exposure appeared to increase the individual effects of SNP rs1800925 T allele and household carpet use on wheeze (OR, 2.0; 95% CI, 1.2–3.6) and late-onset asthma (OR, 3.9; 95% CI, 1.7–9.1). It was suggested that the variant of promoter region, containing a binding site of the nuclear factor of activated T cells (NFAT) transcription factor, regulates IL-13 and IL-4 gene expression [33]. Additionally, our results indicated that children with h1011 haplotype and exposure to carpets may have increased risks for asthma phenotypes (Table 8). We believe that IL-13 genetic variants and exposure to household carpet use may synergistically induce high IL-13 levels to inflammation, which would result in the occurrence of asthma phenotypes in children.
Up to the present, two studies concerning IL-13-environmental interaction on asthma phenotypes in children have been reported. Sadeghnejad and colleagues investigated SNP rs1800925, SNP rs2066960 and SNP rs20541 and demonstrated that the effect of ETS exposure at home was stronger on wheeze with the common IL-13 haplotype compared to those without it [19]. Sorensen et al. reported that children exposed to maternal smoking during pregnancy and with SNP rs20541 C allele had increased risks on wheeze [20]. However, no significant interaction between IL-13 polymorphisms (SNP rs20541 and SNP rs1800925) and ETS exposure at home were noted. In our data, we could not find interactive effects between IL-13 and ETS exposure at home in asthma phenotypes (Table S6). Several reasons, including differences in ethnicity or genotype frequencies, may explain this situation. For example, the genotype frequencies in SNP rs2066960 were 81.5, 17.6% and 0.9% for CC, CA and AA genotypes, respectively, in British population [19]. In SNP rs20541, the genotype frequencies were 61.8∼64.4%, 32.1∼33.1%, and 3.5∼5.1% for CC, CT and TT genotypes, respectively, in European populations [19], [20]. Our study showed distinctly different results: 37.6%, 45.4% and 17.0% for CC, CA and AA genotypes in SNP rs2066960, and 46.7%, 44.4% and 8.9% for CC, CT and TT genotypes in SNP rs20541 (Table 2). All of the results were similar to ethnic Han Chinese in the Beijing population from HapMap (data not shown). Our population did provide evidence for IL-13 genetic variants on asthma phenotypes among Han children.
Age, sex, parental atopic history, maternal smoking during pregnancy, ETS, dampness, incense burning, and pet ownership at home were believed to contribute to asthma and wheeze in childhood [4], [21], [44], [45], [46]. We minimized interference from these confounders by recruiting lifelong non-smokers of similar age, and adjusting potential confounders by regression models. Difference in participation by children with respiratory outcomes who had different carpet exposure histories is unlikely to be significant enough to produce substantial bias, as participation rates in each classroom were high and the characteristics was similar between genotyping and all participants (Table 1). Because the differences in distribution are modest and are probably not associated with the genotypes, it is unlikely that selection of subjects biased the effect estimates in our results. As the study subjects were recruited in an unselected population, unbiased observations of the association between genetic effects and outcomes were expected.
Our definition of household carpet use might not indicate a good quantitative biomarker for measuring allergen levels in the houses. However, higher concentrations of indoor allergens have been reported to be associated with carpet use at home [16], [47]. Exposure assessment from the questionnaire was likely to introduce some misclassification bias shifting the results toward the null. Moreover, the associations between household carpet use and childhood asthma were consistent in another case-control study from our group [18]. Another possible limitation is recall bias of respiratory outcomes. Asthma phenotypes in our study were ascertained by parental-reported questionnaire, so misclassification may have arisen from imperfect parental recall of events. Differential misclassification by IL-13 genetic variants was probably not a major source of bias that accounts for our results, because disease status was defined without the knowledge of genotype. We found that large parts of significant effects were limited on wheeze and effect estimates of wheeze were also stronger than other asthma phenotypes. We believe that wheeze is the most common respiratory symptom in children when occurrence of airway inflammation induced by environmental stimuli. In Taiwan, parents of children with wheeze symptoms might be unlikely to seek medical care, and therefore physician-diagnosed asthma would be underreported. Consistent with previous well-known knowledge, we found the significant genetic association in IL-13 gene, and wheeze is a good predictor for development of asthma in children.
In conclusions, our results showed that genetic variants in IL-13, especially h1011 haplotype, showed adverse effects on respiratory health in children. Household carpet use may influence the severity of diverse allergic inflammatory reactions induced by IL-13 genetic variants. Additional long-term research is necessary to explore the roles played by other genes in determining genetic susceptibility on adverse respiratory outcomes. Identification of gene-environmental interactions in childhood asthma may lead to new and comprehensive insights into asthma pathogenesis and treatment.
Supporting Information
Primer and probe sequences for IL-13 genetic variants.
(DOC)
Pairwise measures of linkage disequilibrium for IL-13 in this study participants.
(DOC)
Association of IL-13 genotypes with asthma phenotypes, by co-dominant and dominant genetic model.
(DOC)
Environmental questions of TCHS questionnaire.
(DOC)
Association of household carpet use and ETS at home with asthma phenotypes among children.
(DOC)
Joint effects of ETS exposure and IL-13 haplotype h1011 on asthma phenotypes among children.
(DOC)
Acknowledgments
We thank all the field workers who supported data collection, the school administrators and teachers, and especially the parents and children who participated in this study.
Funding Statement
This study was supported by the National Science Council (Grant #96-2314-B-006-053, #98-2314-B-002-138-MY3 and #101-2314-B-002-113-MY3). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Spee-van der Wekke J, Meulmeester JF, Radder JJ, Verloove-Vanhorick SP (1998) School absence and treatment in school children with respiratory symptoms in The Netherlands: data from the Child Health Monitoring System. J Epidemiol Community Health 52: 359–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Silverstein MD, Mair JE, Katusic SK, Wollan PC, O'Connell E J, et al. (2001) School attendance and school performance: a population-based study of children with asthma. J Pediatr 139: 278–283. [DOI] [PubMed] [Google Scholar]
- 3. Weiss KB, Sullivan SD (1994) Socio-economic burden of asthma, allergy, and other atopic illnesses. Pediatr Allergy Immunol 5: 7–12. [DOI] [PubMed] [Google Scholar]
- 4. Lee YL, Lin YC, Hwang BF, Guo YL (2005) Changing prevalence of asthma in Taiwanese adolescents: two surveys 6 years apart. Pediatr Allergy Immunol 16: 157–164. [DOI] [PubMed] [Google Scholar]
- 5. Maziak W, Behrens T, Brasky TM, Duhme H, Rzehak P, et al. (2003) Are asthma and allergies in children and adolescents increasing? Results from ISAAC phase I and phase III surveys in Munster, Germany. Allergy 58: 572–579. [DOI] [PubMed] [Google Scholar]
- 6. Martinez FD (2007) Gene-environment interactions in asthma: with apologies to William of Ockham. Proc Am Thorac Soc 4: 26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunninghake GM, Soto-Quiros ME, Lasky-Su J, Avila L, Ly NP, et al.. (2008) Dust mite exposure modifies the effect of functional IL10 polymorphisms on allergy and asthma exacerbations. J Allergy Clin Immunol 122: 93–98, 98 e91–95. [DOI] [PMC free article] [PubMed]
- 8. Wills-Karp M, Chiaramonte M (2003) Interleukin-13 in asthma. Curr Opin Pulm Med 9: 21–27. [DOI] [PubMed] [Google Scholar]
- 9. Zhu Z, Homer RJ, Wang Z, Chen Q, Geba GP, et al. (1999) Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J Clin Invest 103: 779–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wills-Karp M, Luyimbazi J, Xu X, Schofield B, Neben TY, et al. (1998) Interleukin-13: central mediator of allergic asthma. Science 282: 2258–2261. [DOI] [PubMed] [Google Scholar]
- 11. DeMeo DL, Lange C, Silverman EK, Senter JM, Drazen JM, et al. (2002) Univariate and multivariate family-based association analysis of the IL-13 ARG130GLN polymorphism in the Childhood Asthma Management Program. Genet Epidemiol 23: 335–348. [DOI] [PubMed] [Google Scholar]
- 12. Graves PE, Kabesch M, Halonen M, Holberg CJ, Baldini M, et al. (2000) A cluster of seven tightly linked polymorphisms in the IL-13 gene is associated with total serum IgE levels in three populations of white children. J Allergy Clin Immunol 105: 506–513. [DOI] [PubMed] [Google Scholar]
- 13. Hunninghake GM, Soto-Quiros ME, Avila L, Su J, Murphy A, et al. (2007) Polymorphisms in IL13, total IgE, eosinophilia, and asthma exacerbations in childhood. J Allergy Clin Immunol 120: 84–90. [DOI] [PubMed] [Google Scholar]
- 14. Kim HB, Lee YC, Lee SY, Jung J, Jin HS, et al. (2006) Gene-gene interaction between IL-13 and IL-13Ralpha1 is associated with total IgE in Korean children with atopic asthma. J Hum Genet 51: 1055–1062. [DOI] [PubMed] [Google Scholar]
- 15. Busse WW, Lemanske RF Jr (2001) Asthma. N Engl J Med 344: 350–362. [DOI] [PubMed] [Google Scholar]
- 16. Spertini F, Berney M, Foradini F, Roulet CA (2010) Major mite allergen Der f 1 concentration is reduced in buildings with improved energy performance. Allergy 65: 623–629. [DOI] [PubMed] [Google Scholar]
- 17. Tranter DC, Wobbema AT, Norlien K, Dorschner DF (2009) Indoor allergens in Minnesota schools and child care centers. J Occup Environ Hyg 6: 582–591. [DOI] [PubMed] [Google Scholar]
- 18.Chen YC, Tsai CH, Lee YL (2011) Early-Life Indoor Environmental Exposures Increase the Risk of Childhood Asthma. Int J Hyg Environ Health (Accepted). [DOI] [PubMed]
- 19. Sadeghnejad A, Karmaus W, Arshad SH, Kurukulaaratchy R, Huebner M, et al. (2008) IL13 gene polymorphisms modify the effect of exposure to tobacco smoke on persistent wheeze and asthma in childhood, a longitudinal study. Respir Res 9: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sorensen M, Allermann L, Vogel U, Andersen PS, Jespersgaard C, et al. (2009) Polymorphisms in inflammation genes, tobacco smoke and furred pets and wheeze in children. Pediatr Allergy Immunol 20: 614–623. [DOI] [PubMed] [Google Scholar]
- 21. Tsai CH, Huang JH, Hwang BF, Lee YL (2010) Household environmental tobacco smoke and risks of asthma, wheeze and bronchitic symptoms among children in Taiwan. Respir Res 11: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hwang BF, Lee YL (2010) Air Pollution and Prevalence of Bronchitic Symptoms among Children in Taiwan. Chest 138: 956–964. [DOI] [PubMed] [Google Scholar]
- 23. Balding DJ (2006) A tutorial on statistical methods for population association studies. Nat Rev Genet 7: 781–791. [DOI] [PubMed] [Google Scholar]
- 24. de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, et al. (2005) Efficiency and power in genetic association studies. Nature Genetics 37: 1217–1223. [DOI] [PubMed] [Google Scholar]
- 25. Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, et al. (2002) The structure of haplotype blocks in the human genome. Science 296: 2225–2229. [DOI] [PubMed] [Google Scholar]
- 26. Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, et al. (2004) Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet 74: 106–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gill P, Jeffreys AJ, Werrett DJ (1985) Forensic application of DNA ‘fingerprints’. Nature 318: 577–579. [DOI] [PubMed] [Google Scholar]
- 28. Tong IS, Lu Y (2001) Identification of confounders in the assessment of the relationship between lead exposure and child development. Ann Epidemiol 11: 38–45. [DOI] [PubMed] [Google Scholar]
- 29. Greenland S, Finkle WD (1995) A critical look at methods for handling missing covariates in epidemiologic regression analyses. Am J Epidemiol 142: 1255–1264. [DOI] [PubMed] [Google Scholar]
- 30. Stram DO, Leigh Pearce C, Bretsky P, Freedman M, Hirschhorn JN, et al. (2003) Modeling and E-M estimation of haplotype-specific relative risks from genotype data for a case-control study of unrelated individuals. Hum Hered 55: 179–190. [DOI] [PubMed] [Google Scholar]
- 31. Osborne JA (2006) Estimating the false discovery rate using SAS. SAS Users Group International Proceedings 190: 1–10. [Google Scholar]
- 32. Cameron L, Webster RB, Strempel JM, Kiesler P, Kabesch M, et al. (2006) Th2 cell-selective enhancement of human IL13 transcription by IL13-1112C>T, a polymorphism associated with allergic inflammation. J Immunol 177: 8633–8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. van der Pouw Kraan TC, van Veen A, Boeije LC, van Tuyl SA, de Groot ER, et al. (1999) An IL-13 promoter polymorphism associated with increased risk of allergic asthma. Genes Immun 1: 61–65. [DOI] [PubMed] [Google Scholar]
- 34. Hummelshoj T, Bodtger U, Datta P, Malling HJ, Oturai A, et al. (2003) Association between an interleukin-13 promoter polymorphism and atopy. Eur J Immunogenet 30: 355–359. [DOI] [PubMed] [Google Scholar]
- 35. Heinzmann A, Jerkic SP, Ganter K, Kurz T, Blattmann S, et al. (2003) Association study of the IL13 variant Arg110Gln in atopic diseases and juvenile idiopathic arthritis. J Allergy Clin Immunol 112: 735–739. [DOI] [PubMed] [Google Scholar]
- 36. Park HW, Lee JE, Kim SH, Kim YK, Min KU, et al. (2009) Genetic variation of IL13 as a risk factor of reduced lung function in children and adolescents: a cross-sectional population-based study in Korea. Respir Med 103: 284–288. [DOI] [PubMed] [Google Scholar]
- 37. Arima K, Umeshita-Suyama R, Sakata Y, Akaiwa M, Mao XQ, et al. (2002) Upregulation of IL-13 concentration in vivo by the IL13 variant associated with bronchial asthma. J Allergy Clin Immunol 109: 980–987. [DOI] [PubMed] [Google Scholar]
- 38. Ogbuanu IU, Karmaus WJ, Zhang H, Sabo-Attwood T, Ewart S, et al. (2010) Birth order modifies the effect of IL13 gene polymorphisms on serum IgE at age 10 and skin prick test at ages 4, 10 and 18: a prospective birth cohort study. Allergy Asthma Clin Immunol 6: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grunig G, Warnock M, Wakil AE, Venkayya R, Brombacher F, et al. (1998) Requirement for IL-13 independently of IL-4 in experimental asthma. Science 282: 2261–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vojta PJ, Randels SP, Stout J, Muilenberg M, Burge HA, et al. (2001) Effects of physical interventions on house dust mite allergen levels in carpet, bed, and upholstery dust in low-income, urban homes. Environ Health Perspect 109: 815–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chan-Yeung M, Manfreda J, Dimich-Ward H, Lam J, Ferguson A, et al. (1995) Mite and cat allergen levels in homes and severity of asthma. Am J Respir Crit Care Med 152: 1805–1811. [DOI] [PubMed] [Google Scholar]
- 42. Henderson FW, Henry MM, Ivins SS, Morris R, Neebe EC, et al. (1995) Correlates of recurrent wheezing in school-age children. The Physicians of Raleigh Pediatric Associates. Am J Respir Crit Care Med 151: 1786–1793. [DOI] [PubMed] [Google Scholar]
- 43. Sporik R, Holgate ST, Platts-Mills TA, Cogswell JJ (1990) Exposure to house-dust mite allergen (Der p I) and the development of asthma in childhood. A prospective study. N Engl J Med 323: 502–507. [DOI] [PubMed] [Google Scholar]
- 44.Tsai CH, Tung KY, Chen CH, Lee YL (2011) Tumour necrosis factor G-308A polymorphism modifies the effect of home dampness on childhood asthma. Occupational and Environmental Medicine. [DOI] [PubMed]
- 45. Wang IJ, Tsai CH, Chen CH, Tung KY, Lee YL (2011) Glutathione S-transferase, incense burning and asthma in children. Eur Respir J 37: 1371–1377. [DOI] [PubMed] [Google Scholar]
- 46. Takkouche B, Gonzalez-Barcala FJ, Etminan M, Fitzgerald M (2008) Exposure to furry pets and the risk of asthma and allergic rhinitis: a meta-analysis. Allergy 63: 857–864. [DOI] [PubMed] [Google Scholar]
- 47. Mihrshahi S, Marks G, Vanlaar C, Tovey E, Peat J (2002) Predictors of high house dust mite allergen concentrations in residential homes in Sydney. Allergy 57: 137–142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer and probe sequences for IL-13 genetic variants.
(DOC)
Pairwise measures of linkage disequilibrium for IL-13 in this study participants.
(DOC)
Association of IL-13 genotypes with asthma phenotypes, by co-dominant and dominant genetic model.
(DOC)
Environmental questions of TCHS questionnaire.
(DOC)
Association of household carpet use and ETS at home with asthma phenotypes among children.
(DOC)
Joint effects of ETS exposure and IL-13 haplotype h1011 on asthma phenotypes among children.
(DOC)